Abstract
Facioscapulohumeral dystrophy (FSHD) is associated with somatic chromatin relaxation of the D4Z4 repeat array and derepression of the D4Z4-encoded DUX4 retrogene coding for a germline transcription factor. Somatic DUX4 derepression is caused either by a 1–10 unit repeat-array contraction (FSHD1) or by mutations in SMCHD1, which encodes a chromatin repressor that binds to D4Z4 (FSHD2). Here, we show that heterozygous mutations in DNA methyltransferase 3B (DNMT3B) are a likely cause of D4Z4 derepression associated with low levels of DUX4 expression from the D4Z4 repeat and increased penetrance of FSHD. Recessive mutations in DNMT3B were previously shown to cause immunodeficiency, centromeric instability, and facial anomalies (ICF) syndrome. This study suggests that transcription of DUX4 in somatic cells is modified by variations in its epigenetic state and provides a basis for understanding the reduced penetrance of FSHD within families.
Main Text
Facioscapulohumeral dystrophy (FSHD [OMIM: 158900 and 158901]) is a common muscular dystrophy typically manifesting in the second decade and characterized by progressive weakness and atrophy of the facial and upper-extremity muscles. With disease progression, other muscles also become affected.1 A clinical hallmark of the disease is the variability in onset and progression, such that 20% of mutation carriers eventually become wheelchair dependent, and a similar proportion of mutation carriers remain asymptomatic.2
The common form of the disease, FSHD1, is associated with a 1–10 unit contraction of the polymorphic D4Z4 macrosatellite repeat array on chromosome arm 4q (Figure 1A).3, 4 In the healthy control population, this array varies from 8 to 100 units, and 1%–3% of individuals carry an FSHD-sized allele of 8–10 units.5, 6 Each unit of the repeat array contains a copy of the retrogene double homeobox 4 (DUX4 [OMIM: 606009]), which is normally expressed in the testis and silenced in somatic tissue.7 In FSHD1, the epigenetic repression of DUX4 is incomplete in somatic cells, leading to sporadic DUX4 expression in myonuclei.7, 8 Stable DUX4 transcripts are only produced in combination with a polymorphic polyadenylation signal (PAS) immediately distal to the D4Z4 repeat array present in 4qA chromosomal regions, of which two major variants exist (4qA-S and 4qA-L) (Figure 1A).9 Contractions of the highly homologous repeat arrays in 4qB or 4q10 are non-pathogenic because of the absence of a DUX4 PAS.9
Figure 1.
D4Z4 Locus and FSHD2-Affected Families
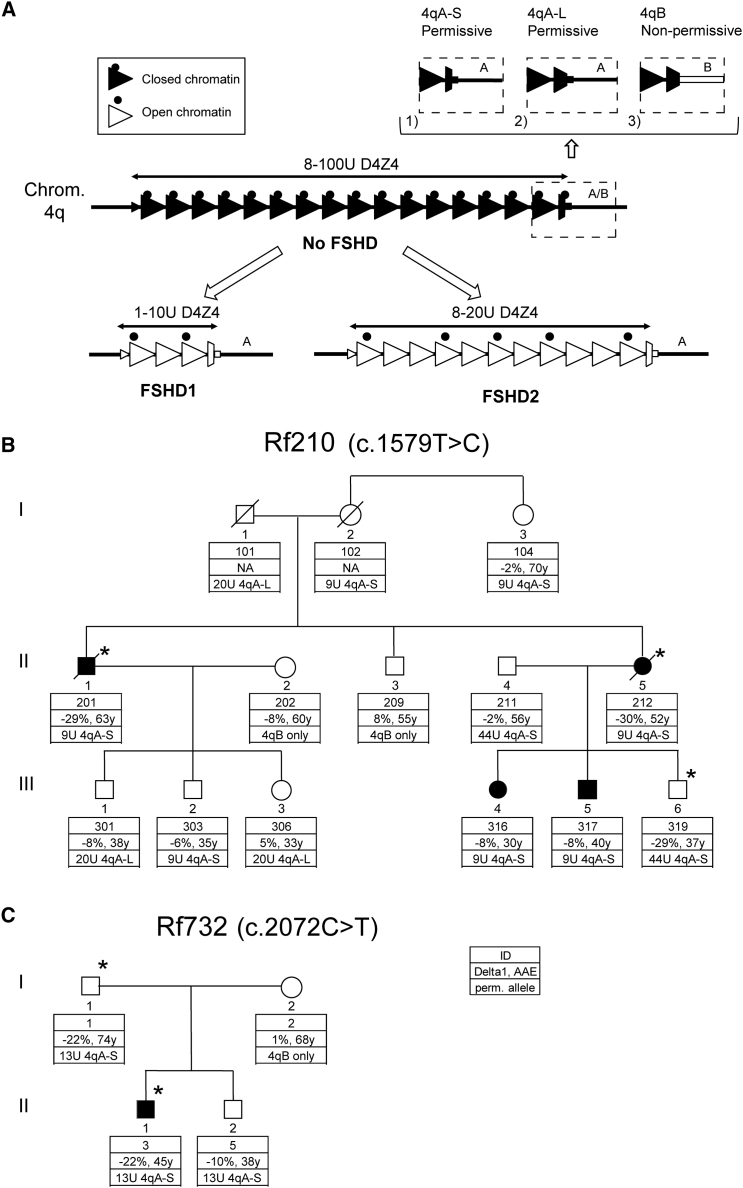
(A) Schematic representation of the D4Z4 locus. In control individuals, the D4Z4 repeat array ranges from 8 to 100 units and shows characteristics of a closed chromatin structure (black triangles) characterized by high CpG methylation, among other things. For both FSHD1 and FSHD2, the chromatin adopts a more open configuration (white triangles) marked by a loss of CpG methylation and other chromatin changes. FSHD1 is caused by a contraction of the D4Z4 repeat to 1–10 units, whereas FSHD2 involves chromatin relaxation due to mutations that affect a chromatin modifier (black dots), most often SMCHD1. The chromatin relaxation must occur in a permissive 4qA (marked by 4qA-S in this figure) or 4qA-L chromosomal region to cause FSHD, given that 4qB chromosomes are non-permissive for FSHD (chromosome 4 variants are displayed in the dashed boxes).9 4qA-S and 4qA-L differ by the length of the last partial D4Z4 unit, and protein studies have demonstrated production of DUX4 from both 4qA variants. The 3′ UTR of DUX4 is missing in 4qB chromosomal regions (white square in dashed box), which makes them non-permissive to DUX4 expression.
(B and C) Pedigrees of families Rf210 (B) and Rf732 (C). Clinically affected individuals are indicated in black. The key shows the family identifier (ID), Delta1 score, age at examination (AAE), and size of the smallest D4Z4 repeat array on a FSHD-permissive allele (4qA-S and 4qA-L). Additionally, it indicates when no permissive allele was present (4qB only). The cDNA position behind the family ID indicates the cDNA position of the DNMT3B mutation (GenBank: NM_006892.3) present in this family. The asterisk indicates individuals carrying the DNMT3B mutation.
Somatic repression of DUX4 requires a combination of epigenetic mechanisms, and D4Z4 hypomethylation has consistently been reported as an aberrant epigenetic feature in FSHD.10, 11, 12, 13 In FSHD1, D4Z4 hypomethylation is restricted to the contracted allele. In the rare FSHD2 type of the disease, D4Z4 hypomethylation is observed on all D4Z4 repeat arrays in the absence of D4Z4 contractions (Figure 1A).14, 15 D4Z4 methylation linearly correlates with the size of the D4Z4 array in control and FSHD-affected individuals.16 FSHD2-affected individuals often carry smaller but normally sized D4Z4 repeat arrays (8–20 units), given that this renders them more susceptible to further D4Z4 hypomethylation.14, 16 Dominant segregation of D4Z4 hypomethylation in FSHD2-affected families was instrumental in identifying mutations in SMCHD1 (structural maintenance of chromosomes flexible hinge domain-containing 1 [OMIM: 614982]) in >85% of these families.17 SMCHD1 is a chromatin repressor involved in the establishment and/or maintenance of CpG methylation at specific loci and binds directly to D4Z4.17, 18, 19 Therefore, the disease presentation in FSHD2 depends on a combination of repeat length and damaging potential of the SMCHD1 mutation.16 Mutations in SMCHD1 have also been reported as modifiers of disease severity in FSHD1-affected families with alleles of 8–10 D4Z4 units.20, 21 Thus, D4Z4 methylation is dependent on repeat-array size and on the activity of the partially characterized D4Z4-repressive mechanisms. Deviations in the expected D4Z4 methylation, expressed as the Delta1 factor, can be diagnostic for the presence of damaging variants in D4Z4-chromatin modifiers. Indeed, Delta1 factors ≤ −22% are generally associated with mutations in SMCHD1.16
Because FSHD2 cannot be explained by SMCHD1 mutations in all affected families, we applied exome sequencing in eight families in whom we found D4Z4 hypomethylation without evidence of an exonic SMCHD1 mutation (Figures 1B and 1C and Figure S1). All samples were obtained in an anonymized manner, and all families gave consent. The study was approved by the medical ethics committees of the Leiden University Medical Center and the Radboud University Medical Center Nijmegen. Whole-exome sequencing (WES) was performed by deCODE Genetics (Reykjavik) in the context of the European Union’s NeurOmics project. To identify variants, we analyzed the WES data by using the deCODE Clinical Sequence Miner. We performed dominant analysis for multiple case and control individuals and annotated gene variants (with moderate to high Variant Effect Predictor consequences) to identify possible dominant mutations. Under these conditions, in two families we identified a potentially damaging variant in DNMT3B (DNA methyltransferase 3B [OMIM: 602900]), encoding a known D4Z4-chromatin modifier. These variants have not been reported previously in dbSNP, the 1000 Genomes Project, the National Heart, Lung, and Blood Institute Exome Sequencing Project (ESP) Exome Variant Server, the Exome Aggregation Consortium (ExAC) Browser, or in-house databases.
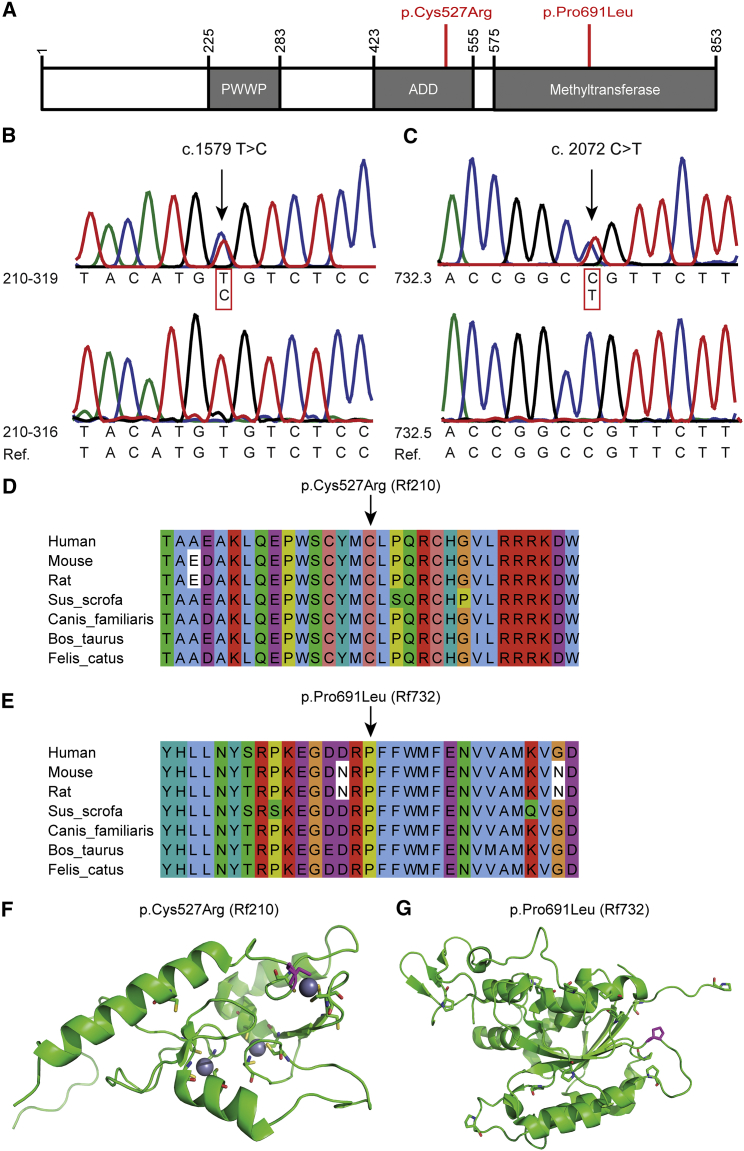
Family Rf210 is a FSHD1-affected family with a 9 unit D4Z4 array in a permissive 4qA chromosomal region (Figure 1B and Table S1). Despite the presence of this disease allele in seven family members, only four of them are clinically affected, whereas one carrier (Rf210.102 [I-2]) could not be clinically examined. D4Z4 methylation at the FseI site was determined by Southern blotting and was expressed as the Delta1 score, which is the observed methylation corrected for the size of the repeat array at the FseI site in D4Z4.16 In Rf210, analysis of D4Z4 methylation identified robust D4Z4 hypomethylation in two severely affected individuals (Rf210.201 [II-1] and Rf210.212 [II-5]) and one clinically unaffected individual (Rf210.319 [III-6]), as evidenced by the strongly reduced Delta1 values. These reduced Delta1 values indicate the involvement of a defective D4Z4-chromatin modifier. Genetic studies excluded the involvement of the SMCHD1 locus (Figure S2), but exome sequencing identified a potentially damaging DNMT3B variant co-segregating with D4Z4 hypomethylation (Figures 2A and 2B and Table S1). This variant (c.1579T>C [p.Cys527Arg] [GenBank: NM_006892.3]) was confirmed by Sanger sequencing and disrupts the C2C2-type zinc-finger motif in the ATRX-DNMT3-DNMT3L (ADD) domain, a highly conserved domain that can be found in several chromatin-associated proteins that play a role in establishing and/or maintaining a normal DNA-methylation pattern (Figures 2B, 2D, and 2F).22, 23 Like SMCHD1, DNMT3B was previously identified as a suppressor of murine metastable epialleles, alleles that display unusual variable expressivity in the absence of genetic heterogeneity depending on their epigenetic state.18, 24, 25 In these Dnmt3b-hypomorphic mice, the ADD domain also seems to be primarily affected.26
Figure 2.
DNMT3B Mutations in FSHD2
(A) Schematic representation of DNMT3B. The amino acid changes (GenBank: NP_008823.1) found in FSHD2-affected families are indicated in red.
(B and C) Sanger sequence confirmation of DNMT3B variants (GenBank: NM_006892.3) in Rf210 and Rf732.
(D and E) Multiple-sequence alignment (MSA) of DNMT3B across distinct species for DNMT3B variants in Rf210 and Rf732. MSA was performed with ClustalOmega, and alignment was viewed in Jalview and colored as in ClustalX.
(F) Ribbon representation of the nuclear-magnetic-resonance structure of the ADD domain of ATRX (PDB: 2JM1).22 The cysteine residues are shown as sticks. Cys527 is shown in magenta. Zinc ions are represented as spheres.
(G) Ribbon representation of the crystallography structure of the C-terminal domain of DNMT3A (chain A [PDB: 2QRV]). The proline residues are shown as sticks. Pro691 is shown in magenta.
In family Rf210, the DNMT3B variant perfectly segregates with D4Z4 hypomethylation, but not with disease presentation. DNMT3B-mutation carrier Rf210.319 (III-6; Figure 1B) might be protected from disease presentation because of the large size of the FSHD-permissive D4Z4 repeat (44 units). This is reminiscent of the situation in SMCHD1-mutation carriers, where individuals with smaller, normally sized D4Z4 repeat arrays (8–20 units) have a greater likelihood of developing FSHD than do individuals with larger repeat arrays.16 The two DNMT3B-variant carriers with a 9 unit D4Z4 array, however, have an age-corrected clinical severity score (ACCS) greater than that of the carriers of only a 9 unit D4Z4 allele. This suggests that the DNMT3B variant acts as a modifier of disease severity in this FSHD1-affected family, similarly to the SMCHD1 mutation in FSHD1-affected families.20 Of the four carriers of a 9 unit D4Z4 array without the DNMT3B variant, two are clinically unaffected (Rf210.104 [I-3] and Rf210.303 [III-2]). This variability in severity is typical for this borderline-FSHD1 repeat-array size. Indeed, 1%–3% of the control population carries an 8–10 unit array on a permissive allele, demonstrating the strongly reduced penetrance of these alleles.5, 6 Penetrance is dependent on age and the degree of D4Z4-chromatin relaxation in somatic tissue, among other things.12, 16, 27
In family Rf732, the index individual (Rf732.3 [II-1]) carries a 13 unit D4Z4 repeat array in a 4qA chromosomal region (Figure 1C and Table S1), and it is also present in his unaffected father and brother. Methylation analysis showed that Rf732.3 (II-1) and his father (Rf732.1 [I-1]) had severe D4Z4 hypomethylation on all four alleles with reduced Delta1 values. Exome sequencing identified a potentially damaging variant affecting a highly conserved residue in the enzymatic domain of DNMT3B (DNMT3B c.2072C>T [p.Pro691Leu] [GenBank: NM_006892.3]) in the index individual and his father; it was confirmed by Sanger sequencing and was absent in the son with normal D4Z4 methylation (Figures 2A, 2C, 2E, and 2G). Although Rf732.1 (I-1) and Rf732.3 (II-1) both carry this DNMT3B variant, have the same Delta1 value, and have a 13 unit FSHD-permissive D4Z4 allele, only Rf732.3 (II-1) is clinically affected. This family emphasizes the reduced penetrance that is typical of FSHD.16, 27 The Delta1 value in this family is low, but not as low as typically found in SMCHD1-mutation carriers.16 This suggests a lesser degree of D4Z4-chromatin relaxation in this family, which might explain why the father has remained unaffected.
Analysis of all coding exons of DNMT3B in 25 additional individuals with a permissive D4Z4 allele and mildly to severely reduced D4Z4 methylation, but not exonic SMCHD1 mutations, did not identify additional mutations in DNMT3B (Tables S2 and S4).
Biallelic DNMT3B mutations have been reported in autosomal-recessive immunodeficiency, centromeric instability, and facial anomalies syndrome type 1 (ICF1 [OMIM: 242860]).28, 29 This primary immunodeficiency syndrome is characterized by hypo- or agammaglobulinemia with B cells and by a distinct facial appearance. There is a progressive decrease in B and T cells during childhood and adolescence.30, 31 The cytogenetic hallmark of ICF syndrome is the presence of chromosome abnormalities involving the juxtacentromeric domains of chromosomes 1, 9, and 16 in metaphase spreads of phytohemagglutinin (PHA)-stimulated cells.30, 32 ICF1-affected individuals show CpG hypomethylation of juxtacentromeric satellite repeat types II and III and the macrosatellite repeats NBL2 and D4Z4.33, 34 ICF1 mutations most often affect the catalytic domain of DNMT3B and are believed to result in strongly reduced DNMT3B activity.31
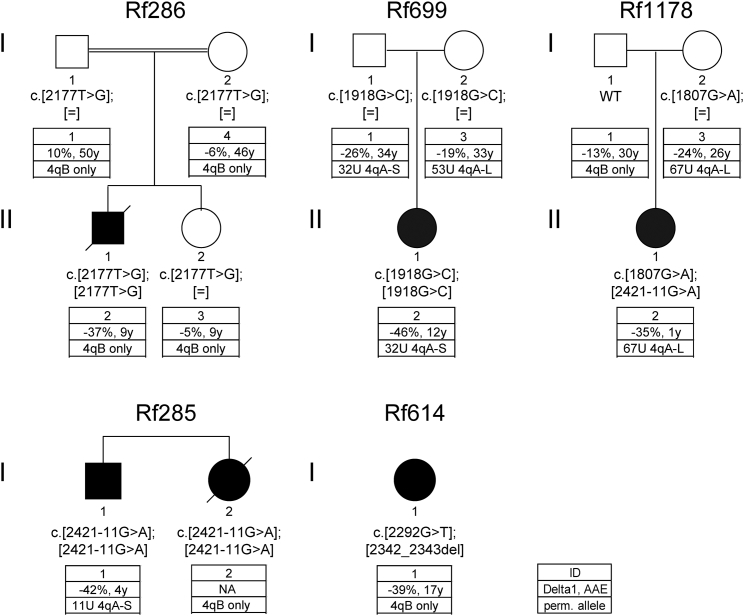
Because our data suggest that FSHD2 and ICF1 can both be caused by DNMT3B mutations—dominant mutations for FSHD2 and recessive mutations for ICF1—we analyzed six ICF1 individuals belonging to five families (Rf285, Rf286, Rf614, Rf699, and Coriell Cell Repositories family 2081, here annotated as Rf1178) for D4Z4 repeat arrays, the presence of a DUX4 PAS, D4Z4 hypomethylation, and DUX4 expression (Figure 3). If possible, we also included unaffected relatives. Table S3 lists all ICF1-affected families with reference to their original description. Consistent with earlier reports,33 methylation analysis showed that all ICF1 individuals tested had severe D4Z4 hypomethylation with Delta1 values varying between −35% and −46% (Figure 3). However, depending on the mutation, some heterozygous carriers (parents of Rf699 and mother of Rf1178) also showed reduced Delta1 values, similar to what we observed in our FSHD2-affected families (−19% to −26%). This not only suggests an additive effect of both DNMT3B mutations in the affected ICF1 children but also puts ICF1-mutation carriers with a reduced Delta1 value at risk of stable DUX4 expression and FSHD if the mutation is combined with a DUX4 PAS. Analysis of D4Z4-repeat sizes, however, showed that about half of the heterozygous DNMT3B carriers in our ICF1-affected families do not carry a FSHD-permissive chromosome. For those who do have D4Z4 repeat arrays on FSHD-permissive chromosomes (containing a DUX4 PAS), the arrays are well beyond the size of what is typically found in FSHD2 individuals (Figure 3). The smallest permissive D4Z4 repeat array found in these heterozygous DNMT3B carriers contained 32 units, suggesting that these individuals might be protected from somatic DUX4 expression because of their long D4Z4 repeat arrays, given that in FSHD2, we already demonstrated a D4Z4-repeat-size-dependent penetrance for SMCHD1 mutations.16 In concordance, to our knowledge, muscle weakness has never been reported in ICF1-mutation carriers.
Figure 3.
Pedigrees of Families Rf286, Rf699, Rf1178, Rf285, and Rf614, Affected by Autosomal-Recessive ICF1
Affected individuals are indicated in black, and DNMT3B mutations (GenBank: NM_006892.3) are shown below each individual. Their clinical phenotypes and DNMT3B mutations have been described before.28, 30, 31, 35, 36 The key description is identical to that in Figure 1.
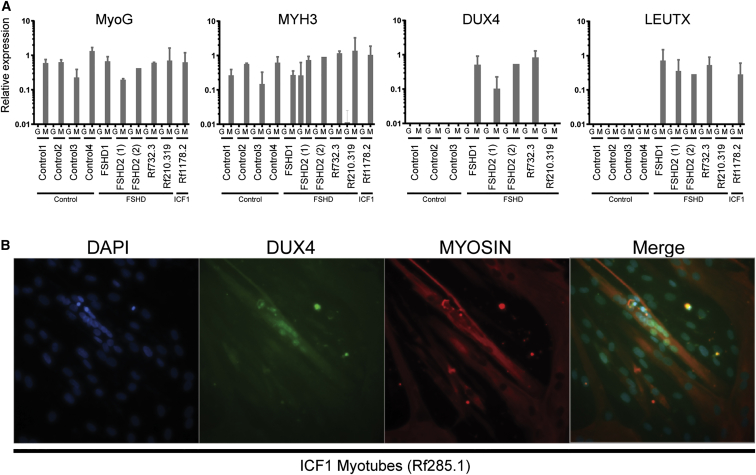
To address the possibility of DUX4 expression in carriers of a single DNMT3B mutation, we trans-differentiated primary fibroblasts of control individuals, FSHD1 and FSHD2 individuals, and unaffected and affected carriers of an FSHD2 mutation in DNMT3B (Rf210.319 [III-6] in Figure 1B and Rf732.3 [II-1] in Figure 1C, respectively) into myotubes by lentiviral MyoD expression. A lentivirus containing GFP or FLAG was used as a control. To examine differentiation, we measured MYOG (OMIM: 159980) and MYH3 (OMIM: 160720) expression levels by qPCR.37, 38 For almost all cell lines, we observed MYOG and MYH3 expression only in the fibroblasts transduced with MyoD, indicating that these cells were trans-differentiated into myogenic cells (Figure 4A). In one FSHD2 cell line (FSHD2-1), MYH3 expression was detected in the GFP-transduced fibroblast as well, possibly because of a technical or biological artifact. We next analyzed the expression of DUX4 and three DUX4 target genes (LEUTX, TRIM43, and PRAMEF2) by qPCR and gel electrophoresis.39 We found expression of DUX4 and DUX4 target genes in MyoD-transduced fibroblasts of FSHD2-affected individual Rf732.3 (II-1, who has a 13 unit D4Z4 repeat array), but not in unaffected individual Rf210.319 (III-6, who has a 44 unit array in a 4qA chromosomal region) (Figure 4A and Figure S3A). No DUX4 expression or upregulated expression of DUX4 target genes was detected in GFP-transduced fibroblasts, and no fibroblasts were available from other FSHD2-affected family members. These data are consistent with the suggestion that heterozygous DNMT3B mutations, only when combined with smaller D4Z4 repeat arrays, can derepress DUX4 in somatic cells and cause FSHD.
Figure 4.
DUX4 Presence in FSHD and ICF1
(A) Expression of MYOG, MYH3, DUX4, and LEUTX (DUX4 target) by qPCR in GFP (G)- or MyoD (M)-lentivirus-transduced fibroblasts from control individuals, FSHD1 and FSHD2 cell lines, and individuals Rf210.319, Rf732.3, and ICF-affected Rf1178.2. All transductions were performed twice for each cell line, except for control individual 4 (1× transduced with GFP and 2× transduced with MyoD) and FSHD2-2 (transduced 1× with GFP and 1× with MyoD). Mean expression values with SDs are shown in relation to those of the reference genes GUSB and RPL27. DUX4 was measured with primers for the most common DUX4-4A-S variant, but the primers did not recognize DUX4-4A-L. The fibroblasts from control individual 4 and Rf1178.2 carry a 4qA-L allele and were therefore excluded from analysis of DUX4 expression. Primers are listed in Table S5.
(B) Immunofluorescent staining for DUX4 and Myosin in fixed ICF1 myotubes from Rf285.1 (Figure 3) shows DUX4 immunoreactivity in a small percentage of myotubes.
To investigate DUX4 expression in ICF1, we trans-differentiated three primary fibroblast cell lines of ICF1 individuals (Rf699.2 [II-1], Rf614.1 [I-1], and Rf1178.2 [II-1]; Figure 3). In Rf699.2 (II-1), who has a 32 unit permissive D4Z4 array, we detected DUX4 in the MyoD-transduced fibroblasts (Figure S3B). DUX4 could not be detected in Rf614.1 (I-1) because she carries two 4qB alleles, which are unable to produce a stable DUX4 transcript (Figure S3B). Our DUX4 primers recognize the most common DUX4-4A-S variant, but not the DUX4-4A-L variant, which is produced from 4qA-L repeats. Because Rf1178.2 (II-1) carries a 4qA-L repeat, we were unable to directly detect DUX4 (Figure S3B). However, the expression of DUX4 target genes was detected in Rf1178.2 (II-1), suggesting that these fibroblasts produce DUX4 (Figure 4A and Figure S3A). These results show that MyoD-transduced fibroblasts in ICF1-affected individuals can produce small amounts of DUX4, indicating that when both DNMT3B alleles are mutated, the epigenetic derepression is sufficient to facilitate DUX4 expression from D4Z4 repeats (Figure S3B). Additionally, myotubes were available from one ICF1 individual from a different family (Rf285.1 [I-1]; Figure 3); this individual has an 11 unit D4Z4 repeat on a FSHD-permissive chromosome 4, and we detected small amounts of DUX4 by immunofluorescent staining (Figure 4B). This ICF1 individual (Rf285.1 [I-1]) might still be too young (15 years) to develop FSHD. Possibly, the short life expectancy of ICF1 individuals in general might obscure the diagnosis of muscle weakness.
Conversely, although ICF1-mutation carriers are reported to be unaffected, we explored the possibility that dominant DNMT3B mutations identified in our FSHD2-affected families might have epigenetic consequences similar to those found in ICF1 or clinical features reminiscent of ICF syndrome. Metaphase analysis of PHA-stimulated peripheral-blood mononuclear cell cultures of FSHD DNMT3B-mutation carrier Rf210.319 (III-6; Figure 1B), but not Rf732.3 (II-1, Figure 1C), indicated a low frequency of formation of multi-branched chromosomes (Figures 5A and 5B). Chromosome decondensations, breaks, and deletions can be found at low frequencies also in ICF1-mutation carriers and control individuals,32 but the formation of multi-branched chromosomes might be specific to the presence of DNMT3B mutations, even in heterozygous carriers. Rf210.319 (III-6) also showed evidence of mild NBL2 hypomethylation in a Southern blot assay, given that the NBL2 repeat is sensitive to digestion by the methylation-sensitive endonuclease Eco52I, albeit to a lesser degree than observed in ICF1 individuals (Figure 5C). Similarly, one heterozygous ICF1-mutation carrier with strongly reduced Delta1 values for D4Z4 (Rf699.1 [I-1]; Figure 3) also showed mild NBL2 hypomethylation (Figure 5C). The fact that not all carriers of the same variant showed NBL2 hypomethylation suggests that heterozygous DNMT3B variants can cause mild and variable NBL2 hypomethylation. Clinically, however, DNMT3B-mutation carrier Rf210.319 (III-6) and his siblings, Rf210.316 (III-4) and Rf210.317 (III-5), do not show signs or features of ICF syndrome and have normal serum immunoglobulin levels and normal numbers of B cells and T cell subsets (Figure S4).
Figure 5.
Metaphase Analysis and NBL2 Southern Blot Analysis of Rf210, Rf732, and ICF1-Affected Families
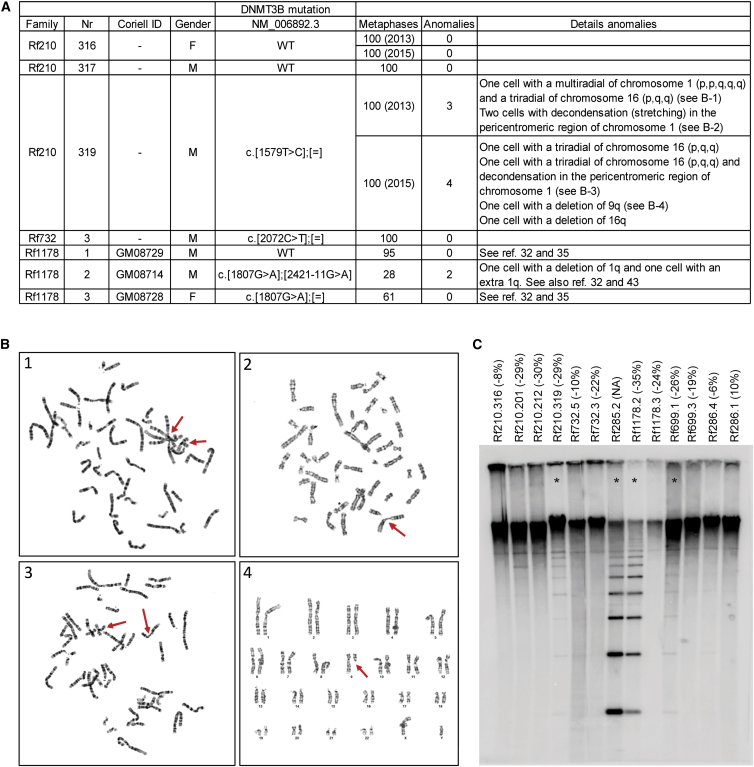
(A) Metaphases were analyzed from three heterozygous DNMT3B-mutation carriers (Rf210.319, Rf732.3, and Rf1178.3), one ICF1 individual (Rf1178.2), and three individuals without a DNMT3B variant (Rf210.316, Rf210.317, and Rf1178.1). Identifiers from Leiden University Medical Center and Coriell, the mutation in DNMT3B (GenBank: NM_006892.3), and the number of analyzed metaphases are indicated. Chromosomal anomalies are listed in the last column.
(B) Four panels show examples of chromosomal anomalies identified in individual Rf210.319. Chromosomal anomalies are indicated with red arrows.
(C) NBL2 Southern blot analysis in Rf210, Rf732, and ICF1-affected families after digestion of 2 μg genomic DNA with the methylation-sensitive endonuclease Eco52I according to previously described protocols.40 Numbers correspond with pedigrees in Figures 1 and 3. Delta1 scores are indicated in brackets. NBL2 was only hypomethylated in the four individuals indicated with an asterisk.
These observations raise the question of why DNMT3B mutations can cause such discordant phenotypes. Mutations that affect the ADD domain of DNMT3B have never been reported in ICF syndrome, but mutations disrupting the ADD domain of DNMT3A have been associated with Tatton-Brown-Rahman syndrome (OMIM: 615879), an overgrowth syndrome with intellectual disability.41 Similarly, mutations that disturb the ADD domain of ATRX have been reported in alpha thalassemia-mental retardation syndrome, X-linked (ATR-X [OMIM: 301040]).22 The ADD domains of ATRX, DNMT3A, and DNMT3B bind to the N terminus of the histone 3 (H3) tail lacking the active lysine 4 (H3K4) methylation mark, where they integrate histone-modification status with DNA methylation.42 Binding of the ADD domain of DNMT3A to the H3 tail stimulates the catalytic activity of this enzyme.43, 44, 45 Likewise, it is possible that the mutation that affects the ADD domain of DNMT3B in family Rf210 also disrupts the DNA-methylation activity of DNMT3B. However, most of the ICF1 mutations, such as the mutation in family Rf732, are located in exons that encode the catalytic domain of DNMT3B. It is not well known why mutations in DNMT3B cause a primary immunodeficiency, but the absence of an immunological phenotype in our FSHD2 families might be explained by the presence of one wild-type DNMT3B allele, given that heterozygous ICF1-mutation carriers also do not present with immunological abnormalities.
Our study implicates that mutations in DNMT3B act as a modifier in FSHD. We propose that, like for SMCHD1, the effect of DNMT3B mutations on DUX4 expression and disease presentation depends on the presence of a DUX4 PAS and on the size of the D4Z4 repeat array. This, combined with the relatively young age at which ICF1 individuals typically succumb to their immunodeficiency, might explain the absence of FSHD in ICF1-affected families. These observations also suggest that FSHD1 and FSHD2 represent polar extremes of a continuous disease mechanism determined by the interaction among D4Z4-repeat size, the presence of a DUX4 PAS, and variations in genes that modify the D4Z4 epigenetic state and provide a firm basis for understanding reduced disease penetrance in the FSHD population.
Acknowledgments
We thank all families for participating in our studies. We thank Marcellus Ubbink (Leiden Institute of Chemistry, Leiden University) for assistance with modeling the mutations and Nisha Verwey (Human Genetics, Leiden University Medical Center) for assistance with the Cellomics platform. Our studies are supported by grants from the NIH National Institute of Neurological Disorders and Stroke (P01NS069539), the Prinses Beatrix Spierfonds (W.OR12-20 and W.OP14-01), the European Union Framework Programme 7 (agreement 2012-305121, NEUROMICS), the FSH Society, Spieren voor Spieren, the FSHD Global Research Foundation, FSHD Stichting, and Friends of FSH Research.
Published: May 5, 2016
Footnotes
Supplemental Data include four figures and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.03.013.
Accession Numbers
The mutations reported in this paper have been deposited in the Leiden Open Variation Database under accession numbers LOVD: 00059205, 00059206, 00059223, 00059224, and 00059225.
Web Resources
1000 Genomes, http://www.1000genomes.org/
Alamut Visual, http://www.interactive-biosoftware.com/alamut-visual/
Exome Aggregation Consortium (ExAC) Browser, http://exac.broadinstitute.org/
Leiden Open Variation Database (LOVD), http://www.lovd.nl/3.0/home
Mutalyzer, https://mutalyzer.nl/
NIGMS Human Genetic Cell Repository, https://catalog.coriell.org/1/NIGMS
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
Richard Fields Center for FSHD Research, https://www.urmc.rochester.edu/fields-center.aspx
Variant Effect Predictor, http://useast.ensembl.org/info/docs/tools/vep/index.html
Supplemental Data
References
- 1.Padberg G.W., Lunt P.W., Koch M., Fardeau M. Diagnostic criteria for facioscapulohumeral muscular dystrophy. Neuromuscul. Disord. 1991;1:231–234. doi: 10.1016/0960-8966(91)90094-9. [DOI] [PubMed] [Google Scholar]
- 2.Tawil R., van der Maarel S.M., Tapscott S.J. Facioscapulohumeral dystrophy: the path to consensus on pathophysiology. Skelet. Muscle. 2014;4:12. doi: 10.1186/2044-5040-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Deutekom J.C., Wijmenga C., van Tienhoven E.A., Gruter A.M., Hewitt J.E., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum. Mol. Genet. 1993;2:2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- 4.Wijmenga C., Hewitt J.E., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet. 1992;2:26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- 5.Lemmers R.J., Wohlgemuth M., van der Gaag K.J., van der Vliet P.J., van Teijlingen C.M., de Knijff P., Padberg G.W., Frants R.R., van der Maarel S.M. Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2007;81:884–894. doi: 10.1086/521986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scionti I., Fabbri G., Fiorillo C., Ricci G., Greco F., D’Amico R., Termanini A., Vercelli L., Tomelleri G., Cao M. Facioscapulohumeral muscular dystrophy: new insights from compound heterozygotes and implication for prenatal genetic counselling. J. Med. Genet. 2012;49:171–178. doi: 10.1136/jmedgenet-2011-100454. [DOI] [PubMed] [Google Scholar]
- 7.Snider L., Geng L.N., Lemmers R.J., Kyba M., Ware C.B., Nelson A.M., Tawil R., Filippova G.N., van der Maarel S.M., Tapscott S.J., Miller D.G. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 2010;6:e1001181. doi: 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tassin A., Laoudj-Chenivesse D., Vanderplanck C., Barro M., Charron S., Ansseau E., Chen Y.W., Mercier J., Coppée F., Belayew A. DUX4 expression in FSHD muscle cells: how could such a rare protein cause a myopathy? J. Cell. Mol. Med. 2013;17:76–89. doi: 10.1111/j.1582-4934.2012.01647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemmers R.J., van der Vliet P.J., Klooster R., Sacconi S., Camaño P., Dauwerse J.G., Snider L., Straasheijm K.R., van Ommen G.J., Padberg G.W. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329:1650–1653. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Overveld P.G., Lemmers R.J., Sandkuijl L.A., Enthoven L., Winokur S.T., Bakels F., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat. Genet. 2003;35:315–317. doi: 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- 11.Huichalaf C., Micheloni S., Ferri G., Caccia R., Gabellini D. DNA methylation analysis of the macrosatellite repeat associated with FSHD muscular dystrophy at single nucleotide level. PLoS ONE. 2014;9:e115278. doi: 10.1371/journal.pone.0115278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones T.I., King O.D., Himeda C.L., Homma S., Chen J.C., Beermann M.L., Yan C., Emerson C.P., Jr., Miller J.B., Wagner K.R., Jones P.L. Individual epigenetic status of the pathogenic D4Z4 macrosatellite correlates with disease in facioscapulohumeral muscular dystrophy. Clin. Epigenetics. 2015;7:37. doi: 10.1186/s13148-015-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartweck L.M., Anderson L.J., Lemmers R.J., Dandapat A., Toso E.A., Dalton J.C., Tawil R., Day J.W., van der Maarel S.M., Kyba M. A focal domain of extreme demethylation within D4Z4 in FSHD2. Neurology. 2013;80:392–399. doi: 10.1212/WNL.0b013e31827f075c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Greef J.C., Lemmers R.J., Camaño P., Day J.W., Sacconi S., Dunand M., van Engelen B.G., Kiuru-Enari S., Padberg G.W., Rosa A.L. Clinical features of facioscapulohumeral muscular dystrophy 2. Neurology. 2010;75:1548–1554. doi: 10.1212/WNL.0b013e3181f96175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balog J., Thijssen P.E., de Greef J.C., Shah B., van Engelen B.G., Yokomori K., Tapscott S.J., Tawil R., van der Maarel S.M. Correlation analysis of clinical parameters with epigenetic modifications in the DUX4 promoter in FSHD. Epigenetics. 2012;7:579–584. doi: 10.4161/epi.20001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemmers R.J., Goeman J.J., van der Vliet P.J., van Nieuwenhuizen M.P., Balog J., Vos-Versteeg M., Camano P., Ramos Arroyo M.A., Jerico I., Rogers M.T. Inter-individual differences in CpG methylation at D4Z4 correlate with clinical variability in FSHD1 and FSHD2. Hum. Mol. Genet. 2015;24:659–669. doi: 10.1093/hmg/ddu486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemmers R.J., Tawil R., Petek L.M., Balog J., Block G.J., Santen G.W., Amell A.M., van der Vliet P.J., Almomani R., Straasheijm K.R. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet. 2012;44:1370–1374. doi: 10.1038/ng.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blewitt M.E., Gendrel A.V., Pang Z., Sparrow D.B., Whitelaw N., Craig J.M., Apedaile A., Hilton D.J., Dunwoodie S.L., Brockdorff N. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat. Genet. 2008;40:663–669. doi: 10.1038/ng.142. [DOI] [PubMed] [Google Scholar]
- 19.Gendrel A.V., Tang Y.A., Suzuki M., Godwin J., Nesterova T.B., Greally J.M., Heard E., Brockdorff N. Epigenetic functions of smchd1 repress gene clusters on the inactive X chromosome and on autosomes. Mol. Cell. Biol. 2013;33:3150–3165. doi: 10.1128/MCB.00145-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sacconi S., Lemmers R.J., Balog J., van der Vliet P.J., Lahaut P., van Nieuwenhuizen M.P., Straasheijm K.R., Debipersad R.D., Vos-Versteeg M., Salviati L. The FSHD2 gene SMCHD1 is a modifier of disease severity in families affected by FSHD1. Am. J. Hum. Genet. 2013;93:744–751. doi: 10.1016/j.ajhg.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsen M., Rost S., El Hajj N., Ferbert A., Deschauer M., Walter M.C., Schoser B., Tacik P., Kress W., Müller C.R. Diagnostic approach for FSHD revisited: SMCHD1 mutations cause FSHD2 and act as modifiers of disease severity in FSHD1. Eur. J. Hum. Genet. 2015;23:808–816. doi: 10.1038/ejhg.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Argentaro A., Yang J.C., Chapman L., Kowalczyk M.S., Gibbons R.J., Higgs D.R., Neuhaus D., Rhodes D. Structural consequences of disease-causing mutations in the ATRX-DNMT3-DNMT3L (ADD) domain of the chromatin-associated protein ATRX. Proc. Natl. Acad. Sci. USA. 2007;104:11939–11944. doi: 10.1073/pnas.0704057104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashimoto H., Vertino P.M., Cheng X. Molecular coupling of DNA methylation and histone methylation. Epigenomics. 2010;2:657–669. doi: 10.2217/epi.10.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rakyan V.K., Blewitt M.E., Druker R., Preis J.I., Whitelaw E. Metastable epialleles in mammals. Trends Genet. 2002;18:348–351. doi: 10.1016/s0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
- 25.Blewitt M.E., Vickaryous N.K., Hemley S.J., Ashe A., Bruxner T.J., Preis J.I., Arkell R., Whitelaw E. An N-ethyl-N-nitrosourea screen for genes involved in variegation in the mouse. Proc. Natl. Acad. Sci. USA. 2005;102:7629–7634. doi: 10.1073/pnas.0409375102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Youngson N.A., Epp T., Roberts A.R., Daxinger L., Ashe A., Huang E., Lester K.L., Harten S.K., Kay G.F., Cox T. No evidence for cumulative effects in a Dnmt3b hypomorph across multiple generations. Mamm. Genome. 2013;24:206–217. doi: 10.1007/s00335-013-9451-5. [DOI] [PubMed] [Google Scholar]
- 27.Ricci G., Scionti I., Sera F., Govi M., D’Amico R., Frambolli I., Mele F., Filosto M., Vercelli L., Ruggiero L. Large scale genotype-phenotype analyses indicate that novel prognostic tools are required for families with facioscapulohumeral muscular dystrophy. Brain. 2013;136:3408–3417. doi: 10.1093/brain/awt226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen R.S., Wijmenga C., Luo P., Stanek A.M., Canfield T.K., Weemaes C.M., Gartler S.M. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc. Natl. Acad. Sci. USA. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu G.L., Bestor T.H., Bourc’his D., Hsieh C.L., Tommerup N., Bugge M., Hulten M., Qu X., Russo J.J., Viegas-Péquignot E. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 30.Hagleitner M.M., Lankester A., Maraschio P., Hultén M., Fryns J.P., Schuetz C., Gimelli G., Davies E.G., Gennery A., Belohradsky B.H. Clinical spectrum of immunodeficiency, centromeric instability and facial dysmorphism (ICF syndrome) J. Med. Genet. 2008;45:93–99. doi: 10.1136/jmg.2007.053397. [DOI] [PubMed] [Google Scholar]
- 31.Weemaes C.M., van Tol M.J., Wang J., van Ostaijen-ten Dam M.M., van Eggermond M.C., Thijssen P.E., Aytekin C., Brunetti-Pierri N., van der Burg M., Graham Davies E. Heterogeneous clinical presentation in ICF syndrome: correlation with underlying gene defects. Eur. J. Hum. Genet. 2013;21:1219–1225. doi: 10.1038/ejhg.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuck-Muller C.M., Narayan A., Tsien F., Smeets D.F., Sawyer J., Fiala E.S., Sohn O.S., Ehrlich M. DNA hypomethylation and unusual chromosome instability in cell lines from ICF syndrome patients. Cytogenet. Cell Genet. 2000;89:121–128. doi: 10.1159/000015590. [DOI] [PubMed] [Google Scholar]
- 33.Kondo T., Bobek M.P., Kuick R., Lamb B., Zhu X., Narayan A., Bourc’his D., Viegas-Péquignot E., Ehrlich M., Hanash S.M. Whole-genome methylation scan in ICF syndrome: hypomethylation of non-satellite DNA repeats D4Z4 and NBL2. Hum. Mol. Genet. 2000;9:597–604. doi: 10.1093/hmg/9.4.597. [DOI] [PubMed] [Google Scholar]
- 34.Jeanpierre M., Turleau C., Aurias A., Prieur M., Ledeist F., Fischer A., Viegas-Pequignot E. An embryonic-like methylation pattern of classical satellite DNA is observed in ICF syndrome. Hum. Mol. Genet. 1993;2:731–735. doi: 10.1093/hmg/2.6.731. [DOI] [PubMed] [Google Scholar]
- 35.Carpenter N.J., Filipovich A., Blaese R.M., Carey T.L., Berkel A.I. Variable immunodeficiency with abnormal condensation of the heterochromatin of chromosomes 1, 9, and 16. J. Pediatr. 1988;112:757–760. doi: 10.1016/s0022-3476(88)80698-x. [DOI] [PubMed] [Google Scholar]
- 36.Okano M., Bell D.W., Haber D.A., Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 37.Larsen J., Pettersson O.J., Jakobsen M., Thomsen R., Pedersen C.B., Hertz J.M., Gregersen N., Corydon T.J., Jensen T.G. Myoblasts generated by lentiviral mediated MyoD transduction of myotonic dystrophy type 1 (DM1) fibroblasts can be used for assays of therapeutic molecules. BMC Res. Notes. 2011;4:490. doi: 10.1186/1756-0500-4-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Racca A.W., Beck A.E., McMillin M.J., Korte F.S., Bamshad M.J., Regnier M. The embryonic myosin R672C mutation that underlies Freeman-Sheldon syndrome impairs cross-bridge detachment and cycling in adult skeletal muscle. Hum. Mol. Genet. 2015;24:3348–3358. doi: 10.1093/hmg/ddv084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yao Z., Snider L., Balog J., Lemmers R.J., Van Der Maarel S.M., Tawil R., Tapscott S.J. DUX4-induced gene expression is the major molecular signature in FSHD skeletal muscle. Hum. Mol. Genet. 2014;23:5342–5352. doi: 10.1093/hmg/ddu251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Greef J.C., Wohlgemuth M., Chan O.A., Hansson K.B., Smeets D., Frants R.R., Weemaes C.M., Padberg G.W., van der Maarel S.M. Hypomethylation is restricted to the D4Z4 repeat array in phenotypic FSHD. Neurology. 2007;69:1018–1026. doi: 10.1212/01.wnl.0000271391.44352.fe. [DOI] [PubMed] [Google Scholar]
- 41.Tatton-Brown K., Seal S., Ruark E., Harmer J., Ramsay E., Del Vecchio Duarte S., Zachariou A., Hanks S., O’Brien E., Aksglaede L., Childhood Overgrowth Consortium Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat. Genet. 2014;46:385–388. doi: 10.1038/ng.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noh K.M., Allis C.D., Li H. Reading between the Lines: “ADD”-ing Histone and DNA Methylation Marks toward a New Epigenetic “Sum”. ACS Chem. Biol. 2016;11:554–563. doi: 10.1021/acschembio.5b00830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li B.Z., Huang Z., Cui Q.Y., Song X.H., Du L., Jeltsch A., Chen P., Li G., Li E., Xu G.L. Histone tails regulate DNA methylation by allosterically activating de novo methyltransferase. Cell Res. 2011;21:1172–1181. doi: 10.1038/cr.2011.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y., Jurkowska R., Soeroes S., Rajavelu A., Dhayalan A., Bock I., Rathert P., Brandt O., Reinhardt R., Fischle W., Jeltsch A. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 2010;38:4246–4253. doi: 10.1093/nar/gkq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo X., Wang L., Li J., Ding Z., Xiao J., Yin X., He S., Shi P., Dong L., Li G. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature. 2015;517:640–644. doi: 10.1038/nature13899. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.