This is the first study in a dimorphic fungal pathogen to investigate the role of a response regulator downstream of two-component signaling systems and its connection to the high-osmolarity glycerol pathway. This study will inspire further research into the downstream components of two-component signaling systems and their role during pathogenic growth.
KEYWORDS: Two-component signaling, dimorphism, morphogenesis, pathogenicity, response regulator
ABSTRACT
For successful infection to occur, a pathogen must be able to evade or tolerate the host’s defense systems. This requires the pathogen to first recognize the host environment and then signal this response to elicit a complex adaptive program in order to activate its own defense strategies. In both prokaryotes and eukaryotes, two-component signaling systems are utilized to sense and respond to changes in the external environment. The hybrid histidine kinases (HHKs) at the start of the two-component signaling pathway have been well characterized in human pathogens. However, how these HHKs regulate processes downstream currently remains unclear. This study describes the role of a response regulator downstream of these HHKs, sskA, in Talaromyces marneffei, a dimorphic human pathogen. sskA is required for asexual reproduction, hyphal morphogenesis, cell wall integrity, osmotic adaptation, and the morphogenesis of yeast cells both in vitro at 37°C and during macrophage infection, but not during dimorphic switching. Comparison of the ΔsskA mutant with a strain in which the mitogen-activated protein kinase (MAPK) of the high-osmolarity glycerol pathway (SakA) has been deleted suggests that SskA acts upstream of this pathway in T. marneffei to regulate these morphogenetic processes. This was confirmed by assessing the amount of phosphorylated SakA in the ΔsskA mutant, antifungal resistance due to a lack of SakA activation, and the ability of a constitutively active sakA allele (sakAF316L) to suppress the ΔsskA mutant phenotypes. We conclude that SskA regulates morphogenesis and osmotic stress adaptation in T. marneffei via phosphorylation of the SakA MAPK of the high-osmolarity glycerol pathway.
IMPORTANCE This is the first study in a dimorphic fungal pathogen to investigate the role of a response regulator downstream of two-component signaling systems and its connection to the high-osmolarity glycerol pathway. This study will inspire further research into the downstream components of two-component signaling systems and their role during pathogenic growth.
INTRODUCTION
The abilities of pathogenic bacteria, parasites, and fungi to invade and survive within mammalian hosts are governed by a complex interaction between the microbe and the host. It is essential for pathogens to evade or tolerate the host’s defense systems in order to allow successful infection to occur. The pathogen is required to recognize the host environment and subsequently signal to elicit changes in gene expression in order to activate its own defense strategies. Two-component signaling systems are utilized in both prokaryotes and eukaryotes to sense and respond to changes in the external environment and are essential for virulence (reviewed in references 1, 2, and 3). In bacteria, two-component signaling systems consist of a membrane-associated sensor histidine kinase (HK) and a cytoplasmic response regulator (RR) protein. An environmental signal triggers the phosphorylation of a conserved histidine residue in the HK kinase domain, and this phosphorylation signal is relayed to a conserved aspartic acid residue in the effector domain of the RR, which mediates changes in gene expression. In fungi, the HK and RR are fused (hybrid HK [HHK]), and additional phosphorelay occurs via a phosphotransmitter protein (HPt) and a second RR. The RR either directly regulates gene expression or activates a mitogen-activated protein kinase (MAPK) pathway that in turn regulates gene expression.
Saccharomyces cerevisiae possesses a single HHK (class VI) and HPt, encoded by SLN1 and YPD1, respectively, and two response regulators, encoded by SSK1 and SKN7 (reviewed in reference 4). In response to oxidative stress, Skn7p binds directly to DNA to regulate changes in gene expression (5). Skn7p binds to DNA regardless of its phosphorylation state; however, expression of SLN1-dependent genes occurs only in response to receiver domain phosphorylation by Sln1p (6, 7). In the absence of stress, Ssk1p negatively regulates the high-osmolarity glycerol (HOG) MAPK signaling pathway. Under hyperosmotic conditions, the kinase domain of Sln1p is inactive, resulting in a cessation of the Sln1p-Ypd1p-Ssk1p phosphorelay. This leads to the rapid dephosphorylation of Ssk1p, which then binds the redundant Ssk2p and Ssk22p MAPKKKs (MAPK kinase kinases), leading to activation of the MAPKK Pbs2p, which activates the MAPK Hog1p (reviewed in reference 8). Pbs2p can also be activated independently of Sln1p by the Sho1p osmosensor via phosphorylation of the Ste11p MAPKKK (9). Ste11p activation is thought to occur by the Ste20p p21-activated protein kinase (PAK) and also involves Cdc42p and Ste50p (reviewed in reference 8). Phosphorylated Hog1p interacts with a number of transcription factors, including Hot1p, Msn2/4p, Sko1p, and Smp1p responsible for the induction of genes required for the response to osmotic stress (10–12). In addition to direct phosphorylation of transcription factors, Hog1p also controls gene expression by chromatin association and by recruiting RNA polymerase II (RNA Pol II) machinery and histone deacetylases to promoters (reviewed in reference 8).
Fungi with more-complex life cycles have a greatly expanded set of HHKs that transmit the phosphorylation signal via a single HPt, orthologous to Ypd1p, and at least two RRs, orthologous to Ssk1p and Skn7p (13). HHKs have been comprehensively investigated in plant pathogens due to mutations in the class III HHKs resulting in reduced pathogenicity and increased resistance to dicarboximide and phenylpyrrole antifungal agents, which act by Hog1-induced glycerol accumulation in the absence of high external osmolarity (14–21). In human pathogens, class III HHKs play conserved roles in fungicide resistance, pathogenicity, and osmotic and oxidative stress adaptation, in addition to regulating other processes required for virulence in a mammalian host such as melanin production, cell wall integrity, mating, dimorphism, and the production of infectious propagules through asexual development (22–24). However, how pathway components downstream of the HHKs act to regulate these processes in human-pathogenic fungi, especially those capable of dimorphic growth, currently remains unclear.
Talaromyces marneffei (formerly called Penicillium marneffei) is a dimorphic human pathogen prevalent in South-East Asia. Like many other human pathogens, T. marneffei exhibits thermally regulated dimorphic switching such that it grows as highly polarized multicellular hyphae at 25°C and switches to a unicellular yeast growth form at 37°C. Deletion of the HHKs encoded by drkA (class III) and slnA (class VI, SLN1 orthologue) has revealed that these HKs have both overlapping and unique roles in T. marneffei, with DrkA playing a more significant role during growth and development. DrkA, and to a lesser extent SlnA, plays a role in asexual development, hyphal morphogenesis, and cell wall integrity (23). SlnA has a unique role during conidial germination, whereas DrkA is essential for yeast growth at 37°C, both in vitro and ex vivo, antifungal resistance, and phosphorylation of the MAPK SakA (orthologous to S. cerevisiae Hog1p) during osmotic stress adaptation (23). This study describes the role of a response regulator downstream of these HHKs, sskA (homologous to S. cerevisiae Ssk1p) in T. marneffei. Like DrkA, sskA is required for the production and viability of asexual spores, hyphal morphogenesis, cell wall integrity, osmotic adaptation, and the morphogenesis of yeast cells both in vitro at 37°C and during macrophage infection, but not during dimorphic switching. Studies using sskA and sakA mutants show that there is a lack of phosphorylated SakA in the ΔsskA mutant. In addition, the ΔsskA mutant is resistant to the dicarboximide and phenylpyrrole classes of antifungals, and a constitutively active sakA allele (sakAF316L [the F-to-L change at position 316 encoded by sakA]) can suppress the ΔsskA mutant phenotypes. These data provide strong evidence that SskA acts upstream of SakA to regulate these processes in T. marneffei.
RESULTS
The T. marneffei genome encodes four putative response regulators.
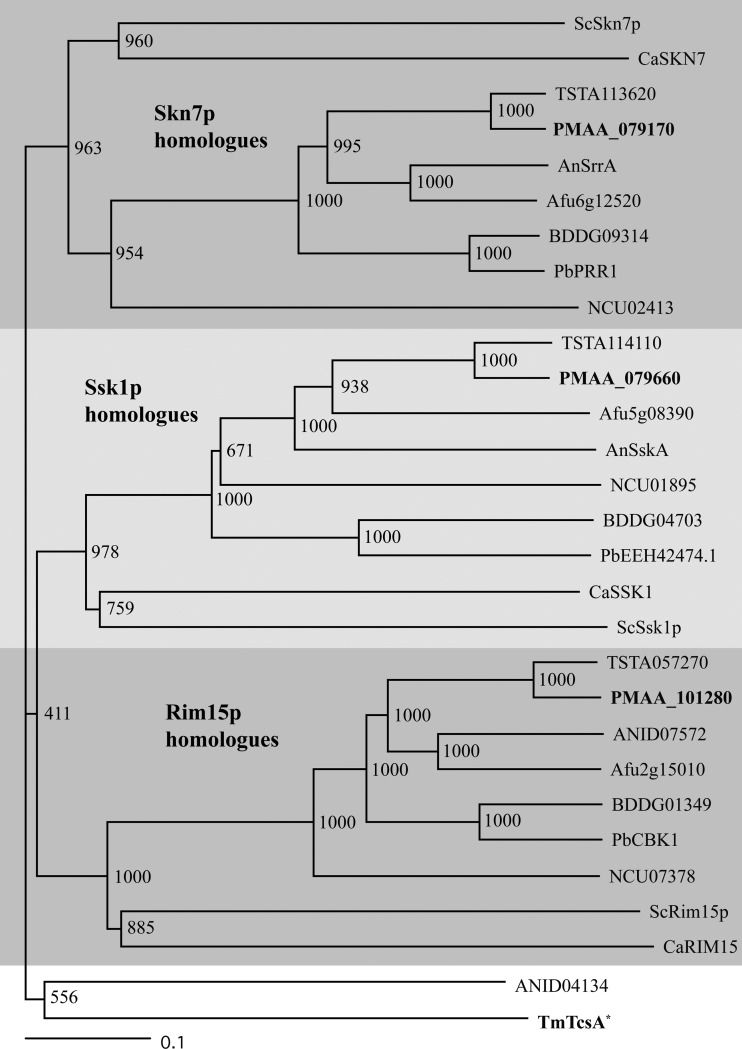
The T. marneffei genome encodes four putative response regulators (RRs) containing response regulator receiver domains (PFam00072) (GenBank accession no. NW_002196663.1, PMAA_079660; NW_002196663.1, PMAA_079170; NW_002196664.1, PMAA_101280; and NW_002196665.1, PMAA_053050). PMAA_079660 and PMAA_079170 show strong sequence homology to S. cerevisiae Ssk1p and Skn7p, respectively (Fig. 1). PMAA_101280 shows sequence homology with S. cerevisiae Rim15p (Fig. 1). Rim15p is a kinase required for cellular proliferation in response to nutrients (25). Rim15p homologues lack the conserved aspartic acid residue in the effector domain found in canonical RRs, such as Ssk1p and Skn7p. The presence of a response regulator receiver domain at residues 220 to 330 in PMAA_053050 initially indicated that this gene encoded a unique response regulator in T. marneffei. However, comparison of the genomic location encompassing PMAA_053050 with that of a close sexual relative Talaromyces stipitatus, revealed the presence of the tcsA HHK homologue (TSTA_019980) in the equivalent genomic location in T. stipitatus. The genomic synteny extended on both sides of this location, and the T. marneffei genome lacked a tcsA homologue elsewhere. Subsequent alignment of PMAA_053050 with homologues of tscA from T. stipitatus, Neurospora crassa, Blastomyces dermatitidis, and Aspergillus nidulans confirmed that PMAA_053050 encodes a truncated form of tcsA lacking the HATPase_c (PF02518) and HisKA (PF00512) histidine kinase domains. Therefore, T. marneffei possesses two canonical RRs homologous to Ssk1p and Skn7p and one noncanonical RR homologous to Rim15p.
FIG 1 .
The T. marneffei genome encodes four putative response regulators. The bootstrapped relatedness tree was generated using ClustalX with protein sequences from Saccharomyces cerevisiae (Sc), Candida albicans (Ca), Talaromyces stipitatus (TSTA), Talaromyces marneffei (Tm), Aspergillus nidulans (An), Aspergillus fumigatus (Afu), Blastomyces dermatitidis (BDD), Paracoccidioides brasiliensis (Pb), and Neurospora crassa (NCU). Skn7p, Ssk1p, and Rim15p homologues are indicated by gray shading. T. marneffei SrrA, SskA, RimA, and TcsA* are highlighted in bold. The asterisk after TmTcsA* indicates that the protein is truncated. The scale bar at bottom indicates the length of branch that represents an amount genetic change of 0.1.
sskA is required for the production and viability of asexual spores.
To investigate the role of sskA in T. marneffei, PMAA_032020 was cloned, and a deletion strain (ΔsskA::pyrG+ [strain G991]) was generated by transforming T. marneffei strain G816 (ΔligD niaD1 pyrG1) (26). To generate a complemented strain, the ΔsskA strain was transformed with an sskA+ fragment targeted to the niaD locus. However, no transformants could be isolated by using this protoplast method, most likely due to osmosensitivity in this strain. Control transformation plates (minus selection) showed a 10-fold reduction in ΔsskA protoplast regeneration. Therefore, a strain was generated in which sskA+ was targeted to niaD (strain G1018). This strain was transformed with the ΔsskA deletion construct to generate ΔsskA sskA+ (strain G1019). This strain exhibited a wild-type phenotype (see Fig. S1A in the supplemental material). To confirm that sskA+ targeting niaD was complementing the ΔsskA phenotype, the ΔsskA sskA+ strain was plated on chlorate to select for the loss of the sskA+ plasmid integrated at niaD. Removal of niaAt sskA+ resulted in a strain displaying the ΔsskA phenotype (Fig. S1B).
Complementation of ΔsskA, ΔmpkA, and ΔmpkB strains. (A) Colonies of a wild-type control strain (ΔligD pyrGt), ΔsskA sskA+, ΔmpkA mpkA+, and ΔmpkB mpkB+ strains grown on ANM plus supplements plus GABA at 25°C for 14 days. (B) The ΔsskA sskA+ strain was plated on chlorate to select for the loss of the sskA+ plasmid integrated at niaD and then grown for 14 days at 25°C on 1% ANM plus GABA plus various stress-inducing agents (calcofluor white [CAL], sodium dodecyl sulfate [SDS], Congo red [CR], sorbitol, and sodium chloride [NaCl]). Removal of niaAt sskA+ resulted in a strain displaying the ΔsskA phenotype. (C) The wild-type, ΔsskA sskA+, ΔmpkA mpkA+, and ΔmpkB mpkB+ strains were grown for 14 days at 25°C on 1% ANM plus GABA plus various stress-inducing agents (calcofluor white [CAL], sodium dodecyl sulfate [SDS], Congo red [CR], hydrogen peroxide [H2O2], sorbitol, potassium chloride [KCl], and sodium chloride [NaCl]). Download Figure S1, TIF file, 7 MB (7.2MB, tif) .
Copyright © 2016 Boyce et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To assess what functions of T. marneffei SskA are independent of MAPK signaling pathways, the phenotype of the ΔsskA mutant was compared to the phenotypes of strains in which the genes encoding one of the three MAPKs was deleted. Deletions of the three MAPKs in T. marneffei, encoded by sakA (GenBank accession no. NW_002196662.1, PMAA_067980), mpkA (NW_002196663.1, PMAA_080160), and mpkB (unannotated in genome sequence) were generated by transforming T. marneffei strain G816 (ΔligD niaD1 pyrG1) (26). Complemented strains (ΔmpkA mpkA+ [strain G959] and ΔmpkB mpkB+ [strain G1022]) were generated by transforming ΔmpkB pyrG− (G958) and ΔmpkB pyrG− (G961) strains with either an mpkA or mpkB fragment targeted to the pyrG locus.
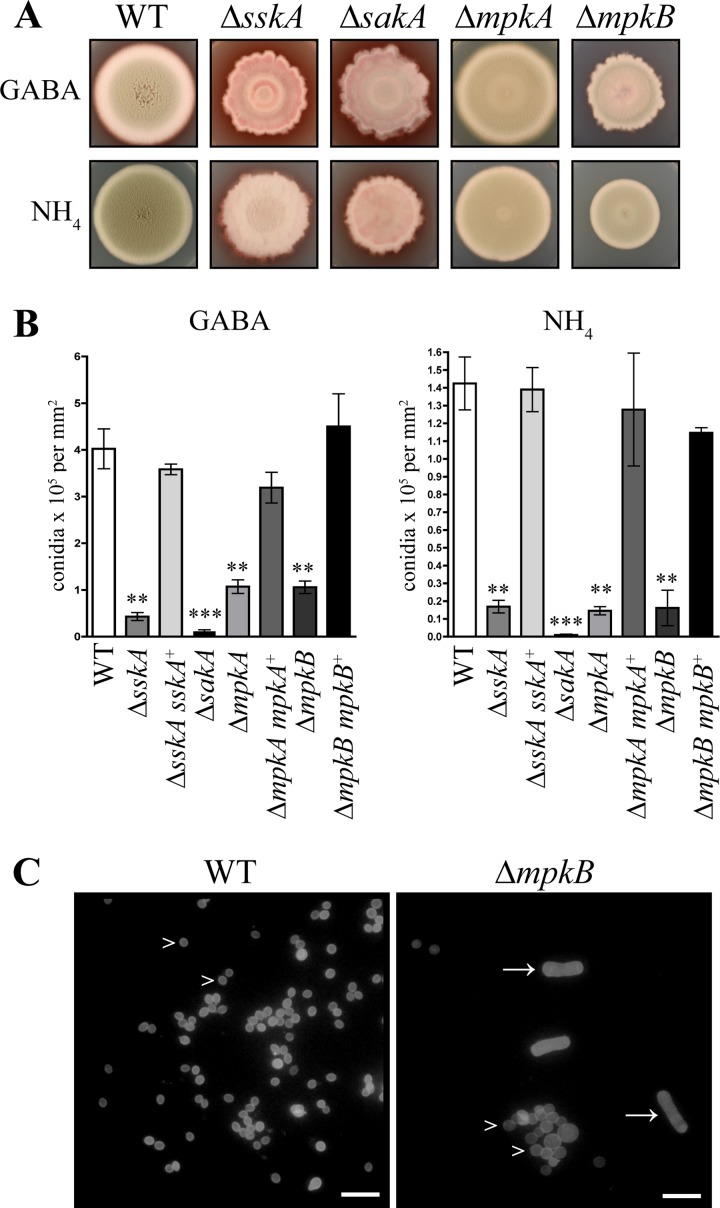
At 25°C, T. marneffei grows as highly polarized vegetative hyphae that form a branched network called a mycelium. Hyphae undergo asexual development, a process that involves cellular differentiation into asexual structures (conidiophores) and culminates in the production of pigmented asexual spores (conidia). After 14 days at 25°C, the ΔsskA strains showed a drastic reduction in the production of asexual development structures. In contrast to the green colonies of the wild type, the ΔsskA colonies appeared pink due to a lack of mature pigmented conidia (Fig. 2A). The ΔsskA strains produced only a small number of conidia in an irregular pattern in the center of the colony and as a ring around the colony center (Fig. 2A). In addition, unlike the circular colonies of the wild type, the ΔsskA mutant colonies exhibited an irregular shape with nonuniform edges (Fig. 2A). The ΔsskA mutant colonies also excreted red pigment into the medium. This phenotype was complemented in the ΔsskA sskA+ strain (see Fig. S1A in the supplemental material). To quantify the decrease in conidiation observed in the ΔsskA mutant, 1 × 105 conidia were spread on an agar plate with either ammonium or gamma-aminobutyric acid (GABA) as the nitrogen source, and the conidial density was calculated after 10 days at 25°C. The ΔsskA mutant showed a statistically significant decrease in conidial density compared to the wild type and the ΔsskA sskA+ strain (Fig. 2B). The ΔsakA mutant colonies showed a phenotype indistinguishable from that of the ΔsskA mutant (Fig. 2A). Similar to the ΔsskA mutant, the ΔsakA mutant also showed a statistically significant decrease in conidial density, but in this case it was even more pronounced (Fig. 2B). Higher osmolarity can often remediate defects in conidiation rate in a number of fungi; however, growth of the ΔsskA and ΔsakA mutants on 0.5 M sorbitol at 25°C for 14 days did not restore visible conidiation (see Fig. 5). Both the ΔmpkA and ΔmpkB mutants also showed a statistically significant decrease in conidiation compared to the wild type and the respective complemented strains, although not to the same extent as the ΔsskA and ΔsakA mutants (Fig. 2A and B). On GABA but not ammonium, the ΔmpkB strain also produced conidia in an irregular pattern in the center of the colony and as a ring around the colony center and showed an irregularly shaped colony with nonuniform edges (Fig. 2A). It was also apparent during the isolation of conidia for this analysis that the ΔmpkB mutant inappropriately produced yeast cells in addition to conidia at 25°C (Fig. 2C). This phenotype was observed only when the ΔmpkB mutant was grown on ammonium as the nitrogen source, not GABA, and did not affect the decrease in conidial density (Fig. 2B). Colonies of the ΔmpkA mpkA+ and ΔmpkB mpkB+ strains were indistinguishable from the wild-type colonies (Fig. S1A).
FIG 2 .
Deletion of sskA and genes encoding MAPKs results in reduced asexual development at 25°C. (A) Colonies of a wild-type (WT) control strain (ΔligD pyrGt) and ΔsskA, ΔsakA, ΔmpkA, and ΔmpkB strains grown on ANM plus supplements plus GABA or ammonium (NH4) at 25°C for 14 days. (B) Quantification of the conidial density (conidia × 105 per mm2) of strains grown on either GABA or ammonium as the sole nitrogen source after 10 days at 25°C. Statistical significance using an unpaired t test and 95% confidence interval is indicated by asterisks as follows: **, P < 0.005; ***, P < 0.001. (C) Conidial preparations of the wild type and the ΔmpkB mutant grown on ammonium as a nitrogen source after 10 days at 25°C. The ΔmpkB mutant inappropriately produces yeast cells (white arrows) in addition to conidia (white arrowheads). Images with epifluorescence to observe calcofluor white-stained fungal cell walls are shown. Bars, 10 µm.
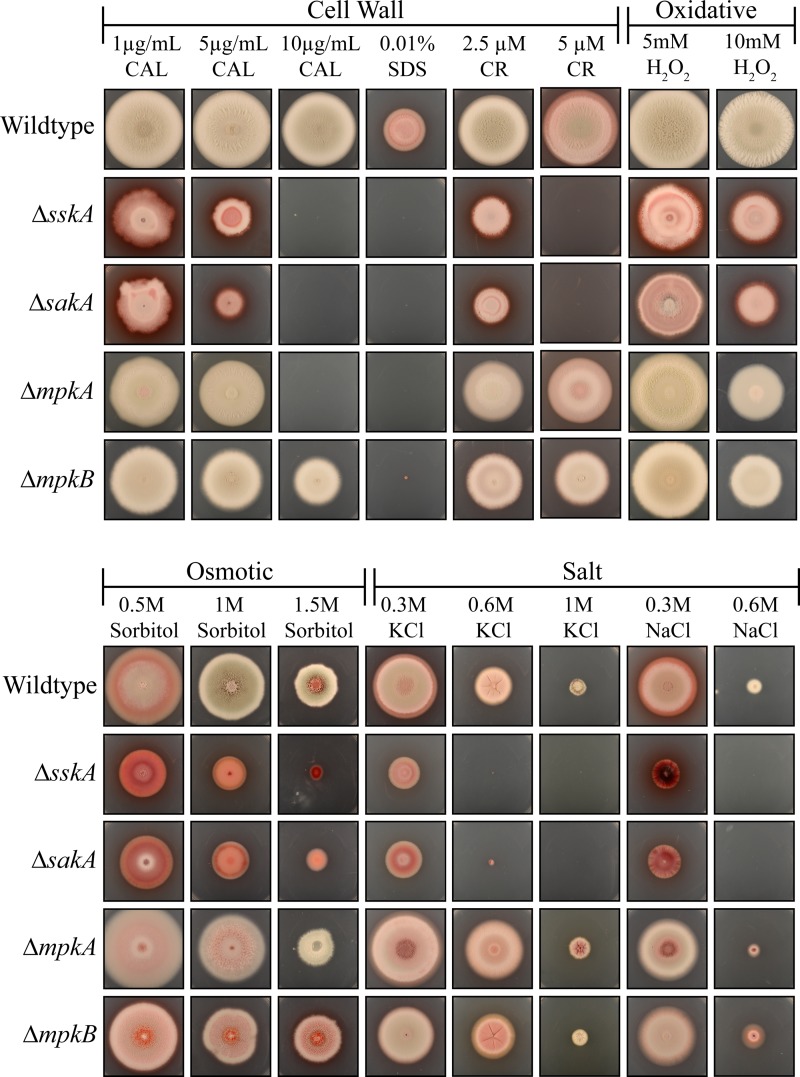
FIG 5 .
sskA and sakA are essential for adaptation to cell wall, osmotic, and salt stress. The wild-type, ΔsskA, ΔsakA, ΔmpkA, and ΔmpkB strains were grown for 14 days at 25°C on 1% ANM plus GABA plus various stress-inducing agents (calcofluor white [CAL], sodium dodecyl sulfate [SDS], Congo red [CR], hydrogen peroxide [H2O2], sorbitol, potassium chloride [KCl], and sodium chloride [NaCl]).
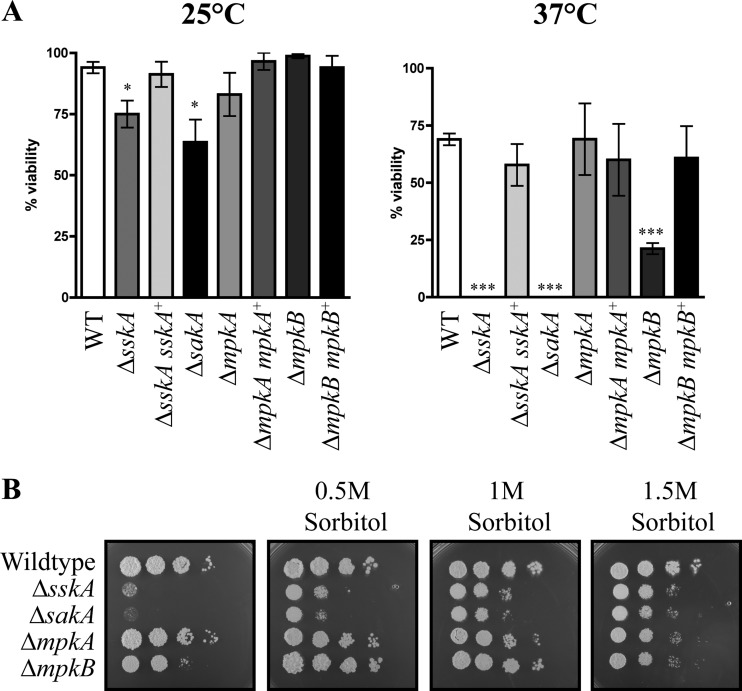
In addition, the viability of conidia was tested at both 25°C and 37°C (Fig. 3A). In contrast to the wild-type, ΔmpkA, and complemented strains, the ΔsskA and ΔsakA conidia showed a small reduction in conidial viability at 25°C and a severe reduction in viability at 37°C (Fig. 3A). These reductions were statistically significant. A statistically significant decrease in conidial viability was also observed in the ΔmpkB strain at 37°C but not 25°C (Fig. 3A). The decreases in viability at 37°C could be remediated by the addition of increasing concentrations of sorbitol (Fig. 3B).
FIG 3 .
sskA and sakA are essential for conidial viability at 37°C. (A) Quantification of the percentage of viable conidia at 25°C and 37°C. Statistical significance using an unpaired t test and 95% confidence interval is indicated by asterisks as follows: *, P < 0.05; ***, P < 0.0001. (B) Conidial suspensions of the wild-type, ΔsskA, ΔsakA, ΔmpkA, and ΔmpkB strains were dropped onto plates containing synthetic dextrose (SD) plus (NH4)2SO4 and 0.5 M, 1 M, or 1.5 M sorbitol and incubated for 5 days at 37°C.
To investigate whether conidiophores possessed wild-type morphology, the wild-type, ΔsskA, ΔsskA sskA+, ΔsakA, ΔmpkA, ΔmpkA mpkA+, ΔmpkB, and ΔmpkB mpkB+ strains were grown on 4 days at 25°C and stained with calcofluor white to visualize cell walls. Wild-type T. marneffei produces conidiophores from a specialized stalk from which differentiated cells are produced sequentially in a budding fashion: metulae bud from the stalk, phialides bud from metulae, and uninucleate conidia bud from phialides. The conidiophores of all strains displayed wild-type morphology (data not shown).
The ΔsskA mutant displays defects in hyphal morphogenesis and cell wall integrity.
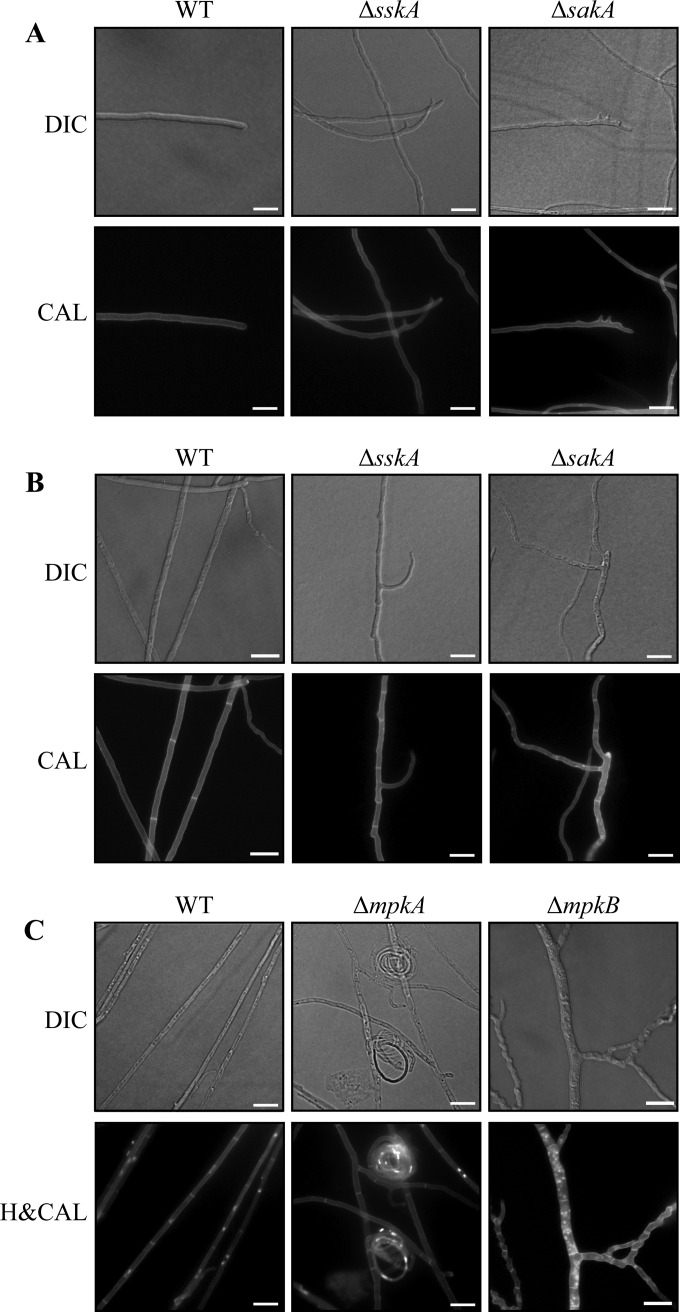
The ΔsskA, ΔsakA, and ΔmpkB mutants exhibited irregularly shaped colonies with nonuniform edges (Fig. 2A). To investigate whether these colonial phenotypes are a result of defects in hyphal morphogenesis, the wild-type, ΔsskA, ΔsskA sskA+, ΔsakA, ΔmpkA, ΔmpkA mpkA+, ΔmpkB, and ΔmpkB mpkB+ strains were grown for 4 days at 25°C and stained with either calcofluor white to visualize cell walls or Hoechst 33258 to observe nuclei. Wild-type T. marneffei grows as septate, branched hyphae (Fig. 4) of which subapical cells are predominately uninucleate, whereas apical cells are multinucleate (Fig. 4C). The ΔsskA and ΔsakA mutants displayed subtle defects in hyphal morphology such as hyphal curling and dichotomous branching in some apical cells (Fig. 4A) as well as abnormal bumps and chitin deposits along hyphae (Fig. 4B) compared to all the other strains. The ΔmpkA mutant produced numerous tightly coiled balls of hyphae, and cell lysis was also evident (Fig. 4C). Interestingly, ΔmpkB hyphae appeared wild type when grown on GABA as the nitrogen source, but hyphae displayed aberrant morphology when grown on ammonium as the nitrogen source (Fig. 4C). Hyphae were swollen and bumpy, exhibited dichotomous branching, and were extremely multinucleate (Fig. 4C). The ΔsskA sskA+, ΔmpkA mpkA+, and ΔmpkB mpkB+ strains were indistinguishable from the wild type (data not shown).
FIG 4 .
Deletion of sskA and genes encoding MAPKs results in defects in hyphal morphology at 25°C. (A to C) The wild-type, ΔsskA, ΔsakA, ΔmpkA, and ΔmpkB strains were grown for 4 days at 25°C on 1% ANM plus GABA (A and B) or (NH4)2SO4 (C) and stained with calcofluor white (CAL) to visualize cell walls and septa (A to C) and Hoechst 33258 to visualize nuclei (C). (A) Apical cells of the ΔsskA and ΔsakA mutants can display dichotomous branching. (B) Subapical hyphal cells of the ΔsskA and ΔsakA mutants display aberrant branching, cellular swellings, and abnormal chitin deposits. (C) Subapical hyphal cells of the ΔmpkA and ΔmpkB mutants. The ΔmpkA mutant exhibited cell lysis and produced tightly coiled balls of hyphae. ΔmpkB hyphae were swollen and bumpy, exhibited dichotomous branching, and were extremely multinucleate. Images were captured using differential interference contrast (DIC) or with epifluorescence to observe fungal cell walls stained with calcofluor white (CAL) or nuclei stained with Hoechst 33258 (H&CAL). Bars, 10 µm.
The abnormal chitin deposits along hyphae of the ΔsskA and ΔsakA mutants and the frequent cell lysis of ΔmpkA mutant suggested defects in cell wall deposition (Fig. 4). To investigate whether the mutants possess cell wall defects, the wild-type, ΔsskA, ΔsskA sskA+, ΔsakA, ΔmpkA, ΔmpkA mpkA+, ΔmpkB, and ΔmpkB mpkB+ strains were plated on increasing concentrations of calcofluor white, Congo red, and sodium dodecyl sulfate (SDS) (Fig. 5). Calcofluor white and Congo red both interfere with the construction and stress response of the cell wall by binding to chitin and beta(1-3)glucan, respectively, whereas SDS disrupts the plasma membrane. Compared to the wild type, the ΔsskA and ΔsakA strains showed increased sensitivity to calcofluor white, SDS, and Congo red (Fig. 5). The ΔmpkA strain was sensitive to calcofluor white and SDS but not Congo red (Fig. 5). The ΔmpkB strain did not show sensitivity to these cell wall stress agents (Fig. 5). The ΔsskA sskA+, ΔmpkA mpkA+, and ΔmpkB mpkB+ strains showed no sensitivity to cell wall stress (see Fig. S1C in the supplemental material).
SskA and SakA are required for adaptation to high osmolarity, salt stress, and oxidative stress.
Deletion of T. marneffei slnA (orthologous to Sln1p) and the gene encoding another hybrid histidine kinase, drkA, in T. marneffei and other filamentous ascomycetes, results in sensitivity to high-osmolarity conditions (14, 15, 18, 20, 21, 23, 27, 28). Deletion of T. marneffei drkA results in increased sensitivity to oxidative stress (23). To investigate whether the deletion of sskA in T. marneffei also results in sensitivity to high-osmolarity conditions, salt stress, and oxidative stress, the wild-type, ΔsskA, ΔsskA sskA+, ΔsakA, ΔmpkA, ΔmpkA mpkA+, ΔmpkB, and ΔmpkB mpkB+ strains were plated on media containing different concentrations of sorbitol, potassium chloride (KCl), sodium chloride (NaCl), and H2O2. In contrast to the wild type and the ΔmpkA and ΔmpkB mutants and complemented strains, the ΔsskA and ΔsakA mutants exhibited sensitivity to high concentrations of sorbitol, KCl, and NaCl (Fig. 5; see Fig. S1C in the supplemental material). Compared to the wild type and the complemented strains, growth of the ΔsskA, ΔsakA, ΔmpkA, and ΔmpkB strains was slightly reduced on 10 mM H2O2 (Fig. 5; Fig. S1C).
SskA acts upstream of SakA during osmotic adaptation.
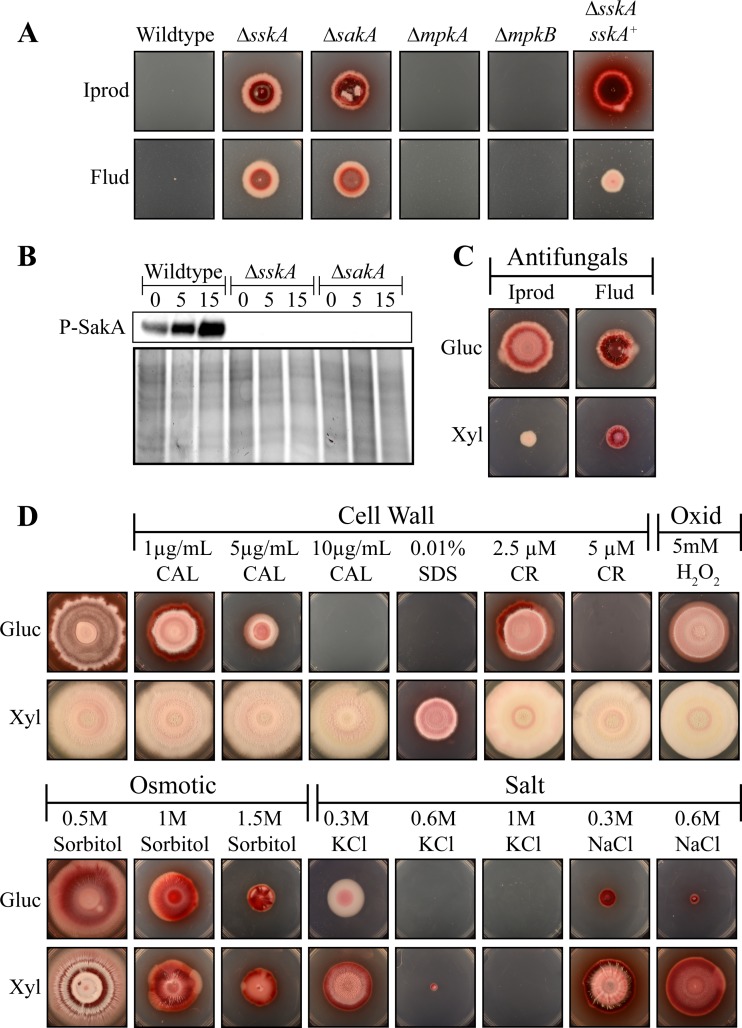
As the phenotypes of the ΔsskA and ΔsakA mutants were indistinguishable, we hypothesized that SskA acts upstream of SakA in T. marneffei. To test whether SskA may be inducing signaling through the SakA pathway, resistance of strains to the dicarboximide and phenylpyrrole classes of antifungals, fludioxonil and iprodione, was tested. The fungicidal activity of these agents is achieved by induction of the HOG MAPK pathway, as glycerol accumulation in the absence of high external osmolarity results in cell death (17). Deletions of HOG1 orthologues (SakA) and their upstream activators confer resistance to these antifungals (1, 15, 18, 20, 22, 28–32). As expected, the T. marneffei ΔsakA mutant is resistant to fludioxonil and iprodione (Fig. 6A). The ΔsskA mutant also shows resistance to fludioxonil and iprodione, suggesting that like drkA, sskA is necessary for induction of the HOG pathway in T. marneffei and is acting upstream of SakA (Fig. 6A) (23). The ΔmpkA and ΔmpkB strains do not show resistance to fludioxonil and iprodione (Fig. 6A). Interestingly, despite showing a wild-type phenotype for all other assays, the ΔsskA sskA+ strain still exhibited resistance to fludioxonil and iprodione (Fig. 6A). This may suggest that wild-type expression is not achieved when sskA is integrated at niaD and that a reduction in sskA expression may be sufficient to result in resistance to these antifungals.
FIG 6 .
SskA acts upstream of SakA during cell wall, oxidative, osmotic, and salt stress adaptation. (A) The wild-type, ΔsskA, ΔsakA, ΔmpkA, ΔmpkB, and ΔsskA sskA+ strains were grown for 14 days at 25°C on phenylpyrrole (iprodione) or dicarboximide (fludioxonil) fungicides. The ΔsskA, ΔsakA, and ΔsskA sskA+ strains show resistance to both iprodione (Iprod) and fludioxonil (Flud). (B) Western blot of phosphorylated SakA (P-SakA) in total denatured protein extracts from the wild-type, ΔsskA, and ΔsakA strains grown for 2 days at 25°C and transferred to media containing 0.3 M NaCl for 0, 5, or 15 min. (C) Growth after 14 days at 25°C of the ΔsskA xylP(p)::sakAF316L strain on dicarboximide (fludioxonil) or phenylpyrrole (iprodione) fungicide and either glucose (noninduced) or xylose (induced). On glucose, the ΔsskA xylP(p)::sakAF316L strain exhibits the ΔsskA phenotype of resistance to both fludioxonil and iprodione. Growth on xylose, and consequent expression of xylP(p)::sakAF316L, partially suppresses this phenotype. (D) Growth after 14 days at 25°C of the ΔsskA xylP(p)::sakAF316L strain on various stress-inducing agents (calcofluor white [CAL], sodium dodecyl sulfate [SDS], Congo red [CR], hydrogen peroxide [H2O2], sorbitol, potassium chloride [KCl], and sodium chloride [NaCl]) and either glucose (noninduced) or xylose (induced).
To confirm that SskA acts upstream of SakA in T. marneffei, the phosphorylation of SakA was assessed in the ΔsskA mutant. Western blot analysis to detect phosphorylated SakA was performed using anti-phospho-p38 antibodies which have been shown to detect phosphorylated SakA homologues in T. marneffei and other fungal species (19, 23, 33–37). In the wild type, phosphorylated SakA was detected at 0 min, and the level of phosphorylated protein increased over time (Fig. 6B). No phosphorylated SakA could be detected in either the ΔsskA or ΔsakA mutant (Fig. 6B).
In addition, the ability of a constitutively active sakA allele (sakAF316L) to suppress the ΔsskA mutant phenotypes was assessed. The equivalent allele in S. cerevisiae Hog1p (F318L) is active, independent of external stimulation by salt and regulation by phosphorylation and the upstream MAPKK Pbs2p (38). As we were unable to successfully transform the ΔsskA mutant, the ΔsskA xylP(p)::sakAF316L strain (strain G1021) was generated by first transforming strain G816 (ΔligD niaD1 pyrG1) with the xylP(p)::sakAF316L construct targeted to niaD (strain G1020) and then transforming this strain with the ΔsskA deletion construct. Growth of the ΔsskA xylP(p)::sakAF316L strain on various stress-inducing agents and either glucose (noninduced) or xylose (induced) was assessed after 14 days of growth at 25°C. On glucose, the ΔsskA xylP(p)::sakAF316L strain exhibits a ΔsskA phenotype (Fig. 6C and D). Expression of xylP(p)::sakAF316L in the ΔsskA mutant (growth on xylose) suppressed the decreased conidiation and irregular shaped colonial phenotype (Fig. 6D). In addition, xylP(p)::sakAF316L expression in the ΔsskA mutant suppressed the sensitivity to cell wall stress, oxidative stress, and NaCl salt stress (Fig. 6D). Growth on xylose did not suppress the ΔsskA sensitivity to osmotic or salt stress caused by KCl and only partially suppressed the resistance to fludioxonil and iprodione (Fig. 6C and D).
sskA is essential for morphogenesis of yeast cells both in vitro and ex vivo at 37°C.
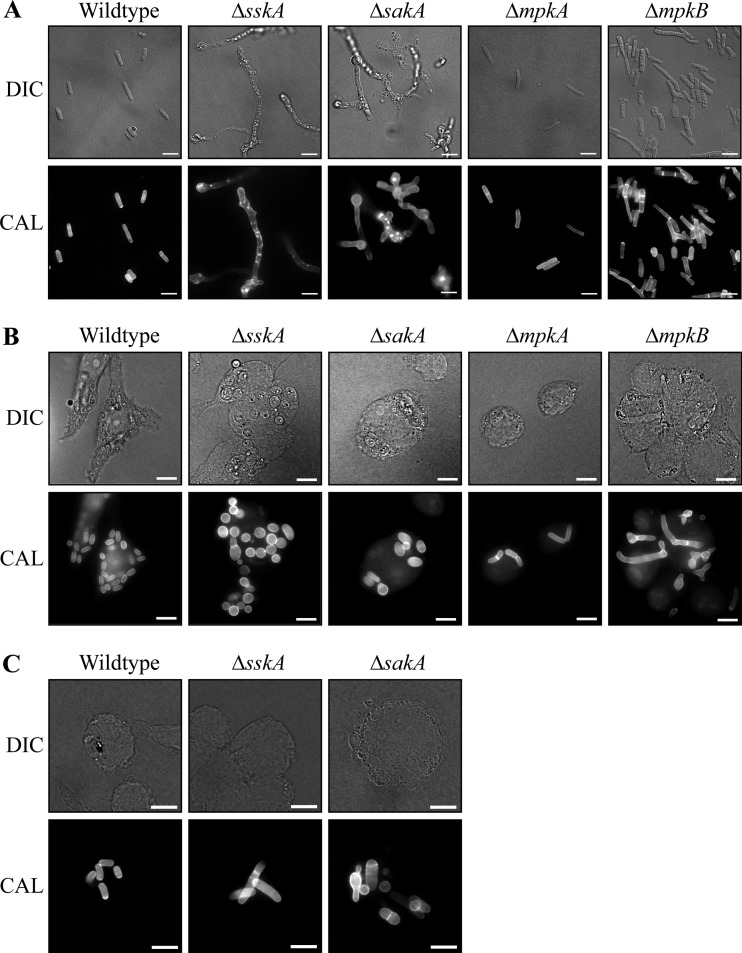
To examine yeast cell morphogenesis in vitro, the wild-type, ΔsskA, ΔsskA sskA+, ΔsakA, ΔmpkA, ΔmpkA mpkA+, ΔmpkB, and ΔmpkB mpkB+ strains were incubated at 37°C (Fig. 7A). After 5 days at 37°C, wild-type conidia have germinated to produce polarized arthroconidiating hyphae that are fragmenting to liberate uninucleate yeast cells. In contrast, the ΔsskA and ΔsakA mutants produced aberrant, swollen cells with few septa and numerous abnormal deposits of chitin (Fig. 7A). No yeast cells were observed in the ΔsskA and ΔsakA strains. The ΔmpkA, ΔmpkB, and complemented strains were indistinguishable from the wild type (Fig. 7A and data not shown).
FIG 7 .
sskA and sakA are essential for morphogenesis of yeast cells both in vitro and ex vivo at 37°C. (A) The wild-type, ΔsskA, ΔsakA, ΔmpkA, and ΔmpkB strains were grown in vitro for 5 days at 37°C on brain heart infusion (BHI) broth. (B) Conidia of the wild-type, ΔsskA, ΔsakA, ΔmpkA, and ΔmpkB strains were used to infect J774 murine macrophages, and growth was assessed 24 h postinfection. (C) Conidia of the wild-type, ΔsskA, and ΔsakA strains were used to infect J774 murine macrophages, and growth was assessed 48 h postinfection. Images were captured using differential interference contrast (DIC) or with epifluorescence to observe fungal cell walls stained with calcofluor white (CAL). Bars, 10 µm.
To assess the roles of sskA, sakA, mpkA, and mpkB during ex vivo growth, conidia of the wild-type, ΔsskA, ΔsskA sskA+, ΔsakA, ΔmpkA, ΔmpkA mpkA+, ΔmpkB, and ΔmpkB mpkB+ strains were used to infect murine J774 macrophages. After 24 h, macrophages infected with wild-type conidia contain numerous yeast cells dividing by fission (Fig. 7B). All strains produced numerous intracellular yeast cells after 24 h. In contrast to the wild type and the ΔsskA sskA+ strain, yeast cells produced by the ΔsskA and ΔsakA strains showed a rounder and more swollen cellular morphology (Fig. 7B). The average size of ΔsskA cells (19.3 ± 0.48 µm2) and ΔsakA (18.6 ± 0.90 µm2) cells significantly increased compared to the wild-type cells (14.6 ± 0.72 µm2). This increase in size is statistically significant (Materials and Methods). Yeast cells produced by the ΔmpkB strain were misshapen and branched but only when conidia used for infections were isolated from plates containing ammonium, not plates containing GABA (Fig. 7B).
After a longer period of incubation (48 h), unlike the wild type, macrophages infected with the ΔsskA and ΔsakA strains contained yeast cells with a mixed population of morphologies, including round swollen cells as at 24 h, in addition to long, aberrantly shaped cells (Fig. 7C). A proportion of ΔsskA and ΔsakA cells also stained very poorly with calcofluor white, and cell lysis was evident (Fig. 7C).
DISCUSSION
Role of two-component signaling during asexual development.
Deletion of three of the seven HHKs encoded by T. marneffei genes, drkA (class III), slnA (class VI), and hhkA (class X), results in decreased asexual development, suggesting that there are numerous environmental signals influencing this process (23, 39). The T. marneffei ΔsskA mutant also showed a reduction in asexual development (conidiation), supporting the hypothesis that it is acting downstream of HHKs to regulate this process. The reduction in conidiation in the ΔsskA mutant was not as severe as previously noted for the ΔdrkA mutant (23), suggesting that the other canonical RR in T. marneffei, SrrA, is also likely to be involved in regulating asexual development. In A. nidulans, both the SSK1 and SKN7 homologues have been shown to regulate asexual development (28). Reduced conidiation in the T. marneffei ΔsskA mutant can be attributed to a lack of SakA phosphorylation, as expression of a constitutively active sakA allele (sakAF316L) in the ΔsskA mutant can suppress this phenotype. This suggests that SskA and SakA regulate the activity of one or more transcription factors required to regulate asexual development in T. marneffei. This could be a homologue to S. cerevisiae Sko1p, Smp1p, or Msn2/4p. Similar to deletion of sskA, deletion of the Msn2/4p homologue in T. marneffei, hgrA, results in an irregular pattern of conidiation in the center of the colony and as a ring around the colony center (41).
In addition, like the ΔdrkA mutant, ΔsskA conidia exhibited reduced viability, especially at 37°C. Deletion of the SskA orthologue in A. nidulans also results in decreased conidial viability, and this is due to a lack of SakA phosphorylation (28, 36). The reduction in viability in T. marneffei is also likely due to a lack of SakA phosphorylation, as the ΔsakA strain showed an identical reduction in conidial viability at both 25°C and 37°C. In A. nidulans, phosphorylated SakA accumulates in the nucleus during normal spore development and is proposed to maintain dormancy and/or prevent germination (42). SakA is dephosphorylated during germination, and constitutive phosphorylation of SakA using fludioxinil treatment results in a failure to germinate (42). Phosphorylated SakA interacts with the AtfA nuclear protein in conidia to regulate the expression of genes required for spore viability (42).
SskA acts upstream of SakA during osmotic adaptation in T. marneffei.
The ΔsskA mutant displays sensitivity to osmotic and salt stress consistent with the postulated role of SskA, acting downstream of drkA and slnA in regulating osmotic adaptation. Deletion of the sskA orthologues in A. nidulans, Cryptococcus neoformans, Cochliobolus Heterostrophus, and Candida albicans also results in sensitivity to osmotic stress (22, 30, 43), albeit only partial sensitivity for C. albicans (27, 44). In C. albicans, SSK1 also appears to have a more important role during adaptation to oxidative stress (45). The resistance of the T. marneffei ΔsskA mutant to the dicarboximide and phenylpyrrole classes of antifungals indicates that SskA is necessary for the phosphorylation of SakA in response to osmotic stress. This was confirmed by showing that no phosphorylated SakA was detected in the ΔsskA mutant. In further support, a constitutively active sakA allele (sakAF316L) can suppress the ΔsskA NaCl-based salt stress sensitivity and partially suppress the resistance to fludioxonil and iprodione. However, this allele was unable to suppress the ΔsskA sensitivity to sorbitol or salt stress caused by KCl. High NaCl concentrations result in ionic imbalance, as Na+ competes with K+ for uptake, resulting in K+ deficiency within the cytosol. In addition, enzymes with K+ binding sites can be inhibited by Na+. High KCl lowers the cellular osmotic potential and restricts the uptake of water. Therefore, the suppression of the NaCl- but not KCl-based salt stress sensitivity suggests that active SskA is directly affecting ionic homeostasis. This may also indicate that SskA interacts with additional factors to regulate the response to high osmolarity or that a cyclic transition of SakA from the phosphorylated to nonphosphorylated state may be crucial for the response to high osmolarity. It is also possible that some functions of T. marneffei SakA are dependent on phosphorylation and interaction with the upstream MAPKK, as the equivalent mutation in S. cerevisiae Hog1p (F318L) is active independent of regulation by phosphorylation and Pbs2p (38).
SskA is necessary for the integrity of the cell wall.
Like the ΔdrkA mutant, the ΔsskA mutant exhibited abnormal chitin deposits along hyphae and showed sensitivity to cell wall stressing agents, indicating defects in cell wall deposition (23). This phenotype is most likely due to a lack of phosphorylated SakA, as the ΔsakA mutant showed a phenotype indistinguishable from the phenotype of the ΔsskA mutant. In addition, the expression of a constitutively active sakA allele could suppress the sensitivity to cell wall stress exhibited by the ΔsskA mutant. In C. albicans, ssk1 is required for the expression of a number of genes encoding cell wall biosynthetic enzymes or proteins that regulate cell wall biosynthesis via Hog1 phosphorylation (45). In S. cerevisiae, the Msn2/4p transcription factors, which are in part responsible for the induction of genes required for osmotic stress, are activated by phosphorylated Hog1p (11). Many filamentous fungi studied thus far do not have a clear Msn2/4p orthologue, and it remains unclear as to whether the general stress response mediated by Msn2/4 in S. cerevisiae is in fact conserved in filamentous fungi (41). The closest homologue in T. marneffei is hgrA which is not required for adaptation to osmotic stress but has roles specific to hyphal growth, including regulating cell wall biosynthesis and cell wall remodeling (41). It is therefore possible that the cell wall defects in the ΔsskA mutant may arise from an inability to activate HgrA via SakA.
It is also possible that SskA is influencing the phosphorylation levels of the MpkA MAPK in response to cell wall stress. However, the sensitivity of the ΔmpkA mutant to calcofluor white and SDS is much milder than those of the ΔsskA and ΔsakA mutants, and no sensitivity to Congo red was observed. In addition, the expression of a constitutively active sakA allele could suppress the sensitivity to cell wall stress exhibited by the ΔsskA mutant, suggesting that it is the lack of SakA phosphorylation that is the main factor contributing to the cell wall sensitivity of the ΔsskA mutant. It is likely that the other canonical RR in T. marneffei, SrrA, is also involved in regulating aspects of the cell wall. In A. nidulans, deletion of srrA results in sensitivity to cell wall stress (28). In addition, putative SrrA binding sites can be identified in the promoters of T. marneffei genes required for α-1,3-glucan synthesis (agsA and agsB), as well as chitin biosynthesis (homologues to S. cerevisiae Chs5p and Chs3p).
Roles of sskA during yeast morphogenesis and infection.
Yeast cells produced by the ΔsskA and ΔsakA strains showed a swollen and aberrant cellular morphology and frequently lysed. This phenotype is predicted to result in an attenuation of virulence in an animal host, although this has yet to be tested. In C. neoformans, virulence correlates directly with the level of activity of the HOG MAPK pathway (33). The hog1 (sakA orthologue) gene is crucial for regulating virulence factors such as capsule size and melanin production in virulent serotypes but not in less virulent serotypes (33). Interestingly, the phosphorylation kinetics and localization pattern of Hog1 differ between virulent serotypes and less virulent serotypes and other yeasts (33). This suggests that small alterations in signaling through the conserved two-component and HOG pathway may provide pathogenic fungi with an opportunity to increase their virulent potential. Ssk1 and Hog1 are also essential for virulence and the colonization of the gastrointestinal tract during infection by C. albicans (40, 44, 46). In C. albicans, the Δssk1 mutant is killed more efficiently by human polymorphonuclear neutrophils (PMNs). This decrease in Δssk1 survival is postulated to be due to increased sensitivity to human defensin-1, a nonoxidative antimicrobial peptide produced by the PMNs, and an increase in the release of PMN cytokines and chemokines during the inflammatory response (47). However, there are likely to be numerous factors contributing to reduced growth during infection. In addition to genes required for osmotic adaptation, Ssk1p and Hog1p in S. cerevisiae independently induce the expression of genes required for hypoxic growth (low oxygen), which also occurs in the host environment (48). Hypoxic conditions lead to a gradual increase in Hog1p phosphorylation, and both Ssk1p and Hog1p are required for induced expression of 33 genes contributing to the maintenance of cell integrity during hypoxia (48).
Interestingly, unlike deletion of drkA, deletion of sskA in T. marneffei did not result in hyphal growth during macrophage infection (23). This suggests that the second canonical RR in T. marneffei, SrrA, is likely responsible for directly regulating the expression of genes required for the dimorphic switch. As the changes in gene expression required for the dimorphic switch remain unknown, this opens up an exciting avenue for future research.
MATERIALS AND METHODS
Molecular techniques and plasmid construction.
T. marneffei genomic DNA was isolated as previously described (49). Southern and Northern blotting was performed with Amersham Hybond N+ membrane and [α-32P]dATP-labeled probe hybridization by standard methods (50). Total denatured proteins were extracted using a FastPrep-24 sample preparation system (MP Biomedicals) by standard methods (50). SDS-polyacrylamide gel electrophoresis using 50 µg of protein per sample was performed with Tris-glycine-buffered 10% Bio-Rad Mini-PROTEAN TGX stain-free precast gels. Western blotting was performed with Immobilon-P polyvinylidene difluoride (PVDF) filters (Millipore) according to the manufacturer’s instructions. Phosphorylated SakA was detected using phospho-p38 MAPK rabbit antibodies (Cell Signaling Technology). Anti-rabbit IgG horseradish peroxidase (HRP)-linked antibody (Cell Signaling Technology) was used as the secondary antibody and was detected using Clarity Western enhanced chemiluminescence (ECL) substrate (Bio-Rad). Western blot signals were detected by using a Bio-Rad ChemiDoc MP imaging system and Image Lab 5.2.
Sequences of the primers are provided in Table S1 in the supplemental material. A PCR product encompassing T. marneffei sskA (PMAA_079660) was generated with primers QQ65 and QQ66 and cloned into pGEM-T Easy to generate pLS7840. To make the pLS7854 deletion construct, a Gateway reaction was performed with pHW7711 (pyrG+) and an inverse PCR-generated product from pLS7840 using the primers QQ67 and QQ68 (51). The complementation construct pKB7937 was generated by cloning a SacII/SpeI fragment from pLS7840 into pHB7615 (niaD targeting pBluescript II SK+). The mpkA gene was cloned into pGEM-T Easy (pKB7779) using primers PP30 and PP31. Then, the XhoI/EcoRV fragment from pKB7779 was cloned into XhoI/StuI-digested pLitmus29, generating pKB7780. The deletion construct pKB7806 was generated by cloning an XbaI/EcoRI pyrG fragment cloned into the XbaI/EcoRI sites of pKB7780. The mpkA gene was also amplified using primers NN67 and NN68 and cloned into pGEM-T Easy, generating pLS7830. The complementation construct pLS7831 was generated by cloning a SpeI/NotI fragment from pLS7830 into pLS7804 (51). The mpkB gene was cloned into pGEM-T Easy (pKB7719) using primers NN47 and NN48. The deletion construct pKB7721 was generated by cloning a BamHI/EcoRV pyrG fragment into BamHI/EcoRV-digested pKB7719. The complementation construct pLS7825 was made by cloning a PCR product generated with primers OO80 and OO81 into pLS7804 (51). The sakA gene was amplified using primers PP26 and PP27 and cloned into pGEM-T Easy, generating pLS7808. The sakAF316L mutation was introduced by performing inverse PCR using primers UU51 and UU52 to generate pKB8002. A PstI/SalI fragment containing the inducible promoter xylP(p) was cloned into PstI/XhoI pHB7615 (niaDt) to generate pKB7996. The PstI/SacII sakAF316L fragment from pKB8002 was cloned into PstI/SacII pKB7996, generating pKB8003 [xylP(p)::sakAF316L niaDt]. The deletion construct was generated by overlap PCR using the primers psak-F1 (F stands for forward) and psak-R2 (R stands for reverse) and PCR products generated from the pGEM-T Easy clone and pALX223 with primers psak-F1 and psakR1 (5′ region), psak-F2 and psak-R2 (3′ region), and pyr-F and pyr-R (pyrG). The deletion cassette was cloned into pGEM_T Easy, generating pCC7859.
Oligonucleotides used in this study Download Table S1, DOC file, 0.04 MB (45.5KB, doc) .
Copyright © 2016 Boyce et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fungal strains and media.
Transformation was performed using the previously described protoplast method (49). Strains used in this study are listed in Table S2 in the supplemental material. The ΔsskA pyrG+ strain (G991), ΔmpkA pyrG+ strain (G1014), ΔmpkB pyrG+ strain (G988), and ΔsakA pyrG+ strain (G990) were generated by transformation of strain G816 (ΔligD niaD1 pyrG1) with linearized deletion constructs pLS7854, pKB7806, pKB7721, and pCC7859 and selecting for PyrG+. The ΔsskA sskA+ strain was generated by transforming strain G816 (ΔligD niaD1 pyrG1) with the pKB7937 plasmid and selecting for niaD+. This strain (G1018) was transformed with the linearized sskA deletion construct pLS7854, generating strain G1019. This strain was plated on 10 mM chlorate to select for the loss of the sskA+ plasmid integrated at niaD. The xylP(p)::sakAF316L ΔsskA strain (G1021) was generated by transforming strain G816 (ΔligD niaD1 pyrG1) with the pKB8003 plasmid and selecting for niaD+. This strain (G1021) was transformed with the linearized sskA deletion construct pLS7854, generating strain G1021. The ΔmpkA pyrG− and ΔmpkB pyrG− strains (Table S2) were generated by plating on medium containing 1 mg/ml 5-fluoroorotic acid (5-FOA) supplemented with 10 mM (NH4)2SO4 and 5 mM uracil to select for the loss of the pyrG marker. These strains are unable to grow in the absence of 5 mM uracil. The ΔmpkA and ΔmpkB deletion mutants were complemented by transformation of the pyrG− strains with pLS7831 (mpkA) and pLS7825 (mpkB) and selecting for pyrG+. Gene deletion and the integration of the complementation constructs at pyrG or niaD was confirmed by Southern blot analysis of genomic DNA.
Strains used in this study Download Table S2, DOC file, 0.05 MB (48.5KB, doc) .
Copyright © 2016 Boyce et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To test the stress resistance of mutants, strains were grown at 25°C and 37°C on A. nidulans minimal medium (ANM) with 1% glucose and 10 mM GABA and supplemented as follows: for salt stress using 0.3, 0.6, and 1 M KCl and 0.3 and 0.6 M NaCl; for osmotic stress using 0.5, 1, and 1.5 M sorbitol; for oxidative stress using 5 and 10 mM H2O2; for cell wall stress using 0.1, 5, and 10 µg/ml calcofluor white, 2.5 and 5 µM Congo red, and 0.01% SDS; for resistance using 30 µg/µl iprodione and 10 µg/µl fludioxonil. All samples were incubated for 14 days.
Macrophage assay.
J774 murine macrophages (1 × 105) were seeded into each well of a six-well microtiter tray containing one sterile coverslip and 2 ml of complete Dulbecco’s modified Eagle medium (complete DMEM) (DMEM, 10% fetal bovine serum, 8 mM l-glutamine, and penicillin-streptomycin). Macrophages were incubated at 37°C for 24 h before activation with 0.1 µg/ml lipopolysaccharide (LPS) from E. coli (Sigma) and incubated a further 24 h at 37°C, washed in phosphate-buffered saline (PBS) and 2 ml of complete DMEM or RPMI 1640 medium containing 1 × 106 conidia was added. A control lacking conidia was also performed. Macrophages were incubated for 2 h at 37°C (to allow conidia to be engulfed), washed once in PBS (to remove free conidia), and incubated a further 24 or 48 h at 37°C. Macrophages were fixed in 4% paraformaldehyde and stained with 1 mg/ml fluorescent brightener 28 (calcofluor white [CAL]) to observe fungal cell walls. Mounted coverslips were examined using differential interference contrast (DIC) and epifluorescence optics for cell wall staining and viewed on a Reichart Jung Polyvar II microscope. Images were captured using a SPOT charge-coupled-device (CCD) camera (Diagnostic Instruments Inc.) and processed in Adobe Photoshop. The numbers of ungerminated conidia, germlings, or yeast cells were recorded in a population of approximately 100 in three independent experiments. Mean and standard error of the mean values were calculated using GraphPad Prism3. Unpaired t tests were performed on the mean and standard error of the mean values to test statistical significance (95% confidence interval).
ACKNOWLEDGMENTS
This work was supported by grant APP1006839 from the National Health and Medical Research Council of Australia to Kylie J. Boyce and Alex Andrianopoulos and a Melbourne Research Fellowship to Kylie J. Boyce from the University of Melbourne. Cunwei Cao was supported by the National Natural Science Foundation of China (grants 81060128, 81271804, and 81571971) and Guangxi Key Laboratory of AIDS Prevention and Treatment.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Jung K, Fried L, Behr S, Heermann R. 2012. Histidine kinases and response regulators in networks. Curr Opin Microbiol 15:118–124. doi: 10.1016/j.mib.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Kruppa M, Calderone R. 2006. Two-component signal transduction in human fungal pathogens. FEMS Yeast Res 6:149–159. doi: 10.1111/j.1567-1364.2006.00024.x. [DOI] [PubMed] [Google Scholar]
- 3.Mascher T. 2014. Bacterial (intramembrane-sensing) histidine kinases: signal transfer rather than stimulus perception. Trends Microbiol 22:559–565. doi: 10.1016/j.tim.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 4.Fassler JS, West AH. 2010. Genetic and biochemical analysis of the SLN1 pathway in Saccharomyces cerevisiae. Methods Enzymol 471:291–317. doi: 10.1016/S0076-6879(10)71016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raitt DC, Johnson AL, Erkine AM, Makino K, Morgan B, Gross DS, Johnston LH. 2000. The Skn7 response regulator of Saccharomyces cerevisiae interacts with HSF1 in vivo and is required for the induction of heat shock genes by oxidative stress. Mol Biol Cell 11:2335–2347. doi: 10.1091/mbc.11.7.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li S, Ault A, Malone CL, Raitt D, Dean S, Johnston LH, Deschenes RJ, Fassler JS. 1998. The yeast histidine protein kinase, Sln1p, mediates phosphotransfer to two response regulators, Ssk1p and Skn7p. EMBO J 17:6952–6962. doi: 10.1093/emboj/17.23.6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li S, Dean S, Li Z, Horecka J, Deschenes RJ, Fassler JS. 2002. The eukaryotic two-component histidine kinase Sln1p regulates OCH1 via the transcription factor, Skn7p. Mol Biol Cell 13:412–424. doi: 10.1091/mbc.01-09-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saito H, Posas F. 2012. Response to hyperosmotic stress. Genetics 192:289–318. doi: 10.1534/genetics.112.140863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Posas F, Saito H. 1997. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science 276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 10.de Nadal E, Casadomé L, Posas F. 2003. Targeting the MEF2-like transcription factor Smp1 by the stress-activated Hog1 mitogen-activated protein kinase. Mol Cell Biol 23:229–237. doi: 10.1128/MCB.23.1.229-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martínez-Pastor MT, Marchler G, Schüller C, Marchler-Bauer A, Ruis H, Estruch F. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J 15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 12.Proft M, Pascual-Ahuir A, de Nadal E, Ariño J, Serrano R, Posas F. 2001. Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J 20:1123–1133. doi: 10.1093/emboj/20.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catlett NL, Yoder OC, Turgeon BG. 2003. Whole-genome analysis of two-component signal transduction genes in fungal pathogens. Eukaryot Cell 2:1151–1161. doi: 10.1128/EC.2.6.1151-1161.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Motoyama T, Kadokura K, Ohira T, Ichiishi A, Fujimura M, Yamaguchi I, Kudo T. 2005. A two-component histidine kinase of the rice blast fungus is involved in osmotic stress response and fungicide action. Fungal Genet Biol 42:200–212. doi: 10.1016/j.fgb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Ochiai N, Fujimura M, Motoyama T, Ichiishi A, Usami R, Horikoshi K, Yamaguchi I. 2001. Characterization of mutations in the two-component histidine kinase gene that confer fludioxonil resistance and osmotic sensitivity in the os-1 mutants of Neurospora crassa. Pest Manag Sci 57:437–442. doi: 10.1002/ps.302. [DOI] [PubMed] [Google Scholar]
- 16.Rispail N, Di Pietro A. 2010. The two-component histidine kinase Fhk1 controls stress adaptation and virulence of Fusarium oxysporum. Mol Plant Pathol 11:395–407. doi: 10.1111/j.1364-3703.2010.00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vetcher L, Menzella HG, Kudo T, Motoyama T, Katz L. 2007. The antifungal polyketide ambruticin targets the HOG pathway. Antimicrob Agents Chemother 51:3734–3736. doi: 10.1128/AAC.00369-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viaud M, Fillinger S, Liu W, Polepalli JS, Le Pêcheur P, Kunduru AR, Leroux P, Legendre L. 2006. A class III histidine kinase acts as a novel virulence factor in Botrytis cinerea. Mol Plant Microbe Interact 19:1042–1050. doi: 10.1094/MPMI-19-1042. [DOI] [PubMed] [Google Scholar]
- 19.Yoshimi A, Kojima K, Takano Y, Tanaka C. 2005. Group III histidine kinase is a positive regulator of Hog1-type mitogen-activated protein kinase in filamentous fungi. Eukaryot Cell 4:1820–1828. doi: 10.1128/EC.4.11.1820-1828.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshimi A, Tsuda M, Tanaka C. 2004. Cloning and characterization of the histidine kinase gene Dic1 from Cochliobolus heterostrophus that confers dicarboximide resistance and osmotic adaptation. Mol Genet Genomics 271:228–236. doi: 10.1007/s00438-003-0974-4. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, Liu K, Zhang X, Song W, Zhao Q, Dong Y, Guo M, Zheng X, Zhang Z. 2010. A two-component histidine kinase, MoSLN1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae. Curr Genet 56:517–528. doi: 10.1007/s00294-010-0319-x. [DOI] [PubMed] [Google Scholar]
- 22.Bahn YS, Kojima K, Cox GM, Heitman J. 2006. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol Biol Cell 17:3122–3135. doi: 10.1091/mbc.E06-02-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyce KJ, Schreider L, Kirszenblat L, Andrianopoulos A. 2011. The two-component histidine kinases DrkA and SlnA are required for in vivo growth in the human pathogen Penicillium marneffei. Mol Microbiol 82:1164–1184. doi: 10.1111/j.1365-2958.2011.07878.x. [DOI] [PubMed] [Google Scholar]
- 24.Nemecek JC, Wüthrich M, Klein BS. 2006. Global control of dimorphism and virulence in fungi. Science 312:583–588. doi: 10.1126/science.1124105. [DOI] [PubMed] [Google Scholar]
- 25.Vidan S, Mitchell AP. 1997. Stimulation of yeast meiotic gene expression by the glucose-repressible protein kinase Rim15p. Mol Cell Biol 17:2688–2697. doi: 10.1128/MCB.17.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bugeja HE, Boyce KJ, Weerasinghe H, Beard S, Jeziorowski A, Pasricha S, Payne M, Schreider L, Andrianopoulos A. 2012. Tools for high efficiency genetic manipulation of the human pathogen Penicillium marneffei. Fungal Genet Biol 49:772–778. doi: 10.1016/j.fgb.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Nagahashi S, Mio T, Ono N, Yamada-Okabe T, Arisawa M, Bussey H, Yamada-Okabe H. 1998. Isolation of CaSLN1 and CaNIK1, the genes for osmosensing histidine kinase homologues, from the pathogenic fungus Candida albicans. Microbiology 144:425–432. doi: 10.1099/00221287-144-2-425. [DOI] [PubMed] [Google Scholar]
- 28.Vargas-Pérez I, Sánchez O, Kawasaki L, Georgellis D, Aguirre J. 2007. Response regulators SrrA and SskA are central components of a phosphorelay system involved in stress signal transduction and asexual sporulation in Aspergillus nidulans. Eukaryot Cell 6:1570–1583. doi: 10.1128/EC.00085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hagiwara D, Matsubayashi Y, Marui J, Furukawa K, Yamashino T, Kanamaru K, Kato M, Abe K, Kobayashi T, Mizuno T. 2007. Characterization of the NikA histidine kinase implicated in the phosphorelay signal transduction of Aspergillus nidulans, with special reference to fungicide responses. Biosci Biotechnol Biochem 71:844–847. doi: 10.1271/bbb.70051. [DOI] [PubMed] [Google Scholar]
- 30.Izumitsu K, Yoshimi A, Tanaka C. 2007. Two-component response regulators Ssk1p and Skn7p additively regulate high-osmolarity adaptation and fungicide sensitivity in Cochliobolus heterostrophus. Eukaryot Cell 6:171–181. doi: 10.1128/EC.00326-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kojima K, Takano Y, Yoshimi A, Tanaka C, Kikuchi T, Okuno T. 2004. Fungicide activity through activation of a fungal signalling pathway. Mol Microbiol 53:1785–1796. doi: 10.1111/j.1365-2958.2004.04244.x. [DOI] [PubMed] [Google Scholar]
- 32.Motoyama T, Ohira T, Kadokura K, Ichiishi A, Fujimura M, Yamaguchi I, Kudo T. 2005. An os-1 family histidine kinase from a filamentous fungus confers fungicide-sensitivity to yeast. Curr Genet 47:298–306. doi: 10.1007/s00294-005-0572-6. [DOI] [PubMed] [Google Scholar]
- 33.Bahn YS, Kojima K, Cox GM, Heitman J. 2005. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol Biol Cell 16:2285–2300. doi: 10.1091/mbc.E04-11-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheetham J, Smith DA, da Silva Dantas A, Doris KS, Patterson MJ, Bruce CR, Quinn J. 2007. A single MAPKKK regulates the Hog1 MAPK pathway in the pathogenic fungus Candida albicans. Mol Biol Cell 18:4603–4614. doi: 10.1091/mbc.E07-06-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du C, Sarfati J, Latge JP, Calderone R. 2006. The role of the sakA (Hog1) and tcsB (sln1) genes in the oxidant adaptation of Aspergillus fumigatus. Med Mycol 44:211–218. doi: 10.1080/13693780500338886. [DOI] [PubMed] [Google Scholar]
- 36.Kawasaki L, Sánchez O, Shiozaki K, Aguirre J. 2002. SakA MAP kinase is involved in stress signal transduction, sexual development and spore viability in Aspergillus nidulans. Mol Microbiol 45:1153–1163. doi: 10.1046/j.1365-2958.2002.03087.x. [DOI] [PubMed] [Google Scholar]
- 37.Rolke Y, Tudzynski P. 2008. The small GTPase Rac and the p21-activated kinase Cla4 in Claviceps purpurea: interaction and impact on polarity, development and pathogenicity. Mol Microbiol 68:405–423. doi: 10.1111/j.1365-2958.2008.06159.x. [DOI] [PubMed] [Google Scholar]
- 38.Bell M, Capone R, Pashtan I, Levitzki A, Engelberg D. 2001. Isolation of hyperactive mutants of the MAPK p38/Hog1 that are independent of MAPK kinase activation. J Biol Chem 276:25351–25358. doi: 10.1074/jbc.M101818200. [DOI] [PubMed] [Google Scholar]
- 39.Wang F, Tao J, Qian Z, You S, Dong H, Shen H, Chen X, Tang S, Ren S. 2009. A histidine kinase PmHHK1 regulates polar growth, sporulation and cell wall composition in the dimorphic fungus Penicillium marneffei. Mycol Res 113:915–923. doi: 10.1016/j.mycres.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 40.Alonso-Monge R, Navarro-García F, Molero G, Diez-Orejas R, Gustin M, Pla J, Sánchez M, Nombela C. 1999. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J Bacteriol 181:3058–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bugeja HE, Hynes MJ, Andrianopoulos A. 2013. HgrA is necessary and sufficient to drive hyphal growth in the dimorphic pathogen Penicillium marneffei. Mol Microbiol 88:998–1014. doi: 10.1111/mmi.12239. [DOI] [PubMed] [Google Scholar]
- 42.Lara-Rojas F, Sánchez O, Kawasaki L, Aguirre J. 2011. Aspergillus nidulans transcription factor AtfA interacts with the MAPK SakA to regulate general stress responses, development and spore functions. Mol Microbiol 80:436–454. doi: 10.1111/j.1365-2958.2011.07581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hagiwara D, Mizuno T, Abe K. 2009. Characterization of NikA histidine kinase and two response regulators with special reference to osmotic adaptation and asexual development in Aspergillus nidulans. Biosci Biotechnol Biochem 73:1566–1571. doi: 10.1271/bbb.90063. [DOI] [PubMed] [Google Scholar]
- 44.Calera JA, Zhao XJ, Calderone R. 2000. Defective hyphal development and avirulence caused by a deletion of the SSK1 response regulator gene in Candida albicans. Infect Immun 68:518–525. doi: 10.1128/IAI.68.2.518-525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chauhan N, Inglis D, Roman E, Pla J, Li D, Calera JA, Calderone R. 2003. Candida albicans response regulator gene SSK1 regulates a subset of genes whose functions are associated with cell wall biosynthesis and adaptation to oxidative stress. Eukaryot Cell 2:1018–1024. doi: 10.1128/EC.2.5.1018-1024.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prieto D, Román E, Correia I, Pla J. 2014. The HOG pathway is critical for the colonization of the mouse gastrointestinal tract by Candida albicans. PLoS One 9:e87128. doi: 10.1371/journal.pone.0087128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du C, Calderone R, Richert J, Li D. 2005. Deletion of the SSK1 response regulator gene in Candida albicans contributes to enhanced killing by human polymorphonuclear neutrophils. Infect Immun 73:865–871. doi: 10.1128/IAI.73.2.865-871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hickman MJ, Spatt D, Winston F. 2011. The Hog1 mitogen-activated protein kinase mediates a hypoxic response in Saccharomyces cerevisiae. Genetics 188:325–338. doi: 10.1534/genetics.111.128322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borneman AR, Hynes MJ, Andrianopoulos A. 2000. The abaA homologue of Penicillium marneffei participates in two developmental programmes: conidiation and dimorphic growth. Mol Microbiol 38:1034–1047. doi: 10.1046/j.1365-2958.2000.02202.x. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning; a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 51.Boyce KJ, Bugeja HE, Weerasinghe H, Payne M, Schreider L, Park C, Woodward T, Andrianopoulos A. 2012. Strategies for the molecular genetic manipulation and visualization of the human fungal pathogen Penicillium marneffei. Fungal Genet Rep 59:1–12. [DOI] [PubMed] [Google Scholar]
- 52.Boyce KJ, McLauchlan A, Schreider L, Andrianopoulos A. 2015. Intracellular growth is dependent on tyrosine catabolism in the dimorphic fungal pathogen Penicillium marneffei. PLoS Pathog 11:e1004790. doi: 10.1371/journal.ppat.1004790. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complementation of ΔsskA, ΔmpkA, and ΔmpkB strains. (A) Colonies of a wild-type control strain (ΔligD pyrGt), ΔsskA sskA+, ΔmpkA mpkA+, and ΔmpkB mpkB+ strains grown on ANM plus supplements plus GABA at 25°C for 14 days. (B) The ΔsskA sskA+ strain was plated on chlorate to select for the loss of the sskA+ plasmid integrated at niaD and then grown for 14 days at 25°C on 1% ANM plus GABA plus various stress-inducing agents (calcofluor white [CAL], sodium dodecyl sulfate [SDS], Congo red [CR], sorbitol, and sodium chloride [NaCl]). Removal of niaAt sskA+ resulted in a strain displaying the ΔsskA phenotype. (C) The wild-type, ΔsskA sskA+, ΔmpkA mpkA+, and ΔmpkB mpkB+ strains were grown for 14 days at 25°C on 1% ANM plus GABA plus various stress-inducing agents (calcofluor white [CAL], sodium dodecyl sulfate [SDS], Congo red [CR], hydrogen peroxide [H2O2], sorbitol, potassium chloride [KCl], and sodium chloride [NaCl]). Download Figure S1, TIF file, 7 MB (7.2MB, tif) .
Copyright © 2016 Boyce et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Oligonucleotides used in this study Download Table S1, DOC file, 0.04 MB (45.5KB, doc) .
Copyright © 2016 Boyce et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains used in this study Download Table S2, DOC file, 0.05 MB (48.5KB, doc) .
Copyright © 2016 Boyce et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.