Whole-genome sequencing (WGS) is quickly becoming a routine method for identifying genes associated with antimicrobial resistance (AR). However, for many microbiologists, the use and analysis of WGS data present a substantial challenge. We developed SSTAR, software with a graphical user interface that enables the identification of known AR genes from WGS and has the unique capacity to easily detect new variants of known AR genes, including truncated protein variants. Current software solutions do not notify the user when genes are truncated and, therefore, likely nonfunctional, which makes phenotype predictions less accurate. SSTAR users can apply any AR database of interest as a reference comparator and can manually add genes that impact resistance, even if such genes are not resistance determinants per se (e.g., porins and efflux pumps).
KEYWORDS: antimicrobial resistance genes, porins, BLAST, SSTAR
ABSTRACT
We present the easy-to-use Sequence Search Tool for Antimicrobial Resistance, SSTAR. It combines a locally executed BLASTN search against a customizable database with an intuitive graphical user interface for identifying antimicrobial resistance (AR) genes from genomic data. Although the database is initially populated from a public repository of acquired resistance determinants (i.e., ARG-ANNOT), it can be customized for particular pathogen groups and resistance mechanisms. For instance, outer membrane porin sequences associated with carbapenem resistance phenotypes can be added, and known intrinsic mechanisms can be included. Unique about this tool is the ability to easily detect putative new alleles and truncated versions of existing AR genes. Variants and potential new alleles are brought to the attention of the user for further investigation. For instance, SSTAR is able to identify modified or truncated versions of porins, which may be of great importance in carbapenemase-negative carbapenem-resistant Enterobacteriaceae. SSTAR is written in Java and is therefore platform independent and compatible with both Windows and Unix operating systems. SSTAR and its manual, which includes a simple installation guide, are freely available from https://github.com/tomdeman-bio/Sequence-Search-Tool-for-Antimicrobial-Resistance-SSTAR-.
IMPORTANCE Whole-genome sequencing (WGS) is quickly becoming a routine method for identifying genes associated with antimicrobial resistance (AR). However, for many microbiologists, the use and analysis of WGS data present a substantial challenge. We developed SSTAR, software with a graphical user interface that enables the identification of known AR genes from WGS and has the unique capacity to easily detect new variants of known AR genes, including truncated protein variants. Current software solutions do not notify the user when genes are truncated and, therefore, likely nonfunctional, which makes phenotype predictions less accurate. SSTAR users can apply any AR database of interest as a reference comparator and can manually add genes that impact resistance, even if such genes are not resistance determinants per se (e.g., porins and efflux pumps).
INTRODUCTION
Antimicrobial resistance (AR) is an ancient phenomenon with a rapid increase in recent years and is a serious worldwide public health threat (1, 2). The emerging carbapenem-resistant microorganisms, like Enterobacteriaceae and Pseudomonas, are especially a concern since they are rendering many treatment options ineffective. The emergence of carbapenem resistance is mediated mainly by acquired carbapenemase genes that encode proteins which inactivate carbapenems and other β-lactam antibiotics (3). Other mechanisms, including modification or loss of outer membrane porin and efflux mechanisms, both of which can be combined with the production of extended-spectrum beta-lactamases (ESBLs) or AmpC-type β-lactamases, have also been described (4). AR genes can be acquired through horizontal gene transfer or can arise due to spontaneous mutations in chromosomal loci. Rapid identification of AR determinants, including prediction of novel alleles, is important for understanding the molecular epidemiology of AR pathogens and informing infection control interventions and may be useful for guiding patient therapy.
In recent years, the cost of DNA sequencing has been decreasing rapidly and the technique has become available for a large community worldwide (5). Whole-genome sequencing (WGS) data are used for strain characterization in the context of outbreak investigation, epidemiological surveillance, and the detection of genes that are associated with antimicrobial resistance (6 – 8). Many online databases containing AR genes are currently available to the public. The two most complete and up-to-date repositories are ResFinder (9) and the Comprehensive Antibiotic Resistance Database (CARD) (10). For these, users upload bacterial genome sequences to a Web service and subsequently retrieve a list of acquired antimicrobial resistance genes. These Web services are relatively slow, which can be a hurdle for users wanting to query multiple genome files.
Offline solutions, including SRST2 (11), ARG-ANNOT (12), and ABRicate (https://github.com/tseemann/abricate), exist as well. In addition to containing acquired resistance genes, the ARG-ANNOT database includes point mutations in chromosomal target genes known to be associated with AR. SRST2, ARG-ANNOT, and ABRicate run locally and give the user more control than Web-based services. SRST2 and ABRicate offer only a command-line interface, while ARG-ANNOT runs on an outdated software package (13). Currently, there is no freely available standalone AR gene detection tool with a “mouse click user interface.” While ARG-ANNOT and ABRicate offer a way to detect putative new AR genes, none of the above-mentioned tools offer an easy way to detect new or truncated alleles or variants of existing AR genes or other genes associated with resistance.
Here, we describe our tool SSTAR (Sequence Search Tool for Antimicrobial Resistance), which combines a modified ARG-ANNOT database, standalone BLAST (14), and an easy-to-use graphical user interface that enables the detection of known AR genes and predicts putative new variants as well as truncated genes in a fast, local, and easily updatable tool.
RESULTS
SSTAR algorithm.
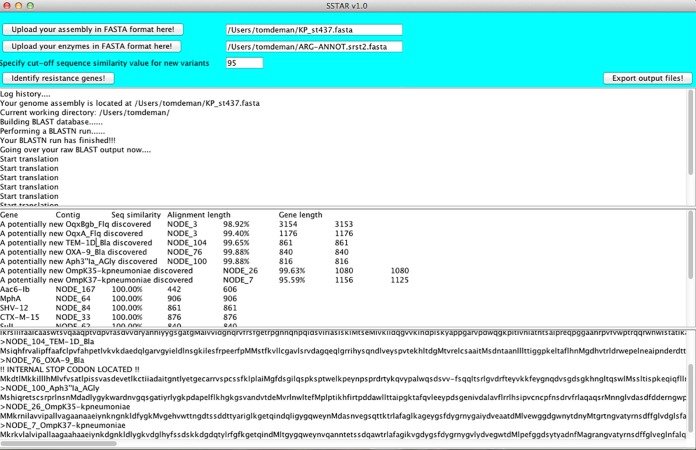
SSTAR accepts two sequence files in FASTA format, one containing the bacterial genome assembly and the other the AR gene collection (Fig. 1). First, the genome assembly is turned into a local nucleotide BLAST database using the BLAST “makeblastdb” utility. Next, AR genes are queried against this reference using BLASTN. AR genes with 100% sequence similarity against 40% or more of a potential gene on the assembly are reported by SSTAR. Reports of more than one partial variant of the same gene located on the same contig suggest an assembly error or artifact, whereas multiple intact variants represent the presence of multiple alleles of the same gene.
FIG 1 .
Graphical user interface of SSTAR. The top two buttons are used for uploading the genome assembly and AR gene collection. The user also fills out a sequence similarity cutoff value for detecting potential new variants.
If all BLASTN hits for a particular clustered AR gene group (e.g., NDM or OXA-48-like) have 95% to 99.99% sequence similarity, SSTAR reports a potentially new variant (PNV) of that gene. The complete region of alignment for each PNV is automatically selected and translated by SSTAR into protein sequence for all three reading frames. PNVs that span less than 80% of an AR gene will not be translated. Resistance determinants with <95% similarity to potential genes on the assembly are not reported.
Although we used a 95% threshold, this value can be set by the user to be more or less stringent for sequence similarity between genes in the AR gene database and those on the genome assembly. Here, we chose 95% since many AR genes within a gene group, like NDM and KPC, differ by only a few nucleotides from each other. When selecting lower similarity values (e.g., 30%), one is more likely to observe false-positive PNVs.
The translated sequence with the fewest internal stop codons is considered the output sequence. If the output protein sequence contains an internal stop codon, it will be marked as truncated. The user should then compare each translated PNV to sequences in the NCBI nonredundant (NR) database using BLASTP. Any potential novel beta-lactamase variant should be sent for confirmation and nomenclature to the NCBI at http://www.ncbi.nlm.nih.gov/projects/pathogens/submit_beta_lactamase.
Resistance genes detected with SSTAR and ARG-ANNOT.
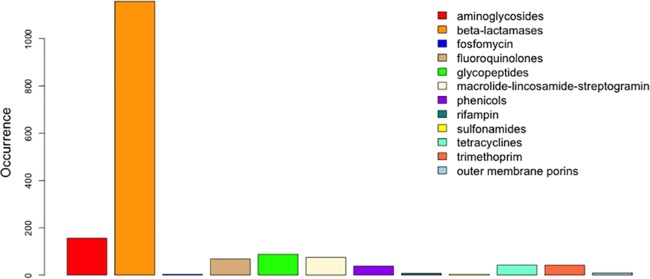
We downloaded the SRST2 version of the ARG-ANNOT database from http://katholt.github.io/srst2/ and manually added several wild-type outer membrane porin sequences (see Materials and Methods). The compositions and distribution of antimicrobial resistance mechanisms in our modified database are illustrated in Fig. 2.
FIG 2 .
Composition of the modified SSTAR SRST2 ARG-ANNOT antimicrobial resistance database. We manually included wild-type outer membrane porin sequences for Escherichia coli, Klebsiella pneumoniae, Enterobacter cloacae, and Enterobacter aerogenes and excluded tetR and blaOXA-30.
Genomic data generated from a Klebsiella pneumoniae ST437 isolate compared to our customized SRST2 ARG-ANNOT database using SSTAR identified numerous resistance mechanisms, including blaNDM-5, blaSHV-12, blaOXA-232, and blaTEM-1, as well as an intact blaampH penicillin binding protein (15, 16) and a truncated blaOXA-9 gene (Table 1). SSTAR recognized an internal stop codon within blaOXA-9, caused by a single nucleotide polymorphism (SNP) and flagged the resultant output with a FASTA-ineligible message. Genes conferring resistance to other classes of antimicrobials, including aminoglycosides, fluoroquinolones, macrolides, sulfonamides, fosfomycin, and trimethoprim, were also identified in K. pneumoniae ST437 (Table 1).
TABLE 1 .
Resistance genes identified in K. pneumoniae ST437 and E. coli ST44 using the ResFinder 2.1 Web service or SSTAR software combined with customized versions of the SRST2 ARG-ANNOT and ResFinder databases
| AR gene detected | Associated resistance |
Presence or absence in K. pneumoniae ST437 |
Presence or absence in E. coli ST44 |
||||
|---|---|---|---|---|---|---|---|
| ResFinder 2.1 | SSTAR/Ad | SSTAR/Re | ResFinder 2.1 | SSTAR/A | SSTAR/R | ||
| aac(6′)-lb-cr | Aminoglycosidesa | – | – | – | + | + | + |
| aac(6′)-lb | Aminoglycosides | + | + | + | – | – | – |
| aph(3′) la | Aminoglycosides | + | + | + | – | – | – |
| aadA1 | Aminoglycosides | + | – | + | – | – | – |
| aadA2 | Aminoglycosides | – | – | – | + | + | + |
| aadA5 | Aminoglycosides | – | – | – | + | + | + |
| strA | Aminoglycosides | – | – | – | + | + | + |
| strB | Aminoglycosides | – | – | – | + | + | + |
| rmtF | Aminoglycosides | + | + | – | – | – | – |
| blaNDM-5 | Beta-lactams | + | + | + | – | – | – |
| blaSHV-12 | Beta-lactams | + | + | + | – | – | – |
| blaCTX-M-15 | Beta-lactams | + | + | + | + | + | + |
| blaTEM-1 | Beta-lactams | + | + | + | – | – | – |
| blaOXA-1 | Beta-lactams | – | – | – | + | + | + |
| blaOXA-9 | Beta-lactams | + | +b | +b | – | – | – |
| blaOXA-232 | Beta-lactams | + | + | + | – | – | – |
| blaampH | Beta-lactams | – | + | – | – | + | – |
| blaampC1 | Beta-lactams | – | – | – | – | + | – |
| blaampC2 | Beta-lactams | – | – | – | – | + | – |
| mph(A) | Macrolides | + | + | + | + | + | + |
| catB4 | Chloramphenicol | – | – | – | + | + | + |
| sul1 | Sulfonamides | + | + | + | + | + | + |
| sul2 | Sulfonamides | – | – | – | + | + | + |
| fosA | Fosfomycin | + | – | + | – | – | – |
| tet(A) | Tetracyclines | – | – | – | + | + | + |
| tet(B) | Tetracyclines | – | – | – | + | + | + |
| dfrA12 | Trimethoprim | – | – | – | + | + | + |
| dfrA14 | Trimethoprim | + | + | + | – | – | – |
| dfrA17 | Trimethoprim | – | – | – | + | + | + |
| oqxA | Fluoroquinolones | + | + | + | – | – | – |
| oqxB | Fluoroquinolones | + | + | + | – | – | – |
| qnrS1 | Fluoroquinolones | + | + | + | – | – | – |
| E. coli ompC (porin)c | Beta-lactams | – | – | – | – | +b | +b |
| E. coli ompF (porin)c | Beta-lactams | – | – | – | – | +b | +b |
| K. pneumoniae ompK35 (porin)c | Beta-lactams | – | + | + | – | – | – |
| K. pneumoniae ompK36 (porin)c | Beta-lactams | – | + | + | – | – | – |
| K. pneumoniae ompK37 (porin)c | Beta-lactams | – | + | + | – | – | – |
And fluoroquinolones.
A truncated variant was identified.
This sequence was manually added to the database.
SSTAR/A, SSTAR with SRST2 ARG-ANNOT.
SSTAR/R, SSTAR with SRST2 ResFinder.
A second analysis was performed on a carbapenem-resistant Escherichia coli ST44 isolate. The SSTAR ARG-ANNOT customized database identified resistance genes, including the beta-lactamase genes blaOXA-1 and blaCTX-M-15, as well as two additional putative cephalosporinase blaampC genes that were present in the ARG-ANNOT database (GenBank accession numbers FN649414 and CP002970) and a blaampH gene (Table 1). As with the K. pneumoniae isolate discussed above, genes conferring resistance to numerous classes of antimicrobials were identified in the genome of E. coli ST44, including aminoglycosides, fluoroquinolones, macrolides, chloramphenicol, sulfonamides, tetracyclines, and trimethoprim (Table 1).
Comparison of resistance genes resulting in the observed resistance phenotype.
MIC values of all carbapenems and cephalosporins tested were elevated for both isolates (Table 2). The NDM carbapenemase identified in K. pneumoniae ST437 explains all beta-lactam resistance observed, with the exception of that to aztreonam, which is not hydrolyzed by metallo-beta-lactamases (17). However, this strain also carries genes that encode two ESBLs, blaSHV-12 and blaCTX-M-15, as well as another class D carbapenemase, blaOXA-232, all of which may contribute to aztreonam resistance. With K. pneumoniae ST437, elevated MICs of tobramycin, amikacin, and gentamicin were also observed, most likely due to the presence of rmtF (18) (Table 1). Resistance to the fluoroquinolones in K. pneumoniae ST437 is likely impacted by the presence of qnrS1 and the oqxA and oqxB efflux pump genes (19, 20). No acquired resistance genes were identified to explain the elevated MICs of chloramphenicol or tetracyclines.
TABLE 2 .
Observed MICs for multidrug-resistant K. pneumoniae ST437 and E. coli ST44a
| Antimicrobial |
K. pneumoniae ST437 |
E. coli ST44 |
||
|---|---|---|---|---|
| MIC (µg/ml) | Interpretation | MIC (µg/ml) | Interpretation | |
| Doripenem | >8 | R | 4 | R |
| Ertapenem | >8 | R | >8 | R |
| Imipenem | 32 | R | 2 | I |
| Meropenem | >8 | R | 8 | R |
| Aztreonam | >64 | R | >64 | R |
| Cefepime | >32 | R | >32 | R |
| Cefotaxime | >64 | R | >64 | R |
| Ceftazidime | >128 | R | >128 | R |
| Ceftriaxone | >32 | R | >32 | R |
| Cefazolin | >8 | R | >8 | R |
| Cefoxitin | >16 | R | >16 | R |
| Ampicillin | >32 | R | >32 | R |
| Amoxicillin-clavulanic acid | >32 | R | >32 | R |
| Piperacillin-tazobactam | >128 | R | >128 | R |
| Colistin | 0.5 | NA | 0.25 | NA |
| Polymyxin B | 0.5 | NA | 0.5 | NA |
| Amikacin | >64 | R | 16 | S |
| Gentamicin | >16 | R | 1 | S |
| Tobramycin | >16 | R | >16 | R |
| Tetracycline | 8 | I | >32 | R |
| Tigecycline | 4 | I | <0.5 | S |
| Trimethoprim-sulfamethoxazole | >8 | R | >8 | R |
| Chloramphenicol | >16 | R | 16 | I |
| Ciprofloxacin | >8 | R | >8 | R |
| Levofloxacin | >8 | R | >8 | R |
R, resistant; I, intermediate; S, susceptible; NA, no breakpoint available.
E. coli ST44 was not susceptible to any of the carbapenems tested and was resistant to all of the extended-spectrum beta-lactams, despite the lack of any known carbapenemase genes; blaCTX-M-15 was the only ESBL identified. E. coli ST44 was resistant to tobramycin but susceptible to amikacin and gentamicin, despite it carrying the aac(6′)-lb-cr aminoglycoside resistance element. This isolate also carried aadA2, aadA5, strA, and strB, which are expected to confer resistance to streptomycin and spectinomycin (21, 22). Escherichia coli ST44 was resistant to both ciprofloxacin and levofloxacin but had no acquired fluoroquinolone resistance determinants except aac(6′)-lb-cr. Genes encoding resistance to chloramphenicol (catB4) and tetracycline (tetA and tetB) were also observed in E. coli ST44, which likely explains its resistance to these two agents.
Porin analysis.
Many phenotypically carbapenem-resistant Enterobacteriaceae do not produce a carbapenemase enzyme but instead harbor a combination of other resistance mechanisms to confer a resistance phenotype. Therefore, we examined the outer membrane porin genes in this organism, which have been described as contributing to carbapenem resistance in organisms that do not produce a carbapenemase (4, 23).
In order to interrogate the chromosomal porin genes in E. coli and K. pneumoniae, wild-type porin gene sequences from each were added to our custom SRST2 ARG-ANNOT database. Using our customized database, SSTAR identified truncations caused by internal stop codons in two major porin genes, ompC and ompF, in E. coli ST44 (Table 1). The T→A mutation at nucleotide position 246 in ompC creates a stop codon and thus a severely truncated protein that is likely nonfunctional. A 1-nucleotide deletion at position 193 in ompF resulted in a frameshift mutation and creation of a stop codon nearby, thereby possibly resulting in a lack of functional OmpF in this isolate. This inferred loss of porin function combined with the presence of blaCTX-M-15 likely explains the carbapenem resistance phenotype in this organism (Table 2).
The K. pneumoniae ST437 isolate carried mutated porin genes in addition to blaNDM-5 and blaOXA-232. Two in-frame insertion sequences were identified in ompK37, at positions 695 and 813, neither of which resulted in internal stop codons. The first insertion sequence generates an extra 4 amino acids in OmpK37 (H-Y-T-H) and the second one a stretch of 6 (S-S-T-N-G-G). The protein sequence similarity between the OmpK37 protein deposited in our modified ARG-ANNOT database and the one detected in K. pneumoniae ST437 was 95%. This modified porin sequence has previously been deposited in the nonredundant (NR) database at the NCBI under accession number WP_002902433 and is not annotated as nonfunctional. Four silent mutations were identified in ompK35 (156T→C, 294G→A, 303G→A, and 786C→T) and are most likely not involved in protein folding or function.
Comparison of results obtained with SSTAR using the ARG-ANNOT database and the ResFinder 2.1 Web service.
In order to test the performance of SSTAR, we compared its results with those of the popular online ResFinder 2.1 Web service. Both SSTAR/ARG-ANNOT and ResFinder 2.1 (https://cge.cbs.dtu.dk//services/ResFinder/) identified blaNDM-5, blaSHV-12, blaOXA-232, blaTEM-1, and blaOXA-9 genes in K. pneumoniae ST437. However, the SSTAR/ARG-ANNOT approach identified an additional beta-lactamase gene, blaampH. This gene is present in the ARG-ANNOT database but not included in the ResFinder repository (Table 1).
The aminoglycoside resistance gene aadA1 was not detected by SSTAR due to several gaps and mismatches between the aadA1 sequence present in ARG-ANNOT and the potential aadA1 gene present in the K. pneumoniae genome. In contrast, the ResFinder database contains numerous aadA1 genes, one of which displayed 100% sequence similarity to a gene in our query genome. A similar phenomenon was observed for fosA, a gene conferring resistance to fosfomycin. There were several fosA genes included in the ResFinder database, one of which was 97.14% similar to our query sequence, but ARG-ANNOT contained only a single fosA gene, which displayed only 74% nucleotide similarity with the identified fosA gene from ResFinder.
ResFinder 2.1 identified blaOXA9 in K. pneumoniae ST437 as having 99.88% sequence similarity to the gene in its database but was not identified as truncated and therefore likely nonfunctional. However, when SSTAR/ARG-ANNOT was used, this previously described mutation was readily identified and highlighted for the user (24).
To further evaluate the SSTAR algorithm, we downloaded the SRST2 ResFinder database (https://github.com/katholt/srst2) and evaluated SSTAR/ResFinder against our K. pneumoniae ST437 isolate. The same AR gene repertoire was identified using the ResFinder database queried with SSTAR and the ResFinder 2.1 tool (Table 1). The inclusion of multiple gene alleles in the ResFinder database allowed SSTAR to annotate the blaTEM-1 gene as blaTEM-1A, whereas it was identified as a possible new variant when the ARG-ANNOT database with only a single blaTEM-1 allele was used as the reference.
When this approach was used with the E. coli ST44 genome, the only observed difference was the detection of blaampC1 and blaampC2, two putative cephalosporinase genes present in the ARG-ANNOT database that are not in the ResFinder collection. blaampH, a weak beta-lactamase, was also identified by SSTAR in E. coli ST44 when ARG-ANNOT was used as a reference, but not with the ResFinder database.
DISCUSSION
SSTAR is an easy-to-use, customizable tool for the very rapid identification of genes and potential variants associated with antimicrobial resistance. In addition, it is stand-alone and contains an intuitive graphical user interface that needs only a few clicks from the user for creating a local BLASTN database, identifying AR genes, as well as porin genes, and presenting these to the user. The average processing time depends heavily on the fragmentation of the genome assembly. A single isolate assembled from high-throughput Illumina short reads, currently the most common and popular sequencing platform, takes approximately 6 s on a 2.3-GHz Intel Core I5 processor laptop with 8 GB of RAM. Genome assemblies containing one long contiguous stretch of DNA, like PacBio or RefSeq sequences, will require considerably longer processing times. The automatic translation of PNVs is convenient and needs only BLASTP verification against a protein database or the beta-lactamase repository at http://www.ncbi.nlm.nih.gov/projects/pathogens/submit_beta_lactamase.
SSTAR’s translation to protein sequence enables detection of potential new variants, truncated enzymes, and porins that otherwise would be missed by examining only to the nucleotide level. This functionality could also be used with chromosomal/nonacquired genes to find new variants that might be involved in conferring resistance, e.g., novel gyrA changes associated with fluoroquinolone resistance (25 – 27). The detection of truncated gene products is especially useful for interrogating carbapenem-resistant microorganisms that do not produce a carbapenemase, where loss or truncation of porins is an important contributor to resistance phenotypes. Even when organisms do produce carbapenemases, porins can have a major role in the phenotypic profile and pathogenic success of particular clones (28).
When an internal stop codon is identified, SSTAR generates an invalid FASTA entry for that gene and therefore forces the user to recognize the “error” and remove the entry from the multi-FASTA file before aligning genes with other AR genes for the detection of PNVs.
Currently available AR detection tools will show the sequence similarity to only the reference gene for blaOXA-9; however, the users are not notified about possible nonfunctional variants. SSTAR identifies an internal stop codon in the blaOXA-9 beta-lactamase gene, resulting in a nonfunctional protein (24). This additional level of information will be increasingly important as genomic information is used to predict organism resistance, as simple gene detection is insufficient to predict phenotypes.
Different AR databases can generate different results due to their included genes. For instance, SSTAR/ARG-ANNOT identified a possible new variant of blaTEM-1; however, when performing the same annotation analyses using SSTAR/ResFinder, we observed a blaTEM-1A wild type. This is due to the fact that ResFinder contains several TEM-1 alleles, whereas ARG-ANNOT contains only one. Thus, it is important that users of these AR databases understand the scope, strengths, and limitations of each repository. By querying multiple databases or creating customized databases, users will gain a more complete understanding of the complement of acquired and well-characterized resistance genes in their isolates of interest. SSTAR/ARG-ANNOT also did not identify the aadA1 and fosA genes that are located in the K. pneumoniae ST437 genome, whereas SSTAR/ResFinder was able to detect those genes because several alleles of both genes were present in the ResFinder database. Using up-to-date AR gene databases is therefore essential for accurately predicting resistance genes, and SSTAR facilitates this option.
This is not meant to imply that genomic data can at this point predict the complete phenotypic profile of an isolate. For instance, both SSTAR/ResFinder and SSTAR/ARG-ANNOT were not able to explain the phenotypic resistance to chloramphenicol or tetracyclines for the K. pneumoniae ST437 isolate or the fluoroquinolone resistance in E. coli ST44. This further emphasizes the need for regular updating of AR databases as new genes are described and customization to include intrinsic mechanisms that strongly correlate with phenotype. Our tool is flexible and can easily be used with different enzyme databases, including any publicly accessible FASTA-formatted gene collection or custom database. When databases other than SRST2 ARG-ANNOT are used, each FASTA header needs to be structured with the CD-HIT-clustered SRST2 sequence header naming structure, which is described in our online manual (https://github.com/tomdeman-bio/Sequence-Search-Tool-for-Antimicrobial-Resistance-SSTAR-).
The SSTAR software is intended for biologists and clinical workers who want to investigate acquired antimicrobial resistance in bacterial genomes. Execution of SSTAR requires only two additional programs for its operation, BLAST+ and Java. This should make usage and installation easy for a broad audience with a limited bioinformatics background. In addition, SSTAR’s capacity for protein translation and identifying mutations that result in premature stop codons is unique among commonly available tools for resistance gene detection and thus provides the user with additional information about genotype-phenotype correlation that might otherwise be missed.
MATERIALS AND METHODS
Database selection.
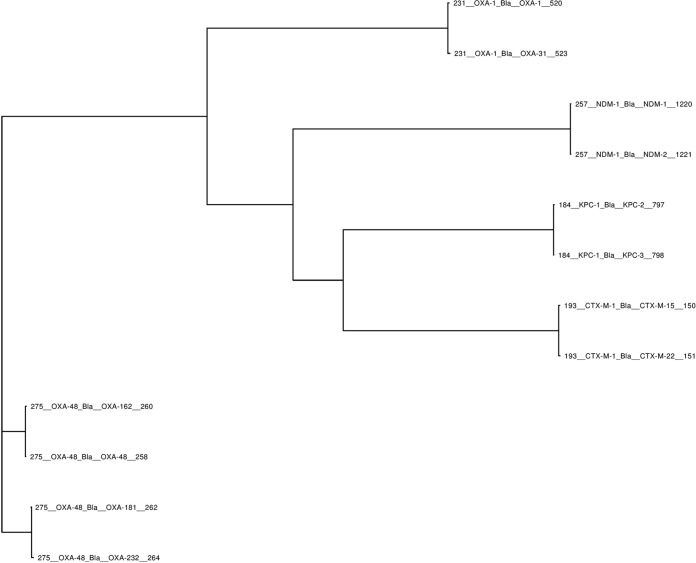
SSTAR can accept any database collection of resistance genes as a comparator for subsequent analysis. For this study, we selected the AR database from ARG-ANNOT, an extensive and curated collection of genes conferring resistance to different antibiotic classes, including aminoglycosides, beta-lactams, fosfomycin, fluoroquinolones, glycopeptides, macrolides, lincosamides, streptogramins, phenicols, rifampin, sulfonamides, tetracyclines, and trimethoprim. A slightly modified version of this database was downloaded from the SRST2 GitHub repository at https://github.com/katholt/srst2/. The AR gene sequences in this version are clustered by means of CD-HIT (29) into gene groups that share >80% sequence similarity. The sequence headers are formatted so that each clustered gene group has a unique number assigned to it. For instance, all New Delhi metallo-beta-lactamase (NDM) alleles fall within the same gene group and therefore start with the same number (i.e., 257). The OXA-type beta-lactamase family, which consists of more-diverse gene groups, contains multiple gene clusters, and each cluster has its own unique number (Fig. 3). SSTAR analyzes each group separately.
FIG 3 .
Several gene groups and associated alleles designated by SRST2 ARG-ANNOT. Each group is assigned a unique number at the beginning of the sequence header; e.g., blaOXA-48 alleles start with 275, and blaKPC alleles start with 184, etc.
We manually excluded tetR from the database since it is a regulatory gene and not a resistance gene and blaOXA-30 because it is 100% identical to blaOXA-1 (30) (http://www.lahey.org/Studies). We also included several wild-type major outer membrane porin sequences from K. pneumoniae (ompK35 [CP011313], ompK36 [FJ577673], and ompK37 [KC195784]), E. coli (ompC [HQ330974] and ompF [CP005930]), Enterobacter cloacae (ompC [AJ316539] and ompF [HM565105]), and Enterobacter aerogenes (omp35 [AY487904] and omp36 [AF336098]).
For comparison, we also used SSTAR to interrogate the genomes in this study against a CD-HIT-clustered version of the ResFinder repository (https://github.com/katholt/srst2/). The complete SSTAR package, along with both databases described above, are available at https://github.com/tomdeman-bio/Sequence-Search-Tool-for-Antimicrobial-Resistance-SSTAR-.
Strains evaluated.
The K. pneumoniae ST437 and E. coli ST44 strains evaluated in this study were isolated from sputum and blood, respectively. These isolates were recognized as being unusually resistant and thus were submitted to the CDC for reference antimicrobial susceptibility testing (AST). AST was performed using the reference broth microdilution method according to the Clinical and Laboratory Standards Institute methodology (31) (Table 2). Additional molecular screening using real-time PCR for the detection of genes encoding KPC, NDM, and OXA-48-like carbapenemases was performed on each isolate as well. Whole-genome sequencing of both specimens, using a MiSeq benchtop sequencer (Illumina, San Diego, CA), was the final step in the process. Sequencing reads were then assembled by means of SPAdes 3.1 (32) and CLC Genomics Workbench v. 7.0.4 (CLC Bio, Aarhus, Denmark). Average genome coverage was 47-fold for K. pneumoniae and 122-fold for E. coli.
Nucleotide sequence accession numbers.
The K. pneumoniae ST437 and E. coli ST44 genome sequences have been deposited in DDBJ/EMBL/GenBank under the accession numbers LART00000000 and LAXC00000000, respectively. The versions used in this paper are LART01000000 and LAXC01000000.
ACKNOWLEDGMENTS
We thank James Kamile Rasheed for his input on antimicrobial resistance mechanisms and Maria Karlsson for naming SSTAR.
This work was made possible through support from the Advanced Molecular Detection (AMD) initiative at the Centers for Disease Control and Prevention.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
REFERENCES
- 1.D’Costa VM, King CE, Kalan L, Morar M, Sung WWL, Schwarz C, Froese D, Zazula G, Calmels F, Debruyne R, Golding GB, Poinar HN, Wright GD. 2011. Antibiotic resistance is ancient. Nature 477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization 2014. Antimicrobial resistance: global report on surveillance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Queenan AM, Bush K. 2007. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev 20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med 18:263–272. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Didelot X, Bowden R, Wilson DJ, Peto TEA, Crook DW. 2012. Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet 13:601–612. doi: 10.1038/nrg3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Köser CU, Ellington MJ, Peacock SJ. 2014. Whole-genome sequencing to control antimicrobial resistance. Trends Genet 30:401–407. doi: 10.1016/j.tig.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleinheinz KA, Joensen KG, Larsen MV. 2014. Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage 4:e27943. doi: 10.4161/bact.27943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Man TJB, Perry KA, Avillan JJ, Rasheed JK, Limbago BM. 2015. Draft genome sequence of a New Delhi metallo-β-lactamase-5 (NDM-5)-producing multidrug-resistant Escherichia coli isolate. Genome Announc 3:e00017–15. doi: 10.1128/genomeA.00017-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O’Brien JS, Pawlowski AC, Piddock LJV, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. 2013. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inouye M, Dashnow H, Raven L-A, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE. 2014. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, Rolain J-M. 2014. Arg-ANNOT (antibiotic resistance gene-ANNOTation), a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall T. BioEdit: biological sequence alignment editor for Win 95/98/NT/2K/XP. Chicago State University, Chicago, IL: http://www.mbioncsu.edu. [Google Scholar]
- 14.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 15.Henderson TA, Young KD, Denome SA, Elf PK. 1997. AmpC and AmpH, proteins related to the class C beta-lactamases, bind penicillin and contribute to the normal morphology of Escherichia coli. J Bacteriol 179:6112–6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González-Leiza SM, de Pedro MA, Ayala JA. 2011. AmpH, a bifunctional dd-endopeptidase and dd-carboxypeptidase of Escherichia coli. J Bacteriol 193:6887–6894. doi: 10.1128/JB.05764-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walsh TR, Toleman MA, Poirel L, Nordmann P. 2005. Metallo-beta-lactamases: the quiet before the storm? Clin Microbiol Rev 18:306–325. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galimand M, Courvalin P, Lambert T. 2012. RmtF, a new member of the aminoglycoside resistance 16S rRNA N7 G1405 methyltransferase family. Antimicrob Agents Chemother 56:3960–3962. doi: 10.1128/AAC.00660-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen LH, Jensen LB, Sørensen HI, Sørensen SJ. 2007. Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J Antimicrob Chemother 60:145–147. doi: 10.1093/jac/dkm167. [DOI] [PubMed] [Google Scholar]
- 20.Domínguez-Herrera J, Velasco C, Docobo-Pérez F, Rodríguez-Martínez JM, López-Rojas R, Briales A, Pichardo C, Díaz-de-Alba P, Rodríguez-Baño J, Pascual A, Pachón J. 2013. Impact of qnrA1, qnrB1 and qnrS1 on the efficacy of ciprofloxacin and levofloxacin in an experimental pneumonia model caused by Escherichia coli with or without the GyrA mutation Ser83Leu. J Antimicrob Chemother 68:1609–1615. doi: 10.1093/jac/dkt063. [DOI] [PubMed] [Google Scholar]
- 21.Hollingshead S, Vapnek D. 1985. Nucleotide sequence analysis of a gene encoding a streptomycin/spectinomycin adenylyltransferase. Plasmid 13:17–30. doi: 10.1016/0147-619X(85)90052-6. [DOI] [PubMed] [Google Scholar]
- 22.Sandvang D. 1999. Novel streptomycin and spectinomycin resistance gene as a gene cassette within a class 1 integron isolated from Escherichia coli. Antimicrob Agents Chemother 43:3036–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wozniak A, Villagra NA, Undabarrena A, Gallardo N, Keller N, Moraga M, Román JC, Mora GC, García P. 2012. Porin alterations present in non-carbapenemase-producing Enterobacteriaceae with high and intermediate levels of carbapenem resistance in Chile. J Med Microbiol 61:1270–1279. doi: 10.1099/jmm.0.045799-0. [DOI] [PubMed] [Google Scholar]
- 24.Poirel L, Héritier C, Tolün V, Nordmann P. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother 48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson JR, Tchesnokova V, Johnston B, Clabots C, Roberts PL, Billig M, Riddell K, Rogers P, Qin X, Butler-Wu S, Price LB, Aziz M, Nicolas-Chanoine M-H, Debroy C, Robicsek A, Hansen G, Urban C, Platell J, Trott DJ, Zhanel G, Weissman SJ, Cookson BT, Fang FC, Limaye AP, Scholes D, Chattopadhyay S, Hooper DC, Sokurenko EV. 2013. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J Infect Dis 207:919–928. doi: 10.1093/infdis/jis933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong L-C, Gao D, Gao Y-H, Liu S-M, Ma H-X. 2014. Fluoroquinolone resistance mechanism of clinical isolates and selected mutants of Pasteurella multocida from bovine respiratory disease in China. J Vet Med Sci 76:1655–1657. doi: 10.1292/jvms.14-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willby M, Sikes RD, Malik S, Metchock B, Posey JE. 2015. Correlation between GyrA substitutions and ofloxacin, levofloxacin, and moxifloxacin cross-resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 59:5427–5434. doi: 10.1128/AAC.00662-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowers JR, Kitchel B, Driebe EM, MacCannell DR, Roe C, Lemmer D, de Man T, Rasheed JK, Engelthaler DM, Keim P, Limbago BM. 2015. Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae pandemic. PLoS One 10:e0133727. doi: 10.1371/journal.pone.0133727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zankari E. 2014. Comparison of the Web tools ARG-ANNOT and ResFinder for detection of resistance genes in bacteria. Antimicrob Agents Chemother 58:4986–4986. doi: 10.1128/AAC.02620-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clinical and Laboratory Standards Institute 2015. Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement. Document M100-S25. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 32.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]