Abstract
Methylmalonic acid (MMA) is a by-product of propionic acid metabolism through the vitamin B12 (cobalamin)-dependent enzyme methylmalonyl CoA mutase. Elevated MMA concentrations are a hallmark of several inborn errors of metabolism and indicators of cobalamin deficiency in older persons. In a genome-wide analysis of 2,210 healthy young Irish adults (median age 22 years) we identified a strong association of plasma MMA with SNPs in 3-hydroxyisobutyryl-CoA hydrolase (HIBCH, p = 8.42 × 10−89) and acyl-CoA synthetase family member 3 (ACSF3, p = 3.48 × 10−19). These loci accounted for 12% of the variance in MMA concentration. The most strongly associated SNP (HIBCH rs291466; c:2T>C) causes a missense change of the initiator methionine codon (minor-allele frequency = 0.43) to threonine. Surprisingly, the resulting variant, p.Met1?, is associated with increased expression of HIBCH mRNA and encoded protein. These homozygotes had, on average, 46% higher MMA concentrations than methionine-encoding homozygotes in young adults with generally low MMA concentrations (0.17 [0.14–0.21] μmol/L; median [25th–75th quartile]). The association between MMA levels and HIBCH rs291466 was highly significant in a replication cohort of 1,481 older individuals (median age 79 years) with elevated plasma MMA concentrations (0.34 [0.24–0.51] μmol/L; p = 4.0 × 10−26). In a longitudinal study of 185 pregnant women and their newborns, the association of this SNP remained significant across the gestational trimesters and in newborns. HIBCH is unique to valine catabolism. Studies evaluating flux through the valine catabolic pathway in humans should account for these variants. Furthermore, this SNP could help resolve equivocal clinical tests where plasma MMA values have been used to diagnose cobalamin deficiency.
Introduction
Methylmalonic acid (MMA) is a by-product of the reaction catalyzed by methylmalonylCoA mutase (MUT), one of two enzymes in mammals that use vitamin B12 (cobalamin) as a cofactor.1 This mitochondrial enzyme plays an important role in the metabolism of odd-chain fatty acids and the branched-chain amino acids valine, leucine, and isoleucine. Catabolism of these molecules produces methylmalonylCoA which is converted to succinylCoA via MUT activity. If MUT does not function effectively, the MMA concentration in blood increases. Rare high-penetrance mutations in MUT (MIM: 609058) or in enzymes affecting the transport or synthesis of 5′adenosylcobalamin cause a number of inborn errors of metabolism characterized by methylmalonic acidemia.2 However, there are currently no data to suggest that genetic variation in MUT determines plasma MMA levels outside of rare inborn errors of metabolism. Furthermore, there are no data on the relative contributions of branched-chain amino acid or fatty acid oxidation pathways to circulating MMA concentrations in healthy individuals. Transient or persistently elevated circulating MMA levels are observed in individuals with severe mutations in ALDH6A1 (MIM: 603178), which encodes methylmalonate semialdehyde dehydrogenase,3 and ACSF3 (MIM: 614245), which encodes a mitochondrial AcylCoA synthetase considered to have malonylCoA and methylmalonyl CoA activities,4, 5 and in mitochondrial depletion diseases such as succinate Co-A ligase deficiency due to mutations of mitochondrial SUCLG1 (MIM: 611224).6
In contrast to diseases of newborns that result in elevated MMA concentration, cobalamin deficiency is relatively common among the elderly; estimates of deficiency range from 6% to over 40% in this population.7, 8, 9, 10, 11 Untreated cobalamin deficiency can lead to megaloblastic anemia or irreversible neurological damage.12 The MUT enzyme is extremely sensitive to cobalamin deficiency, and circulating MMA concentration is regarded as a sensitive and specific marker of cobalamin status, such that plasma concentrations within the normal reference range (<0.27 μmol/L) exclude cobalamin deficiency and concentrations >0.5 μmol/L almost invariably indicate deficiency.13, 14 However, MMA concentration is influenced by age and renal impairment.15, 16, 17 Many older persons have marginally elevated MMA concentrations in the range of 0.27–0.37μmol/L.18 This elevation is not associated with the status of other cobalamin biomarkers, such as the serum total cobalamin or plasma homocysteine. Circulating MMA concentrations in excess of 0.37 μmol/L are used to define inadequate cobalamin status;14, 19, 20, 21 nevertheless, in one study of older subjects, known determinants of MMA levels (age, plasma creatinine, and plasma total cobalamin) accounted for less than 17% of the variation in this trait.16 Several reports have demonstrated transiently high MMA concentration in plasma and urine of neonates up to 6 months old,22 but it remains unclear whether this is related to low cobalamin status or whether other processes involved in MMA handling that are independent of vitamin B12 function are not fully developed.23 Identification of genetic factors that influence circulating MMA concentration in healthy individuals would greatly help in interpreting MMA levels that are elevated but of unknown clinical significance.
To identify loci that influence the plasma MMA concentration in healthy individuals, we conducted a genome-wide association study (GWAS) in a cohort of healthy young Irish adults aged between 18 and 28 years. Our study of this ethnically and environmentally homogeneous group identified variants that were also associated with circulating MMA levels in two Irish replication cohorts (older adults and pregnant women). We present evidence that the functional variant underlying the strongest association signal acts at the level of transcription.
Material and Methods
Recruitment of the Subjects for the GWAS
The Trinity Student Study (TSS) population has been described previously.24, 25, 26 The cohort was established over one academic year (2003–2004) for the purpose of exploring the heritability of quantitative traits. A total of 2,524 eligible students attending the University of Dublin, Trinity College (TCD) were invited to participate in the study. Eligibility criteria included being between 18 and 28 years old, having no current serious medical condition, and being of Irish ethnicity based on geographic origins of grandparents. All participants gave a non-fasting 30 mL venous blood sample and completed a health and lifestyle questionnaire in which information on age, gender, height, weight, medical history and medications, smoking, alcohol intake, and dietary habits was collected. Detailed supplement and fortified food intake data were additionally collected as part of the questionnaire. Participants were paid a modest honorarium for completing the study. Fifteen students did not return a questionnaire and were excluded. One student was found to have participated twice and the second blood sample and questionnaire were excluded. Samples from 2,508 participants with questionnaire data went forward for metabolite analysis and DNA extraction. Ethical approval was obtained from the Dublin Federated Hospitals Research Ethics Committee, which is affiliated with TCD, and was reviewed by the Office of Human Subjects Research at the NIH. Written informed consent was obtained from the participants when they enrolled.
Recruitment of the Older Adult Replication Cohort
The replication cohort consisted of 5,186 subjects recruited as part of the Trinity, Ulster, Department of Agriculture (TUDA) Study, a study of older Irish adults designed to investigate nutritional factors, related gene-nutrient interactions, and a range of health and lifestyle factors in the development of chronic diseases of aging in non-institutionalized adults. Recruitment was carried out in Dublin and in Northern Ireland between September 2008 and December 2012 from either hospital outpatient clinics or the community, and subjects were deemed eligible if they were aged over 60 years, of Irish parents, and without a prior diagnosis of dementia. Detailed health, lifestyle, and clinical information, including medication use, smoking status, physical activity, and vitamin supplement usage, was obtained for all subjects. Further details of the cohort have been published previously.27 Cobalamin status was assessed with both serum total cobalamin and serum holotranscobalamin (holoTC) measurements on all subjects. Plasma MMA concentration was measured on a subset of 1,511 individuals, based on their cobalamin status. All individuals who had either serum cobalamin ≤ 148 pmol/L or holoTC ≤ 30 pmol/L (n = 856) and a random (generated from subject ID) selection of individuals with both serum cobalamin and holoTC above the respective cut-offs (n = 655) were assigned to plasma MMA analysis in order to perform a quantitative trait analysis of plasma MMA concentration on the pooled samples. The study was conducted according to the guidelines in the Declaration of Helsinki, and ethical approval was granted by the relevant authorities in each jurisdiction: the Research Ethics Committee of St. James’s Hospital and The Adelaide and Meath Hospital, Dublin, and the Office for Research Ethics Committees, Northern Ireland (ORECNI; reference 08/NI/RO3113) with corresponding approvals from The Northern and Western Health and Social Care Trusts, Northern Ireland. Participants in all studies provided written informed consent at the time of enrollment.
Recruitment of the Longitudinal Pregnancy Cohort
A total of 201 women were recruited between 2004 and 2005 at their first ante-natal visit to the Coombe Women and Infants University Hospital in Dublin, Ireland. They agreed to participate in a longitudinal study involving collection of non-fasting blood samples at 14 weeks, 24 weeks, and 34 weeks of pregnancy and a cord blood sample upon delivery. Although time points are missing for some women, and cord blood samples were only available from 117 deliveries, all available samples were retained for analysis. All participants had uncomplicated pregnancies and gave birth to infants free of any major health issues. Participants’ blood samples were used to extract genomic DNA for genotyping and to measure the concentrations of circulating metabolites in plasma. All participants gave written informed consent, and ethical approval was granted by the Coombe Women and Infants University Hospital Research Ethics Committee and the Office of Human Research Subjects at the NIH.
Samples for the TSS GWAS discovery cohort and for both replication cohorts were rendered anonymous prior to analysis.
Metabolite Analysis
Non-fasting blood samples were collected into EDTA, lithium heparin, and clotting tubes. Samples for MMA, cobalamin, holoTC, and total homocysteine (tHcy) analysis were processed within 3 hr of collection and were stored below −80°C, until analyzed. Plasma MMA concentration was measured by Bevital, Bergen, with gas chromatography mass spectroscopy.28 Plasma MMA measures were obtained for 2,483 of 2,508 participants. Missing plasma MMA values were due to an insufficient sample for the initial measure or its repeat in the case of a high coefficient of variation. Serum cobalamin was measured with colistin-resistant L. delbreuckii by microbiological assay.29 HoloTC and plasma homocysteine were measured via automated AxSym-based immunofluorescence methods.30, 31 Inter-assay CVs for all methods were as follows: MMA, 8.1%; serum total cobalamin, 10.6%; serum holoTC, 9.4%; plasma homocysteine, 2.2%. Circulating MMA concentrations are reported to be stable during long-term storage below −40°C and are not affected by recent food intake.32
Blood DNA Extraction and Genotyping of the TSS
We extracted genomic DNA from all samples by using QIAGEN QIAamp DNA Blood Mini Kits. In the TSS, eighteen samples with low quantity DNA were excluded and 2,490 samples were sent for genotyping. Genome-wide SNP genotyping was conducted at the Center for Inherited Disease Research (CIDR) in Baltimore with Illumina 1M HumanOmni1-Quad_v1-0_B chips. High-quality genotypes were obtained on 2,438 study samples, 14 randomly selected duplicates, and HapMap controls. The blind duplicates had a concordance rate of 99.997%, and the HapMap samples had a 99.71% concordance rate.
Genotyping Quality Control and Data Preparation for GWAS
In a preliminary analysis for anomalies, ten subjects were excluded because of gender discrepancy between self-report and genotypes (n = 7) and abnormal sex chromosomes (n = 3; one XYY male and two XX/XO mosaic females). 16 subjects with incomplete anthropometric data were removed. Siblings from 77 families reported entry into the study. These included two siblings from 74 families and three siblings from three families. Pair-wise tests for relatedness and population stratification within all the genotyped samples were performed with PLINK v.2,33 and a relatively independent but otherwise random subset of 25,718 SNPs (all selected SNPs had pair-wise intermarker linkage disequilibrium [LD] r2 < 0.04 for both short and long-range blocks of SNPs, minor-allele frequency [MAF] > 0.1 and genotype call rates > 0.99). These analyses confirmed the self-reported sibling relationships, and identified additional pairs of individuals who appeared closely related, to the level of 1st cousins. In order to reduce effects of relatedness, only one individual from each family was retained. In total, 175 individuals were removed (80 siblings and 95 1st cousins). Finally, principal components (PC) analysis was performed with the EIGENSOFT 3.0 software,34 and this same low-LD subset of 25,718 SNPs to detect potential population stratification. Scores from the first two PCs (PC1 and PC2) were examined, and two samples were detected as outliers from the main-population cluster of samples, suggesting that their ancestry might be different than that of the remainder of the cohort (6 SD away from the mean). These two samples were excluded from further analyses. Otherwise, the sample set was remarkably homogeneous for genetic variation, consistent with accurate self-report of participants and their Irish ethnicity. No significant evidence of population stratification (no significant principal components) was observed in these data after dropping the two outlier samples. As such, no adjustment was made for stratification in the association analyses. The full data preparation process resulted in a final dataset of 2,235 study samples in the GWAS association analysis.
Quality control assessment was performed based on genotyping data for 1,008,829 SNPs. SNPs and/or samples were excluded in two stages. In stage 1, SNPs that had less than 95% call rates were dropped and then samples with less than 97% call rates or that showed cryptic relatedness were dropped (57 samples total). In stage 2, SNPs were dropped that had (1) less than 98% call rate, (2) Mendelian errors when we used Hapmap trios, (3) discordant markers when we used Hapmap controls, and (4) discordant markers from one or more pairs based on study duplicates. SNPs with extreme deviation from the Hardy-Weinberg equilibrium (p < 1 × 10−4) were flagged for future reference and remained in the analysis. We also measured LD in our dataset and examined LD plots by using the HaploView program.35 The data were checked for batch effects with several methods from published GWAS quality control guidelines.36 No significant batch effects were detected. These methods included (1) descriptive statistics per batch (average MAF, average genotyping call rate across all SNPs for each plate) and (2) basic allelic chi-square association test (1 df) using the “--loop-assoc” command in PLINK; results compared each batch to the others and at random (median p value, number of significant results at p value thresholds of < 1 × 10−3 and < 1 × 10−4). The final dataset contained 758,443 genotyped SNPs (3,512 of these SNPs had Hardy-Weinberg equilibrium p < 10−4) after the exclusion of monomorphic and low MAF SNPs (MAF < 0.01) for 2,235 samples. TSS genotype and phenotype data have been deposited in dbGAP ([dbGAP: phs000789.v1.p1] Collaborative Study of Genes, Nutrients and Metabolites [CSGNM]).
Imputation of SNPs in the TSS
The MACH1.0 program37 was used with a set of 757,533 post-quality-control autosomal SNPs to impute ungenotyped SNPs with the HapMap II CEU (CEPH; Utah residents with ancestry from northern and western Europe) reference panel, resulting in 2,350,751 imputed genotypes per person after quality control, such that imputed SNPs were only retained for analysis if the squared correlations between imputed and actual genotypes for a set of masked genotypes (R2) was greater than 0.5. We used 200 individuals for estimating model parameters with 100 iterations for the Monte Carlo procedure by using all available haplotypes for each update, and then we imputed all the SNPs in the HapMap II CEU reference panel.
GWAS Statistical Analysis
Plasma MMA results were available for 2,210 of the 2,235 samples that passed the SNP genotype-based quality control analyses, so the discovery analysis for plasma MMA concentration relates to these 2,210 individuals. Standard descriptive statistics, including frequency distribution of MMA values, median, mean, theoretical versus sample quantile Q-Q plots, and measurements of skewness and kurtosis, were performed on the observed MMA measures both with and without transformation. The distribution of the MMA measurements in the TSS study participants deviated significantly from a normal distribution, and a log10 transformation was used to normalize the MMA values for the linear regression analyses. Therefore, for the GWAS, genotyped and imputed autosomal SNPs that passed quality control were tested for association with the log10-transformed MMA values, adjusted for age and sex. Other factors known to influence MMA levels (renal function, serum cobalamin) were not adjusted for given that renal function and cobalamin absorption are not compromised in this young, healthy cohort. In the simple linear regression model, the association test of each marker was performed with PLINK v.2 under the assumption of an additive genetic model (i.e., number of minor alleles, 0, 1, and 2). The genomic control lambda was 0.991 for both the untransformed trait and the transformed trait (Table S1), and results using the transformed trait are reported here. (See Figure 1 and Figures S1–S3 for results using the transformed MMA levels in the analysis of the genotyped SNPs and in the complete set of genotyped plus imputed SNPs, and see Table S2 for a list of SNPs for which missense or nonsense variants were significantly associated with MMA levels.) The contributed variation of each marker to MMA concentration was calculated by regression R squared (R2). Manhattan plots and Q-Q plots were generated with R scripts written within our institution. To further evaluate the distribution of p values obtained in this GWAS, Q-Q plots without the markers on chromosomes 2 and 16 were also generated, to remove the effect on the plots of these extremely significant SNPs (Figures S1 and S3). Genomic-control lambda values were calculated for each analysis (Table S2).
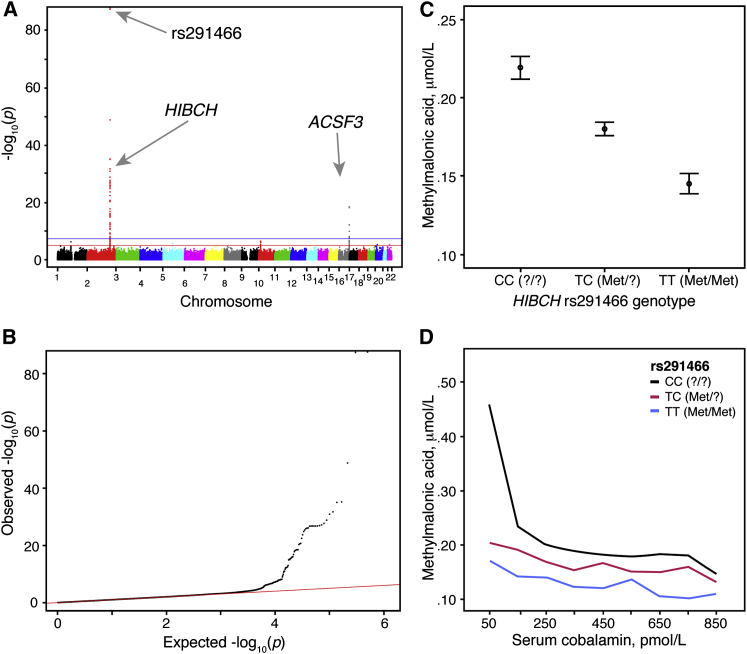
Figure 1.
Genome-wide Analysis of Plasma MMA Levels in the TSS Cohort
At the genome-wide level, HIBCH rs291466 is the SNP most significantly associated with log10-transformed plasma MMA concentration in the TSS cohort of 2,210 young, healthy Irish adults.
(A) A Manhattan plot of the log10-transformed MMA GWAS of 758,443 genotyped SNPs. The horizontal blue line indicates the genome-wide significance threshold (p = 5 × 10−8). The horizontal red line indicates the suggestive significance threshold (p = 1 × 10−05). The two strongest signals are found in HIBCH and ACSF3.
(B) Q-Q plot of p values obtained for the log10-transformed MMA GWAS.
(C) Distribution of MMA plasma levels for each HIBCH rs291466 genotype group. Means and 95% confidence intervals for each genotype group’s MMA levels are shown (ANOVA p < 0.0001).
(D) Values for plasma MMA plotted by serum cobalamin for each HIBCH rs291466 genotype group of young adults. Cobalamin data were categorized by groups in ascending order of 100 pmol/L. Median MMA values were calculated within each grouped category. Line graphs were plotted as median MMA against median cobalamin within each group.
Genotyping in the Replication Cohorts and Statistical Analysis of Metabolites in the TSS, TUDA, and Longitudinal Pregnancy Cohorts
All 5,186 TUDA samples were genotyped for HIBCH (MIM: 610690) rs291466 (GenBank: NM_014362.3:c.2T>C) and ACSF3 rs1054747 ([GenBank: NM_001127214.3] c.∗231G>A) by LGC Genomics (Herts, UK) with KASP genotyping chemistry. The call rates were 98.9% and 98.0%, respectively. Duplicate samples (2%) were included, as well as the addition of a single control sample to every plate. Concordance for all duplicates was >99%. In this sample, measures of plasma MMA data were available for 1,511 individuals; 1,481 of these had genotype data for HIBCH rs291466 and 1,458 of these had genotype data for ACSF3 rs1054747. The replication analysis therefore relates to these 1,481 individuals. All 201 pregnant mothers and 117 cord bloods were genotyped for HIBCH rs291466 and ACSF3 rs1054747 by detection of allele-specific extension products via matrix-assisted laser desorption/ionization and time-of-flight (MALDI-TOF) mass spectrometry (Sequenom). For both SNPs, the call rates were 94% for the mothers and ≥96% for the newborns, and genotypes were 100% concordant for the >12% of samples that were re-plated and independently typed. The final replication analyses involved 185 mothers and 110 newborns with valid HIBCH rs291466 genotype and MMA data, and 182 mothers and 113 newborns with valid ACSF3 rs1054747 genotype and MMA data.
In the TSS, direct genotyping of HIBCH rs291466 yielded calls for 2,208 of the 2,210 subjects with MMA data who were included in the GWAS. Similar to the TSS data, distribution of the MMA measurements in the TUDA study participants and in the longitudinal pregnancy cohort did not approximate normal distributions, and a log10 transformation was used to approximately normalize the MMA values for the linear regression analyses. HIBCH rs291466 was tested for association with the transformed unadjusted MMA values in the TUDA cohort and in the longitudinal pregnancy cohort. These association tests were performed with PLINK v.2 by simple linear regression and under the assumption of an additive genetic model as described above for the TSS GWAS.
Data for cobalamin biomarkers in the TSS and the replication datasets were not normally distributed. For Table 1, Table 2, Table 3, all data are presented as medians with 25th and 75th percentile values (interquartile intervals; IQI). Comparisons of data across the three HIBCH rs291466 genotype groups were carried out with the non-parametric Kruskal-Wallis one-way ANOVA by ranks for three or more independent groups. p values < 0.05 were considered significant and represent a difference across the three genotype groups, without specifying exactly which groups are different.
Table 1.
Subject Characteristics of the TSS Discovery Cohort, with Participants Grouped by HIBCH rs291466 Genotype
| Characteristic |
Alla |
CC (?/?) |
CT (?/Met) |
TT (Met/Met) |
Kruskal-Wallis Testcp Value |
|---|---|---|---|---|---|
| Median (IQIb) | Median (IQIb) | Median (IQIb) | Median (IQIb) | ||
| No. of Individuals | 2,208 | 721 | 1,065 | 422 | – |
| Sex (M/F) | 907/1,301 | 299/422 | 442/623 | 166/256 | – |
| Age (years) | 22.0 (21.0–24.0) | 22.0 (21.0–23.0) | 22.0 (21.0–24.0) | 23.0 (21.0–24.0) | 0.008 |
| Body Mass Index (kg/m2) | 22.6 (21.0–24.4) | 22.5 (21.0–24.5) | 22.7 (21.1–24.5) | 22.5 (20.8–24.4) | 0.454 |
| Creatinine (μmol/L) | 64.8 (56.4–74.6) | 65.1 (56.7–75.4) | 64.7 (56.4–74.4) | 64.1 (56.3–74.1) | 0.586 |
| Hemoglobin (g/dL) | 14.1 (13.1–15.2) | 14.1 (13.2–15.3) | 14.1 (13.1–15.2) | 14.1 (13.1–15.4) | 0.677 |
| Urea (mmol/L) | 4.30 (3.70–5.10) | 4.40 (3.70–5.13) | 4.30 (3.60–5.10) | 4.20 (3.50–5.00) | 0.030 |
| Plasma cobalamin (pmol/L) | 306 (231–411) | 307 (231–421) | 305 (232–405) | 305 (230–411) | 0.675 |
| holoTC (pmol/L) | 53.7 (39.1–72.2) | 54.1 (39.5–72.8) | 53.2 (38.9–71.4) | 55.4 (39.5–73.3) | 0.388 |
| Homocysteine (μmol/L) | 8.13 (7.00–9.62) | 8.17 (6.99–9.61) | 8.17 (7.08–9.61) | 7.92 (6.83–9.75) | 0.319 |
| MMA (μmol/L) | 0.17 (0.14–0.21) | 0.19 (0.16–0.24) | 0.16 (0.14–0.20) | 0.13 (0.11–16) | < 0.0001d |
M, male; F, female.
Valid HIBCH rs291466 genotypes were obtained for 2,208 of the 2,210 TSS subjects in the GWAS study.
IQI (inter-quartile interval) represents the 25th–75th percentile.
The non-transformed data were compared with the Kruskal-Wallis one-way ANOVA by ranks for three or more independent groups; p < 0.05 was considered significant.
The association test for this SNP was also performed by simple linear regression with PLINK v.2 and under the assumption of an additive genetic model (number of minor alleles: 0, 1, and 2). p = 1.95 × 10−87 was obtained for the unadjusted log10-transformed data.
Table 2.
Subject Characteristics of the TUDA Replication Cohort, with Participants Grouped by HIBCH rs291466 Genotype
| Characteristic |
Alla |
CC (?/?) |
CT (?/Met) |
TT (Met/Met) |
Kruskal-Wallis Testcp Value |
|---|---|---|---|---|---|
| Median (IQIb) | Median (IQIb) | Median (IQIb) | Median (IQIb) | ||
| No. of Individuals | 1,481 | 460 | 723 | 298 | – |
| Sex (M/F) | 493/991 | 168/293 | 231/494 | 94/204 | – |
| Age (years) | 79.0 (72.6–83.9) | 79.5 (72.8–84.7) | 78.6 (72.0–83.2) | 78.9 (73.2–84.1) | 0.285 |
| Body Mass Index (kg/m2) | 27.1 (23.6–30.8) | 27.3 (23.6–31.0) | 27.0 (23.8–30.6) | 26.8 (23.6–30.6) | 0.742 |
| Creatinine (μmol/L) | 82.0 (69–101) | 82.0 (69.0–101) | 82.0 (69.5–101) | 81.0 (71.0–103) | 0.878 |
| Hemoglobin (g/dL) | 12.8 (11.7–13.8) | 12.8 (11.7–14.0) | 12.8 (11.7–13.8) | 12.7 (11.7–13.8) | 0.630 |
| Urea (mmol/L) | 6.7 (5.4–8.7) | 6.6 (5.4–8.8) | 6.8 (5.4–8.7) | 6.8 (5.5–8.7) | 0.726 |
| Serum cobalamin (pmol/L) | 185 (131–287) | 177 (128–274) | 187 (135–286) | 194 (128–307) | 0.270 |
| holoTC (pmol/L) | 39.0 (25.8–57.0) | 38.4 (25.6–55.0) | 38.8 (25.9–57.5) | 39.9 (25.8–60.9) | 0.550 |
| Homocysteine (μmol/L) | 16.2 (13.0–20.5) | 15.9 (12.5–19.7) | 16.3 (13.2–20.9) | 16.2 (13.3–20.3) | 0.097 |
| MMA (μmol/L) | 0.34 (0.24–0.51) | 0.42 (0.29–0.62) | 0.33 (0.24–0.50) | 0.26 (0.19–0.37) | < 0.0001c |
M, male; F, female.
IQI (inter-quartile interval) represents the 25th–75th percentile.
The non-transformed data were compared with the Kruskal-Wallis one-way ANOVA by ranks for three or more independent groups; p<0.05 was considered significant.
The association test for this SNP was also performed by simple linear regression with PLINK v.2 and under the assumption of an additive genetic model (number of minor alleles: 0, 1, and 2). p = 4.01 × 10−26 was obtained for the unadjusted log10-transformed data.
Table 3.
Subject Characteristics of the Longitudinal Pregnancy Replication Cohort at First Antenatal Clinic Visit, with Participants Grouped by HIBCH rs291466 Genotype
| Characteristic |
All |
CC (?/?) |
CT (?/Met) |
TT (Met/Met) |
Kruskal-Wallis Testbp value |
|---|---|---|---|---|---|
| Median (IQIa) | Median (IQIa) | Median (IQIa) | Median (IQIa) | ||
| No. of Individuals | 185 | 70 | 80 | 35 | – |
| Age (y) | 30.0 (25.0–33.0) | 30.0 (26.5–34.0) | 28.0 (25.0–31.0) | 30.0 (25.0–32.0) | 0.044 |
| Body Mass Index (kg/m2) | 24.8 (21.9–28.2) | 25.9 (22.5–28.2) | 24.3 (21.0–27.6) | 24.8 (21.5–28.5) | 0.292 |
| Gestation (weeks) | 14.2 (12.9–15.4) | 14.0 (12.4–15.6) | 14.1 (12.8–15.5) | 13.9 (13.3–14.7) | 0.914 |
| Creatinine (μmol/L) | 46.6 (42.4–51.3) | 46.7 (42.4–51.9) | 46.2 (42.4–50.7) | 48.1 (42.9–51.3) | 0.700 |
| Serum cobalamin (pmol/L) | 171 (120–215) | 180 (133–225) | 156 (108–205) | 184 (140–217) | 0.054 |
| holoTC (pmol/L) | 33.6 (24.7–43.7) | 36.4 (27.5–46.4) | 31.1 (21.7–39.4) | 36.7 (25.2–43.7) | 0.034 |
| Homocysteine (μmol/L) | 6.29 (5.35–7.34) | 6.06 (5.06–7.27) | 6.50 (5.51–7.63) | 6.01 (5.27–7.47) | 0.097 |
IQI (inter-quartile interval) represents the 25th–75th percentile.
The non-transformed data were compared with the Kruskal-Wallis one-way ANOVA by ranks for three or more independent groups; p < 0.05 was considered significant.
GWAS Catalog Query
To determine whether the lead SNPs identified in this study might have been previously associated with any disease phenotype or other trait, we searched the National Human Genome Research Institute and the European Bioinformatics Institute (NHGRI-EBI) GWAS Catalog.38 No entries were found for HIBCH rs291466 or ACSF3 rs1054747, or for SNPs in LD (r2 > 0.8, 1000 Genomes EUR super population) with them (n = 40 and n = 84, respectively).
Analysis of mRNA Abundance
Because HIBCH rs291466 and ACSF3 rs1054747 are coding SNPs, RNA-seq data can be used to quantitate transcripts carrying each allele. Two approaches were used to evaluate whether mRNA abundance is influenced by the SNPs associated with plasma MMA concentration. First, RNa-seq data from heterozygotes were used to determine how much HIBCH or ACSF3 transcript was expressed from each allele. Second, individuals were divided into groups based on HIBCH rs291466 or ACSF3 rs1054747 genotypes to compare the overall abundance of these transcripts.
First, to calculate the relative ratios of the T (Met) and C (?) rs291466 alleles, the raw BAM files for the 211 HIBCH rs291466 heterozygous individuals in the Geuvadis RNA Sequencing Project39 were downloaded. Samtools’ mpileup was used to find the number of reads with T (Met) or C (?) alleles expressed in the lymphoblastoid cell lines generated from each individual and the data were plotted in R. To determine the statistical significance of the T (Met) and C (?) allele abundance in the RNA-seq data, a Pearson’s chi-square test was performed.
Second, we checked relative mRNA expression between the HIBCH rs291466 genotype groups by using RPKM (reads per kilobase of transcript per million reads mapped) scores from the GD462.GeneQuantRPKM.50FN.samplename.resk10.txt.gz file39 and plotting in R (v.2.15.1). Significance was calculated with anova() from the base package in R. To confirm these results, Genotype-Tissue Expression (GTEx, v.4) data were similarly analyzed.40 HIBCH microarray expression scores in lymphoblastoid tissue were re-plotted by HIBCH rs291466 genotype group with ggplot() from the ggplot2 package (0.9.2.1) in R. For display purposes, HapMap CEU (n = 60), YRI (Yoruba in Ibadan, Nigeria; n = 60), and JPT/CHB (Japanese in Tokyo, Japan/Han Chinese in Beijing, China; n = 89) individuals were combined into a single group for analysis. Significance was calculated with anova() from the base package in R. These analyses were repeated for ACSF3 rs1054747.
Western Blotting and Subcellular Localization of HIBCH
To determine whether HIBCH rs291466 grossly alters protein levels, western blotting of whole-cell extracts was performed. Five different Coriell cell lines (GM18621, GM18961, GM17443, GM17449, GM17454) with the TT (Met/Met) genotype and four different Coriell cell lines (GM12814, GM17438, GM17456, GM17460) with the CC (?/?) genotype were analyzed. Whole-cell extracts were prepared by lysing cells in RIPA buffer (10 mM Tris [pH 7.4]; 150 mM NaCl; 5 mM EDTA [pH = 8]; 1% Triton X-100; 0.1% sodium deoxycholate; 0.1% SDS) supplemented with protease inhibitor cocktail (Sigma). Western blotting was performed with the Odyssey LI-COR Imaging system. In brief, nitrocellulose membranes were blotted in Odyssey blocking buffer for 1 hr, followed by 1 hr incubation with HIBCH Prestige antibody (Sigma), HIBCH antibody (Proteintech), or proliferating cell nuclear antigen (PCNA [MIM: 176740]) antibody (Santa Cruz), which was used as a loading control. After extensive washing, the membranes were incubated with Odyssey donkey anti-rabbit IRDye-800 or Odyssey donkey anti-mouse IRDye-680 for 0.5 hr. Western blot membranes were quantified on an Odyssey LI-COR Imaging system. HIBCH signal (green channel) was normalized using PCNA signal (red channel) as an internal control. Both HIBCH and PCNA signals were derived from a single western blot. Three independent experiments were performed. A representative western blot is shown in Figure 3D.
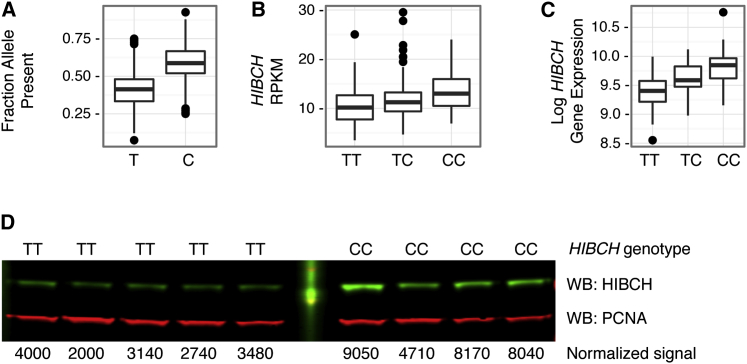
Figure 3.
Association of HIBCH rs291466 Genotype with HIBCH mRNA and Protein Levels
(A) Relative amounts of HIBCH rs291466 mRNA isoforms expressed in heterozygous individuals. The fraction of RNA-seq reads for each HIBCH rs291466 allele was calculated for 211 heterozygotes using genotype and expression data from the GEUVADIS RNA Sequencing Project, based on the 1000 Genomes EUR samples. Distributions for each allele (T [Met] versus C [?]) are shown.
(B) HIBCH mRNA expression by rs291466 genotype (TT [Met/Met], TC [Met/?], and CC [?/?]). Distribution of HIBCH RPKM (reads per kilobase of transcript per million reads mapped) scores for each HIBCH rs291466 genotype group (100 TT [Met/Met], 211 TC [Met/?], and 151 CC [?/?]), based on 1000 Genomes EUR samples, is shown.
(C) Distribution of HIBCH expression scores by HIBCH rs291466 genotype groups (TT [Met/Met], TC [Met/?], and C [?/?]) for 219 individuals (CEU, YRI, JPT, CHB), based on data from the GTEx Project 3, v.2.
(D) Western blot analysis of HIBCH in HIBCH homozygotes (TT ([Met/Met] and CC [?/?]). Whole-cell lysates of nine independent Coriell cell lines were immunoblotted for HIBCH as well as PCNA as a control (WB, western blot). Normalized signals (HIBCH/PCNA) for each cell line are shown below the blot.
Similarly, to determine whether HIBCH rs291466 influences the subcellular localization of HIBCH, western blotting of cytoplasmic, mitochondrial, and nuclear fractions was performed. Mitochondrial import was assessed via western blot after performance of subcellular fractionation (Qproteome Cell Compartment Kit, QIAGEN). Fraction purity was assessed with a control antibody specific for the mitochondrial compartments.
Results
GWAS Discovery Cohort
We identified two regions of genome-wide significant association with log10-transformed MMA values (Figures 1A and 1B). The strongest association was observed with HIBCH rs291466 (p = 8.42 × 10−89), a directly typed SNP on chromosome 2 accounting for 9.9% of the total variance. This SNP is a non-synonymous polymorphism (c.2T>C) resulting in a methionine to a presumed threonine change in the translational start codon of the encoded protein, p.Met1? (note: the coding impact of this SNP in the initiator methionine is unknown and we therefore refer to this isoform as “?”). Although T (Met) is the ancestral allele (based on the allele present in the great apes), the C (?) allele is more common in the studied population. The median plasma MMA concentration for the corresponding CC (?/?) homozygotes was 46% higher than the TT (Met/Met) homozygotes (Table 1). This effect was independent of biochemical markers of cobalamin status, including serum total cobalamin, holoTC, and plasma homocysteine (Figures 1C and 1D and Table 1).
The second region of genome-wide significance was in the ACSF3 region on chromosome 16, consistent with previous studies showing that severe mutations in ACSF3 affect MMA concentrations. Patients with these mutations were found to have combined malonic and methylmalonic aciduria (CMAMMA [MIM: 614265]).4, 5 The described patients are homozygous or compound heterozygous for rare coding changes in ACSF3 and have plasma MMA levels an order of magnitude higher than normal. The strongest ACSF3 signal among the genotyped SNPs (rs1054747, MAF = 0.05, p = 3.48 × 10−19) fell in the 3′ UTR of the gene and accounted for 2.1% of the total variance. Just three homozygotes carrying the minor A allele were detected, but GA heterozygotes had a higher plasma MMA concentration than GG homozygotes (0.22 μM versus 0.18 μM). This SNP is linked (D′ = 1, r2 > 0.92) to SNPs in two adjacent non-coding RNAs (LINC00304, LOC400558) and the nearby cadherin 15 (CDH15) gene, but ACSF3 remained the best candidate gene for containing the causal variant because of its biological relevance. The 40 SNPs in high LD (r2 ≥ 0.95) with ACSF3 rs1054747 are intronic or intergenic, and the functional variant responsible for influencing MMA concentration remains unidentified. No regions of significance were observed in chromosome 6, the location of MUT. Next, we focused our efforts on performing replication analyses in two independent populations and undertaking functional characterization studies.
Older Adult Replication Cohort
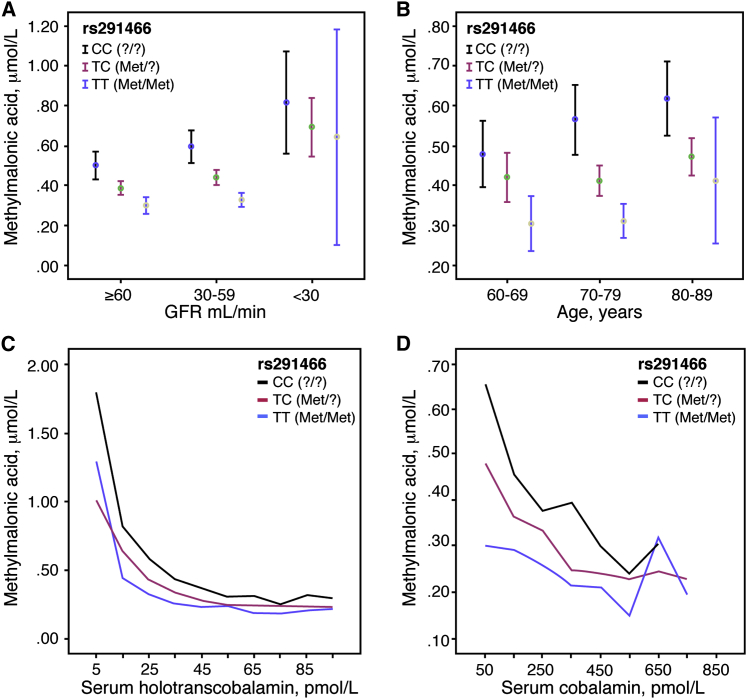
Because MMA concentration is known to increase significantly with age, it was possible that the impact of this genotype would disappear in the elderly. To test this hypothesis, we genotyped HIBCH rs291466 in 1,481 older individuals in whom we had measured plasma MMA levels and other biochemical markers of cobalamin status (Table 2). As in our young adults, the genotype effect for HIBCH rs291466 on the transformed MMA data was highly significant (p = 4.01 × 10−26), and the genotype effect was discernible even when these subjects were categorized into three groups by age or by glomerular filtration rate (GFR, a measure of renal function; Figures 2A and 2B). Of note, this effect was independent of cobalamin status using either holoTC or total cobalamin as the serum indicators (Figures 2C and 2D) and was also independent of plasma homocysteine, a metabolic biomarker of cobalamin deficiency and a metabolite that, like MMA, is sensitive to reduced renal function.
Figure 2.
Replication of the HIBCH rs291466 Association with Plasma MMA Concentration in the TUDA Cohort
The HIBCH rs291466 association with plasma MMA concentrations was confirmed in 1,481 older Irish adults in the TUDA cohort.
(A) Plasma MMA concentration (mean and 95% confidence interval) for each HIBCH rs291466 genotype group in older Irish adults sorted by glomerular filtration rate (GFR), a measure of renal function. GFR ≤ 29 mL/min indicates severely impaired renal function, GFR 30–59 mL/min indicates moderately impaired function, and GFR ≥ 60 mL/min indicates normal function.
(B) Plasma MMA concentration (mean and 95% confidence interval) for each HIBCH rs291466 genotype group in older Irish adults sorted by age group.
(C) Plasma MMA levels plotted as a function of serum holoTC levels in each HIBCH rs291466 genotype group. HoloTC data were categorized by groups in ascending order of 10 pmol/L. Median MMA values were calculated within each grouped category. Line graphs were plotted as median MMA against median holoTC within each group.
(D) Plasma MMA levels plotted as function of serum cobalamin levels in each HIBCH rs291466 genotype group. Cobalamin data were categorized by groups in ascending order of 100 pmol/L. Median MMA values were calculated within each grouped category. Line graphs were plotted as median MMA concentration against median cobalamin concentration within each group.
The effect of ACSF3 rs1054747 on log10-transformed MMA levels was similarly assessed in this population of older Irish adults, resulting in replication of the originally observed effect (p = 0.0003).
Longitudinal Pregnancy Replication Cohort
MMA concentrations are altered during pregnancy and in newborns;41, 42 therefore, replication of the influence of HIBCH rs291466 on MMA concentration was tested in samples from a longitudinal study of pregnant women and their newborn offspring (Table 3). MMA concentration distributions by HIBCH rs291466 genotype show significantly different effects for each of three trimesters of pregnancy and for cord bloods (p < 8.0 × 10−05 for each maternal time point; p = 0.0005 for cord bloods, Table 4). TT (Met/Met) homozygotes have the lowest median MMA concentration, whereas CC (?/?) homozygotes have the highest median MMA concentration, consistent with the direction of effect originally observed in the discovery cohort of young Irish adults.
Table 4.
Plasma MMA Concentrations in the Longitudinal Pregnancy Replication Cohort and in Cord Blood with Participants Grouped by HIBCH rs291466 Genotype
| Gestation Time Point |
All |
CC (?/?) |
CT (?/Met) |
TT (Met/Met) |
Replication testbp Value |
|---|---|---|---|---|---|
| Median (IQIa) | Median (IQIa) | Median (IQIa) | Median (IQIa) | ||
| 14 weeks (μmol/L) | 0.15 (0.11–0.21), n = 184 | 0.18 (0.13–0.25), n = 69 | 0.15 (0.12–0.21), n = 80 | 0.10 (0.08–0.14), n = 35 | 1.95 × 10−8 |
| 24 weeks (μmol/L) | 0.18 (0.14–0.25), n = 132 | 0.23 (0.17–0.29), n = 49 | 0.18 (0.15–0.25), n = 53 | 0.14 (0.12–0.17), n = 30 | 1.05 × 10−5 |
| 34 weeks (μmol/L) | 0.21 (0.16–0.31), n = 129 | 0.25 (0.19–0.36), n = 48 | 0.21 (0.17–0.31), n = 57 | 0.15 (0.14–0.20), n = 24 | 7.39 × 10−5 |
| Cord blood, delivery (μmol/L) | 0.40 (0.30–0.58), n = 110 | 0.47 (0.30–0.67), n = 43 | 0.40 (0.34–0.58), n = 49 | 0.26 (0.23–0.3), n = 18 | 0.0005 |
IQI (inter-quartile interval) represents the 25th–75th percentile.
Replication was performed by simple linear regression of the unadjusted log10-transformed data via PLINK v.2 and under the assumption of an additive genetic; p < 0.05 was considered significant.
Association of ACSF3 rs1054747 with log10-transformed MMA values was not observed at any time point for these pregnant women (14 weeks, p = 0.721; 24 weeks, p = 0.671; 34 weeks, p = 0.795). Interestingly, replication in the cord blood samples yielded a borderline association (p = 0.051) with mean MMA levels increasing with the number of A alleles at this locus (0.61 μM versus 0.46 μM for AG and GG genotype groups, respectively), as originally observed.
Functional Studies
Because the signal associated with HIBCH rs291466 was driven by a common allele in a gene previously unknown to influence MMA and had the largest impact on MMA levels, we pursued functional characterization studies. Alterations of initiator methionine residues have been associated with loss-of-function alleles.43, 44, 45 We assessed the impact of this missense change on HIBCH (see below). To determine whether HIBCH rs291466 influences the expression of HIBCH mRNA, we analyzed RNA-seq data generated from lymphoblastoid lines.39, 46 In 211 HIBCH rs291466 heterozygous individuals, 59% of the reads contained the C (?) allele and 41% contained the T (Met) allele, (p < 2.2 × 10−12; Figure 3A). We then examined whether absolute HIBCH mRNA expression changed depending on HIBCH rs291466 genotype. The median mRNA expression scores of TT (Met/Met), TC (Met/?), and CC (?/?) genotype groups increased from 10.2 to 11.3 to 13.0, respectively (p < 5.02 × 10−11, Figure 3B). No other gene in a 1-megabase window around HIBCH had this rs291466-dependent effect (data not shown). An independent expression dataset40 confirmed these results (p < 3.87 × 10−14; Figure 3C), indicating that the loss of the initiating methionine is associated with an allele-specific increase in HIBCH mRNA expression.
To test whether HIBCH levels differ between individuals with HIBCH rs291466 homozygous genotypes, we obtained lymphoblast cell lines derived from nine individuals with the TT (Met/Met) genotype or the CC (?/?) genotype. Immunoblot analysis, used to quantify HIBCH levels, showed that cell lines with the CC (?/?) genotype have higher steady-state levels of HIBCH than cell lines with the TT (Met/Met) genotype (Figure 3D).
Discussion
Three aspects of this GWAS of plasma MMA concentration are particularly noteworthy. First, we report the strong association of HIBCH rs291466 with MMA levels in a young, healthy, ethnically homogeneous cohort from Ireland, as well as its replication in two ethnically matched Irish cohorts representing significantly different stages of life. Second, the value of GWASs is emphasized given that this study revealed that the largest genetic influence on MMA levels is not related to cobalamin metabolism and is instead related to a little-studied pathway involved in valine catabolism. Last, we note the potential complexity of determining the mode of function of any phenotype-associated variant, as shown by the unexpected apparent mechanism through which the HIBCH rs291466 (p.Met1?) association signal achieves its effect on MMA levels. In this case the variant associated with the strongest GWAS signal would be expected to produce a null or hypomorphic allele, i.e., one that produces a protein that is not properly targeted to the mitochondrion. Our experiments reveal the opposite; the missense allele associated with higher levels of MMA produces more mature enzyme and acts at the level of mRNA. It is unclear whether this allele directly influences transcription or whether this activity resides with another linked SNP.
The clinical utility of MMA concentration as a confirmatory biomarker of cobalamin deficiency makes this metabolite an important candidate for GWASs. Recent GWAS results for cobalamin have reported a strong association with fucosyltransferase 2 (FUT2 [MIM: 182100]), and in several of these studies, MUT was also identified as a significant determinant of cobalamin at a genome-wide level.47, 48, 49, 50 We observed no such associations in our GWAS of MMA concentration, and the top MMA GWAS result is in fact unrelated to cobalamin metabolism. This is consistent with two mechanisms for elevated levels of circulating MMA. First, a reduction in cobalamin reduces activity of the MUT enzyme, resulting in accumulation of MMA. Second, a cobalamin-independent mechanism of increased flux through the valine catabolism pathway (e.g., via increased levels of HIBCH due to the C [?] allele of rs291466) can result in increased production of MMA. The effect of the latter was not anticipated, but is very strong.
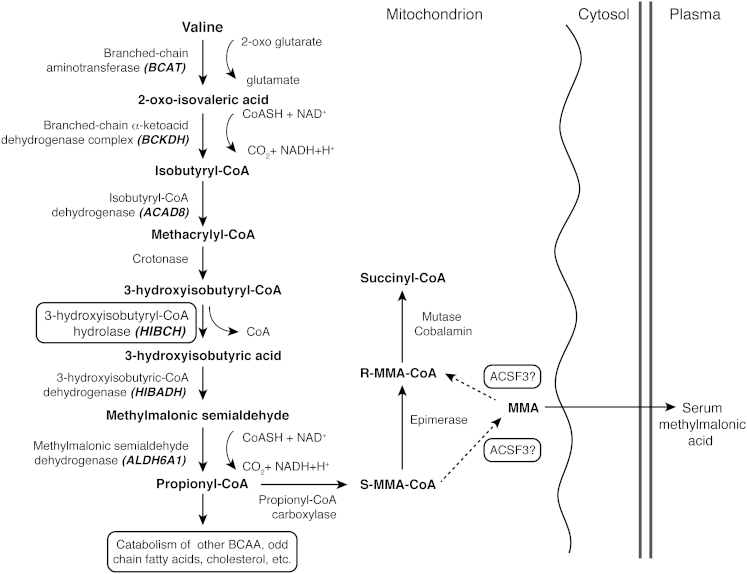
Although HIBCH rs291466 is the more common variant (frequency of allele associated with elevated MMA: 0.57 versus 0.05) with greater impact on plasma MMA concentration (contribution to variance: 9.1% versus 2.2%), we also genotyped the top ACSF3 polymorphism (rs1054747) in our replication cohorts. ACSF3 rs1054747 was significantly associated with MMA levels in the sample of ∼1,400 older Irish adults, but not in our longitudinal pregnancy cohort (Table 4). The combination of the low MAF and the substantially smaller sample size (∼180 pregnant women) most likely diminished our power to detect the effect of this SNP on MMA levels. In support of this, we note that their newborns did show a borderline association (Table 4). ACSF3 rs1054747 was not found to be associated with mRNA levels of ACSF3 by examination of the Geuvadis RNA-seq data (Figure S5) or by querying the GTEx Portal (over 30 tissues, v.6). This SNP, or the causal variant linked to it, does not seem to act at the level of transcription. However, there are pre-existing data and biological arguments supporting the role of ACSF3 in MMA metabolism. First, the genome-wide signal in ACSF3 confirms recent observations of mutations in this gene as a cause of CMAMMA.4, 5 Second, our observations on this poorly understood member of the acyl CoA synthetase family of enzymes reinforce the notion of a reversible MMA shunt to provide R-MMACoA for the MUT reaction through the deacetylation of S-MMACoA and subsequent reacetylation of the free R-MMA, as described by Montgomery et al.51 and supported by Alfares et al.4 Our data further indicate that spontaneous racemization of free MMA could make a more substantial contribution to the pathway than hitherto supposed (see Figure 4) and adds to the debate on whether a functional MMACoA epimerase enzyme is critical for metabolism down this pathway.51, 52 Our data and these biological observations confirm the connection between ACSF3 activity and steady-state levels of MMA in plasma.
Figure 4.
The Valine Catabolism Pathway
All branched-chain amino acids (BCAAs) are first transaminated to the 2-oxo derivative via branched-chain aminotransferase (BCAT). The oxo-derivatives then undergo irreversible oxidative decarboxylation to a CoA ester derivative. In the case of valine, this produces isobutyryl-CoA, which is converted to methacrylyl-CoA, an intermediate that is regarded as highly reactive and potentially toxic within mitochondria. Next, methacrylyl CoA is hydrated via crotonase (methacrylyl-CoA hydratase) to form 3-hydroxyisobutyryl-CoA. In the following step, unique to valine metabolism, the CoA is removed to form 3-hydroxyisobutyric acid, a molecule that is easily diffusible across the mitochondrial membrane. The reaction is catalyzed by HIBCH. 3-hydroxyisobutyric acid is subsequently converted to methylmalonic semialdehyde, and in the next step, CoA is reinstated in the pathway by the oxidative decarboxylation of methylmalonic semi-aldehyde to propionyl-CoA. Further catabolism of propionyl-CoA through cobalamin-dependent methylmalonyl-CoA mutase (MUT) is common to other BCAAs, odd-chain fatty acids, cholesterol, etc. An epimerase enzyme converts S-methylmalonyl-CoA (S-MMA-CoA) to R-MMA-CoA, the epimer used in the MUT reaction. A MMA shunt has been proposed whereby a deacetylation- reacetylation step, catalyzed by the malonyl and MMA-specific AcylCoA synthetase ACSF3 produces free MMA, which can undergo spontaneous racemization and be re-acetylated in the R-MMA-CoA form.
We also confirmed the association of MMA concentration and HIBCH rs291466. This SNP was not only the top genome-wide association signal in a young, healthy Irish cohort, it was also significant in an older Irish cross-sectional cohort and in a longitudinal cohort of pregnant Irish women and their newborns. The persistence of this genetic effect is remarkable considering the known potential for altered MMA levels at each of these stages of life. Maternal circulating cobalamin decreases over the course of pregnancy as a result of hemodilution and other pregnancy related factors.41 This is accompanied by changes in maternal plasma MMA concentration throughout pregnancy and in neonatal blood41, 42 that could have obscured the replication signal (Table 4). Similarly, in the sample of older adults, the effect of HIBCH rs291466 on MMA concentration was detected despite the noise introduced by the expected and observed (Table 2) age-related increase in MMA levels as a result of factors such as impaired cobalamin absorption and decreased renal function. The ability to detect the GWAS signal of HIBCH rs291466 in both replication cohorts speaks to the strength of effect of this variant in the face of a phenotype with many known but unaccounted confounders.
Our findings demonstrate that HIBCH rs291466 is an important determinant of MMA status, regardless of age or renal function and, as such, is relevant to clinical determination of cobalamin deficiency. Low serum cobalamin concentrations are common in older persons, and it is difficult to assess their clinical significance because the classic symptoms of cobalamin deficiency are often absent or are similar to a variety of disorders in elderly individuals. MMA concentration is regarded as the most reliable biochemical indicator of cobalamin deficiency and is used as a confirmatory test when serum cobalamin or holoTC concentrations are indeterminate and clinical symptoms are unreliable.14, 20, 53 In our older replication cohort, the median MMA concentration in CC (?/?) individuals was 0.42 μmol/L (Table 2). By the standard definition of abnormally elevated MMA concentration (> 0.37 μmol/L), 58% of CC (?/?) homozygotes would be considered cobalamin deficient. Although holoTC concentration has been suggested to be a more reliable marker of cobalamin deficiency than serum total cobalamin,54, 55 only half of these high-MMA individuals (52%) would be regarded as having a possible deficiency (holoTC < 30 pmol/l). Thus, 28% of all CC (?/?) individuals might be falsely classified as cobalamin deficient when using MMA values alone. In contrast, among TT (Met/Met) individuals, far fewer (25%) had MMA concentrations above 0.37 μmol/L, and the majority of those with high MMA levels also had low cobalamin status (73%) (holoTC < 30 pmol/L), consistent with a true cobalamin deficiency. Choosing other thresholds for total cobalamin (110 pmol/L) or holoTC (35 pmol/L) changed the proportion of individuals within deficient or sufficient categories but did not remove the strong genotype effect. Thus, this common HIBCH polymorphism is a cobalamin-independent determinant of plasma MMA concentration that influences its effectiveness as a cobalamin status indicator in older adults when applying current clinical cut-offs.
The unanticipated discovery of a variant in HIBCH as a strongly significant contributor to the variance of plasma MMA concentration highlights this little-studied enzyme of branched-chain amino acid metabolism that for several decades has been cited as an unusual feature within the valine oxidation pathway. β-oxidation of odd-chain fatty acids, catabolism of branched-chain amino acids, and metabolism of cholesterol side-chain moieties all follow pathways that generate methylmalonyl-CoA, with coenzyme A (CoA) being added early in the catabolic process. Unlike leucine and isoleucine, valine degradation contains a unique intermediate step, carried out by mitochondrial HIBCH, whereby CoA is removed from 3-hydroxyisobutyryl-CoA, producing 3-hydroxyisobutyric acid, an easily diffusible free acid. Two steps later, CoA is reintroduced when methylmalonic semialdehyde is converted to propionyl-CoA (Figure 4). HIBCH is reported to be abundant in human liver and might be important to remove the preceding toxic intermediate, methacrylyl-CoA.56, 57 Animal studies also suggest that release of the HIBCH product, 3-hydroxybutyric acid, might be a mechanism to conserve the gluconeogenic potential of valine.58 Severe mutations in HIBCH are extremely rare and all reported affected children have died early in life.59, 60
What are the consequences of this common but previously unstudied polymorphism? It is possible that the reduced amount of HIBCH enzyme produced by the methionine-encoding allele causes a decreased flux of 3-hydroxyisobutyryl-CoA through HIBCH, perhaps leading to higher mitochondrial concentrations of upstream intermediates and moderately altered C4-hydroxy carnitines in the blood of TT (Met/Met) individuals. This model is consistent with what is observed in subjects with rare severe mutations in this enzyme.59, 61 Methacrylyl-CoA, an intermediate compound in valine catabolism, is highly reactive and forms toxic conjugates with cysteine.62 The concentration of this compound might be increased when HIBCH activity is low (such as T [Met] homozygotes compared to C [?] homozygotes). Higher concentrations of methacrylyl-CoA within mitochondria could inhibit enzymes involved in oxidative phosphorylation.59 It is also possible that the methionine-encoding allele results in a less efficient gluconeogenic system. We did not measure these metabolites, and we did not detect any consistent changes in liver function tests by genotype in our discovery or our replication cohort of older adults (Tables 1 and 2). Additional studies of the phenotypic and metabolic consequences of this polymorphism are warranted. No other phenotypes have been reported to be associated with HIBCH or ACSF3 in the GWAS catalog. Recently, Raffler and colleagues reported an association between variation in HIBCH and an unidentified compound in urine.63 We anticipate that this compound will be a cleared derivative of valine catabolism. The absence of any known associations between these two loci and reported phenotypes might mean that elevated MMA levels in general do not directly contribute to disease, but rather report on its presence or its potential to develop.
Beyond the impact of this SNP, the mechanism of action of the HIBCH rs291466 signal requires elucidation. Although T (Met) is the minor allele for HIBCH rs291466 in this Irish population, phylogenetic analysis suggests that it is likely to be the ancestral allele. T (Met) is also the major allele in the publicly genotyped HapMap CHB, JPT, and YRI samples. The derived C (?) allele results in higher MMA concentrations. Given that MMA is a downstream product of valine catabolism, our metabolic results suggest that the resulting enzyme is more active or more abundant. Because this polymorphism is located at the start of the mitochondrial leader sequence, it might alter the efficiency at which the protein is imported into mitochondria. We tested this model in cell culture studies. Both isoforms of the enzyme were equally efficiently translocated into the mitochondrial matrix (Figure S4). These results suggest that a downstream methionine residue is used to initiate translation and the shortened leader efficiently directs mitochondrial import. Lymphoblastoid RNA-seq data and expression quantitative trait loci (eQTL) analysis demonstrated that, compared to the T (Met) allele, the C (?) allele is associated with a significant increase of HIBCH mRNA levels, presumably resulting in the increased enzyme concentrations that we observed in cultured lymphoblastoid cells. This effect is observed in eQTL analysis (p < 5.5 × 10−06) of more than 30 tissue types, including liver (GTEx Portal, v.6). On the basis of the genetic code alone, it is predicted that the loss of the initiating methionine would alter HIBCH function via translation or at the level of the produced protein. However, our data support the conclusion that it is likely that this SNP or another variant linked to it produces the metabolic phenotype by influencing mRNA transcription or stability. There are 33 SNPs in strong LD with HIBCH rs291466 (r2 > 0.95 in the EUR cohort), all of which are intronic or 5′ of the transcriptional start site and reside in an area containing chromatin marks consistent with active transcription. Further study is required to determine the identity and mechanism of the functional variant underlying the HIBCH rs291466 association with MMA levels.
In conclusion, we have identified two genetic loci that significantly affect the plasma concentration of MMA. These loci provide important data underpinning the current understanding of the pathways leading to MMA production. Our data suggest that genetic variation in HIBCH might be especially relevant in situations where protein catabolism is elevated, i.e., states of acute disease and in the elderly.
Acknowledgments
The authors acknowledge the contributions made by the participants in the Trinity Student Study, the Trinity Ulster Department of Agriculture study, and the longitudinal pregnancy study. The TSS GWAS work was supported in part by the Intramural Research Programs of the National Human Genome Research Institute (NHGRI), the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the NIH, and the Health Research Board, Dublin. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the NIH (https://hpc.nih.gov/systems). C.D.C. is the recipient of an NHGRI Health Disparities Research Fellowship. The TUDA study was funded by the Irish Department of Agriculture, Food and the Marine’s Food Institutional Research Measure initiative and the Northern Ireland Department for Employment and Learning under the Cross-Border Research and Development Programme “Strengthening the all-Island Research Base.” Holotranscobalamin and methylmalonic acid measurements in the TUDA cohort were sponsored by Axis-Shield Diagnostics.
Published: April 28, 2016
Footnotes
Supplemental Data include five figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.03.005.
Contributor Information
Anne M. Molloy, Email: amolloy@tcd.ie.
Lawrence C. Brody, Email: lbrody@mail.nih.gov.
Accession Numbers
The accession number for the TSS genotype and phenotype data reported in this paper is dbGAP: phs000789.v1.p1.
Web Resources
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
Geuvadis RNA Sequencing Project, http://www.geuvadis.org/web/geuvadis/RNAseq-project
GWAS Catalog v.1.0.1, accessed February 2, 2016, http://www.ebi.ac.uk/gwas/
International HapMap GTEx Portal, http://www.gtexportal.org/home/
Internation HapMap Project, http://hapmap.ncbi.nlm.nih.gov/
OMIM, http://www.omim.org/
Supplemental Data
References
- 1.Ludwig M.L., Matthews R.G. Structure-based perspectives on B12-dependent enzymes. Annu. Rev. Biochem. 1997;66:269–313. doi: 10.1146/annurev.biochem.66.1.269. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee R., Chowdhury S. Methylmalonyl-CoA mutase. In: Banerjee R., editor. Chemistry and Biochemistry of B12. John Wiley & Sons, Inc; New York: 1999. pp. 707–729. [Google Scholar]
- 3.Marcadier J.L., Smith A.M., Pohl D., Schwartzentruber J., Al-Dirbashi O.Y., Majewski J., Ferdinandusse S., Wanders R.J., Bulman D.E., Boycott K.M., FORGE Canada Consortium Mutations in ALDH6A1 encoding methylmalonate semialdehyde dehydrogenase are associated with dysmyelination and transient methylmalonic aciduria. Orphanet J. Rare Dis. 2013;8:98. doi: 10.1186/1750-1172-8-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alfares A., Nunez L.D., Al-Thihli K., Mitchell J., Melançon S., Anastasio N., Ha K.C., Majewski J., Rosenblatt D.S., Braverman N. Combined malonic and methylmalonic aciduria: exome sequencing reveals mutations in the ACSF3 gene in patients with a non-classic phenotype. J. Med. Genet. 2011;48:602–605. doi: 10.1136/jmedgenet-2011-100230. [DOI] [PubMed] [Google Scholar]
- 5.Sloan J.L., Johnston J.J., Manoli I., Chandler R.J., Krause C., Carrillo-Carrasco N., Chandrasekaran S.D., Sysol J.R., O’Brien K., Hauser N.S., NIH Intramural Sequencing Center Group Exome sequencing identifies ACSF3 as a cause of combined malonic and methylmalonic aciduria. Nat. Genet. 2011;43:883–886. doi: 10.1038/ng.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valayannopoulos V., Haudry C., Serre V., Barth M., Boddaert N., Arnoux J.B., Cormier-Daire V., Rio M., Rabier D., Vassault A. New SUCLG1 patients expanding the phenotypic spectrum of this rare cause of mild methylmalonic aciduria. Mitochondrion. 2010;10:335–341. doi: 10.1016/j.mito.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Baik H.W., Russell R.M. Vitamin B12 deficiency in the elderly. Annu. Rev. Nutr. 1999;19:357–377. doi: 10.1146/annurev.nutr.19.1.357. [DOI] [PubMed] [Google Scholar]
- 8.Allen L.H. How common is vitamin B-12 deficiency? Am. J. Clin. Nutr. 2009;89:693S–696S. doi: 10.3945/ajcn.2008.26947A. [DOI] [PubMed] [Google Scholar]
- 9.Johnson M.A., Hawthorne N.A., Brackett W.R., Fischer J.G., Gunter E.W., Allen R.H., Stabler S.P. Hyperhomocysteinemia and vitamin B-12 deficiency in elderly using Title IIIc nutrition services. Am. J. Clin. Nutr. 2003;77:211–220. doi: 10.1093/ajcn/77.1.211. [DOI] [PubMed] [Google Scholar]
- 10.Lindenbaum J., Rosenberg I.H., Wilson P.W., Stabler S.P., Allen R.H. Prevalence of cobalamin deficiency in the Framingham elderly population. Am. J. Clin. Nutr. 1994;60:2–11. doi: 10.1093/ajcn/60.1.2. [DOI] [PubMed] [Google Scholar]
- 11.Pennypacker L.C., Allen R.H., Kelly J.P., Matthews L.M., Grigsby J., Kaye K., Lindenbaum J., Stabler S.P. High prevalence of cobalamin deficiency in elderly outpatients. J. Am. Geriatr. Soc. 1992;40:1197–1204. [PubMed] [Google Scholar]
- 12.Reynolds E. Vitamin B12, folic acid, and the nervous system. Lancet Neurol. 2006;5:949–960. doi: 10.1016/S1474-4422(06)70598-1. [DOI] [PubMed] [Google Scholar]
- 13.Stabler S.P., Lindenbaum J., Allen R.H. The use of homocysteine and other metabolites in the specific diagnosis of vitamin B-12 deficiency. J. Nutr. 1996;126(4, Suppl):1266S–1272S. doi: 10.1093/jn/126.suppl_4.1266S. [DOI] [PubMed] [Google Scholar]
- 14.Savage D.G., Lindenbaum J., Stabler S.P., Allen R.H. Sensitivity of serum methylmalonic acid and total homocysteine determinations for diagnosing cobalamin and folate deficiencies. Am. J. Med. 1994;96:239–246. doi: 10.1016/0002-9343(94)90149-x. [DOI] [PubMed] [Google Scholar]
- 15.Morris M.S., Jacques P.F., Rosenberg I.H., Selhub J. Elevated serum methylmalonic acid concentrations are common among elderly Americans. J. Nutr. 2002;132:2799–2803. doi: 10.1093/jn/132.9.2799. [DOI] [PubMed] [Google Scholar]
- 16.Vogiatzoglou A., Oulhaj A., Smith A.D., Nurk E., Drevon C.A., Ueland P.M., Vollset S.E., Tell G.S., Refsum H. Determinants of plasma methylmalonic acid in a large population: implications for assessment of vitamin B12 status. Clin. Chem. 2009;55:2198–2206. doi: 10.1373/clinchem.2009.128678. [DOI] [PubMed] [Google Scholar]
- 17.Bailey R.L., Durazo-Arvizu R.A., Carmel R., Green R., Pfeiffer C.M., Sempos C.T., Carriquiry A., Yetley E.A. Modeling a methylmalonic acid-derived change point for serum vitamin B-12 for adults in NHANES. Am. J. Clin. Nutr. 2013;98:460–467. doi: 10.3945/ajcn.113.061234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey R.L., Carmel R., Green R., Pfeiffer C.M., Cogswell M.E., Osterloh J.D., Sempos C.T., Yetley E.A. Monitoring of vitamin B-12 nutritional status in the United States by using plasma methylmalonic acid and serum vitamin B-12. Am. J. Clin. Nutr. 2011;94:552–561. doi: 10.3945/ajcn.111.015222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sobczyńska-Malefora A., Gorska R., Pelisser M., Ruwona P., Witchlow B., Harrington D.J. An audit of holotranscobalamin (“Active” B12) and methylmalonic acid assays for the assessment of vitamin B12 status: application in a mixed patient population. Clin. Biochem. 2014;47:82–86. doi: 10.1016/j.clinbiochem.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 20.Clarke R., Refsum H., Birks J., Evans J.G., Johnston C., Sherliker P., Ueland P.M., Schneede J., McPartlin J., Nexo E., Scott J.M. Screening for vitamin B-12 and folate deficiency in older persons. Am. J. Clin. Nutr. 2003;77:1241–1247. doi: 10.1093/ajcn/77.5.1241. [DOI] [PubMed] [Google Scholar]
- 21.Hølleland G., Schneede J., Ueland P.M., Lund P.K., Refsum H., Sandberg S. Cobalamin deficiency in general practice. Assessment of the diagnostic utility and cost-benefit analysis of methylmalonic acid determination in relation to current diagnostic strategies. Clin. Chem. 1999;45:189–198. [PubMed] [Google Scholar]
- 22.Molloy A.M., Kirke P.N., Brody L.C., Scott J.M., Mills J.L. Effects of folate and vitamin B12 deficiencies during pregnancy on fetal, infant, and child development. Food Nutr. Bull. 2008;29:S101–S115. doi: 10.1177/15648265080292S114. [DOI] [PubMed] [Google Scholar]
- 23.Bjørke Monsen A.L., Ueland P.M. Homocysteine and methylmalonic acid in diagnosis and risk assessment from infancy to adolescence. Am. J. Clin. Nutr. 2003;78:7–21. doi: 10.1093/ajcn/78.1.7. [DOI] [PubMed] [Google Scholar]
- 24.Desch K.C., Ozel A.B., Siemieniak D., Kalish Y., Shavit J.A., Thornburg C.D., Sharathkumar A.A., McHugh C.P., Laurie C.C., Crenshaw A. Linkage analysis identifies a locus for plasma von Willebrand factor undetected by genome-wide association. Proc. Natl. Acad. Sci. USA. 2013;110:588–593. doi: 10.1073/pnas.1219885110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mills J.L., Carter T.C., Scott J.M., Troendle J.F., Gibney E.R., Shane B., Kirke P.N., Ueland P.M., Brody L.C., Molloy A.M. Do high blood folate concentrations exacerbate metabolic abnormalities in people with low vitamin B-12 status? Am. J. Clin. Nutr. 2011;94:495–500. doi: 10.3945/ajcn.111.014621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carter T.C., Pangilinan F., Molloy A.M., Fan R., Wang Y., Shane B., Gibney E.R., Midttun Ø., Ueland P.M., Cropp C.D. Common Variants at Putative Regulatory Sites of the Tissue Nonspecific Alkaline Phosphatase Gene Influence Circulating Pyridoxal 5′-Phosphate Concentration in Healthy Adults. J. Nutr. 2015;145:1386–1393. doi: 10.3945/jn.114.208769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laird E., McNulty H., Ward M., Hoey L., McSorley E., Wallace J.M., Carson E., Molloy A.M., Healy M., Casey M.C. Vitamin D deficiency is associated with inflammation in older Irish adults. J. Clin. Endocrinol. Metab. 2014;99:1807–1815. doi: 10.1210/jc.2013-3507. [DOI] [PubMed] [Google Scholar]
- 28.Windelberg A., Arseth O., Kvalheim G., Ueland P.M. Automated assay for the determination of methylmalonic acid, total homocysteine, and related amino acids in human serum or plasma by means of methylchloroformate derivatization and gas chromatography-mass spectrometry. Clin. Chem. 2005;51:2103–2109. doi: 10.1373/clinchem.2005.053835. [DOI] [PubMed] [Google Scholar]
- 29.Kelleher B.P., Walshe K.G., Scott J.M., O’Broin S.D. Microbiological assay for vitamin B12 with use of a colistin-sulfate-resistant organism. Clin. Chem. 1987;33:52–54. [PubMed] [Google Scholar]
- 30.Brady J., Wilson L., McGregor L., Valente E., Orning L. Active B12: a rapid, automated assay for holotranscobalamin on the Abbott AxSYM analyzer. Clin. Chem. 2008;54:567–573. doi: 10.1373/clinchem.2007.096784. [DOI] [PubMed] [Google Scholar]
- 31.Lonati S., Novembrino C., Ippolito S., Accinni R., Galli C., Troonen H., Campolo J., Della Noce C., Lunghi G., Catena F.B. Analytical performance and method comparison study of the total homocysteine fluorescence polarization immunoassay (FPIA) on the AxSYM analyzer. Clin. Chem. Lab. Med. 2004;42:228–234. doi: 10.1515/CCLM.2004.041. [DOI] [PubMed] [Google Scholar]
- 32.Rasmussen K., Møller J., Ostergaard K., Kristensen M.O., Jensen J. Methylmalonic acid concentrations in serum of normal subjects: biological variability and effect of oral L-isoleucine loads before and after intramuscular administration of cobalamin. Clin. Chem. 1990;36:1295–1299. [PubMed] [Google Scholar]
- 33.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patterson N., Price A.L., Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 36.Laurie C.C., Doheny K.F., Mirel D.B., Pugh E.W., Bierut L.J., Bhangale T., Boehm F., Caporaso N.E., Cornelis M.C., Edenberg H.J., GENEVA Investigators Quality control and quality assurance in genotypic data for genome-wide association studies. Genet. Epidemiol. 2010;34:591–602. doi: 10.1002/gepi.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lappalainen T., Sammeth M., Friedländer M.R., ’t Hoen P.A., Monlong J., Rivas M.A., Gonzàlez-Porta M., Kurbatova N., Griebel T., Ferreira P.G., Geuvadis Consortium Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Consortium G., GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy M.M., Molloy A.M., Ueland P.M., Fernandez-Ballart J.D., Schneede J., Arija V., Scott J.M. Longitudinal study of the effect of pregnancy on maternal and fetal cobalamin status in healthy women and their offspring. J. Nutr. 2007;137:1863–1867. doi: 10.1093/jn/137.8.1863. [DOI] [PubMed] [Google Scholar]
- 42.Bjørke Monsen A.L., Ueland P.M., Vollset S.E., Guttormsen A.B., Markestad T., Solheim E., Refsum H. Determinants of cobalamin status in newborns. Pediatrics. 2001;108:624–630. doi: 10.1542/peds.108.3.624. [DOI] [PubMed] [Google Scholar]
- 43.Akizu N., Shembesh N.M., Ben-Omran T., Bastaki L., Al-Tawari A., Zaki M.S., Koul R., Spencer E., Rosti R.O., Scott E. Whole-exome sequencing identifies mutated c12orf57 in recessive corpus callosum hypoplasia. Am. J. Hum. Genet. 2013;92:392–400. doi: 10.1016/j.ajhg.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramalho A.S., Lewandowska M.A., Farinha C.M., Mendes F., Gonçalves J., Barreto C., Harris A., Amaral M.D. Deletion of CFTR translation start site reveals functional isoforms of the protein in CF patients. Cell. Physiol. Biochem. 2009;24:335–346. doi: 10.1159/000257426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Hove J.L., Saenz M.S., Thomas J.A., Gallagher R.C., Lovell M.A., Fenton L.Z., Shanske S., Myers S.M., Wanders R.J., Ruiter J. Succinyl-CoA ligase deficiency: a mitochondrial hepatoencephalomyopathy. Pediatr. Res. 2010;68:159–164. doi: 10.1203/PDR.0b013e3181e5c3a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hazra A., Kraft P., Selhub J., Giovannucci E.L., Thomas G., Hoover R.N., Chanock S.J., Hunter D.J. Common variants of FUT2 are associated with plasma vitamin B12 levels. Nat. Genet. 2008;40:1160–1162. doi: 10.1038/ng.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka T., Scheet P., Giusti B., Bandinelli S., Piras M.G., Usala G., Lai S., Mulas A., Corsi A.M., Vestrini A. Genome-wide association study of vitamin B6, vitamin B12, folate, and homocysteine blood concentrations. Am. J. Hum. Genet. 2009;84:477–482. doi: 10.1016/j.ajhg.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grarup N., Sulem P., Sandholt C.H., Thorleifsson G., Ahluwalia T.S., Steinthorsdottir V., Bjarnason H., Gudbjartsson D.F., Magnusson O.T., Sparsø T. Genetic architecture of vitamin B12 and folate levels uncovered applying deeply sequenced large datasets. PLoS Genet. 2013;9:e1003530. doi: 10.1371/journal.pgen.1003530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin X., Lu D., Gao Y., Tao S., Yang X., Feng J., Tan A., Zhang H., Hu Y., Qin X. Genome-wide association study identifies novel loci associated with serum level of vitamin B12 in Chinese men. Hum. Mol. Genet. 2012;21:2610–2617. doi: 10.1093/hmg/dds062. [DOI] [PubMed] [Google Scholar]
- 51.Montgomery J.A., Mamer O.A., Scriver C.R. Metabolism of methylmalonic acid in rats. Is methylmalonyl-coenzyme a racemase deficiency symptomatic in man? J. Clin. Invest. 1983;72:1937–1947. doi: 10.1172/JCI111158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gradinger A.B., Bélair C., Worgan L.C., Li C.D., Lavallée J., Roquis D., Watkins D., Rosenblatt D.S. Atypical methylmalonic aciduria: frequency of mutations in the methylmalonyl CoA epimerase gene (MCEE) Hum. Mutat. 2007;28:1045. doi: 10.1002/humu.9507. [DOI] [PubMed] [Google Scholar]
- 53.Carmel R. Biomarkers of cobalamin (vitamin B-12) status in the epidemiologic setting: a critical overview of context, applications, and performance characteristics of cobalamin, methylmalonic acid, and holotranscobalamin II. Am. J. Clin. Nutr. 2011;94:348S–358S. doi: 10.3945/ajcn.111.013441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Valente E., Scott J.M., Ueland P.M., Cunningham C., Casey M., Molloy A.M. Diagnostic accuracy of holotranscobalamin, methylmalonic acid, serum cobalamin, and other indicators of tissue vitamin B12 status in the elderly. Clin. Chem. 2011;57:856–863. doi: 10.1373/clinchem.2010.158154. [DOI] [PubMed] [Google Scholar]
- 55.Herrmann W., Obeid R., Schorr H., Geisel J. The usefulness of holotranscobalamin in predicting vitamin B12 status in different clinical settings. Curr. Drug Metab. 2005;6:47–53. doi: 10.2174/1389200052997384. [DOI] [PubMed] [Google Scholar]
- 56.Taniguchi K., Nonami T., Nakao A., Harada A., Kurokawa T., Sugiyama S., Fujitsuka N., Shimomura Y., Hutson S.M., Harris R.A., Takagi H. The valine catabolic pathway in human liver: effect of cirrhosis on enzyme activities. Hepatology. 1996;24:1395–1398. doi: 10.1002/hep.510240614. [DOI] [PubMed] [Google Scholar]
- 57.Shimomura Y., Murakami T., Fujitsuka N., Nakai N., Sato Y., Sugiyama S., Shimomura N., Irwin J., Hawes J.W., Harris R.A. Purification and partial characterization of 3-hydroxyisobutyryl-coenzyme A hydrolase of rat liver. J. Biol. Chem. 1994;269:14248–14253. [PubMed] [Google Scholar]
- 58.Letto J., Brosnan M.E., Brosnan J.T. Valine metabolism. Gluconeogenesis from 3-hydroxyisobutyrate. Biochem. J. 1986;240:909–912. doi: 10.1042/bj2400909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ferdinandusse S., Waterham H.R., Heales S.J., Brown G.K., Hargreaves I.P., Taanman J.W., Gunny R., Abulhoul L., Wanders R.J., Clayton P.T. HIBCH mutations can cause Leigh-like disease with combined deficiency of multiple mitochondrial respiratory chain enzymes and pyruvate dehydrogenase. Orphanet J. Rare Dis. 2013;8:188. doi: 10.1186/1750-1172-8-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loupatty F.J., Clayton P.T., Ruiter J.P., Ofman R., Ijlst L., Brown G.K., Thorburn D.R., Harris R.A., Duran M., Desousa C. Mutations in the gene encoding 3-hydroxyisobutyryl-CoA hydrolase results in progressive infantile neurodegeneration. Am. J. Hum. Genet. 2007;80:195–199. doi: 10.1086/510725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peters H., Ferdinandusse S., Ruiter J.P., Wanders R.J., Boneh A., Pitt J. Metabolite studies in HIBCH and ECHS1 defects: Implications for screening. Mol. Genet. Metab. 2015;115:168–173. doi: 10.1016/j.ymgme.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 62.Brown G.K., Hunt S.M., Scholem R., Fowler K., Grimes A., Mercer J.F., Truscott R.M., Cotton R.G., Rogers J.G., Danks D.M. beta-hydroxyisobutyryl coenzyme A deacylase deficiency: a defect in valine metabolism associated with physical malformations. Pediatrics. 1982;70:532–538. [PubMed] [Google Scholar]
- 63.Raffler J., Friedrich N., Arnold M., Kacprowski T., Rueedi R., Altmaier E., Bergmann S., Budde K., Gieger C., Homuth G. Genome-Wide Association Study with Targeted and Non-targeted NMR Metabolomics Identifies 15 Novel Loci of Urinary Human Metabolic Individuality. PLoS Genet. 2015;11:e1005487. doi: 10.1371/journal.pgen.1005487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.