Abstract
To identify clinically important molecular subtypes of prostate cancer (PCa), we characterized the somatic landscape of aggressive tumors via deep, whole-genome sequencing. In our discovery set of ten tumor/normal subject pairs with Gleason scores of 8–10 at diagnosis, coordinated analysis of germline and somatic variants, including single-nucleotide variants, indels, and structural variants, revealed biallelic BRCA2 disruptions in a subset of samples. Compared to the other samples, the PCa BRCA2-deficient tumors exhibited a complex and highly specific mutation signature, featuring a 2.88-fold increased somatic mutation rate, depletion of context-specific C>T substitutions, and an enrichment for deletions, especially those longer than 10 bp. We next performed a BRCA2 deficiency-targeted reanalysis of 150 metastatic PCa tumors, and each of the 18 BRCA2-mutated samples recapitulated the BRCA2 deficiency-associated mutation signature, underscoring the potent influence of these lesions on somatic mutagenesis and tumor evolution. Among all 21 individuals with BRCA2-deficient tumors, only about half carried deleterious germline alleles. Importantly, the somatic mutation signature in tumors with one germline and one somatic risk allele was indistinguishable from those with purely somatic mutations. Our observations clearly demonstrate that BRCA2-disrupted tumors represent a unique and clinically relevant molecular subtype of aggressive PCa, highlighting both the promise and utility of this mutation signature as a prognostic and treatment-selection biomarker. Further, any test designed to leverage BRCA2 status as a biomarker for PCa must consider both germline and somatic mutations and all types of deleterious mutations.
Introduction
Prostate cancer (PCa [MIM: 176807]) is the most common non-cutaneous malignancy and second leading cause of cancer deaths in American men, with approximately 220,800 diagnoses and 27,540 deaths projected in 2015.1 The 5-year survival for local disease is nearly 100%, compared to only 28% for metastatic disease.2 This outcome disparity frames the major clinical challenge associated with PCa: distinguishing those men who are likely to get metastatic disease, which might be prevented by specific and early therapy, while minimizing the iatrogenic morbidity associated with overtreatment of indolent disease. Though clinical measures including Gleason score and quantification of prostate-specific antigen have prognostic utility, the current risk stratification framework misclassifies a critical subset of tumors. Consequently, a great deal of PCa research is focused on finding molecular and genetic biomarkers that facilitate early and accurate identification of men with potentially high-risk tumors.
Whole-exome sequencing (WES) and whole-genome sequencing (WGS) studies have provided a window into the biology that drives oncogenesis and progression of PCa tumors by enabling unbiased exploration of somatic mutations in prostate tumors that span the spectrum of aggressiveness disease.3, 4, 5, 6, 7, 8, 9, 10 WES-based studies of tumors have highlighted genes that are recurrently mutated,3, 4, 6, 8 and WGS efforts defined a prominent role for structural rearrangements in tumor evolution.5, 7 These findings suggest that the genome-wide interplay between somatic single-nucleotide variants (sSNVs), indels, and structural variants (SVs) is important for understanding the repertoire of genomic aberrations that contribute to PCa. This hypothesis was confirmed by a recent study that reported different variant types combining to knock out both copies of recurrently mutated genes in metastatic PCa tumors.8 In spite of these findings, considerable work remains to understand the relationship between somatic genomic alterations and tumor aggressiveness.
Our initial approach used deep WGS in a discovery set of ten high-Gleason-grade prostate tumor/normal subject pairs from the Mayo Clinic to search for drivers of PCa aggressiveness. Via combined analysis of germline and somatic SNVs, indels, and SVs, we uncovered biallelic loss of BRCA2 (MIM: 600185) in three of the ten sequenced tumors. Although BRCA2 mutations or larger chromosome13 deletions have been reported to affect a small percentage of PCa tumors,3, 8, 9, 10 the effect of these mutations on the PCa tumor genome has not been elucidated. As such, although the clinical importance of BRCA2 deficiency might be inferred, we sought to explicitly define the genome-wide consequences of biallelic BRCA2 loss in PCa tumors and thereby solidify the clinical importance of BRCA2 defects in PCa.
Breast, ovarian, pancreatic, and gastric tumors with germline and/or somatic BRCA2 defects have a distinctive somatic mutation profile that results from the inability of cells to repair double-strand DNA breaks via the high-fidelity homologous recombination (HR) pathway.11, 12, 13, 14, 15, 16 These tumors exhibited an elevated mutation rate and also had characteristic substitution and indel patterns, evidence that BRCA2 loss produces a powerful, pervasive effect on the cancer genome. We hypothesized that if BRCA2 mutations are crucial drivers of PCa tumor evolution, then samples with biallelic loss of the gene should exhibit a somatic mutation profile that mirrors the BRCA2 deficiency from other tumor types. Our WGS characterization of the three discovery set tumors from the Mayo Clinic, as well as our BRCA2 deficiency-targeted reanalysis of 150 metastatic tumors, including 18 with BRCA2 defects, supports this hypothesis. Furthermore, we show that PCa tumors with purely somatic disruption of BRCA2 not only have the same mutation signature, but occur at the same frequency as tumors with germline plus somatic mutations. Thus, our analyses suggest that tumor BRCA2 status and the associated somatic mutation signature represent a clinically relevant molecular biomarker in PCa.
Material and Methods
Sequencing, Variant Calling, and Analysis of WGS Samples
The discovery set of ten Mayo Clinic tumor samples was selected for sequencing based on high Gleason score and availability of both peripheral blood DNA and fresh frozen prostatectomy samples. Subjects had a mean age at diagnosis of 63.6 years (range 54–77 years) and all were of European descent (Tables 1 and S1). The Mayo Clinic IRB approved the study design, and consent was obtained at the time of sample collection. For each frozen tumor sample, an initial H&E-stained slide was reviewed by a pathologist. An area was marked that contained more than 70% tumor cells and a Gleason score greater or equal to 8. The block was then serially cut into six 10-μm thick sections immediately after the marked H&E slide. With the H&E-stained slide as a guide, the marked area was macro-dissected from unstained sections on dry ice and placed into a tube for DNA extraction. The DNA was extracted via the Puregene Tissue Kit (QIAGEN) and the corresponding genomic DNA was extracted from white blood cells via the AutoGen FLEX STAR system (AutoGen). Sequencing libraries were prepared with the Illumina TruSeq v2 kit (Illumina) and then sequenced with the Illumina HiSeq 2000 Genome Analyzer platform to generate 100-base paired-end reads, according to manufacturer’s protocols. Tumor DNA was sequenced to a genome-wide average depth of 95.3× and average germline coverage was 48.3× (Tables 1 and S2).
Table 1.
The Whole Genomes of Nine Highly Aggressive, Treatment-Naive Primary Prostate Tumors and One Nodal Metastasis Were Sequenced
| Sample | Gleason Score | Age at Diagnosis | PSA at Diagnosis (ng/dL) | Germline Cov. | Tumor Cov. | Est. Purity (%)a | Est. Ploidy |
|---|---|---|---|---|---|---|---|
| MC-1T | 9 | 60 | 5.1 | 52.1 | 101.5 | 49 | 2.14 |
| MC-2T | 8 | 68 | 7.1 | 48.1 | 87.5 | 51 | 1.96 |
| MC-3T | 9 | 54 | 17.5 | 48.4 | 98.5 | 56 | 2.1 |
| MC-4T | 9 | 77 | 17.7 | 50.5 | 97.7 | 50 | 2.15 |
| MC-5T | 9 | 62 | 4.4 | 52.2 | 100.3 | 79 | 2.08 |
| MC-6T | 10 | 76 | 15.2 | 46.9 | 103.5 | 50 | 2.19 |
| MC-7T | 9 | 56 | 7.93 | 45.5 | 96.2 | 62 | 2.22 |
| MC-8T | 9 | 66 | 11.14 | 48.2 | 82.5 | 59 | 2.25 |
| MC-9T | 8 | 54 | 7.68 | 45.7 | 92.2 | 75 | 1.96 |
| MC-10M | 7b | 63 | 22.3 | 45.1 | 93.1 | 75 | 2.51 |
| MEAN | – | 63.6 | 11.6 | 48.3 | 95.3 | 60.6 | 2.2 |
Additional clinical information and sequencing metrics can be found in Tables S1 and S2. Purity and ploidy were estimated with Patchwork.24
All tumors were macrodissected to target at least 70% Gleason grade 4+ cytoarchitecture, and the purity estimate reported here is the actual purity calculated from sequencing data.
For MC-10M, the Gleason score refers to the primary tumor, while the sequenced DNA was isolated from a nodal metastasis.
After sequencing data were generated, both the discovery set samples and the WGS reads from the 50 tumors spanning the aggressiveness spectrum described by Baca et al.5 were processed via an identical analysis pipeline. Reads were aligned to the NCBI GRCh37 human reference genome with BWA v.0.7.2,17 PCR duplicates were marked with Picard v.1.87, and GATK v.3.2 was used for local realignment around indels and base quality score recalibration.18
We performed SNV and indel discovery, genotyping, and variant quality score recalibration in all tumor and germline samples simultaneously, according to the GATK HaplotypeCaller v.3.2 best practices recommendations.19 Indels that were multiallelic or found in any germline sample were subtracted from each tumor, and remaining indels with VQSLOD > −0.8209 (Tranche 98.0) were retained for somatic analyses. Indels were classified as either deletions or insertions, and the length of each variant was calculated (Table S3). We used MuTect to compare tumor and normal bam files for first pass sSNV detection,20 then removed any variants that were (1) present in the HaplotypeCaller genotypes from germline samples, (2) present in dbSNP but not the COSMIC database, or (3) powered at less than 0.9 for either the somatic or germline sample. Somatic substitution mutational signatures were created with the reference allele, alternate allele, and adjacent bases in the reference, as previously described.12 The predicted effect of indels and sSNVs were calculated with the Variant Effect Predictor21 and annotated with further functional predictions with CADD v.1.2.22 Somatic and germline BRCA2 indels were confirmed with Sanger sequencing. For MC-6T, Haploview was used to evaluate linkage disequilibrium between the germline indel and variants inside the deleted region,23 and phased haplotypes from the 1000 Genomes Project was used to project which chromosomes were affected by the rare germline indel and the somatic deletion.
Genomic architecture analysis was conducted with the R package Patchwork, which leverages normalized coverage and allelic imbalance to detect clonal and subclonal changes in copy number in 10-kb windows across the genome.24 Patchwork plots for MC-1T were excluded from further analyses due to high variability in normalized coverage between the tumor and germline samples. Local somatic allelic imbalance was detected using plots of SNV sites that (1) were genotyped as heterozygous in the germline sample, (2) were present in dbSNP v.137 and therefore had a higher likelihood of being true germline variant positions, and (3) had a reference allele fraction in the normal sample between 0.45 and 0.55 (to remove positions with systematic allelic bias). Patchwork was also used to estimate tumor purity and ploidy.
We used DELLY v.0.5.5 to identify deletions (DEL), tandem duplications (DUP), inversions (INV), and translocations (TRA) in all tumor and normal genomes.25 Raw somatic DELLY calls were filtered to remove probable false positives by excluding SVs that had a size <1 kb or where either SV breakpoint fell within 100 kb from the start or end of a chromosome. Candidate SVs were merged into a single event when both breakpoints were ±200 bp for INV, DEL, DUP and ±500 bp for TRA. SVs were also filtered if split-read support (SR) < 1 and paired-end read support (PE) < 5 or PE < 1 and SR < 5. As with the SNVs and indels, we employed a panel of normals approach to remove germline SVs or systematic errors. We excluded any variant of the same subtype that was present in any germline genome within the same margin of errors used for merging. A catalog of high-confidence sSNVs, indels, and SVs that are predicted to disrupt the coding region of genes and were found in the discovery set and the tumors unselected for aggressiveness are in Table S4.
BRCA2 Deficiency-Targeted Reanalysis of Metastatic WES Tumors
We performed a BRCA2 deficiency-targeted reevaluation of all variants documented in 150 metastatic PCa samples from Table S3 of the publication by Robinson et al.8 to determine whether the HR-deficient mutation signature was also present in this dataset, which included 18 samples with biallelic BRCA2 mutations, one sample with biallelic hits in BRCA1 (MIM: 113705), and three samples with biallelic aberrations in DNA mismatch repair genes. The trinucleotide sequence context of each substitution was extracted from the hg19 reference sequence using a bespoke python script, and mutation signatures were created for each tumor, as above. Deletion-to-insertion ratios and indel length distributions were tabulated for each tumor (Table S3). All variants were annotated with CADD, and raw scores were used to compare groups of samples, as recommended by the authors of that program.22 Samples with mutations in BRCA1 or mismatch repair (MMR) genes were excluded from all comparisons of mutation rates or characteristics. Instead, the 18 BRCA2-disrupted tumors were compared to the 128 samples without BRCA1, BRCA2, or MMR defects. The single BRCA1-mutated tumor, as well as the three MMR-deficient samples, were evaluated independently.
Statistical Methods
Hierarchical clustering was performed on the substitution signatures with the complete linkage method using R hclust package, which supported grouping the discovery set tumors by BRCA2 status for downstream analysis. For all comparisons of tumor subgroups, observed values were log transformed and p values were fitted with a logistic regression model using the R glm package.26 Two-by-two tables were evaluated for significant associations using Fisher’s exact test.
Pathways Analysis
Pathways analysis for the metastatic tumors was performed on a gene list comprising truncating sSNVs and indels plus genes affected by fusions using the online portal for the Gene Set Enrichment Analysis (GSEA) program.27 For each tumor, we tested for enrichment of mutated genes in canonical pathways using the list of genes with truncating mutations, in-frame indels, fusion genes, or missense changes that had a PHRED-scaled CADD score22 >20. BRCA2 mutations were masked to avoid bias related to the fact that these aberrations were present in all tumors in the HR-deficient group. All pathways with FDR q value < 0.05 were retained. For each pathway, we then compared the number of BRCA2-mutated tumors enriched for that pathway versus the number of tumors without BRCA1, BRCA2, or MMR mutations.
Results
To access the genomic landscape of aggressive, treatment-naive prostate tumors, we initially performed deep, paired-end tumor/normal whole-genome sequencing of nine fresh frozen prostatectomy samples with Gleason scores of 8–10 (MC1-9T), plus one nodal metastasis from a tenth individual (MC-10M), all from the Mayo Clinic (Tables 1 and S1). Mean tumor coverage was 95.3× and mean normal sample coverage was 48.3× (Tables 1 and S2). In each matched tumor and normal sample, we genotyped SNVs, indels, and SVs, and then used quality filters and a panel of normal samples to remove germline variants and systematic errors.
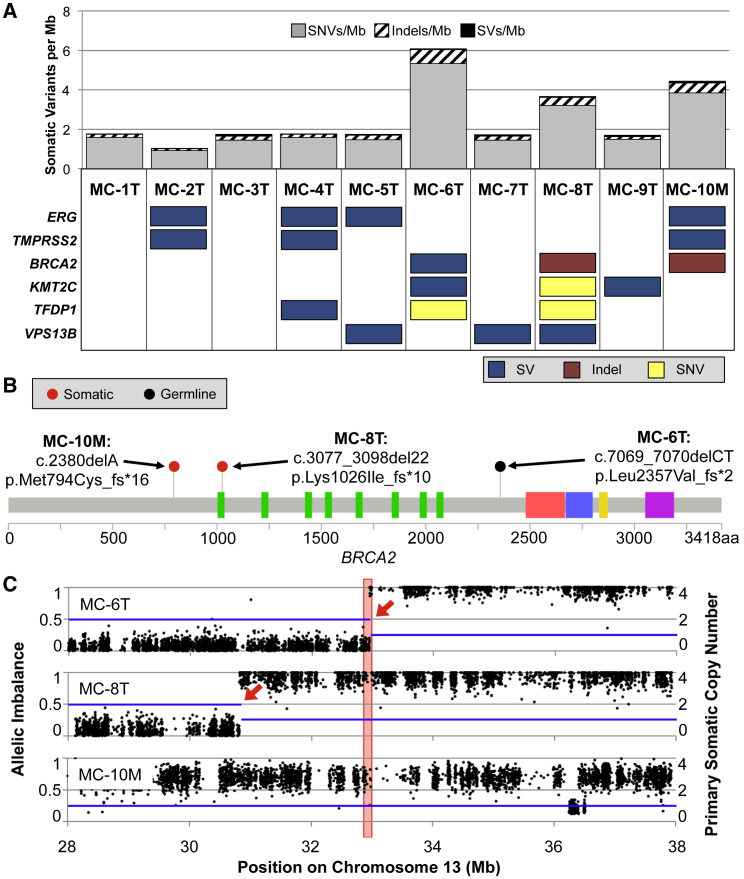
Inspection of somatic mutation counts revealed that three tumors (MC-6T, MC-8T, and MC-10M) had an elevated somatic mutation rate compared to the other samples in this discovery set (Figure 1A). For all variant types combined, these samples had a 2.88-fold greater mutation rate compared to the other seven aggressive prostate tumors, with means of 4.73 ± 0.55 per Mb versus 1.64 ± 0.27 per Mb, respectively (Figure 1A, p value = 4.74 × 10−4). Although this elevated mutation rate was significant for both somatic SNVs and small indels (Figure S1, p value = 4.74 × 10−4 for both), SVs did not contribute to this signal, either in aggregate (1.03-fold enrichment, p value = 0.818) or among any of the assessed SV subtypes, including large deletions, translocations, duplications, and inversions (somatic mutation metrics for all samples are in Table S3).
Figure 1.
Inherited or Acquired Mutations Combine to Cause Biallelic Loss of BRCA2 in Three of the Ten Discovery Set PCa Tumors
(A) Three samples had a significantly elevated mutation rate compared to the other seven tumors. Six genes harbored somatic mutations in three or more tumors. For this purpose, SVs were counted only when the breakpoints were within a gene and the orientation or integrity of at least one exon was interrupted. BRCA2 was the only gene that was mutated in all three tumors with an elevated mutation rate.
(B) Truncating indels, all confirmed via Sanger sequencing, were detected in each of the affected tumors. Nucleotide and protein changes are reported with respect to RefSeq accession numbers GenBank: NM_000059.3 and NP_000050.2, respectively.
(C) All three tumors also have copy loss at the locus. The blue line indicates somatic copy number, with SV breakpoints marked by red arrows. Black dots represent allelic imbalance for heterozygous dbSNP 137 SNVs that had variant allele fraction between 45% and 55% in the normal DNA. The red box indicates the position of BRCA2.
There were six genes that harbored somatic mutations in at least three tumors: ERG (MIM: 165080), TMPRSS2 (MIM: 602060), BRCA2, KMT2C (MIM: 606833), TFDP1 (MIM: 189902), and VPS13B (MIM: 607817) (Figure 1A). The majority of the variants in recurrently mutated genes were SVs, accounting for 14 of 19 mutations overall. In addition, the aggressive tumor discovery set had two samples with mutations in other genes that have been found to be recurrently mutated in PCa, including FOXA1 (MIM: 602294), PARK2 (MIM: 602544), PTEN (MIM: 601728), SPOP (MIM: 602650), and TP53 (MIM: 191170) (Table S4).
Discovery Set Samples with Biallelic BRCA2 Mutations Had a Distinctive Somatic Mutation Landscape
We then investigated whether the tumors with an elevated total mutation rate had mutations in the same gene or pathway and found that all three tumors had biallelic disruption of BRCA2 (Figure 1). One tumor (MC-6T) had one germline and one somatic mutation, whereas the other two (MC-8T and MC-10M) each had two deleterious somatic mutations. In all three, one SV and one indel combined to abolish both copies of BRCA2 (Figure 1). In two samples (MC-8T and the metastatic lesion MC-10M) we observed that complex structural rearrangements caused a single copy loss of BRCA2, and somatic frameshift deletions in exon 11 disrupted the other allele (Figures 1B and 1C). In MC-10M, follow-up Sanger sequencing showed that the somatic indel was present in the primary tumor, in addition to the metastatic lesion. MC-6T had a somatic 39.1-Mb deletion with a breakpoint between exons 21 and 22 that excised the last six exons of the gene, and also a pathogenic germline frameshift deletion in exon 14 (Figures 1B and 1C). These two truncating variants probably affect both chromosomes, as determined by 1000 Genomes-based phasing of a common SNP on the same sequencing reads as the germline indel, and hemizygous SNPs after the somatic deletion breakpoint (Figure S2 and Table S5). Purity-corrected allelic ratios of the frameshift indels, together with Patchwork24 analysis of both allelic imbalance and normalized tumor sequencing depth for SVs (Figure S3), indicated that biallelic BRCA2 loss was clonal in all three tumors.
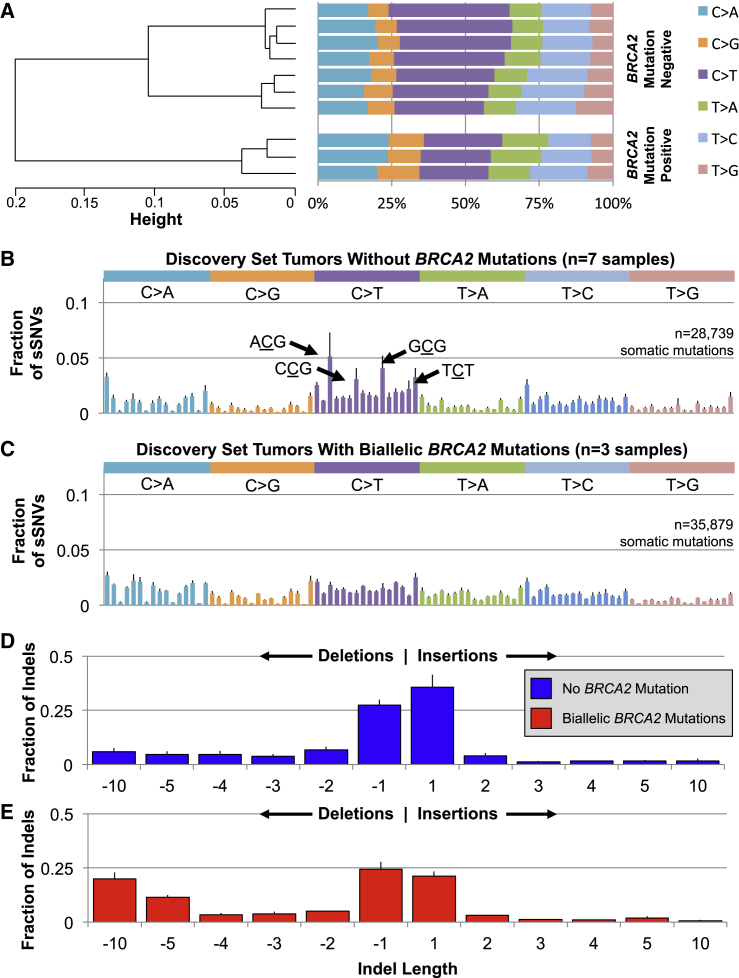
BRCA2-disrupted tumors not only harbored 2.91-fold more sSNVs (Figures 1A and S1A) but also exhibited a distinct somatic substitution profile. These three samples had a significantly lower transition to transversion ratio than the other tumors (p value = 4.74 × 10−4; Figure S1B). Indeed, the seven BRCA2-intact tumors were dominated by C>T transitions, which accounted for an average of 35.6% of all substitutions in these samples (Figures 2A and S4). Placed into their trinucleotide sequence context, these C>T substitutions were especially common in NpCpG triplets (Figure 2B). Tumors with biallelic BRCA2 mutations, in contrast, showed closer to equal representation of each substitution, with C>T mutations amounting to only an average of 24.5% of sSNVs (Figures 2A and S4). Within their trinucleotide sequence context, this lack of a single dominant substitution was further manifest as a flattened mutation signature, without the characteristic context-specific transitions seen in the other tumors (Figure 2C).
Figure 2.
In the BRCA2-Deficient Tumors, both sSNVs and Indels Had Distinctive Characteristics
(A) Tumors without biallelic BRCA2 mutations had nearly twice as many C>T sSNVs than any other substitution. C>T transitions represented a significantly smaller proportion of the substitutions in BRCA2-deficient tumors (Student’s t test p value = 0.0015).
(B) This signal is driven by a predilection in BRCA2-intact tumors for C>T substitutions within specific trinucleotide sequence contexts, particularly ACG, CCG, CCG, or TCT, where “C” is the substituted base.
(C) This pattern was absent in the three BRCA2-mutated tumors, which instead had closer to equal likelihood of each substitution and each trinucleotide sequence context.
(D) HR-competent tumors acquired somatic insertions and deletions at roughly the same rate, and these were overwhelmingly short, with 63.1% spanning a single base.
(E) In the setting of BRCA2 deficiency, deletions were more common than insertions and long deletions were common, accounting for more than 30% of all somatic indels in these tumors.
For all panels with error bars, these represent the standard deviation from the mean of all samples in that category.
Biallelic loss of BRCA2 was also associated with a 3.13-fold increase in the somatic indel rate (Figures 1A and S1C; p value = 4.74 × 10−4), and, like the sSNVs, the characteristics of these mutations were divergent from the other tumors. This elevated somatic indel rate was driven by both insertions (p value = 2.49 × 10−3) and deletions (p value = 4.74 × 10−4), but the magnitude of enrichment was greater for deletions (4.15-fold versus 1.96-fold, Table S3). Tumors without BRCA2 mutations acquired deletions and insertions at nearly the same rate, leading to a mean ratio of 1.18 × 0.22 (Figures 2D and S1D). In contrast, samples with biallelic BRCA2 loss had more than twice as many deletions as insertions, with a mean ratio of 2.43 ± 0.18 (Figures 2E and S1D, p value = 4.74 × 10−4). In addition, loss of BRCA2 yielded longer deletions (Figures 2D and 2E). On average, 28.2% of deletions spanned ten or more bases in these tumors, compared to only 10.8% in the remaining samples (Figures 2D, 2E, and S1E, p value = 4.74 × 10−4).
BRCA2-Deficient Tumors Were Outliers Compared to PCa Samples Spanning the Aggressiveness Spectrum
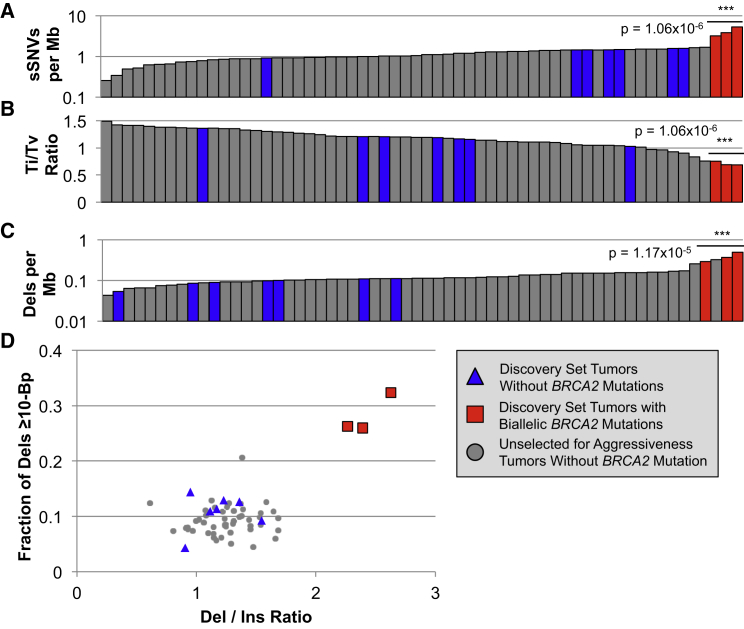
We next sought to place BRCA2 disruption-associated somatic mutation rate and patterns into the context of other prostate tumors from across the clinical aggressiveness spectrum. Using the same pipeline established for the discovery set of tumors, we regenotyped somatic variants in the WGS of 50 prostate tumors, which were made publically available from a recent publication by Baca et al.5 These tumors were treatment naive and were not specifically selected for aggressive disease with 35 Gleason ≤ 7, 11 Gleason 8–10, and 4 tumors without a published Gleason score. The average WGS sequencing depth was 61× for tumors and 34× for matched normal. Among these 50 additional tumors, none harbored monoallelic or biallelic BRCA2 mutations. Strikingly, when considered within the context of the discovery set tumors plus those that were unselected for aggressiveness, the three discovery set samples with biallelic BRCA2 mutations remained clear outliers by all measures that defined the distinctive BRCA2-deficient molecular subtype. In comparison with all tumors from both datasets, as well as only those with Gleason score of eight or greater, the BRCA2-deficient tumors had the three highest sSNV rates (Figure 3A, p value = 1.06 × 10−6) and also the three lowest transition-transversion ratios (Figure 3B, p value = 1.06 × 10−6). The divergent BRCA2-disrupted indel trends also persisted when placed into the context of all 60 tumors. The three BRCA2 mutation-carrying tumors had a significantly elevated deletion mutation rate (Figure 3C, p value = 1.17 × 10−5), in addition to being the only samples with a preponderance of both deletions over insertions plus enrichment for long deletions (Figure 3D). It is unlikely that systematic differences between the studies played any role in the persistence of these BRCA2 mutation-associated patterns, because the seven discovery set tumors without BRCA2 disruptions were largely indistinguishable from the samples that were unselected for aggressiveness (Figure 3, p values = 1.06 × 10−6 for both observations).
Figure 3.
The Somatic Mutation Patterns Found in the Three Discovery Set Tumors with Biallelic BRCA2 Loss Were Maintained when Compared to 50 Additional PCa Tumor WGS5 that Spanned the Range of Clinical Aggressiveness
Samples are color coded by BRCA2 status and study of origin, as indicated in the legend. As observed in the discovery set alone, biallelic BRCA2 loss was associated with (A) an elevated sSNV mutation rate, (B) a depressed transition-transversion ratio, and (C) enrichment for somatic deletions. Strikingly, the three BRCA2-deficient tumors were the only samples with elevations of both the deletion to insertion ratio and the fraction of deletions exceeding 10 bp (D).
Metastatic Tumors with Biallelic BRCA2 Mutations Recapitulated the Somatic Mutation Profile
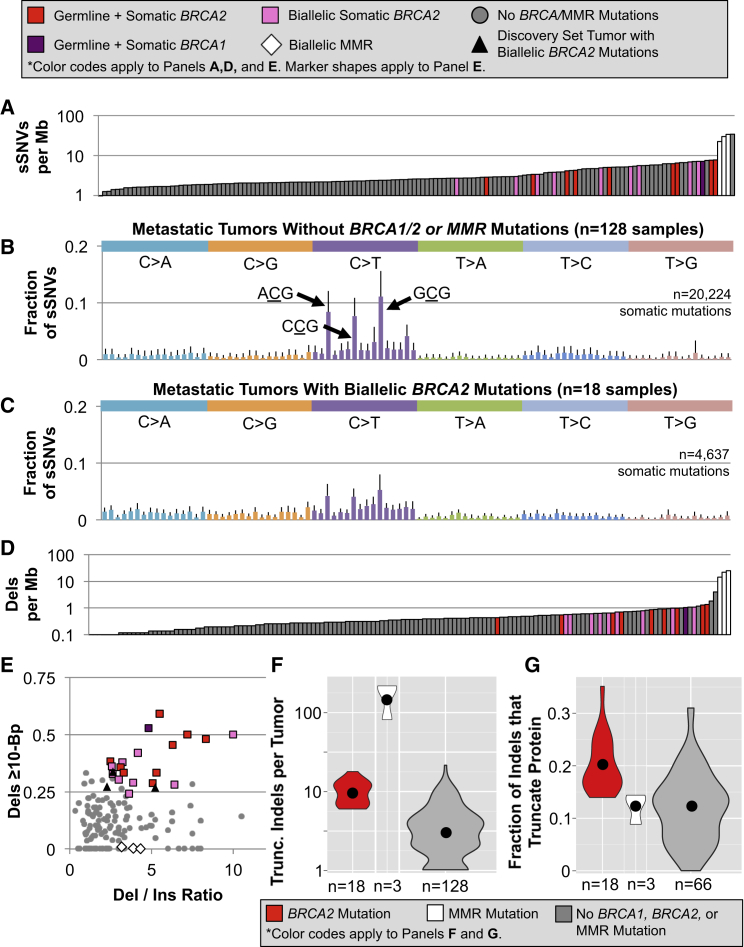
We next tested the reproducibility of the BRCA2-mutation-associated somatic genomic signature by performing a BRCA2 deficiency-targeted reanalysis of 150 metastatic tumors that were recently characterized by Robinson et al.8 Although the other two datasets contained metastatic tumors, the Robinson et al. dataset is subsequently referred to as “metastatic tumors” because of the selection criteria used for data collection. In deep WES and copy-number assessment of primary tumors and metastases from 18 metastatic PCa cases, 12.0% had biallelic mutations in BRCA2 and one tumor (0.7%) had biallelic loss of BRCA1. As in the BRCA2-mutated tumors from the discovery set, complex combinations of both germline and somatic lesions contributed to the loss of BRCA2: nine samples (6.0%) had both germline and somatic BRCA2 mutations, and nine tumors (6.0%) had purely somatic SNVs, indels, and SVs that combined to yield biallelic loss of BRCA2 (Table S6). The single sample with biallelic BRCA1 mutations also had one germline and one somatic mutation.
Like the BRCA2-mutated discovery set samples, we found that the metastatic tumors with BRCA2 mutations had a significantly elevated sSNV mutation rate (Figure 4A, p value = 1.24 × 10−5; somatic mutation metrics for all samples are in Table S3). Within the 18 BRCA2-deficient samples, we did not detect a significant difference in mutation rate between those with purely somatic biallelic mutations versus a combination of germline and somatic variants (Figure 4A, p value = 0.469). The single tumor with biallelic loss of BRCA1 had a similar sSNV rate to the BRCA2-mutated samples.
Figure 4.
Nearly Identical Somatic Mutation Signatures Were Detected in Our BRCA2 Deficiency-Targeted Reanalysis of 150 Metastatic Prostate Tumors,8 18 with Biallelic Loss of BRCA2, 1 with Biallelic Mutations in BRCA1, and 3 with Biallelic MMR Defects
(A) Germline and somatic defects shared a statistically indistinguishable propensity to cause a modest increase in the sSNV mutation rate.
(B) C>T substitutions were most frequent in metastatic HR-competent PCa, exhibiting even greater enrichment than observed in the discovery set tumors.
(C) In contrast, C>T transitions were not dramatically overrepresented in the BRCA2-deficient tumors. Much of the difference is derived from a paucity of substitutions within the triplets ACG, CCG, and GCG, which were characteristic in their HR-intact counterparts.
(D) As seen in these tumors among sSNVs, germline and somatic HR defects led to a similar rise in the indel mutation rate.
(E) Samples with biallelic HR deficiency had enrichment for both deletions over insertions and long deletions, recapitulating the trend observed in the discovery set tumors.
(F) The number of truncating indels was partially determined by differences in the somatic indel rate.
(G) BRCA2-mutated tumors had a higher percentage of indels that were predicted to truncate the encoded protein.
For all panels with error bars, these represent the standard deviation from the mean of all samples in that category.
Although the BRCA2-disrupted tumors had an elevated mutation rate compared to the average metastatic sample, it was notable that four tumors exhibited an even higher sSNV mutation rate. Three of these had biallelic somatic disruptions of MLH1 (MIM: 120436) or MSH2 (MIM: 609309), mismatch repair (MMR) genes that when mutated produce a classic hypermutator phenotype.28 Compared to these hypermutated MMR-deficient samples, the sSNV mutation rate in BRCA2-mutated tumors was significantly lower, with means of 28.72 ± 5.82 per Mb and 5.05 ± 1.70 per Mb, respectively (Figure 4A, p value = 3.32 × 10−5).
Upon examination of the somatic substitutions and their trinucleotide sequence contexts, the metastatic samples mirrored the overarching trends seen in their discovery set counterparts. The 128 tumors without BRCA1, BRCA2, or MMR mutations exhibited even more dramatic enrichment for C>T substitutions than was observed in the seven discovery set non-BRCA2 mutated tumors, accounting for 51.9% of all sSNVs, and these were especially common within the triplets ACG, CCG, and GCG (Figure 4B). In contrast, the 18 samples with biallelic BRCA2 mutations had a significantly lower proportion of C>T substitutions, accounting for just 37.4% (Figure 4C, p value = 8.90 × 10−9). It was also notable that the tumor with biallelic BRCA1 mutations (Figure S5A) exhibited a pattern that closely resembled the BRCA2 deficiency-associated pattern (Figure 4C). In contrast, the substitution context signature in MMR-mutated tumors (Figure S5B) was indistinguishable from the other 128 samples (Figure 4B).
To determine whether these disparate C>T substitution patterns influence the severity of the predicted effect on the encoded protein, we annotated coding sSNVs with CADD.22 Although BRCA2-deficient tumors had more mutations at all CADD scores spanning the severity spectrum (Figure S6A), we observed no enrichment of highly damaging mutations after adjusting for the mutation rate (Figure S6B).
The indels identified in the BRCA2-disrupted metastatic tumors also recapitulated the molecular signature observed in the discovery set BRCA2-mutated tumors. In this dataset, small deletions occurred at a significantly higher rate in BRCA2-mutated samples than in the BRCA1-, BRCA2-, and MMR-intact tumors (Figure 4D, p value = 8.38 × 10−8). Similar to the sSNVs, the sample with biallelic BRCA1 mutations had an indel mutation rate that was consistent with the BRCA2-deficient tumors. However, the BRCA2-mutated deletion rate was 26.3-fold lower than in the three tumors with biallelic MMR lesions (Figure 4D, p value = 3.32 × 10−5). As we found for the sSNV mutation rate, germline versus somatic origins of BRCA2 defects did not significantly impact the somatic deletion rate (Figure 4D, p value = 0.531). Although there were other tumors in the metastatic dataset that had either an elevated deletion to insertion ratio or enrichment of long deletions, the 18 BRCA2- and 1 BRCA1-mutated tumors were the only samples that were outliers for both characteristics (Figure 4E, p values = 5.17 × 10−5 and < 2.2 × 10−6, respectively). The MMR-deficient tumors were similar to one another, with an elevated deletion to insertion ratio, but in contrast to the BRCA2-mutated samples, they were dominated by very short deletions.
To determine whether biallelic BRCA2 mutation status influenced the predicted severity of indels, we compared the number and rate of truncating indels in BRCA2-mutated samples against the other tumors. The MMR- and BRCA2-deficient metastatic tumors had a higher mean number of truncating indels per sample than the other tumors (Figure 4F). Although this was partially explained by the elevated mutation rate, further inspection of the 87 tumors with at least 25 somatic indels revealed that the BRCA2-disrupted tumors (p value = 4.94 × 10−4), but not the MMR-mutated samples, had a higher mean proportion of variants that were predicted to truncate the protein (Figure 4G).
Pathways Analysis Highlights the Unique Evolution of BRCA2-Mutated Tumors
In the metastatic tumor dataset, biallelic BRCA2 deficiency was also associated with somatic mutation patterns in other genes and pathways (Figure S7). Somatic PTEN mutations were significantly underrepresented in BRCA2-mutated tumors, as they were found in 44.5% of samples without BRCA1, BRCA2, or MMR aberrations, but only 16.7% of BRCA2-deficient samples (OR = 0.25, p value = 0.020). Other recurrently mutated genes, APC (MIM: 611731), RB1 (MIM: 614041), and ATM (MIM: 607585), were also underrepresented to a lesser extent in tumors without BRCA2 defects, though the mutation rates did not meet the significance threshold for these genes. Notably, ETS fusions, damaging mutations to TP53, and AR (MIM: 313700) activating alterations occurred at approximately the same rate, irrespective of BRCA2 status (OR = 0.71, 1.21, and 0.94, respectively). PVT1 (MIM: 165140) (OR = 8.86, p value = 0.0085) and OPHN1 (MIM: 300127) (OR = 4.29, p value = 0.036) mutations were more common in the setting of BRCA2 deficiency. CHD1 (MIM: 602118) and KMT2C were also enriched in that scenario, with ORs of 4.07 and 3.46, respectively.
Gene Set Enrichment Analysis27 for each sample did not point to a single pathway driving difference beyond BRCA2 mutations. However, tumors without BRCA1, BRCA2, or MMR mutations were enriched for mutations in canonical cancer driver pathways, including those related to telomerase, TP53, RB, the cell cycle, and receptor tyrosine kinases (Table S7). The KEGG pathway database includes gene sets for multiple specific cancers, with each set comprised of numerous, often overlapping oncogenic drivers. These malignancy-specific pathways were frequently implicated in samples without BRCA1, BRCA2, or MMR deficiency, with 53/128 (41.4%) samples significantly enriched for at least one KEGG cancer pathway, compared to only 4/18 (22.2%) of BRCA2-mutated tumors (OR = 0.40, 95% CI = 0.13–1.29, p value = 0.128) (Table S7). Focal adhesion, collagen formation, integrins, and other cell-cell interaction networks were among the pathways that were more commonly enriched in BRCA2-deficient samples (Table S7).
Discussion
Our initial WGS of ten high-Gleason-grade tumors from the Mayo Clinic revealed three samples with biallelic BRCA2 mutations, and these lesions were associated with a highly specific somatic mutation signature. When compared with the remaining samples, the BRCA2-deficient tumors had a modest elevation in the somatic mutation rate, a reduction in the rate of C>T substitutions, and an increased rate of long somatic deletions. The somatic SNV and indel mutation signature remained specific to BRCA2-deficient tumors when compared to 50 additional PCa tumors across the tumor aggressiveness spectrum. Furthermore, the mutation pattern was reproducible in our BRCA2 deficiency-targeted reanalysis of 150 metastatic tumors, where 18 tumors with biallelic BRCA2 loss and a single tumor with biallelic BRCA1 mutations mirrored the somatic mutation signature observed in the discovery set of BRCA2-mutated tumors, but diverged from the 128 tumors without loss of BRCA1, BRCA2, or MMR. The PCa BRCA2-deficient mutation signature closely recapitulated that found in breast, ovarian, pancreatic, and gastric cancers with mutations in BRCA1 or BRCA2.11, 12, 13, 14, 15 The pattern was seen in all PCa samples with biallelic deleterious mutations in the gene, which suggests that this mutational process is a key driver of somatic mutations in affected tumors and defines a clinically important molecular subtype of PCa, irrespective of primary versus metastatic origins.
In a recent characterization of the trinucleotide substitution profile across 36 cancer types, including 520 PCa tumors, Alexandrov et al. found no prostate tumors with the flattened mutation signature associated with BRCA2 deficiency,14 leading the authors to conclude that biallelic HR mutations do not play an important role in PCa. However, we speculate that the 520 tumors were unselected for disease aggressiveness, meaning only a small portion are likely to be Gleason 8–10. Our study uncovered no HR mutations in 39 non-aggressive tumors, but, in marked contrast, biallelic loss and the associated mutation signature were present in 22/171 (12.9%) of aggressive and metastatic tumors in our study, suggesting that loss of BRCA2, and possibly BRCA1, can play a crucial driver role in these more clinically important tumors.
The observed mutation signature has been attributed to the fact that HR-deficient cells cannot engage in double strand break repair (DSBR) via the high-fidelity HR pathway, but instead rely on error-prone strategies. Non-homologous end joining (NHEJ) is one alternative repair mechanism that leverages local microhomology near the breakpoints to reapproximate the free DNA ends, followed by excision of the overlapping microhomologous DNA.29 Because the length of sequence identity determines the number of bases deleted, DSBR via NHEJ probably explains why HR-deficient breast and prostate tumors are dominated by long deletions.11 Though BRCA2 participates in many aspects of the DNA damage response, reliance upon non-HR mechanisms for DSBR might also contribute to the characteristic sSNV profile, because alternative DSBR pathways induce mutations via error-prone religation and gap-filling strategies.30 One possibility is that the more equal representation of each substitution observed in HR-deficient samples probably reflects the fact that DSBR-related substitutions arise as a function of their proximity to the DNA break, as opposed to the context-specific substitution pattern found in other prostate tumors, where the recurrent C>T transitions probably arose from deamination of 5-methyl-cytosine at XpCpG trinucleotides.
In the PCa tumors analyzed in our study, HR-inactivating mutations exert a prominent effect on the mutational landscape of prostate tumors, irrespective of whether BRCA2 disruptions are inherited or purely somatic. There was no apparent difference in the somatic mutation rates (Figures 4A and 4D) or the nature of sSNVs and indels (Figures 4B, 4C, and 4E). The equivalent effects of germline and somatic BRCA2 defects, paired with the highly similar somatic mutation profiles of BRCA2-deficient tumors from multiple tumor types, underscore the importance of BRCA2 deficiency as a driver of somatic mutagenesis. Thus, BRCA2 defects drive tumor evolution without regard for the tissue type or origins of the mutation, and both germline and somatic mutations appear to represent important molecular biomarkers in PCa.
Our analysis supports previous observations that PCa in subjects with germline BRCA2 risk alleles leads to a higher rate of tumor aggressiveness, rapid progression, and poor outcomes.31, 32, 33 Inherited BRCA2 alleles have also been linked to a higher attributable risk in early-onset cases and high-risk PCa families.34, 35, 36 Across all the samples assessed in this study, 10/171 (5.8%) of subjects with aggressive tumors had a germline BRCA2 mutation. Our findings indicate that truncating germline alleles are somewhat more common in aggressive primary tumors with Gleason scores ≥8 (1/21 samples, 4.6%), and especially in metastatic cases (9/150, 6.0%), compared to case subjects from the general population.37, 38 The fact that our study design explicitly targeted tumors with aggressive clinical behavior probably underlies our observed enrichment of germline BRCA2-truncating alleles, given the increased risk of disease and aggressiveness associated with these mutations. Furthermore, the stronger association between both PCa risk and aggressiveness for BRCA2 compared to BRCA139 might explain our finding that BRCA2 was mutated more often than BRCA1 in aggressive and metastatic PCa. Our findings bolster targeted screening paradigms suggested previously, including prostate cancer screening in carriers of germline BRCA2 risk alleles40 as well as BRCA2 screening for men diagnosed with aggressive or metastatic PCa,8 particularly when there is a family history of BRCA2-associated malignancies.
Historically, oncogenesis in BRCA2-deficient cells has largely been attributed to genomic rearrangements that result from incompetent HR,41, 42, 43 a hypothesis supported by observations that non-PCa tumors that are HR deficient have more SVs than other tumors of the same type.15, 44 In our analysis of PCa tumors, however, BRCA2-mutated tumors did not have significantly more somatic SVs than other tumors, perhaps due to the fact that PCa tumors spanning the aggressiveness spectrum have a high SV burden, which has been suggested as a key driver of prostate tumor evolution.5 Taken together, other genomic derangements resulting from HR deficiency must contribute to the more aggressive phenotype observed in BRCA2-disrupted tumors. We found comparatively more sSNVs in tumors with biallelic loss of BRCA2, but mutations resulting from the distinctive HR-deficient substitution profile were not more damaging than those that occur in the other 128 tumors (Figure S6). For indels, we observed both a higher mutation rate in BRCA2-deficient samples and found that those indels are more likely to cause predicted protein truncations (Figures 4F and 4G), probably due to the fact that the longer deletions in these tumors have a greater chance of overlapping key genomic features. Based on these findings, BRCA2 loss can contribute to tumor aggressiveness via an increased sSNV and indel mutation rate, as well as the enhanced deleteriousness of the characteristic long deletions, and not necessarily via the genomic instability that has been suggested as an oncogenic mechanism in other tumors.
Somatic mutation patterns in BRCA2-mutated versus other samples suggest that HR defects influence PCa tumor evolution via different mechanisms than other cancers. PTEN disruptions were common in metastatic samples without BRCA2 mutations (57/128, 44.5%), but not in samples with biallelic BRCA2 loss (3/18, 16.7%). This pattern differs from HR-deficient breast and ovarian cancers, where depletion of the PTEN protein is thought to play an important role in tumor development.45, 46, 47 BRCA2-deficient tumors also exhibit an aggressive clinical phenotype, in spite of the fact that somatic PTEN loss has been suggested as a key mediator of prostate tumor progression, especially in conjunction with ETS fusions48 or suppression of the JNK pathway.49 Although TP53 is thought to play a key role in oncogenesis in breast and ovarian cancers with HR defects,47, 50 TP53 mutations were present in only 10/18 (55.6%) of the BRCA2-deficient samples in our study. Our data indicate that PCa tumors with BRCA2 defects proceed through oncogenesis and progression via different mechanisms than other BRCA1- or BRCA2-associated malignancies. Instead, PVT1, CDH1, or other genes that are more frequently mutated in samples with BRCA2 loss might influence evolution of PCa tumors with BRCA2 defects. Larger studies of HR-deficient tumors would enable further dissection of interactions between these genes in PCa.
Each example of biallelic BRCA2 loss in our study resulted from a complex combination of variants, including 10/21 subjects with germline truncating alleles, so any effort to identify clinically actionable variants in PCa tumors must be sensitive to all variant types, germline and somatic. In the 21 deficient tumors across the discovery set and metastatic PCa studies, sSNVs represented just 7.1% (3/42) of the damaging alleles, whereas indels were 40.5% (17/42) of the total and SVs accounted for the remaining 52.4% (22/42). The overwhelming contribution of difficult-to-detect variant types, paired with the fact that tumor sequencing studies often do not systematically assess germline alleles, might underlie the low reported frequency of BRCA2 mutations in earlier PCa sequencing studies, including those that explicitly targeted more aggressive tumors.3, 4, 5, 6, 7, 8, 9, 10
Our analysis emphasizes that in order to leverage BRCA2 status as a clinical biomarker in PCa, both germline and somatic mutations, as well as all types of deleterious mutations, must be considered. Given the mutation types identified in our analysis, genotyping with multiple approaches would probably be necessary to achieve the highest possible sensitivity for clinically relevant mutations in PCa. In the near term, economical detection of BRCA2 mutation status could be accomplished via exon sequencing plus regional copy-number detection, though this approach might miss balanced structural rearrangements, epigenetic silencing, or disruptions elsewhere in the HR pathway. As sequencing-based diagnostics are assimilated into clinical testing protocols, incorporation of additional genome-wide metrics like the mutational signature demonstrated here (Figure 4) could yield more sensitive detection of HR defects. Future studies are needed in order to determine which approach most accurately identifies tumors with HR defects. Indeed, the recent PCa clinical trial of a PARP inhibitor showed dramatic response rates in tumors with defects spanning the HR pathway, concluding that targeted sequencing is needed for all DNA-repair pathway genes.51 However, given the difficulty in identifying all possible HR-disrupting somatic genomic aberrations, the molecular signature might provide the most comprehensive way to identify this specific subgroup of PCa tumors.
Notably, the three MMR-deficient tumors in the metastatic tumor set also had a dramatic, distinguishing hypermutation phenotype (Figure 4), which has been described previously in prostate tumors and other cancers with MMR inactivation.6, 8, 28, 52 In contrast to BRCA2-deficient tumors, the MMR-deficient tumors exhibited an elevated deletion-to-insertion ratio with an enrichment of short deletions. The MMR tumors also had a consistent C to T sSNV substitution pattern that mirrored the overarching trend among HR-competent prostate tumors in our study, as well as MMR-deficient tumors from multiple tissues of origin in a pan-cancer analysis.12 In prostate tumors, these genes are often inactivated via complex genomic rearrangements,52 lesions that can be difficult to detect with WES. Thus, the somatic hypermutation with characteristic indel and sSNV signatures might be useful for identifying MMR-deficient tumors as clinical sequencing becomes more widely adopted.
Biallelic BRCA2 mutations were relatively common in aggressive and metastatic PCa tumors. Defects in this gene are a potent driver of mutagenic processes in tumors with germline or somatic variants, defining a unique molecular subtype of PCa. BRCA2-deficient PCa tumors do not have more SVs than other tumors, suggesting that the aggressive nature of these cancers is not solely due to structural rearrangements. Instead, our data suggest that the higher rate of sSNVs and indels, as well as the increased deleteriousness of the indels, might contribute substantially to tumor evolution and progression in BRCA2-deficient PCa tumors. Our analysis also highlights the need to evaluate all variant types for both germline and somatic mutations in BRCA2, because half of the deficient tumors have somatic-only disruptions. Finally, our work strongly implicates BRCA2 status and the HR-deficiency-associated molecular signature as potentially important prognostic and treatment selection biomarkers in aggressive and metastatic PCa.
Acknowledgments
This work was supported by Intramural Research Program of the National Human Genome Research Institute (E.A.O., B.D., D.M.K., B.W.D., E.K.), Prostate Cancer SPORE Grant # CA17205 (S.N.T., L.S.T.), and the NIH-Cambridge Scholars Program (B.D.). We thank the Mayo Clinic Pathology Core for sample processing. This work utilized the NIH Biowulf high-performance computing cluster. Finally, we thank the many men who provided samples for this study.
Published: April 14, 2016
Footnotes
Supplemental Data include seven figures and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.03.003.
Accession Numbers
Sequencing data for the Mayo Clinic discovery set tumors are deposited in dbGAP under accession number phs001105.v1.p1.
Web Resources
The URLs for data presented herein are as follows:
Supplemental Data
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2015. CA Cancer J. Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis C.E., Lin C.C., Mariotto A.B., Siegel R.L., Stein K.D., Kramer J.L., Alteri R., Robbins A.S., Jemal A. Cancer treatment and survivorship statistics, 2014. CA Cancer J. Clin. 2014;64:252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 3.Grasso C.S., Wu Y.-M., Robinson D.R., Cao X., Dhanasekaran S.M., Khan A.P., Quist M.J., Jing X., Lonigro R.J., Brenner J.C. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbieri C.E., Baca S.C., Lawrence M.S., Demichelis F., Blattner M., Theurillat J.-P., White T.A., Stojanov P., Van Allen E., Stransky N. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baca S.C., Prandi D., Lawrence M.S., Mosquera J.M., Romanel A., Drier Y., Park K., Kitabayashi N., MacDonald T.Y., Ghandi M. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar A., White T.A., MacKenzie A.P., Clegg N., Lee C., Dumpit R.F., Coleman I., Ng S.B., Salipante S.J., Rieder M.J. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proc. Natl. Acad. Sci. USA. 2011;108:17087–17092. doi: 10.1073/pnas.1108745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger M.F., Lawrence M.S., Demichelis F., Drier Y., Cibulskis K., Sivachenko A.Y., Sboner A., Esgueva R., Pflueger D., Sougnez C. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson D., Van Allen E.M., Wu Y.-M., Schultz N., Lonigro R.J., Mosquera J.-M., Montgomery B., Taplin M.-E., Pritchard C.C., Attard G. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beltran H., Yelensky R., Frampton G.M., Park K., Downing S.R., MacDonald T.Y., Jarosz M., Lipson D., Tagawa S.T., Nanus D.M. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur. Urol. 2013;63:920–926. doi: 10.1016/j.eururo.2012.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abeshouse A., Ahn J., Akbani R., Ally A., Amin S., Andry C.D., Annala M., Aprikian A., Armenia J., Arora A., Cancer Genome Atlas Research Network The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nik-Zainal S., Alexandrov L.B., Wedge D.C., Van Loo P., Greenman C.D., Raine K., Jones D., Hinton J., Marshall J., Stebbings L.A., Breast Cancer Working Group of the International Cancer Genome Consortium Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A.J.R., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.-L., Australian Pancreatic Cancer Genome Initiative. ICGC Breast Cancer Consortium. ICGC MMML-Seq Consortium. ICGC PedBrain Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang D., Khan S., Sun Y., Hess K., Shmulevich I., Sood A.K., Zhang W. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. JAMA. 2011;306:1557–1565. doi: 10.1001/jama.2011.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexandrov L.B., Nik-Zainal S., Siu H.C., Leung S.Y., Stratton M.R. A mutational signature in gastric cancer suggests therapeutic strategies. Nat. Commun. 2015;6:8683. doi: 10.1038/ncomms9683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waddell N., Pajic M., Patch A.-M., Chang D.K., Kassahn K.S., Bailey P., Johns A.L., Miller D., Nones K., Quek K., Australian Pancreatic Cancer Genome Initiative Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patch A.-M., Christie E.L., Etemadmoghadam D., Garsed D.W., George J., Fereday S., Nones K., Cowin P., Alsop K., Bailey P.J., Australian Ovarian Cancer Study Group Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–494. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 17.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLaren W., Pritchard B., Rios D., Chen Y., Flicek P., Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 24.Mayrhofer M., DiLorenzo S., Isaksson A. Patchwork: allele-specific copy number analysis of whole-genome sequenced tumor tissue. Genome Biol. 2013;14:R24. doi: 10.1186/gb-2013-14-3-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rausch T., Zichner T., Schlattl A., Stütz A.M., Benes V., Korbel J.O. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R Team . R Foundation for Statistical Computing; Vienna: 2013. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 27.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunter C., Smith R., Cahill D.P., Stephens P., Stevens C., Teague J., Greenman C., Edkins S., Bignell G., Davies H. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66:3987–3991. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore J.K., Haber J.E. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malkova A., Haber J.E. Mutations arising during repair of chromosome breaks. Annu. Rev. Genet. 2012;46:455–473. doi: 10.1146/annurev-genet-110711-155547. [DOI] [PubMed] [Google Scholar]
- 31.Narod S.A., Neuhausen S., Vichodez G., Armel S., Lynch H.T., Ghadirian P., Cummings S., Olopade O., Stoppa-Lyonnet D., Couch F., Hereditary Breast Cancer Study Group Rapid progression of prostate cancer in men with a BRCA2 mutation. Br. J. Cancer. 2008;99:371–374. doi: 10.1038/sj.bjc.6604453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitra A., Fisher C., Foster C.S., Jameson C., Barbachanno Y., Bartlett J., Bancroft E., Doherty R., Kote-Jarai Z., Peock S., IMPACT and EMBRACE Collaborators Prostate cancer in male BRCA1 and BRCA2 mutation carriers has a more aggressive phenotype. Br. J. Cancer. 2008;98:502–507. doi: 10.1038/sj.bjc.6604132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castro E., Goh C., Leongamornlert D., Saunders E., Tymrakiewicz M., Dadaev T., Govindasami K., Guy M., Ellis S., Frost D. Effect of BRCA mutations on metastatic relapse and cause-specific survival after radical treatment for localised prostate cancer. Eur. Urol. 2015;68:186–193. doi: 10.1016/j.eururo.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 34.Breast Cancer Linkage Consortium Cancer risks in BRCA2 mutation carriers. J. Natl. Cancer Inst. 1999;91:1310–1316. doi: 10.1093/jnci/91.15.1310. [DOI] [PubMed] [Google Scholar]
- 35.Gayther S.A., de Foy K.A., Harrington P., Pharoah P., Dunsmuir W.D., Edwards S.M., Gillett C., Ardern-Jones A., Dearnaley D.P., Easton D.F., The Cancer Research Campaign/British Prostate Group United Kingdom Familial Prostate Cancer Study Collaborators The frequency of germ-line mutations in the breast cancer predisposition genes BRCA1 and BRCA2 in familial prostate cancer. Cancer Res. 2000;60:4513–4518. [PubMed] [Google Scholar]
- 36.Kote-Jarai Z., Leongamornlert D., Saunders E., Tymrakiewicz M., Castro E., Mahmud N., Guy M., Edwards S., O’Brien L., Sawyer E., UKGPCS Collaborators BRCA2 is a moderate penetrance gene contributing to young-onset prostate cancer: implications for genetic testing in prostate cancer patients. Br. J. Cancer. 2011;105:1230–1234. doi: 10.1038/bjc.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agalliu I., Karlins E., Kwon E.M., Iwasaki L.M., Diamond A., Ostrander E.A., Stanford J.L. Rare germline mutations in the BRCA2 gene are associated with early-onset prostate cancer. Br. J. Cancer. 2007;97:826–831. doi: 10.1038/sj.bjc.6603929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sinclair C.S., Berry R., Schaid D., Thibodeau S.N., Couch F.J. BRCA1 and BRCA2 have a limited role in familial prostate cancer. Cancer Res. 2000;60:1371–1375. [PubMed] [Google Scholar]
- 39.Attard G., Parker C., Eeles R.A., Schröder F., Tomlins S.A., Tannock I., Drake C.G., de Bono J.S. Prostate cancer. Lancet. 2016;387:70–82. doi: 10.1016/S0140-6736(14)61947-4. [DOI] [PubMed] [Google Scholar]
- 40.Bancroft E.K., Page E.C., Castro E., Lilja H., Vickers A., Sjoberg D., Assel M., Foster C.S., Mitchell G., Drew K., IMPACT Collaborators Targeted prostate cancer screening in BRCA1 and BRCA2 mutation carriers: results from the initial screening round of the IMPACT study. Eur. Urol. 2014;66:489–499. doi: 10.1016/j.eururo.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia F., Taghian D.G., DeFrank J.S., Zeng Z.C., Willers H., Iliakis G., Powell S.N. Deficiency of human BRCA2 leads to impaired homologous recombination but maintains normal nonhomologous end joining. Proc. Natl. Acad. Sci. USA. 2001;98:8644–8649. doi: 10.1073/pnas.151253498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu V.P., Koehler M., Steinlein C., Schmid M., Hanakahi L.A., van Gool A.J., West S.C., Venkitaraman A.R. Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 2000;14:1400–1406. [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng L., Li S., Boyer T.G., Lee W.H. Lessons learned from BRCA1 and BRCA2. Oncogene. 2000;19:6159–6175. doi: 10.1038/sj.onc.1203968. [DOI] [PubMed] [Google Scholar]
- 44.Tutt A., Gabriel A., Bertwistle D., Connor F., Paterson H., Peacock J., Ross G., Ashworth A. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr. Biol. 1999;9:1107–1110. doi: 10.1016/s0960-9822(99)80479-5. [DOI] [PubMed] [Google Scholar]
- 45.Press J.Z., De Luca A., Boyd N., Young S., Troussard A., Ridge Y., Kaurah P., Kalloger S.E., Blood K.A., Smith M. Ovarian carcinomas with genetic and epigenetic BRCA1 loss have distinct molecular abnormalities. BMC Cancer. 2008;8:17. doi: 10.1186/1471-2407-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saal L.H., Gruvberger-Saal S.K., Persson C., Lövgren K., Jumppanen M., Staaf J., Jönsson G., Pires M.M., Maurer M., Holm K. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat. Genet. 2008;40:102–107. doi: 10.1038/ng.2007.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martins F.C., De S., Almendro V., Gönen M., Park S.Y., Blum J.L., Herlihy W., Ethington G., Schnitt S.J., Tung N. Evolutionary pathways in BRCA1-associated breast tumors. Cancer Discov. 2012;2:503–511. doi: 10.1158/2159-8290.CD-11-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carver B.S., Tran J., Gopalan A., Chen Z., Shaikh S., Carracedo A., Alimonti A., Nardella C., Varmeh S., Scardino P.T. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat. Genet. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hübner A., Mulholland D.J., Standen C.L., Karasarides M., Cavanagh-Kyros J., Barrett T., Chi H., Greiner D.L., Tournier C., Sawyers C.L. JNK and PTEN cooperatively control the development of invasive adenocarcinoma of the prostate. Proc. Natl. Acad. Sci. USA. 2012;109:12046–12051. doi: 10.1073/pnas.1209660109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jonkers J., Meuwissen R., van der Gulden H., Peterse H., van der Valk M., Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 51.Mateo J., Carreira S., Sandhu S., Miranda S., Mossop H., Perez-Lopez R., Nava Rodrigues D., Robinson D., Omlin A., Tunariu N. DNA-repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pritchard C.C., Morrissey C., Kumar A., Zhang X., Smith C., Coleman I., Salipante S.J., Milbank J., Yu M., Grady W.M. Complex MSH2 and MSH6 mutations in hypermutated microsatellite unstable advanced prostate cancer. Nat. Commun. 2014;5:4988. doi: 10.1038/ncomms5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.