Abstract
Energy production by chloroplasts and mitochondria causes constant oxidative damage. A functioning photosynthetic cell requires quality-control mechanisms to turn over and degrade chloroplasts damaged by reactive oxygen species (ROS). Here, we generated a conditionally lethal Arabidopsis mutant that accumulated excess protoporphyrin IX in the chloroplast and produced singlet oxygen. Damaged chloroplasts were subsequently ubiquitinated and selectively degraded. A genetic screen identified the plant U-box 4 (PUB4) E3 ubiquitin ligase as being necessary for this process. pub4-6 mutants had defects in stress adaptation and longevity. Thus, we have identified a signal that leads to the targeted removal of ROS-overproducing chloroplasts.
In chloroplasts, an electron transport chain allows energy from sunlight to be used. When the chloroplast’s capacity to transfer electrons is exceeded (owing to high irradiance, drought, or extreme temperatures) reactive oxygen species (ROS) (e.g., singlet oxygen (1O2) or peroxides) are generated (1). ROS can then damage DNA, proteins, lipids, and other cellular components. Thus, safeguards have evolved that allow for rapid cellular responses to these injuries.
Information about ROS damage is relayed by chloroplast-to-nucleus (retrograde) signals that broadly regulate nuclear genes involved in chloroplast function and stress adaptation (2, 3). Retrograde signals involve chloroplast-localized tetrapyrroles (hemes and chlorophylls) (4), chloroplast gene expression (5), chloroplast-produced ROS (6, 7), and metabolites (8–10). These signals are assumed to affect the function of all 50 to 100 chloroplasts in a cell. Here, we looked for other quality-control mechanisms that could work at the level of the individual chloroplast.
Plastid ferrochelatases 1 and 2 (FC1 and FC2) are conserved enzymes at the heme-chlorophyll branch point of the chloroplast-localized tetrapyrrole biosynthetic pathway (fig. S1). They convert protoporphyrin IX (Proto) to heme and may play a role in the quality control of individual chloroplasts (11–13). To test this hypothesis, we monitored Arabidopsis thaliana fc1 and fc2 mutants (fig. S2, A to C) during de-etiolation. De-etiolation involves development of nongreen plastids into mature chloroplasts and requires chloroplast signaling. When this process is disrupted, photoautotrophic growth can be inhibited (14). Seedlings were grown in the dark for 4 days and then moved to light. After 1 day of light, wild-type (wt) and fc1-1 plants became green, whereas fc2-1 and fc2-2 mutants failed to do so (Fig. 1, A and B), and photosynthetic cells in cotyledons died (Fig. 1C).
Fig. 1. fc2 mutants suffer from chloroplast degradation and photooxidative stress during dark-to-light transitions.
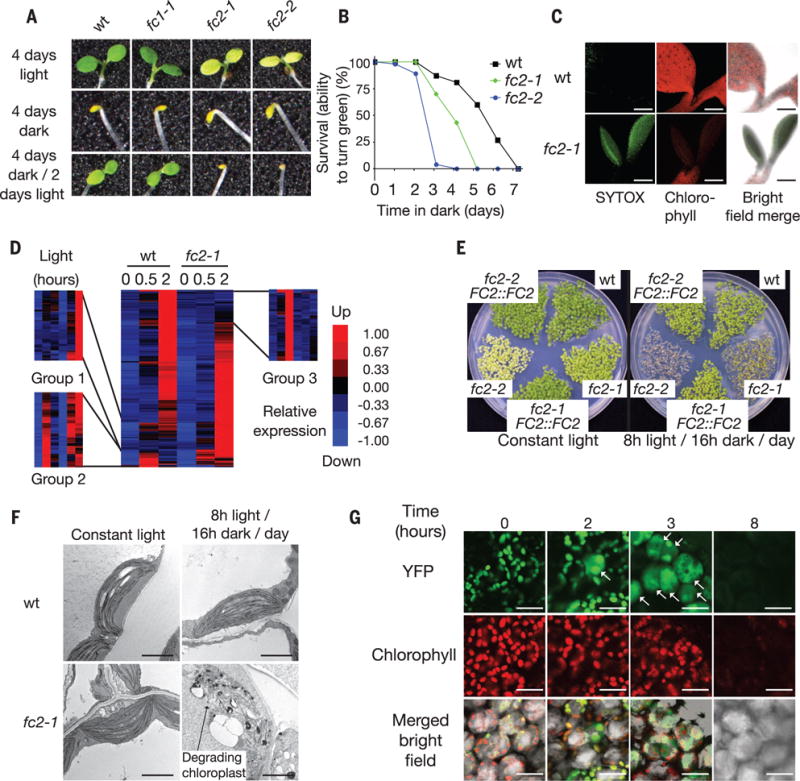
(A) fc2 mutant seedlings are unable to turn green after 4 days of growth in the dark. (B) The survival rate (the ability to turn green in 48 hours of light) of 100 seedlings first grown for the indicated number of days in the dark. (C) fc2-1 cotyledons undergo cell death, as shown by SYTOX green staining of dead nuclei. Scale bars, 200 μm. (D) A microarray analysis of transcripts induced (at least twofold, P ≤ 0.05) in 4-day-old dark grown seedlings during the first 2 hours of de-etiolation. (E) Five-day-old seedling phenotypes under different day lengths. (F) Representative TEM micrographs of chloroplasts in 3-day-old cotyledon mesophyll cells 2 hours after dawn. Scale bars, 2 μm. (G) Confocal images of 3-day-old fc2-1 seedling cotyledons grown in 8 hours of light/day expressing plastid-targeted YFP (RBCS-YFP) before (time 0) or after light exposure. White arrows indicate cells with cytoplasmic YFP. Scale bars, 20 μm.
To determine the cause of this phenotype, we profiled the transcriptomes of wt and fc2-1 seedlings at the start of de-etiolation. RNA profiles were similar between these genotypes before the dark-to-light shift (Fig. 1D, table S1), but diverged after 2 hours of light. Genes enriched for plastid function (group 3) were repressed, whereas genes enriched for heat stress (group 1) and chitin response (group 2) (table S2) were more highly expressed in fc2-1 than in wt. Thus, the dark-to-light transition appeared to cause photooxidative stress in the fc2-1 mutant, possibly within chloroplasts.
To test this, we grew seedlings in 24-hour light/dark cycling periods (Fig. 1E and fig. S3, A and B). Wt and fc2 seedlings turned green when grown in constant light (24 hours) or 16 hours of light (8 hours of dark)/day. In shorter light periods [8 hours of light (16 hours of dark)/day or 4 hours of light (20 hours of dark)/day], fc2 seedlings, unlike wt, did not become green; transgenic complementation confirmed this was an fc2 mutant defect. For plants provided with 8 hours of light/day, the third dawn blocked their ability to turn green, and stress marker genes were induced (fig. S3, C and D, and table S3). This condition was used for most of the subsequent seedling experiments. These phenotypes were not dependent on EXE1, which mediates chloroplast-induced cell death (7) (fig. S4, A to I, and supplementary text).
We used transmission electron microscopy (TEM) to monitor the development of fc2 chloroplasts. In constant light conditions, the ultrastructure of fc2-1 chloroplasts was similar to that of wt (Fig. 1F). In 8 hours of light/day, fc2-1 chloroplasts were severely damaged, with swollen thylakoid membranes, large plastoglobule structures, and disrupted outer envelope membranes. This loss of chloroplast integrity was confirmed using fc2-1 seedlings expressing chloroplast-localized yellow fluorescent protein (YFP) (Fig. 1G and fig. S3H). When these lines were grown in 8 hours of light/day, YFP leaked into the cytoplasm after 1 hour. After 8 hours, YFP and chlorophyll fluorescence were barely detectable. Thus a chloroplast stress signal induced fast chloroplast degradation.
Because tetrapyrroles can cause photooxidative damage (15), we monitored the levels of five chlorophyll intermediates. In 8 hours of light/day growth conditions, Proto was the only one that accumulated in fc2 mutants (Fig. 2A and fig. S5A). Genetic complementation experiments of fc2 mutants showed that expression of FC1 could not prevent this accumulation (Fig. 2A), but did restore wt levels of heme (Fig. 2B) and chlorophyll (Fig. 2C and fig. S5B) in constant light and protochlorophyllide (fig. S5C) in the dark. In 8 hours of light/day growth conditions, FC1 expression did not restore the ability to turn green (Fig. 2C and fig. S5B), block chloroplast degradation (fig. S4I), or block stress marker gene induction (fig. S4H) in fc2-1 plants. Expressing maize FC1 or a catalytically dead variant of FC2 (H295A) (12) also did not restore an ability to turn green to fc2-1 plants (figs. S6 and S7, A and B). Finally, reducing excess tetrapyrrole accumulation in fc2-1 by introducing hema1 and hema2 mutations that reduce the first step of tetrapyrrole synthesis (fig. S1) allowed seedlings to avoid chloroplast degradation when provided with 8 hours of light/day (Fig. 2C and fig. S4I). Together these findings suggested that Proto accumulation correlated with chloroplast degradation and confirmed that FC2 has a specialized function in plastids (11, 12, 16).
Fig. 2. fc2 mutants accumulate Proto and 1O2.
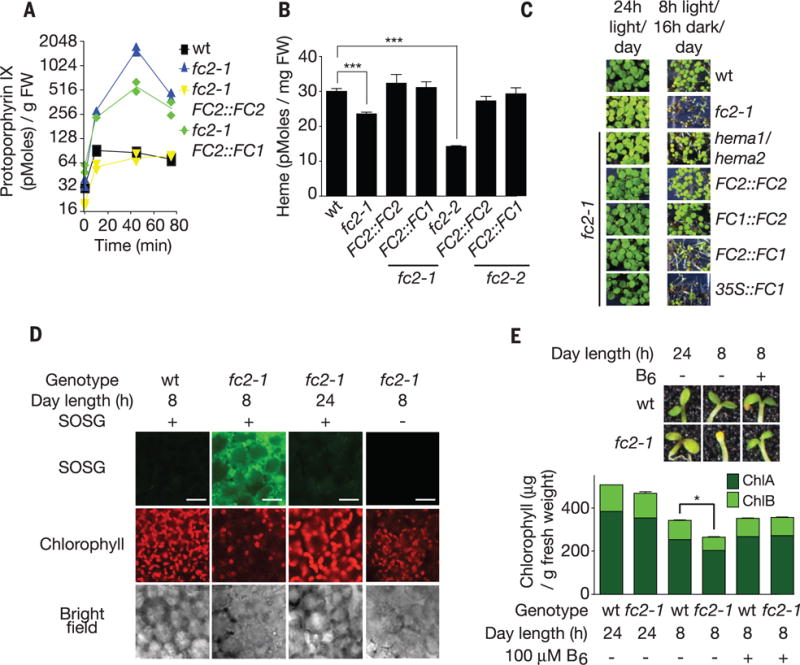
(A) Time course (starting at dawn) of Proto levels in 3-day-old seedlings provided with 8 hours of light/day (n ≥ 2 replicates). (B) Free heme levels of seedlings (n = 3 replicates). (C) Phenotypes of transgenic seedlings grown for 5 days in the indicated conditions. (D) Representative confocal images of singlet oxygen (as shown by singlet oxygen sensor green) accumulation in cotyledon mesophyll cells (2 hours after dawn) of 3-day-old seedlings provided with 8 hours of light/day. Scale bars, 20 μm. (E) Effect of feeding vitamin B6 on chlorophyll levels (n = 3 replicates) and survival. All values are means T SEM. *P ≤ 0.05, ***P ≤ 0.001, two-tailed Student’s t test.
Proto is a photosensitizing molecule that generates 1O2 in cells (17). As expected, fc2-1 mutants provided with 8 hours of light/day accumulated 1O2 (Fig. 2D) and induced 1O2 and oxidative stress-responsive genes within 2 hours (fig. S8A). Feeding the 1O2 scavenger vitamin B6 (18) blocked chloroplast degradation in fc2-1 seedlings and restored their ability to become green (Fig. 2E and fig. S8B). Thus, a burst of 1O2, owing to Proto accumulation in fc2 mutants, was likely to be responsible for chloroplast degradation and an inability to turn green.
To identify genes required for 1O2-induced chloroplast degradation, we screened for second site mutations in fc2-1 mutants that restored a wt ability to green when provided with 4 hours of light/day. By one-step mapping and whole-genome sequencing, we identified 24 ferrochelatase-two-suppressor (fts) mutants affecting 17 independent loci (Fig. 3A). Four of these loci were characterized; three were genes that affect tetrapyrrole and 1O2 accumulation (fig. S9A; fig. S10, A to E; and table S4) (19).
Fig. 3. A mutant screen to identify genes involved in chloroplast stress and degradation.
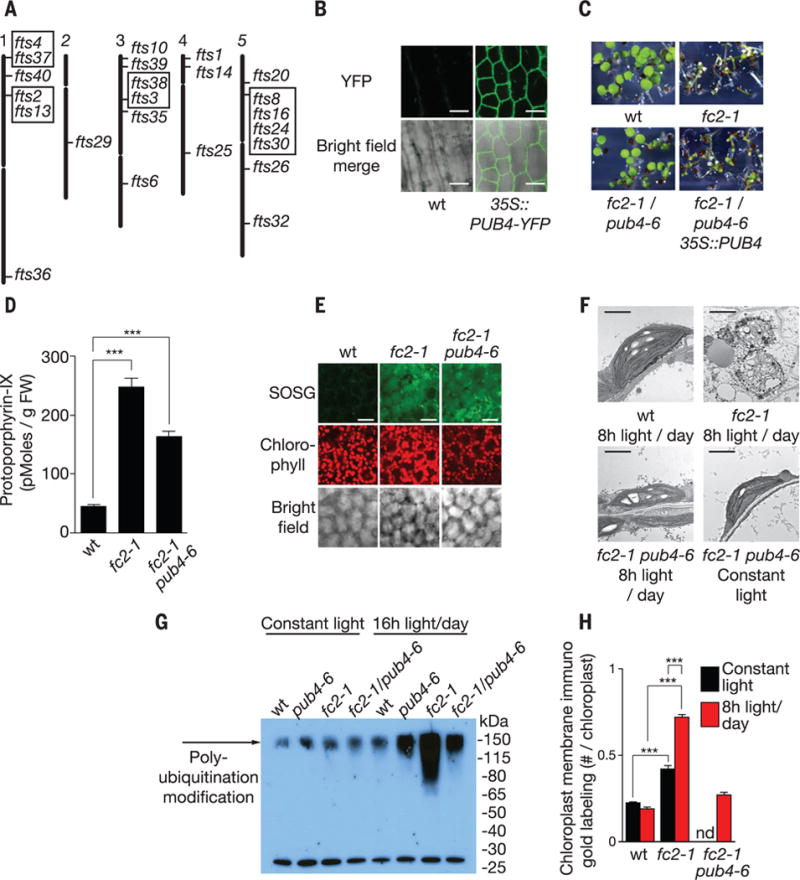
(A) Locations of 24 independent fts (ferrochelatase two suppressors) mutations representing 17 loci. Boxed mutants are allelic. (B) Subcellular localization of stably expressed PUB4-YFP in hypocotyl cells. (C) Complementation of the fts29 (pub4-6) phenotype with a wt copy of PUB4 grown for 5 days in 8 hours of light/day. (D) Proto levels in 3-day-old seedlings provided 8 hours of light/day 10 min after dawn (n = 3 replicates). (E) Representative confocal images of 1O2 accumulation in cotyledon mesophyll cells (2 hours after dawn) of 3-day-old seedlings provided with 8 hours of light/day. Scale bars, 20 μm. (F) Representative TEM micrographs of chloroplasts in 3-day-old cotyledon mesophyll cells 6 hours after dawn. Scale bars, 2 μm. (G) Antiubiquitin immunoblot of whole chloroplast fractions isolated from leaves 3 hours after dawn. Plants were grown for 2 weeks in constant light and then transferred to 16 hours of light/day or kept in constant light for 2 days. (H) Quantification of chloroplast envelope-associated ubiquitin by immunoelectron microscopy (gold labeling) (19) in 3-day-old seedlings (n = 3 seedlings, 10 cells each) 1 hour after dawn. All values are means T SEM. ***P ≤ 0.001, two-tailed Student’s t test.
The fourth loci, fts29 (called pub4-6 hereafter), had a missense mutation in Plant U-Box 4 (PUB4) E3 ubiquitin ligase, a broadly expressed gene encoding an active cytoplasmic-localized (Fig. 3B and fig. S11) E3 ubiquitin ligase involved in cell death and development (20, 21). E3 ubiquitin ligases catalyze the transfer of ubiquitin from an E2 ubiquitin–conjugating enzyme to a protein substrate. A wt copy of PUB4 complemented the pub4-6 phenotype (Fig. 3C and fig. S12, A and B) and pub4-6 was allelic with pub4 T-DNA lines (fig. S12, C to D). fc2-1/pub4-6 had no reduction in tetrapyrrole synthesis [protochlorophyllide, chlorophyll, and ALA levels remained elevated (fig. S10, A to C)]. When provided with 8 hours of light/day, Proto and 1O2 (Fig. 3, D and E) accumulated in fc2-1/pub4-6, but chloroplasts were not degraded. Instead, chloroplasts appeared stressed with irregular shapes and angular membranes (Fig. 3F). fc2-1/pub4-6 mutants still induced many nuclear stress-associated genes (fig. S13), which suggested that chloroplast degradation was not because of 1O2 damage per se but was because of a 1O2-generated signal involving PUB4.
Because E3 ligases ubiquitinate protein substrates, we tested whether the ubiquitination of chloroplast proteins was linked to chloroplast degradation. Chloroplast fractions of fc2-1 plants provided with 16 hours of light/day exhibited a PUB4-dependent increase in polyubiquitinated protein(s) compared with wt (Fig. 3G and fig. S14, A and B). Immuno-electron microscopy also showed a PUB4-dependent increase of ubiquitin on the chloroplast envelopes in fc2-1 mutants (Fig. 3H and fig. S14C) (19), which suggested that ubiquitination was responsible for the chloroplast degradation in fc2-1 plants.
Next, we tested if chloroplast degradation happened under permissive conditions without cell death. In constant light conditions, chloroplast degradation occurred in wt, but fc2 mutants had more chloroplasts being degraded (Fig. 4, A to E) (19). These aberrant chloroplasts often had starch granules (indicating that they had been photosynthetic) (Fig. 4D), large plastoglobules, and compressed grana/thylakoid membranes (Fig. 4E and fig. S15, A and B). Sometimes these chloroplasts were observed to be interacting with a globular vacuole (Fig. 4, B and C, and fig. S15, C and D). Nearby organelles appeared normal, which suggested that specific chloroplasts had been selected for degradation. Degradation depended on PUB4; pub4-6 and fc2-1/pub4-6 plants had less chloroplast degradation than their parent lines wt and fc2-1, respectively (Fig. 4A). pub4-6 plants also accumulated more chloroplasts/cell area and chlorophyll (Fig. 4, F and G, and fig. S16, A to C). Thus, PUB4 may control chloroplast degradation in healthy cells.
Fig. 4. PUB4 is part of a chloroplast quality-control pathway.
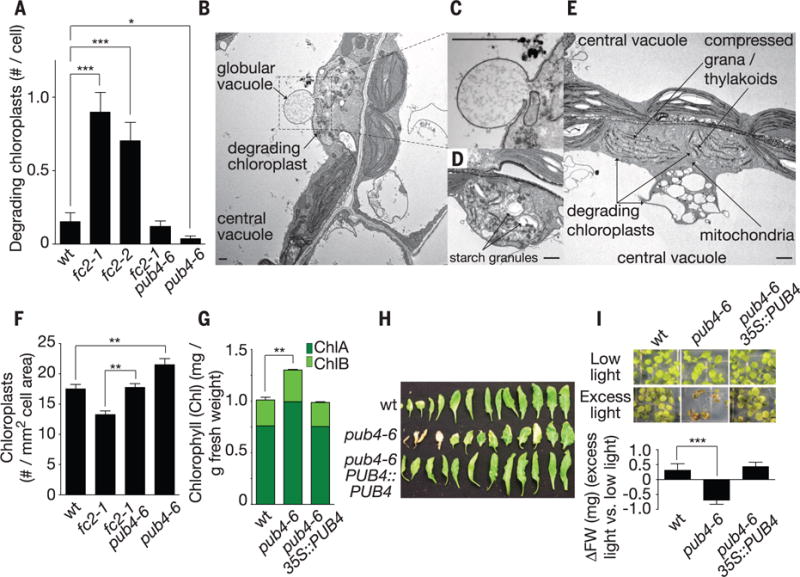
(A) Quantification of degrading chloroplasts using scanning electron microscopy micrographs (19) of cotyledon mesophyll cells of 5-day-old seedlings grown in constant light (n ≥ 54 cells). (B to E) TEM images of chloroplasts undergoing various levels of degradation in fc2-1 seedlings. Scale bars, 1 μm. (F) Quantification of chloroplasts in cotyledon palisade mesophyll cells (n ≥ 6 seedlings) and (G) chlorophyll levels (n = 3 replicates) of 6-day-old seedlings grown in constant light. (H) Senescing rosette leaves of 5-week-old plants grown in constant light. (I) Phenotypes and fresh weight (FW) of seedlings (n = 7 groups of 10) grown for 1 week in low light (30 μmol · m−2 · s−1 at 22°C) and then transferred to excess light (275 μmol · m−2 · s−1 at 15°C) for 1 week. All values are means T SEM. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, two-tailed Student’s t test.
To test if chloroplast degradation is important for long-term maintenance of photosynthetic cells during stress (22, 23), we characterized the phenotypes of pub4-6 single mutants. pub4-6 mutants senesced early (Fig. 4H and fig. S16, D and E) and were stunted in excess light that causes long-term chloroplast oxidative damage (Fig. 4I). Thus, a ubiquitination-dependent chloroplast degradation pathway and/or PUB4 may be important for stress adaptation and longevity.
Indiscriminate chloroplast degradation can occur during osmotic stress (24) or used to recycle nutrients during starvation or senescence (25, 26). Here, we found that plant cells also use a selective chloroplast degradation system to remove ROS-damaged chloroplasts. When chloroplasts accumulated 1O2, proteins in their envelope membranes became ubiquitinated by the direct or indirect action of PUB4, and were degraded (fig. S17). 1O2 is the major ROS species generated during photosynthesis (27), but its short half-life (~4 μs) ensures that it is confined only to chloroplasts in which it was generated (28) so that healthy chloroplasts are not accidently degraded.
Supplementary Material
Acknowledgments
We thank S. Orchard for photographs of plants, C. Lanz for assistance with sequencing, X. Wang for help with SHORE analysis, C. Procko, E. Lee, and A. Tsang for technical assistance, and D. O’Keefe for useful discussions about the manuscript. J.D.W., A.S., J.G. and J.C. acknowledge long-term support from the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy through grant DE-FG02-04ER15540 and from the Howard Hughes Medical Institute. J.G. was supported by an NIH Kirschstein fellowship (1F32GM096610). M.S.J. and J.A.J.F. received funding from the Waitt Advanced Biophotonics Center, NCI P30 Cancer Center Support Grant CA014195 (JAJF), NINDS P30 Neuroscience Center Core Grant NS072031 and the W.M. Keck Foundation. P.S. and D.W. were supported by a Gottfried Wilhelm Leibniz Award to the Deutsche Forschungsgemeinschaft (D.F.G.), and by the Max Planck Society. We thank J. Callis for sharing the anti-ubiquitin antibody and R. Tsien and L. Gross for use of their HPLC. Microarray data are deposited at Gene Expression Omnibus (GSE71764). All biological materials in this study are available from J.C. under a materials transfer agreement with the Salk Institute for Biological Studies/HHMI.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/350/6259/450/suppl/DC1 Materials and Methods
Supplementary Text
Figs. S1 to S17
Tables S1 to S9
References (29–63)
REFERENCES AND NOTES
- 1.Asada K. Plant Physiol. 2006;141:391–396. doi: 10.1104/pp.106.082040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chi W, Sun X, Zhang L. Annu Rev Plant Biol. 2013;64:559–582. doi: 10.1146/annurev-arplant-050312-120147. [DOI] [PubMed] [Google Scholar]
- 3.Leister D. Front Plant Sci. 2012;3:135. doi: 10.3389/fpls.2012.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. Proc Natl Acad Sci USA. 2001;98:2053–2058. doi: 10.1073/pnas.98.4.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gray JC, Sullivan JA, Wang JH, Jerome CA, MacLean D. Philos Trans R Soc Lond B Biol Sci. 2003;358:135–144. doi: 10.1098/rstb.2002.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramel F, et al. Plant Cell. 2013;25:1445–1462. doi: 10.1105/tpc.113.109827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner D, et al. Science. 2004;306:1183–1185. doi: 10.1126/science.1103178. [DOI] [PubMed] [Google Scholar]
- 8.Estavillo GM, et al. Plant Cell. 2011;23:3992–4012. doi: 10.1105/tpc.111.091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramel F, et al. Proc Natl Acad Sci USA. 2012;109:5535–5540. doi: 10.1073/pnas.1115982109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao Y, et al. Cell. 2012;149:1525–1535. doi: 10.1016/j.cell.2012.04.038. [DOI] [PubMed] [Google Scholar]
- 11.Scharfenberg M, et al. Plant Cell Environ. 2015;38:280–298. doi: 10.1111/pce.12248. [DOI] [PubMed] [Google Scholar]
- 12.Woodson JD, Perez-Ruiz JM, Chory J. Curr Biol. 2011;21:897–903. doi: 10.1016/j.cub.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woodson JD, Perez-Ruiz JM, Schmitz RJ, Ecker JR, Chory J. Plant J. 2013;73:1–13. doi: 10.1111/tpj.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruckle ME, DeMarco SM, Larkin RM. Plant Cell. 2007;19:3944–3960. doi: 10.1105/tpc.107.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka R, Tanaka A. Annu Rev Plant Biol. 2007;58:321–346. doi: 10.1146/annurev.arplant.57.032905.105448. [DOI] [PubMed] [Google Scholar]
- 16.Papenbrock J, et al. Plant J. 2001;28:41–50. doi: 10.1046/j.1365-313x.2001.01126.x. [DOI] [PubMed] [Google Scholar]
- 17.Kennedy JC, Pottier RH. J Photochem Photobiol B. 1992;14:275–292. doi: 10.1016/1011-1344(92)85108-7. [DOI] [PubMed] [Google Scholar]
- 18.Bilski P, Li MY, Ehrenshaft M, Daub ME, Chignell CF. Photochem Photobiol. 2000;71:129–134. doi: 10.1562/0031-8655(2000)071<0129:sipvbp>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 19.Materials and Methods are available as supplementary materials on Science online.
- 20.Kinoshita A, et al. Development. 2015;142:444–453. doi: 10.1242/dev.113167. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, et al. Plant J. 2013;74:511–523. doi: 10.1111/tpj.12146. [DOI] [PubMed] [Google Scholar]
- 22.Kusaba M, Tanaka A, Tanaka R. Photosynth Res. 2013;117:221–234. doi: 10.1007/s11120-013-9862-x. [DOI] [PubMed] [Google Scholar]
- 23.Li Z, Wakao S, Fischer BB, Niyogi KK. Annu Rev Plant Biol. 2009;60:239–260. doi: 10.1146/annurev.arplant.58.032806.103844. [DOI] [PubMed] [Google Scholar]
- 24.Wang S, Blumwald E. Plant Cell. 2014;26:4875–4888. doi: 10.1105/tpc.114.133116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michaeli S, Honig A, Levanony H, Peled-Zehavi H, Galili G. Plant Cell. 2014;26:4084–4101. doi: 10.1105/tpc.114.129999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiba A, Ishida H, Nishizawa NK, Makino A, Mae T. Plant Cell Physiol. 2003;44:914–921. doi: 10.1093/pcp/pcg118. [DOI] [PubMed] [Google Scholar]
- 27.Triantaphylidès C, et al. Plant Physiol. 2008;148:960–968. doi: 10.1104/pp.108.125690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogilby PR. Chem Soc Rev. 2010;39:3181–3209. doi: 10.1039/b926014p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


