Antibiotics alter the gastrointestinal microbiota, allowing for Clostridium difficile infection, which is a significant public health problem. Changes in the structure of the gut microbiota alter the metabolome, specifically the production of secondary bile acids. Specific bile acids are able to initiate C. difficile spore germination and also inhibit C. difficile growth in vitro, although no study to date has defined physiologically relevant bile acids in the gastrointestinal tract. In this study, we define the bile acids C. difficile spores encounter in the small and large intestines before and after various antibiotic treatments. Antibiotics that alter the gut microbiota and deplete secondary bile acid production allow C. difficile colonization, representing a mechanism of colonization resistance. Multiple secondary bile acids in the large intestine were able to inhibit C. difficile spore germination and growth at physiological concentrations and represent new targets to combat C. difficile in the large intestine.
KEYWORDS: Clostridium difficile, bile acids, antibiotics, microbiota, colonization resistance
ABSTRACT
It is hypothesized that the depletion of microbial members responsible for converting primary bile acids into secondary bile acids reduces resistance to Clostridium difficile colonization. To date, inhibition of C. difficile growth by secondary bile acids has only been shown in vitro. Using targeted bile acid metabolomics, we sought to define the physiologically relevant concentrations of primary and secondary bile acids present in the murine small and large intestinal tracts and how these impact C. difficile dynamics. We treated mice with a variety of antibiotics to create distinct microbial and metabolic (bile acid) environments and directly tested their ability to support or inhibit C. difficile spore germination and outgrowth ex vivo. Susceptibility to C. difficile in the large intestine was observed only after specific broad-spectrum antibiotic treatment (cefoperazone, clindamycin, and vancomycin) and was accompanied by a significant loss of secondary bile acids (deoxycholate, lithocholate, ursodeoxycholate, hyodeoxycholate, and ω-muricholate). These changes were correlated to the loss of specific microbiota community members, the Lachnospiraceae and Ruminococcaceae families. Additionally, physiological concentrations of secondary bile acids present during C. difficile resistance were able to inhibit spore germination and outgrowth in vitro. Interestingly, we observed that C. difficile spore germination and outgrowth were supported constantly in murine small intestinal content regardless of antibiotic perturbation, suggesting that targeting growth of C. difficile will prove most important for future therapeutics and that antibiotic-related changes are organ specific. Understanding how the gut microbiota regulates bile acids throughout the intestine will aid the development of future therapies for C. difficile infection and other metabolically relevant disorders such as obesity and diabetes.
IMPORTANCE Antibiotics alter the gastrointestinal microbiota, allowing for Clostridium difficile infection, which is a significant public health problem. Changes in the structure of the gut microbiota alter the metabolome, specifically the production of secondary bile acids. Specific bile acids are able to initiate C. difficile spore germination and also inhibit C. difficile growth in vitro, although no study to date has defined physiologically relevant bile acids in the gastrointestinal tract. In this study, we define the bile acids C. difficile spores encounter in the small and large intestines before and after various antibiotic treatments. Antibiotics that alter the gut microbiota and deplete secondary bile acid production allow C. difficile colonization, representing a mechanism of colonization resistance. Multiple secondary bile acids in the large intestine were able to inhibit C. difficile spore germination and growth at physiological concentrations and represent new targets to combat C. difficile in the large intestine.
INTRODUCTION
Clostridium difficile infection (CDI) is a significant public health problem, associated with increasing morbidity, mortality, and health care-related costs in the United States and around the globe (1). Current treatment for patients with CDI includes the antibiotics vancomycin and metronidazole; however, even after successful treatment, this therapy is associated with more than 20% of cases relapsing (2–5). Even though antibiotics are the first line of treatment, they are also key risk factors in the pathogenesis of CDI (6, 7). Antibiotics alter the resident gut microbiota, decreasing resistance against C. difficile colonization (8–10). However, the exact mechanism for colonization resistance is still unknown.
By altering the gut microbiota, antibiotics ultimately change the gut metabolome (11, 12), specifically the composition and concentration of bile acids (11, 13–15). Bile acids are synthesized by hepatic enzymes from cholesterol and are important for lipoprotein, glucose, drug, and energy metabolism (16, 17). Mice synthesize two primary bile acids, cholate (CA) and β-muricholate (βMCA), whereas humans synthesize CA and chendeoxycholate (CDCA) (18). Bile acids are further conjugated with taurine and glycine (19). Once made in the liver, 95% of primary bile acids, both unconjugated and conjugated, are absorbed in the terminal ileum and through the hepatic system (16, 18). Primary bile acids that make it to the large intestine are biotransformed by members of the gut microbiota via two enzymatic reactions, deconjugation and dehydroxylation, into secondary bile acids, including ω-muricholate (ωMCA), hyodeoxycholate (HDCA), ursodeoxycholate, (UDCA), lithocholate (LCA), and deoxycholate (DCA) (16, 20, 21).
C. difficile is a spore-forming organism that requires specific bile acids for maximal germination into a metabolically active vegetative cell, where it can grow to high cell density and produce toxins (22–24). However, many microbially derived secondary bile acids inhibit C. difficile growth (22, 25, 26). Specific bile acids are able to enhance or inhibit C. difficile spore germination and vegetative cell outgrowth in vitro, and this is thought to be important for colonization resistance against C. difficile. Wilson et al. first suggested this concept in 1983, and more recent data support this hypothesis (25, 27–30). Multiple studies showing that restoration of secondary bile acids by members of the gut microbiota help restore colonization resistance against C. difficile in humans and in mice have been published recently (12, 28, 29). However, most studies defining the dynamics between C. difficile and bile acids have been done in vitro, and it is not clear how physiologically relevant this is in vivo (22). Based on our previous work and work by others, we hypothesize that antibiotics associated with decreased colonization resistance against C. difficile in the gut not only alter the gut microbiota but also decrease secondary bile acid pools, allowing for C. difficile spore germination and outgrowth. To test this hypothesis, we used a variety of antibiotics to create distinct microbial and metabolic (bile acid) environments in the murine gut and directly tested their ability to support or inhibit C. difficile spore germination and outgrowth ex vivo.
Here we show that susceptibility to C. difficile spore germination and outgrowth occurs in murine small intestinal content (ileum) regardless of antibiotic perturbation. Susceptibility to C. difficile spore germination and outgrowth in the large intestine (cecum) was present only after specific broad-spectrum antibiotic treatment (cefoperazone, clindamycin, and vancomycin) and was accompanied by a loss of secondary bile acids and significant changes to the gut microbiota. In vivo concentrations of secondary bile acids present during C. difficile resistance were able to inhibit spore germination and outgrowth in vitro. This study illustrates how antibiotics associated with increased risk of CDI are able to alter the gut microbiota, which more importantly results in a loss of secondary bile acid production, allowing for C. difficile colonization. Understanding how the gut microbiota regulates bile acids in both the small and large intestines is vital for designing future therapies to restore colonization resistance against C. difficile and for other metabolic disorders, including obesity and diabetes.
RESULTS
Small intestinal content supports C. difficile spore germination and outgrowth before and after antibiotic treatment.
Groups of C57BL/6 mice were treated with various antibiotics (cefoperazone, clindamycin, vancomycin, metronidazole, and kanamycin), and some groups were allowed to recover off of the antibiotic cefoperazone for up to 6 weeks in order to create different microbial and metabolic environments in the small intestine (ileum) and the large intestine (cecum) (Fig. 1). At the time of necropsy (depicted by red circles in Fig. 1), ileal and cecal contents were collected for ex vivo C. difficile spore germination and outgrowth assays. After a 6-h incubation, C. difficile VPI 10463 spores (approximately 104 spores/ml) were able to germinate and initiate outgrowth in ileal content from non-antibiotic-treated mice and almost all antibiotic-treated mice (Fig. 2A). Spores added to phosphate-buffered saline (PBS [negative control]) did not germinate or outgrow due to the lack of germinant present. The only two metabolic environments from ileal content that were able to initiate germination and outgrowth of spores but failed to reach significance were from mice allowed 3 and 4 weeks of recovery after cefoperazone.
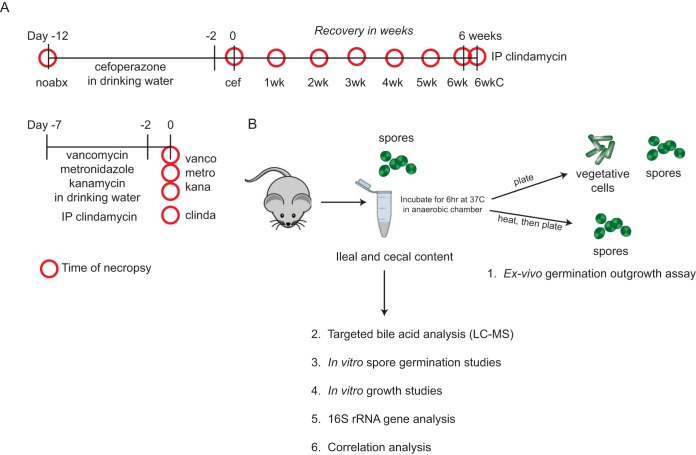
FIG 1 .
Antibiotic treatment scheme and experimental design. (A) C57BL/6 mice were treated with various antibiotics that would result in different microbial and metabolic environments. Red circles represent the time of necropsy for each group (n = 7 to 10 mice per group). (B) At the time of necropsy, ileal and cecal contents were collected. Ex vivo germination and outgrowth of C. difficile spores were measured in paired ileal and cecal contents from all treatment groups as well as by targeted bile acid analysis. In vitro spore germination and growth studies were done using relevant in vivo ileal and cecal bile acid concentrations. Microbiome analysis was also done to understand the relationship between gut bacteria and bile acids using correlation analysis. Abbreviations: noabx, no antibiotic; cef, cefoperazone; 1wk to 6wk, number of weeks off cefoperazone; 6wkC, 6 weeks off cefoperazone plus an intraperitoneal administration (IP) of clindamycin; vanco, vancomycin; metro, metronidazole; kana, kanamycin; clinda, clindamycin.
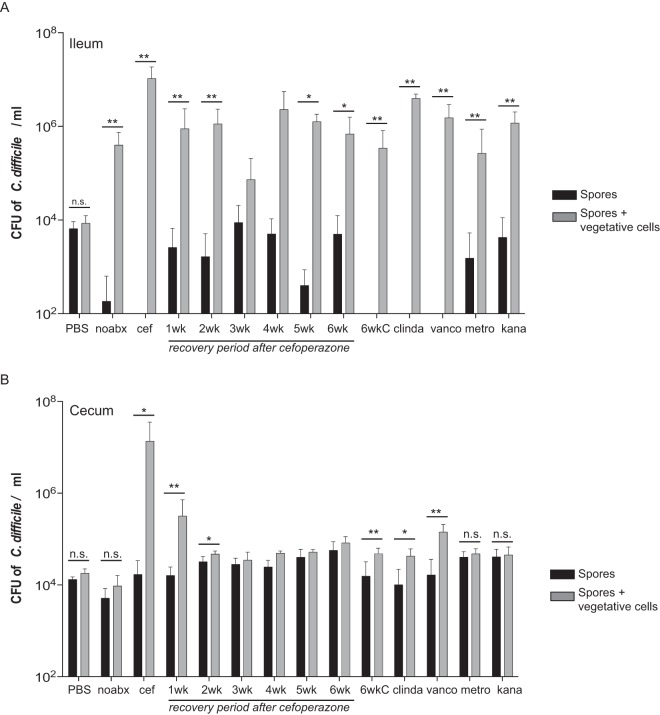
FIG 2 .
C. difficile ex vivo spore germination and outgrowth assays in murine ileal and cecal contents. Ex vivo germination and outgrowth of C. difficile spores were done in ileal (A) and cecal (B) contents collected from mice treated with various antibiotics. C. difficile VPI 10463 spores inoculated into the ileal contents of mice treated with or without antibiotics (noabx) allowed for spore germination and outgrowth after a 6-h period, whereas spores in non-antibiotic-treated cecal content did not. Only specific antibiotic treatments (cefoperazone [cef], 1 or 2 weeks off cefoperazone, 6 weeks off cefoperazone plus an intraperitoneal administration of clindamycin [6wkC], clindamycin [clinda], and vancomycin [vanco]) in the cecum supported spore germination and outgrowth. Black bars represent spores only, and gray bars represent spores and vegetative cells. Significance between groups was determined by Mann-Whitney nonparametric t test. Error bars represent the mean ± standard error of the mean (SEM) (*, P < 0.05; **, P < 0.01). n.s. not significant.
Large intestinal content supports C. difficile spore germination and outgrowth only after antibiotic treatment.
Unlike ileal content, C. difficile spores were only able to germinate and outgrow in cecal content from mice treated with specific antibiotics (Fig. 2B). We previously demonstrated non-antibiotic-treated mouse cecum does not support spore germination or outgrowth ex-vivo, whereas cefoperazone-treated mouse cecum does (12, 31). The only metabolic environments that allowed for significant spore germination and outgrowth in the mouse cecum ex vivo were directly after cefoperazone, 1 and 2 weeks of recovery after cefoperazone, 6 weeks of recovery after cefoperazone plus clindamycin, clindamycin alone, and vancomycin.
Resistance to C. difficile germination and outgrowth is associated with secondary bile acids.
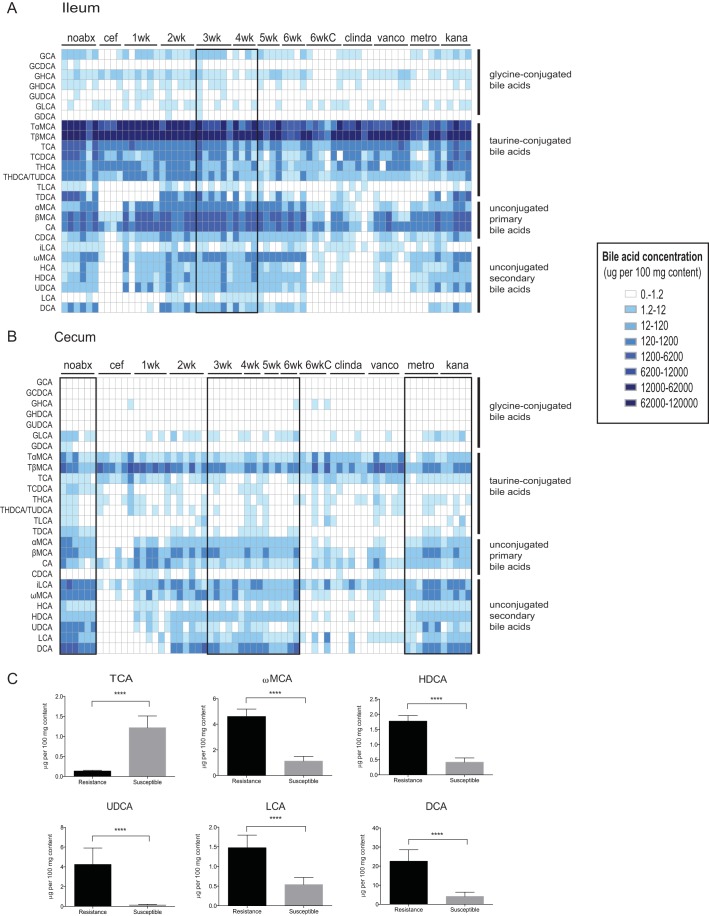
Since bile acids are required for C. difficile spore germination and are also able to inhibit outgrowth (22, 32), we defined the bile acids present in paired ileal and cecal samples from ex vivo germination and outgrowth assays. Prior to antibiotics, ileal content contains mostly primary bile acids, specifically taurine-conjugated and unconjugated primary bile acids, with a lesser concentration of unconjugated secondary bile acids (Fig. 3A). Primary bile acids taurocholate (TCA) and cholate (CA), both germinants of C. difficile spores, are always present at high concentrations in the ileum (average concentrations in vivo, TCA, 0.03%, and CA, 0.05%). This is in contrast to the cecum, where unconjugated secondary bile acids make up the majority of bile acids prior to antibiotics (Fig. 3B). Antibiotic treatments that supported C. difficile spore germination and outgrowth ex vivo in Fig. 2B (directly after cefoperazone, 1 and 2 weeks of recovery after cefoperazone, 6 weeks of recovery after cefoperazone plus clindamycin, clindamycin alone, and vancomycin) resulted in significantly more TCA and less secondary bile acids in the cecum (Fig. 3B and susceptible in panel C; see Fig. S1 and S2 in the supplemental material). Murine cecal contents that did not support C. difficile spore germination and outgrowth are highlighted in the black boxes in Fig. 3B and are associated with an increase in unconjugated secondary bile acids (resistant in Fig. 3C) (average concentrations in vivo, ωMCA, 0.004%, HDCA, 0.002%, UDCA, 0.004%, LCA, 0.001%, and DCA, 0.023%).
FIG 3 .
Targeted bile acid metabolomics of murine ileal and cecal contents. Bile acids were analyzed by LC-MS from paired ileal (A) and cecal (B) contents from ex vivo spore germination and outgrowth assays. A heat map shows the bile acid concentration present in micrograms per 100 mg of gut content, ranging from 1 to 120,000. The black boxes represent samples that did not reach significance in supporting spore germination and outgrowth of C. difficile spores in Fig. 2. (C) Bile acids present in cecal content that did not support spore germination and outgrowth in the black bars (resistance) or were able to support germination and outgrowth (susceptible) are compared. Significance between groups was determined by Mann-Whitney nonparametric t test. Error bars represent the mean ± SEM (****, P < 0.0001).
Targeted bile acid metabolomics from murine ileal content. Bile acids were analyzed by LC-MS from paired ileal content from ex vivo spore germination and outgrowth assays. Bile acid concentrations are represented in micrograms per 100 mg of gut content from each treatment group. Antibiotic abbreviations: noabx, no antibiotic; cef, cefoperazone; 1 week to 6 weeks, weeks off of cefoperazone; 6wkC, 6 weeks off of cefoperazone plus an intraperitoneal administration of clindamycin; vanco, vancomycin; metro, metronidazole; kana, kanamycin; clinda, clindamycin. Download Figure S1, EPS file, 1.9 MB (2MB, eps) .
Copyright © 2016 Theriot et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Targeted bile acid metabolomics from murine cecal content. Bile acids were analyzed by LC-MS from paired cecal contents from ex vivo spore germination and outgrowth assays. Bile acid concentrations are represented in micrograms per 100 mg of gut content from each treatment group. Download Figure S2, EPS file, 1.7 MB (1.8MB, eps) .
Copyright © 2016 Theriot et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We previously demonstrated using untargeted metabolomics that directly after cefoperazone treatment there was a significant decrease in unconjugated secondary bile acids in the murine cecum, and 6 weeks later, the levels returned to baseline or prior to antibiotic treatment (12). Here we show using a more sensitive targeted bile acid assay that secondary bile acids start to return to baseline levels within 2 weeks of recovery of cefoperazone, and by 6 weeks of recovery, they return to baseline levels (see Fig. S3 in the supplemental material).
Antibiotics alter murine cecal bile acids. (A) Nonmetric multidimensional scaling (NMDS) illustrates dissimilarity indices via Horn distances between the bile acid profiles of mouse cecal samples labeled by antibiotic treatment. (B) Paired intestinal samples were used in ex vivo assays to determine if C. difficile spores were able to germinate and outgrow. Bile acid profiles that supported C. difficile germination and outgrowth are susceptible (red), and those that did not support germination and outgrowth are resistant (black). Download Figure S3, EPS file, 1.5 MB (1.5MB, eps) .
Copyright © 2016 Theriot et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Secondary bile acids associated with resistance to C. difficile inhibit spore germination.
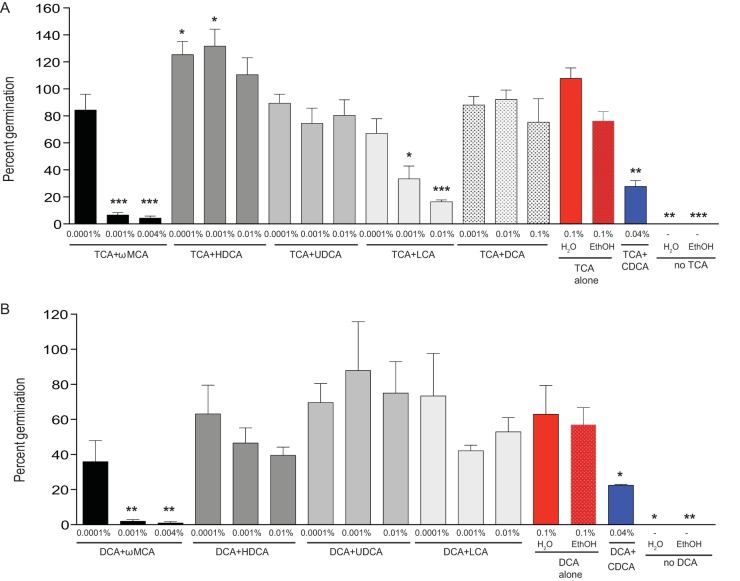
To define the relationship between relevant in vivo levels of secondary bile acids and colonization resistance to C. difficile in Fig. 2, we tested the ability of these bile acids to inhibit or enhance the first step of C. difficile colonization, spore germination. Previous studies have shown TCA is required for maximal spore germination, and chendeoxycholate (CDCA) is able to inhibit this interaction (26). Figure 4A shows maximal spore germination with TCA (0.1%) alone and significant inhibition of spore germination with the addition of CDCA (0.04%). Since most bile acids are soluble in ethanol (ωMCA, HDCA, UDCA, and LCA) a positive control of TCA made with ethanol was used for these comparisons. Secondary bile acids that significantly interfered with TCA-mediated spore germination in a concentration-dependent manner were ωMCA (0.001% and 0.004%) and LCA (0.001% and 0.01%), whereas HDCA (0.0001% and 0.001%) enhanced spore germination at lower concentrations.
FIG 4 .
Secondary bile acids present during C. difficile resistance inhibit spore germination. In vitro spore germination inhibition assays were performed with C. difficile strain VPI 10463 to assess if the secondary bile acids ω-muricholate (ωMCA), hyodeoxycholate (HDCA), ursodeoxycholate (UDCA), lithocholate (LCA), and deoxycholate (DCA) were able to inhibit spore germination with known germinants TCA and DCA. Spores were incubated for 30 min in (A) BHI plus TCA (0.1%) or (B) BHI plus DCA (0.1%) supplemented with a range of secondary bile acids at relevant in vivo concentrations. Positive controls include BHI plus TCA (0.1%) or DCA (0.1%) alone with mock H2O and mock ethanol (EthOH) (red bars). A negative control was also used: BHI plus TCA supplemented with CDCA (0.04%) or BHI plus DCA supplemented with CDCA (0.04%), a known inhibitor of spore germination (blue bar). Negative controls include BHI alone without the addition of TCA or DCA. The data presented represent the mean ± standard deviation (SD) from triplicate experiments and were significant compared to the positive controls (A) TCA alone or (B) DCA alone (Student’s t test, *, P < 0.05; **, P < 0.01; ***, P < 0.001).
Since the concentration of TCA (average concentration in vivo TCA, 0.0001%) is not high enough to support spore germination in the murine cecum during resistance to C. difficile, we turned our attention to the secondary bile acid DCA, which is highly abundant during resistance to C. difficile (average concentration in vivo DCA, 0.023%) and is a known germinant of C. difficile spores (12, 22). In Fig. 4B, spore germination with DCA (0.1%) alone does not reach the same maximal level of spore germination as TCA (0.1%) alone (Fig. 4A). The addition of CDCA (0.04%) significantly interfered with DCA-mediated spore germination as well as supplementation with ωMCA (0.001% and 0.004%). Secondary bile acids present in the cecum during resistance to C. difficile (ωMCA and LCA) were able to inhibit germination of C. difficile spores.
Secondary bile acids associated with resistance to C. difficile inhibit growth.
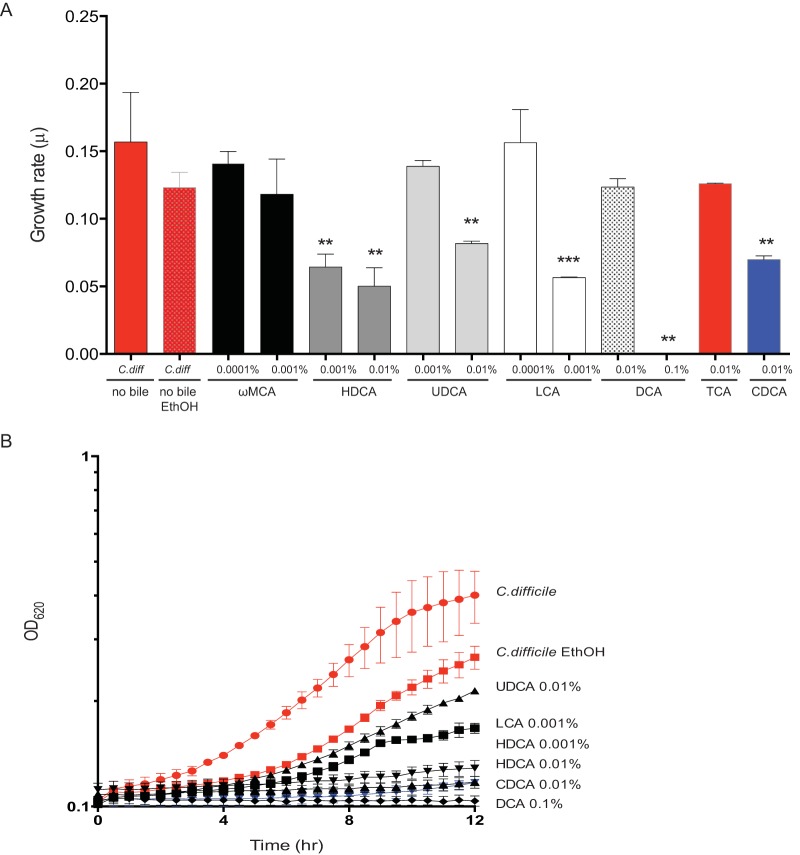
To further characterize the dynamics between the pathogen and secondary bile acids, we evaluated growth of C. difficile over a 24-h period in vitro in the presence of relevant in vivo concentrations of secondary bile acids. In media supplemented with secondary bile acids HDCA, UDCA, LCA, and DCA, C. difficile exhibited decreased growth rates compared to controls (C. difficile alone and C. difficile with ethanol added and no bile) without the addition of secondary bile acids. (Ethanol was added to a C. difficile culture to account for the ethanol in the bile acids.) C. difficile growth curves with secondary bile acids that significantly inhibited the growth rate from Fig. 5A are displayed in Fig. 5B (see Fig. S4 in the supplemental material). Secondary bile acids present during resistance to C. difficile were able to inhibit growth of C. difficile.
FIG 5 .
Secondary bile acids present during C. difficile resistance inhibit growth of C. difficile. (A) Growth rates (per hour) are shown. C. difficile was grown in BHI medium supplemented with various secondary bile acids with a range of in vivo concentrations (0.001% to 0.1%) present during resistance to C. difficile. The data presented represent the means ± SD from triplicate experiments and were significant compared to positive controls C. difficile and C. difficile with mock ethanol (EthOH) without bile acids (Students t test, **, P < 0.05; ***, P < 0.01). (B) Growth curve of representative secondary bile acids that significantly decreased the growth rate of C. difficile in panel A.
C. difficile growth curves supplemented with secondary bile acids. (A) In vitro growth of C. difficile strain VPI 10463 was done in BHI medium over a 12-h period anaerobically. BHI medium was supplemented with various secondary bile acids (black line), with a range of in vivo concentrations (0.001% to 0.1%) present during resistance to C. difficile. The data presented represent the mean ± SD from triplicate experiments. Positive controls include C. difficile (red line) and C. difficile with mock ethanol (EthOH) (red line) without bile acids. Download Figure S4, EPS file, 1.3 MB (1.3MB, eps) .
Copyright © 2016 Theriot et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Members of the Lachnospiraceae and Ruminococcaceae families in the large intestinal microbiota are positively correlated with secondary bile acids.
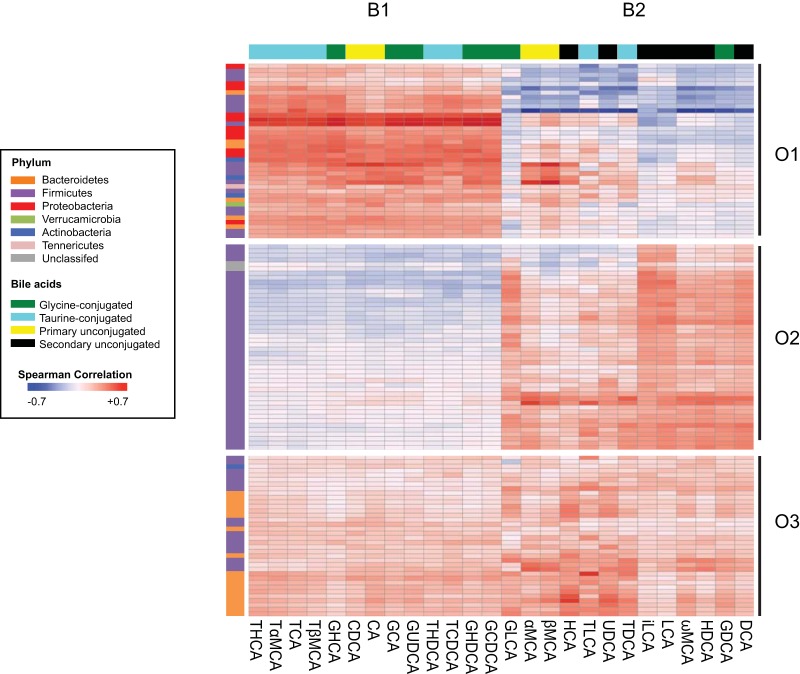
Since many secondary bile acids shape resistance to C. difficile, and members of the gut microbiota are responsible for the biotransformation of primary bile acids to secondary bile acids, we defined the microbiome of each ileal and cecal sample (see Fig. S5 and S6 in the supplemental material). To examine the relationship between members of the gut microbiota and bile acids, we calculated the Spearman’s rank correlation coefficient for all operational taxonomic units (OTU) within the microbiome and bile acids using data from the mouse ileum and cecum (Fig. 6). To visualize these correlations, we performed unsupervised clustering of OTU and bile acids from all groups, which revealed three distinct OTU clusters (O1 to O3 in Table S1 in the supplemental material) and two bile acid clusters (B1 and B2). The organization of the correlation revealed distinctive relationships between OTU and bile acids in the different groups. OTU in the first OTU cluster (O1) were positively correlated with bile acids in the first bile acid cluster (B1) and negatively correlated with most of the bile acids in cluster B2, which contains all of the secondary bile acids (black boxes in Fig. 6). The OTU in cluster O1 include many members from the Proteobacteria and Firmicutes phyla, more specifically from the Enterobacteriaceae and Lactobacillaceae families (see Table S1). Many of these bacteria were present in samples that supported C. difficile germination and outgrowth. This is in contrast to the relationship between clusters O2 and B2, which has a positive correlation. O2 is made up of members from the Firmicutes phylum: specifically, these Lachnospiraceae and Ruminococcaceae family members are positively correlated with all secondary bile acids (see Table S1). Many of these bacteria were present in samples that provided resistance against C. difficile germination and outgrowth. Cluster O3 is made up of many members from the Bacteroidetes phylum, from the Porphyromonadaceae family (see Table S1) and is positively correlated with all bile acids from clusters B1 and B2.
FIG 6 .
Correlation analysis of the gut microbiome and bile acids. Spearman’s correlation analysis was done with all 121 OTU (i.e., OTU that were greater than 1% of the total bacterial population) in the microbiome, color coded by phylum and grouped based on unsupervised clustering. All 26 bile acids detected were similarly clustered and are color coded based on structure. There were three distinct clusters of OTU (O1 to O3) and two distinct clusters of bile acids (B1 and B2). The heat map scale ranges from positively correlated, +0.7, to negatively correlated, −0.7.
Structural changes to the murine ileal and cecal microbiome. A heat map of the top 121 operational taxonomic units (OTU) (i.e., OTU that were >1% of the total bacterial population) in the same orientation as Fig. 6. The bacterial phyla are listed by color, and the scale ranges from 0 to 100% relative OTU abundance. The black boxes represent samples that did not reach significance in supporting spore germination and outgrowth of C. difficile spores in Fig. 2. Download Figure S5, JPG file, 0.9 MB (959.5KB, jpg) .
Copyright © 2016 Theriot et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Structural changes to the murine ileal and cecal microbiome. OTU are listed in order from Fig. 6 (and see Fig. S5 in the supplemental material) as displayed by taxonomic classification. Download Table S1, XLS file, 0.05 MB (49.5KB, xls) .
Copyright © 2016 Theriot et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Using a targeted metabolomics approach, we defined the in vivo concentrations of bile acids before and after various antibiotic treatments in the murine small and large intestines. C. difficile spores were able to germinate and outgrow in most ileal content and cecal content that was depleted of secondary bile acids and had significant alterations to the microbiome. To further define the mechanism of colonization resistance against C. difficile, we conducted in vitro studies to show that in vivo concentrations of secondary bile acids were able to inhibit spore germination and growth in the large intestine. Previous in vitro studies looking at the interaction between bile acids and C. difficile spore germination and growth observed inhibition of germination with ωMCA, LCA, and UDCA and inhibition of growth with ωMCA, LCA, and DCA (12, 22, 25, 26, 28, 32, 33). However, they were not based on in vivo bile acid levels in the gastrointestinal (GI) tract. Newer, more sensitive mass spectrometric technology has allowed us to define the real-time physiological concentrations in vivo as opposed to untargeted metabolomic approaches, which only yield relative abundance (34). Defining the composition and concentration of bile acids that are able to inhibit or enhance C. difficile spore germination and outgrowth in the gut is critical for the development of targeted bacterial therapeutics to prevent C. difficile.
Interestingly, we observed that C. difficile spores were able to germinate in ileal content of the mouse before and after antibiotic treatment. Germination has been demonstrated in the small intestine in other rodent models (31, 35, 36). Although primary bile acids are absorbed in the small intestine, it is likely that sufficient levels are present to allow some level of spore germination. We detected consistently high levels of the germinants TCA and CA, even in the absence of antibiotic treatment, a finding that has been reported previously (14). It is hard to say what the concentration of primary bile acids is in the human small intestine because collection of such samples is difficult, although we know that human serum is rich in primary bile acids, which are mostly absorbed in the small intestine (37).
It is also challenging to say what the oxygen content is in the small intestine. Multiple studies measuring oxygen tension in the small intestine of rats, sheep, ducks, and mice show a spatial distribution, where the mucosa is more oxygen rich and the lumen is more anaerobic (38–40). More recent literature using sensitive oxygen-measuring imaging found the ileum in a mouse, which is relevant to our model, to be quite anaerobic and to resemble that of the colon (38). Similarly, the oxygen concentrations in the duck small intestinal lumen measured by microelectrodes were 25 mm Hg closest to the villi and <0.5 mm Hg in the center of the lumen (39). This further supports that the luminal content of the ileum is anaerobic. This study and our previous work also suggest that spores will always germinate to some degree in small intestinal content (31). Therefore, prevention of C. difficile from growing in the cecum and large intestine will be critical.
In the cecum, we observed that specific antibiotic treatments (cefoperazone, vancomycin, and clindamycin) altered the gut microbiome and decreased the secondary bile acid load, allowing C. difficile to germinate and outgrow. Some of these antibiotics are associated with susceptibility to CDI, with the highest risk associated with cephalosporin, clindamycin, penicillin, and fluoroquinolones (7, 41, 42). Interestingly, vancomycin is the preferred treatment for CDI but has been shown to alter the microbiome, bile acid metabolism, and host physiology in both mice and humans (13, 15, 43–45).
In our study, bacterial members from the Firmicutes phylum, specifically the Lachnospiraceae and Ruminococcaceae families, were positively correlated with secondary bile acids in the cecum and resistance to C. difficile. A small subset of spore-forming, anaerobic members of the class Clostridia are able to perform enzymatic reactions on conjugated bile acids, including deconjugation and 7α-dehydroxylation, which is a multistep biotransformation from CA to DCA and CDCA to LCA (16, 17, 20, 46). Well-characterized bacteria that have 7α-dehydroxylation activity are from the Clostridium species and include Clostridium scindens, Clostridium hiranonis, Clostridium hylemonae, and Clostridium sordellii, which belong to the Blautia, Ruminococcaceae, and Lachnospiraceae families (46–48). Most recently there is renewed interest in C. scindens because of its high 7α-dehydroxylation activity and its presence in patients’ resistant to C. difficile (25, 28). Future therapies to restore colonization resistance against C. difficile could potentially include targeted bacterial cocktails that are able to restore the level of secondary bile acids in the large intestine to inhibit C. difficile (28, 49). Much attention has focused on increasing DCA levels in the gut. However, increased levels of DCA are associated with an increased risk of colon cancer (50, 51). This study identifies new secondary bile acid targets (LCA, UDCA, HDCA, and ωMCA) at inhibitory concentrations, which could be produced by bacterial therapies to inhibit C. difficile.
Bacterial members from the Proteobacteria and Firmicutes phyla, more specifically from the Enterobacteriaceae and Lactobacillaceae families, were negatively correlated with secondary bile acids in this study. This is consistent with another study in which blooms of proinflammatory Enterobacteriaceae were found in cirrhotic patients and corresponded with a decrease in fecal bile acid levels (52). It is not known whether members from the Proteobacteria phylum are sensitive to the antimicrobial properties of secondary bile acids, but it has been suggested that they may limit their growth in the gut. Members of the Lactobacillaceae family include many Lactobacillus species, and they contain potent bile salt hydrolases, which are able to deconjugate glycine- or taurine-conjugated bile acids into unconjugated bile acids (53, 54). The lack of bacteria able to make secondary bile acids after antibiotics would cause a buildup of primary bile acids like TCA and CA, which are germinants of C. difficile spores.
More recently, human studies have shown recovery of fecal secondary bile acids, and members from the class Clostridia, including members from the Lachnospiraceae family, were associated with successful fecal transplantation in patients with recurrent CDI (29, 55). Even though this is promising, spore germination and colonization occurs upstream of the feces. The current standard of measuring the gut microbiota structure is via feces collection, and it is difficult to collect samples from the more relevant upper human GI tract for studying C. difficile. Although the bile acid profile of mice differs from that of humans, an animal model of CDI allows for the assessment of bile acid concentrations throughout the GI tract (31). More promising is the observation that more sensitive bile acid metabolomics, such as liquid chromatography-tandem mass spectrometry (LC-MS/MS) assays, are seeing similar bile acid species in both human and mouse sera (37), suggesting the usefulness of this mouse model.
Finally, there are potential limitations to the present study, including the use of C. difficile VPI 10463, which is still used in many studies today, although it is not a clinically relevant strain (8, 28, 31). This strain still provides us with a reproducible model of C. difficile infection in the mouse. Moving forward, these observations need to be validated with clinically relevant strains as they use a wide range of bile acids for germination (56, 57). The ex vivo approach we used is powerful, but it does not account for transit time in the GI tract, as it is a static sample. Even though bile acids are able to inhibit spore germination and the growth rate of C. difficile in the gut, bile acids do not represent the sole mechanism for colonization resistance against C. difficile. Other factors that could contribute to colonization resistance in the gut and in our ex vivo samples include competition for nutrients by other members of the gut microbiota.
Since alterations in the gut microbiome and bile acid metabolism are associated with many diseases, including diabetes, obesity, cancer, and metabolic syndrome (21, 50, 51, 58), further investigation is needed to understand the complex relationship between the gut microbiota, bile acids, and host physiology.
MATERIALS AND METHODS
Ethics statement.
The University Committee on the Use and Care of Animals (UCUCA) at the University of Michigan approved this study. The University of Michigan laboratory animal care policies follow the Public Health Service Policy on Humane Care and Use of Laboratory Animals (59). Animals were assessed twice daily for physical condition and behavior, and those assessed as moribund were humanely euthanized by CO2 asphyxiation. Trained animal technicians performed animal husbandry in an AAALAC-accredited facility.
Animals and housing.
Five- to 8-week-old C57BL/6 wild-type (WT) mice (male or female) were obtained from a breeding colony at the University of Michigan that was originally established using animals purchased from Jackson Laboratories (Bar Harbor, ME). Mice were housed with autoclaved food, bedding, and water. Cage changes were performed in a laminar flow hood. Mice had a 12-h cycle of light and darkness.
Antibiotic treatment of animals and sample collection.
Groups of C57BL/6 WT mice (male and female) ranging from 5 to 8 weeks in age were treated with a variety of antibiotics (Fig. 1). Groups of mice (n = 7 to 10 per group) were given cefoperazone (0.5 mg/ml) in sterile drinking water for 10 days followed by a recovery period of 2 days and 1, 2, 3, 4, 5, and 6 weeks with regular drinking water. Three additional groups of mice were treated with a 5-day course of vancomycin (0.5 mg/ml), metronidazole (0.5 mg/ml), or kanamycin (0.5 mg/ml) in their drinking water followed by a 2-day recovery period with regular drinking water. Mice treated with clindamycin were given a subcutaneous injection of 1 mg/ml clindamycin solution so that each mouse received 10 mg/kg of body weight followed by a 1-day recovery. An additional group of mice was given an injection of clindamycin after 6 weeks of recovery from a 10-day treatment with cefoperazone (0.5 mg/ml) followed by a 1-day recovery after the clindamycin injection. Non-antibiotic-treated controls for this experiment included mice given sterile drinking water during the duration of these experiments. After each treatment, mice were euthanized, and at the time of necropsy, ileal and cecal contents were collected, flash-frozen in liquid nitrogen, and stored at −80°C until further analysis.
Bile acid analysis of mouse ileal and cecal contents. (i) Sample preparation.
A two-step extraction of gut content with ethanol and 1:1 chloroform-methanol was done with isotope-labeled internal standards spiked in. Extraction solvent was added to gut samples for homogenization with a probe sonicator. Homogenized samples were kept on ice for 10 min and centrifuged at 45°C at 13000 rpm for 10 min. Supernatants containing metabolites were transferred to another tube, and pellets were extracted with 1:1 chloroform-methanol. Supernatants from the two steps were combined and dried under vacuum at 45°C by using a vacuum centrifuge. Dried samples were reconstituted in 50:50 methanol-water for LC-MS analysis. A series of calibration standards were prepared and analyzed along with samples to quantify metabolites.
(ii) LC-MS analysis.
An Agilent 1290 liquid chromatograph (Agilent, Santa Clara, CA) and waters Acquity BEH C18 column with 1.7-µm particle size (2.1 by 100 mm; Waters, Milford, MA) was used for chromatographic separation. Mobile phase A (MPA) was 0.1% formic acid in LC-MS-grade water. Mobile phase B (MPB) was 0.1% formic acid in acetonitrile. The mobile phase gradient consisted of 0 to 0.5 min of 5% MPB, 3 min of 25% MPB, 17 min of 40% MPB, and 19 to 21 min of 95% MPB and was run on an Agilent 6490 triple-quadrupole mass spectrometer (Agilent, Santa Clara, CA). Data were processed by MassHunter workstation software, version B.06. Table 1 lists the bile acids detected by name and abbreviation as well as their MRM transitions (i.e., parent m/z→daughter m/z).
TABLE 1 .
Bile acids identified by LC-MS analysis
| Name | Abbreviation | MRM transitiona |
|---|---|---|
| α-Muricholate | αMCA | 407.3→407.3 |
| β-Muricholate | βMCA | 407.3→407.3 |
| ω-Muricholate | ωMCA | 407.3→407.3 |
| Hyocholate (γ-muricholate) | HCA (γ-MCA) | 407.3→407.3 |
| Cholate | CA | 407.3→407.3 |
| Chenodeoxycholic acid | CDCA | 391.3→391.3 |
| Deoxycholic acid | DCA | 391.3→391.3 |
| Ursodeoxycholic acid | UDCA | 391.3→391.3 |
| Hyodeoxycholate | HDCA | 391.3→391.3 |
| Glycocholic acid | GCA | 464.3→74.0 |
| Glycochenodeoxycholate | GCDCA | 448.3→74.0 |
| Glycodeoxycholate | GDCA | 448.3→74.0 |
| Glycoursodeoxycholate | GUDCA | 448.3→74.0 |
| Glycohyodeoxycholate | GHDCA | 448.3→74.0 |
| Glycolithocholate | GLCA | 432.3→74.0 |
| Lithocholic acid | LCA | 375.3→375.3 |
| Taurocholic acid | TCA | 514.3→80.0 |
| Taurochenodeoxycholate | TCDCA | 498.3→80.0 |
| Taurodeoxycholate | TDCA | 498.3→80.0 |
| Tauroursodeoxycholate | TUDCA | 498.3→80.0 |
| Taurohyodeoxycholate/tauroursodeoxycholate | THDCA/TUDCA | 498.3→80.0 |
| Taurolithocholate | TLCA | 482.3→80.0 |
| Tauro-β-muricholic acid | T-βMCA | 514.3→80.0 |
| Taurohyocholate | THCA | 514.3→80.0 |
| Tauro-α-muricholic acid | TαMCA | 514.3→80.0 |
| Glycohyocholic acid | GHCA | 464.3→74.0 |
Parent m/z→daughter m/z.
C. difficile spore preparation.
C. difficile VPI 10463 spores were prepared as described in previous studies (12, 60).
Ex vivo spore germination and outgrowth studies in mouse ileal and cecal contents.
C. difficile spores were subjected to heat treatment (65°C for 20 min) prior to use for these studies. Mouse ileal and cecal contents (non-antibiotic treated and antibiotic treated) were weighed and passed into an anaerobic chamber, diluted at a 1:1 ratio with PBS, and spun down for 5 s in a microcentrifuge. Approximately 103 C. difficile spores (104 spores/ml) were added to diluted ileal content, cecal content, and a PBS-only control at a final volume of 0.1 ml and then incubated at 37°C anaerobically for 6 h. After incubation, bacterial enumeration was done on taurocholate-cycloserine-cefoxitin-fructose agar (TCCFA) selective medium (12). The remaining ileal and cecal contents were heat treated at 65°C for 20 min to kill off any vegetative cells, and a subsequent bacterial enumeration was done. Samples plated prior to heating represent spores and vegetative cells (germination and outgrowth). Samples plated after heat treatment represent the spores remaining.
C. difficile in vitro germination assays with secondary bile acids.
Spores were subjected to heat treatment (65°C for 20 min) prior to use for these studies. Sterile bile acid solutions dissolved in either water or ethanol were prepared and passed into an anaerobic chamber and added to brain heart infusion (BHI) plus 100 mg/liter l-cysteine broth with either 0.1% taurocholate (TCA) or 0.1% deoxycholate (DCA) as a positive control. C. difficile spores were added to broth and allowed to incubate for 30 min at 37°C anaerobically. Bacterial enumeration of the samples was performed on both BHI plates (vegetative cells) and BHI agar supplemented with 0.1% TCA (total CFU). Since 0.04% chendeoxycholate (CDCA) is a known inhibitor of TCA-mediated spore germination, this was used as a negative control in this assay (32). TCA at 0.1% and DCA at 0.1% made in water or ethanol (10%) were used as positive controls of spore germination. The percentage of germination of the positive controls TCA and DCA alone was calculated as [(CFU on BHI)/(CFU on BHI + TCA or DCA)] × 100. Inhibition of germination with TCA with the addition of secondary bile acids ωMCA, HDCA, UDCA, LCA, and DCA was calculated as [(CFU on BHI + TCA + secondary bile acids)/(CFU on BHI + TCA alone)] × 100. Inhibition of germination with DCA with the addition of secondary bile acids ωMCA, HDCA, UDCA, and LCA was calculated as [(CFU on BHI + DCA + secondary bile acids)/(CFU on BHI + DCA alone)] × 100. Germination under each condition was performed in triplicate. Controls were always used and compared to the proper bile acid backgrounds of water or ethanol.
C. difficile in vitro growth curves with secondary bile acids.
C. difficile VPI 10463 was cultured overnight at 37°C in BHI plus 100 mg/liter·l-cysteine broth in an anaerobic growth chamber (Coy labs). After 14 h of growth, C. difficile was subcultured 1:10 into BHI plus 100 mg/liter·l-cysteine and allowed to grow for 3 h anaerobically at 37°C. The culture was then diluted in fresh BHI so that the starting optical density at 620 nm (OD620) was 0.01. The cell suspension was added to a 96-well plate at a final volume of 0.2 ml. Sterile bile acid solutions were added to the wells in triplicate. Cell density (OD620) was monitored every 30 min for 24 h, shaking the plate for 90 s before each reading, in a Tecan plate reader (reference no. 16039400) inside an anaerobic chamber. The specific growth rate was determined as μ = ln(X/Xo)/T, where X is the OD620 value during the linear portion of growth and T is time in hours. Values given are representative of the mean μ values from two independent cultures done in triplicate. The growth curve data in Fig. S5 in the supplemental material start at an OD of 0.1 because BHI medium alone has a starting OD620 of 0.1.
The bile acids used in all in vitro assays include taurocholic acid sodium salt hydrate (TCA) (Sigma-Aldrich, CAS no. 345909-26-4), sodium deoxycholate (DCA) (Sigma-Aldrich, CAS no. 302-9504), chenodeoxycholic acid (CDCA) (Alfa Aesar, CAS no. 474-25-9), lithocholic acid (LCA) (Sigma-Aldrich, CAS no. 434-13-9), ursodeoxycholic acid (UDCA) (Sigma-Aldrich, CAS no. 128-13-2), ω-muricholic acid (ωMCA) (Steraloids, catalog ID no. C1888-000), hyocholic acid (HCA) (Steraloids, CAS no. 547-75-1), and hyodeoxycholic acid (HDCA) (Steraloids, CAS no. 83-49-8). TCA and DCA were soluble in water, and the rest of the bile acids were soluble in ethanol.
Illumina MiSeq sequencing of bacterial communities.
Microbial DNA was extracted from mouse ileal and cecal tissue snips that also included luminal content using the PowerSoil-htp 96-well soil DNA isolation kit (Mo Bio Laboratories, Inc.). The V4 region of the 16S rRNA gene was amplified from each sample using a dual-indexing sequencing strategy (61). Each 20-µl PCR mixture contained 2 µl of 10× Accuprime PCR buffer II (Life Technologies), 0.15 µl of Accuprime high-fidelity Taq (catalog no. 12346094) high-fidelity DNA polymerase (Life Technologies), 2 µl of a 4.0 µM primer set, 1 µl DNA, and 11.85 µl sterile double-distilled water (ddH2O) (free of DNA, RNase, and DNase contamination). The template DNA concentration was 1 to 10 ng/µl for a high bacterial DNA/host DNA ratio. PCR was performed under the following conditions: 2 min at 95°C, followed by 30 cycles of 95°C for 20 s, 55°C for 15 s, and 72°C for 5 min, followed by 72°C for 10 min. For low-biomass samples (ileal content), a “touchdown PCR” protocol was performed. Each 20-µl PCR mixture contained 2 µl of 10× Accuprime PCR buffer II (Life Technologies), 0.15 µl of Accuprime high-fidelity Taq (catalog no. 12346094) high-fidelity DNA polymerase (Life Technologies), 2 µl of 4.0 µM primer set, 1 µl DNA, and 11.85 µl sterile ddH2O (free of DNA, RNase, and DNase contamination). The template DNA concentration was 1 to 10 ng/µl for a high bacterial DNA/host DNA ratio. PCR was performed under the following conditions: 2 min at 95°C, followed by 20 cycles of 95°C for 20 s, 60°C for 15 s, and 72°C for 5 min (with a 0.3°C increase of the 60°C annealing temperature each cycle), followed by 20 cycles of 95°C for 20 s, 55°C for 15 s, and 72°C for 5 min, followed by 72°C for 10 min. Libraries were normalized using a Life Technologies SequalPrep normalization plate kit (catalog no. A10510-01) following the manufacturer’s protocol. The concentration of the pooled samples was determined using the Kapa Biosystems library quantification kit for Illumina platforms (KapaBiosystems KK4854). The sizes of the amplicons in the library were determined using the Agilent Bioanalyzer high-sensitivity DNA analysis kit (catalog no. 5067-4626). The final library consisted of equal molar amounts from each of the plates, normalized to the pooled plate at the lowest concentration.
Sequencing was done on the Illumina MiSeq platform, using a MiSeq reagent kit V2 with 500 cycles (catalog no. MS-102-2003) according to the manufacturer’s instructions, with modifications (61). Libraries were prepared according to Illumina’s protocol for preparing libraries for sequencing on the MiSeq (part 15039740 Rev. D) for 2 or 4 nM libraries. The final load concentration was 4 pM (but it can be up to 8 pM) with a 10% PhiX spike to add diversity. Sequencing reagents were prepared according to Illumina’s protocol for 16S sequencing with the Illumina MiSeq personal sequencer (61). (Updated versions of this protocol can be found at http://www.mothur.org/wiki/MiSeq_SOP.) Custom read 1, read 2, and index primers were added to the reagent cartridge, and FASTQ files were generated for paired-end reads.
Microbiome analysis.
Analysis of the V4 region of the 16S rRNA gene was done using mothur (version 1.33.3) (61, 62). Briefly, the standard operating procedure (SOP) at http://www.mothur.org/wiki/MiSeq_SOP was followed to process the MiSeq data. The paired-end reads were assembled into contigs and then aligned to the SILVA 16S rRNA sequence database (63, 64) and were classified to the mothur-adapted RDP training set v9 (65) using the Wang method and an 80% bootstrap minimum to the family taxonomic level. All samples with <500 sequences were removed. Chimeric sequences were removed using UCHIME (66). Sequences were clustered into operational taxonomic units (OTU) using a 3% species-level definition. The OTU data were then filtered to include only those OTU that made up 1% or more of the total sequences. The percentage of relative abundance of bacterial phyla and family members in each sample was calculated. A cutoff of 0.03 (97%) was used to define operational taxonomic units (OTU) and used to calculate the inverse Simpson index as a measure for diversity. Standard packages in R were used to create average heat maps per mouse treatment group and changes in the inverse Simpson index per treatment group (see Fig. S5 and S6 in the supplemental material).
Correlation analysis.
We computed the Spearman’s rank correlation coefficients (r) for all pairs among the 26 bile acids and 121 OTU (Fig. 6) using “method = ‘spearman,’ use = ‘pairwise.complete.obs’” in R package as seen in reference 12.
Statistical analysis.
Statistical tests were performed using Prism version 6.00d for Mac Os X (GraphPad Software, La Jolla, CA, USA). Significance between groups of ex vivo spore germination and outgrowth studies was done by Mann-Whitney nonparametric t test (Fig. 2). Significance between controls and all other groups in in vitro germination and growth studies were calculated by Student’s parametric t test (Fig. 4 and 5). The Kruskal-Wallis one-way analysis of variance test followed by Dunn’s multiple comparison test was used to calculate the significance of bacterial diversity of non-antibiotic-treated controls compared to all other groups in Fig. S6 in the supplemental material. Statistical significance was set at a P value of <0.05 for all analyses.
Bacterial diversity of the murine ileal and cecal microbiome. Bacterial diversity of the (A) ileal and (B) cecal microbiota of all treatment groups measured by the inverse Simpson index. All treatment groups were compared to non-antibiotic-treated controls by Kruskal-Wallis one-way analysis of variance test followed by Dunn’s multiple comparison test. Each bar represents the mean ± SEM (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Download Figure S6, EPS file, 1.5 MB (1.5MB, eps) .
Copyright © 2016 Theriot et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
We thank Kathy Wozniak for assisting with the animal studies of C. difficile. We thank Jun Li and Anna Seekatz for discussions on bioinformatics analysis and data visualization. We would also like to thank Judy Opp, April Cockburn, and Harriette Carrington for constructing the DNA libraries and conducting the MiSeq run. We thank Chunhai Ruan, Steve Brown, Chuck Burant, and Maureen Kachman at the Metabolomics Core Services (supported by grant U24 DK097153 of the NIH Common Funds Project to the University of Michigan) for the development and execution of the targeted metabolomic analysis.
This research was supported in part by The University of Michigan Medical School Host Microbiome Initiative. This work was funded by NIH grant U19AI090871 (V.B.Y.) and by Career Development Award in Metabolomics grant K01GM109236 (C.M.T.) by NIGMS, NIH.
C.M.T. and V.B.Y. conceived and designed the experiments. C.M.T. and A.A.B. performed the experiments. C.M.T. and A.A.B. performed the analysis. C.M.T., A.A.B., and V.B.Y. wrote and edited the manuscript.
REFERENCES
- 1.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC. 2015. Burden of Clostridium difficile infection in the United States. N Engl J Med 372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Figueroa I, Johnson S, Sambol SP, Goldstein EJC, Citron DM, Gerding DN. 2012. Relapse versus reinfection: recurrent Clostridium difficile infection following treatment with fidaxomicin or vancomycin. Clin Infect Dis 55(Suppl 2):S104–S109. doi: 10.1093/cid/cis357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louie TJ, Miller MA, Mullane KM, Weiss K, Lentnek A, Golan Y, Gorbach S, Sears P, Shue Y. 2011. Fidaxomicin versus vancomycin for Clostridium difficile infection. N Engl J Med 364:422–431. doi: 10.1056/NEJMoa0910812. [DOI] [PubMed] [Google Scholar]
- 4.Zar FA, Bakkanagari SR, Moorthi KM, Davis MB. 2007. A comparison of vancomycin and metronidazole for the treatment of Clostridium difficile-associated diarrhea, stratified by disease severity. Clin Infect Dis 45:302–307. doi: 10.1086/519265. [DOI] [PubMed] [Google Scholar]
- 5.Cohen SH, Gerding DN, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox M, Society for Healthcare Epidemiology of America, Infectious Diseases Society of America . 2010. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect Control Hosp Epidemiol 31:431–455. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 6.McFee RB. 2009. Clostridium difficile: emerging public health threat and other nosocomial or hospital acquired infections. Introduction. Dis Mon 55:419–421. doi: 10.1016/j.disamonth.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 7.Pepin J, Saheb N, Coulombe MA, Alary ME, Corriveau MP, Authier S, Leblanc M, Rivard G, Bettez M, Primeau V, Nguyen M, Jacob CE, Lanthier L. 2005. Emergence of fluoroquinolones as the predominant risk factor for Clostridium difficile-associated diarrhea: a cohort study during an epidemic in Quebec. Clin Infect Dis 41:1254–1260. doi: 10.1086/496986. [DOI] [PubMed] [Google Scholar]
- 8.Reeves AE, Theriot CM, Bergin IL, Huffnagle GB, Schloss PD, Young VB. 2011. The interplay between microbiome dynamics and pathogen dynamics in a murine model of Clostridium difficile infection. Gut Microbes 2:145–158. doi: 10.4161/gmic.2.3.16333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, Viale A, Ubeda C, Xavier J, Pamer EG. 2012. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun 80:62–73. doi: 10.1128/IAI.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bassis CM, Theriot CM, Young VB. 2014. Alteration of the murine gastrointestinal microbiota by tigecycline leads to increased susceptibility to Clostridium difficile infection. Antimicrob Agents Chemother 58:2767–2774. doi: 10.1128/AAC.02262-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antunes LC, Han J, Ferreira RB, Lolic P, Borchers CH, Finlay BB. 2011. The effect of antibiotic treatment on the intestinal metabolome. Antimicrob Agents Chemother 55:1494–1503. doi: 10.1128/AAC.01664-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theriot CM, Koenigsknecht MJ, Carlson PE Jr, Hatton GE, Nelson AM, Li B, Huffnagle GB, Li JZ, Young VB. 2014. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun 5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vrieze A, Out C, Fuentes S, Jonker L, Reuling I, Kootte RS, van Nood E, Holleman F, Knaapen M, Romijn JA, Soeters MR, Blaak EE, Dallinga-Thie GM, Reijnders D, Ackermans MT, Serlie MJ, Knop FK, Holst JJ, van der Ley C, Kema IP, Zoetendal EG, de Vos WM, Hoekstra JB, Stroes ES, Groen AK, Nieuwdorp M. 2014. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J Hepatol 60:824–831. doi: 10.1016/j.jhep.2013.11.034. [DOI] [PubMed] [Google Scholar]
- 14.Sayin S, Wahlström A, Felin J, Jäntti S, Marschall H, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, Bäckhed F. 2013. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Yap IKS, Li JV, Saric J, Martin F, Davies H, Wang Y, Wilson ID, Nicholson JK, Utzinger J, Marchesi JR, Holmes E. 2008. Metabonomic and microbiological analysis of the dynamic effect of vancomycin-induced gut microbiota modification in the mouse. J Proteome Res 7:3718–3728. doi: 10.1021/pr700864x. [DOI] [PubMed] [Google Scholar]
- 16.Ridlon JM, Kang D-J, Hylemon PB. 2006. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Chiang JYL. 2009. Bile acids: regulation of synthesis. J Lipid Res 50:1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russell DW. 2003. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem 72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- 19.Claus SP, Ellero SL, Berger B, Krause L, Bruttin A, Molina J, Paris A, Want EJ, de Waziers I, Cloarec O, Richards SE, Wang Y, Dumas ME, Ross A, Rezzi S, Kochhar S, Van Bladeren P, Lindon JC, Holmes E, Nicholson JK. 2011. Colonization-induced host-gut microbial metabolic interaction. mBio 2:e00271–e00210. doi: 10.1128/mBio.00271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Midtvedt T. 1974. Microbial bile acid transformation. Am J Clin Nutr 27:1341–1347. [DOI] [PubMed] [Google Scholar]
- 21.Kuipers F, Bloks VW, Groen AK. 2014. Beyond intestinal soap—bile acids in metabolic control. Nat Rev Endocrinol 10:488–498. doi: 10.1038/nrendo.2014.60. [DOI] [PubMed] [Google Scholar]
- 22.Sorg JA, Sonenshein AL. 2008. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol 190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson KH, Kennedy MJ, Fekety FR. 1982. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J Clin Microbiol 15:443–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlsson S, Lindberg A, Norin E, Burman LG, Akerlund T. 2000. Toxins, butyric acid, and other short-chain fatty acids are coordinately expressed and down-regulated by cysteine in Clostridium difficile. Infect Immun 68:5881–5888. doi: 10.1128/IAI.68.10.5881-5888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorg JA, Sonenshein AL. 2010. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol 192:4983–4990. doi: 10.1128/JB.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Francis MB, Allen CA, Sorg JA. 2013. Muricholic acids inhibit Clostridium difficile spore germination and growth. PLoS One 8:e73653. doi: 10.1371/journal.pone.0073653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sorg J. 2014. Microbial bile acid metabolic clusters: the bouncers at the bar. Cell Host Microbe 16:551–552. doi: 10.1016/j.chom.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MR, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weingarden AR, Chen C, Bobr A, Yao D, Lu Y, Nelson VM, Sadowsky MJ, Khoruts A. 2014. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am J Physiol Gastrointest Liver Physiol 306:G310–G319. doi: 10.1152/ajpgi.00282.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson KH. 1983. Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J Clin Microbiol 18:1017–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koenigsknecht MJ, Theriot CM, Bergin IL, Schumacher CA, Schloss PD, Young VB. 2015. Dynamics and establishment of Clostridium difficile infection in the murine gastrointestinal tract. Infect Immun 83:934–941. doi: 10.1128/IAI.02768-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sorg JA, Sonenshein AL. 2009. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J Bacteriol 191:1115–1117. doi: 10.1128/JB.01260-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howerton A, Ramirez N, Abel-Santos E. 2011. Mapping interactions between germinants and Clostridium difficile spores. J Bacteriol 193:274–282. doi: 10.1128/JB.00980-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hagio M, Matsumoto M, Fukushima M, Hara H, Ishizuka S. 2009. Improved analysis of bile acids in tissues and intestinal contents of rats using LC/ESI-MS. J Lipid Res 50:173–180. doi: 10.1194/jlr.D800041-JLR200. [DOI] [PubMed] [Google Scholar]
- 35.Giel JL, Sorg JA, Sonenshein AL, Zhu J. 2010. Metabolism of bile salts in mice influences spore germination in Clostridium difficile. PLoS One 5:e8740. doi: 10.1371/journal.pone.0008740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson KH, Sheagren JN, Freter R. 1985. Population dynamics of ingested Clostridium difficile in the gastrointestinal tract of the Syrian hamster. J Infect Dis 151:355–361. doi: 10.1093/infdis/151.2.355. [DOI] [PubMed] [Google Scholar]
- 37.Han J, Liu Y, Wang R, Yang J, Ling V, Borchers CH. 2015. Metabolic profiling of bile acids in human and mouse blood by LC-MS/MS in combination with phospholipid-depletion solid-phase extraction. Anal Chem 87:1127–1136. doi: 10.1021/ac503816u. [DOI] [PubMed] [Google Scholar]
- 38.He G, Shankar RA, Chzhan M, Samouilov A, Kuppusamy P, Zweier JL. 1999. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc Natl Acad Sci U S A 96:4586–4591. doi: 10.1073/pnas.96.8.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crompton DW, Shrimpton DH, Silver IA. 1965. Measurements of the oxygen tension in the lumen of the small intestine of the domestic duck. J Exp Biol 43:473–478. [DOI] [PubMed] [Google Scholar]
- 40.Rogers WP. 1949. On the relative importance of aerobic metabolism in small nematode parasites of the alimentary tract: oxygen tensions in the normal environment of the parasites. Aust J Biol Sci 2:157–165. [Google Scholar]
- 41.Owens R, Donskey C, Gaynes R, Loo V, Muto C. 2008. Antimicrobial-associated risk factors for Clostridium difficile infection. Clin Infect Dis 46(Suppl 1):S19–S31. doi: 10.1086/521859. [DOI] [PubMed] [Google Scholar]
- 42.Freeman J, Wilcox MH. 1999. Antibiotics and Clostridium difficile. Microbes Infect 1:377–384. doi: 10.1016/S1286-4579(99)80054-9. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, Limaye PB, Renaud HJ, Klaassen CD. 2014. Effect of various antibiotics on modulation of intestinal microbiota and bile acid profile in mice. Toxicol Appl Pharmacol 277:138–145. doi: 10.1016/j.taap.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis BB, Buffie CG, Carter RA, Leiner I, Toussaint NC, Miller LC, Gobourne A, Ling L, Pamer EG. 2015. Loss of microbiota-mediated colonization resistance to Clostridium difficile infection with oral vancomycin compared with metronidazole. J Infect Dis 212:1656–1665. doi: 10.1093/infdis/jiv256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schubert AM, Sinani H, Schloss PD. 2015. Antibiotic-induced alterations of the murine gut microbiota and subsequent effects on colonization resistance against Clostridium difficile. mBio 6:e00974. doi: 10.1128/mBio.00974-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wells JE, Hylemon PB. 2000. Identification and characterization of a bile acid 7alpha-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7alpha-dehydroxylating strain isolated from human feces. Appl Environ Microbiol 66:1107–1113. doi: 10.1128/AEM.66.3.1107-1113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mallonee DH, White WB, Hylemon PB. 1990. Cloning and sequencing of a bile acid-inducible operon from Eubacterium sp. strain vpi 12708. J Bacteriol 172:7011–7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ridlon JM, Kang D, Hylemon PB. 2010. Isolation and characterization of a bile acid inducible 7alpha-dehydroxylating operon in Clostridium hylemonae TN271. Anaerobe 16:137–146. doi: 10.1016/j.anaerobe.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, Goulding D, Rad R, Schreiber F, Brandt C, Deakin LJ, Pickard DJ, Duncan SH, Flint HJ, Clark TG, Parkhill J, Dougan G. 2012. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog 8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bernstein H, Bernstein C, Payne C, Dvorakova K, Garewal H. 2005. Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res 589:47–65. doi: 10.1016/j.mrrev.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 51.McGarr SE, Ridlon JM, Hylemon PB. 2005. Diet, anaerobic bacterial metabolism, and colon cancer: a review of the literature. J Clin Gastroenterol 39:98–109. [PubMed] [Google Scholar]
- 52.Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, Takei H, Muto A, Nittono H, Ridlon JM, White MB, Noble NA, Monteith P, Fuchs M, Thacker LR, Sikaroodi M, Bajaj JS. 2013. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol 58:949–955. doi: 10.1016/j.jhep.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elkins CA, Moser SA, Savage DC. 2001. Genes encoding bile salt hydrolases and conjugated bile salt transporters in Lactobacillus johnsonii 100–100 and other Lactobacillus species. Microbiology 147:3403–3412. doi: 10.1099/00221287-147-12-3403. [DOI] [PubMed] [Google Scholar]
- 54.Jones BV, Begley M, Hill C, Gahan CGM, Marchesi JR. 2008. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A 105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hamilton MJ, Weingarden AR, Unno T, Khoruts A, Sadowsky MJ. 2013. High-throughput DNA sequence analysis reveals stable engraftment of gut microbiota following transplantation of previously frozen fecal bacteria. Gut Microbes 4:125–135. doi: 10.4161/gmic.23571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carlson PE Jr, Walk ST, Bourgis AE, Liu MW, Kopliku F, Lo E, Young VB, Aronoff DM, Hanna PC. 2013. The relationship between phenotype, ribotype, and clinical disease in human Clostridium difficile isolates. Anaerobe 24:109–116. doi: 10.1016/j.anaerobe.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heeg D, Burns DA, Cartman ST, Minton NP. 2012. Spores of Clostridium difficile clinical isolates display a diverse germination response to bile salts. PLoS One 7:e32381. doi: 10.1371/journal.pone.0032381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kohli R, Myronovych A, Tan BK, Salazar-Gonzalez R, Miles L, Zhang W, Oehrle M, Sandoval DA, Ryan KK, Seeley RJ, Setchell KDR. 2015. Bile acid signaling: mechanism for bariatric surgery, cure for NASH? Dig Dis 33:440–446. doi: 10.1159/000371699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.National Institutes of Health 2002. Public Health Service policy on humane care and use of laboratory animals. Office of Laboratory Animal Welfare, National Institutes of Health, Bethesda, MD. [Google Scholar]
- 60.Theriot CM, Koumpouras CC, Carlson PE, Bergin II, Aronoff DM, Young VB. 2011. Cefoperazone-treated mice as an experimental platform to assess differential virulence of Clostridium difficile strains. Gut Microbes 2:326–334. doi: 10.4161/gmic.19142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. 2007. Silva: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Targeted bile acid metabolomics from murine ileal content. Bile acids were analyzed by LC-MS from paired ileal content from ex vivo spore germination and outgrowth assays. Bile acid concentrations are represented in micrograms per 100 mg of gut content from each treatment group. Antibiotic abbreviations: noabx, no antibiotic; cef, cefoperazone; 1 week to 6 weeks, weeks off of cefoperazone; 6wkC, 6 weeks off of cefoperazone plus an intraperitoneal administration of clindamycin; vanco, vancomycin; metro, metronidazole; kana, kanamycin; clinda, clindamycin. Download Figure S1, EPS file, 1.9 MB (2MB, eps) .
Copyright © 2016 Theriot et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Targeted bile acid metabolomics from murine cecal content. Bile acids were analyzed by LC-MS from paired cecal contents from ex vivo spore germination and outgrowth assays. Bile acid concentrations are represented in micrograms per 100 mg of gut content from each treatment group. Download Figure S2, EPS file, 1.7 MB (1.8MB, eps) .
Copyright © 2016 Theriot et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Antibiotics alter murine cecal bile acids. (A) Nonmetric multidimensional scaling (NMDS) illustrates dissimilarity indices via Horn distances between the bile acid profiles of mouse cecal samples labeled by antibiotic treatment. (B) Paired intestinal samples were used in ex vivo assays to determine if C. difficile spores were able to germinate and outgrow. Bile acid profiles that supported C. difficile germination and outgrowth are susceptible (red), and those that did not support germination and outgrowth are resistant (black). Download Figure S3, EPS file, 1.5 MB (1.5MB, eps) .
Copyright © 2016 Theriot et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
C. difficile growth curves supplemented with secondary bile acids. (A) In vitro growth of C. difficile strain VPI 10463 was done in BHI medium over a 12-h period anaerobically. BHI medium was supplemented with various secondary bile acids (black line), with a range of in vivo concentrations (0.001% to 0.1%) present during resistance to C. difficile. The data presented represent the mean ± SD from triplicate experiments. Positive controls include C. difficile (red line) and C. difficile with mock ethanol (EthOH) (red line) without bile acids. Download Figure S4, EPS file, 1.3 MB (1.3MB, eps) .
Copyright © 2016 Theriot et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Structural changes to the murine ileal and cecal microbiome. A heat map of the top 121 operational taxonomic units (OTU) (i.e., OTU that were >1% of the total bacterial population) in the same orientation as Fig. 6. The bacterial phyla are listed by color, and the scale ranges from 0 to 100% relative OTU abundance. The black boxes represent samples that did not reach significance in supporting spore germination and outgrowth of C. difficile spores in Fig. 2. Download Figure S5, JPG file, 0.9 MB (959.5KB, jpg) .
Copyright © 2016 Theriot et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Structural changes to the murine ileal and cecal microbiome. OTU are listed in order from Fig. 6 (and see Fig. S5 in the supplemental material) as displayed by taxonomic classification. Download Table S1, XLS file, 0.05 MB (49.5KB, xls) .
Copyright © 2016 Theriot et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial diversity of the murine ileal and cecal microbiome. Bacterial diversity of the (A) ileal and (B) cecal microbiota of all treatment groups measured by the inverse Simpson index. All treatment groups were compared to non-antibiotic-treated controls by Kruskal-Wallis one-way analysis of variance test followed by Dunn’s multiple comparison test. Each bar represents the mean ± SEM (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Download Figure S6, EPS file, 1.5 MB (1.5MB, eps) .
Copyright © 2016 Theriot et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.