Abstract
4,5-Diazafluoren-9-one (DAF) has been identified as a highly effective ligand in a number of Pd-catalyzed oxidation reactions, but the mechanistic basis for its utility has not been elucidated. Here, we present the complex coordination chemistry of DAF and palladium(II) carboxylate salts. Multiple complexes among an equilibrating mixture of species have been characterized by 1H and 15N NMR spectroscopy and X-ray crystallography. These complexes include monomeric and dimeric PdII species, with monodentate (κ1), bidentate (κ2) and bridging (μ:κ1:κ1) DAF coordination modes. Titration studies of DAF and Pd(OAc)2 reveal the formation of two dimeric DAF/Pd(OAc)2 complexes at low [DAF] and four monomeric species at higher [DAF]. The dimeric complexes feature two bridging acetate ligands together with either a bridging or non-bridging-κ1 DAF ligand coordinated to each PdII center. The monomeric structures consist of three isomeric Pd(κ1-DAF)2(OAc)2 complexes, together with Pd(κ2-DAF)(OAc)2 in which the DAF exhibits a traditional bidentate coordination mode. Replacing DAF with the structurally related, but more-electron-rich derivative 9,9-dimethyl-4,5-diazafluorene (Me2DAF) simplifies the equilibrium mixture to two complexes, a dimeric species in which the Me2DAF bridges the two Pd centers and a monomeric species with a traditional κ2-Me2DAF coordination mode. The use of DAF in combination with other carboxylate ligands (CF3CO2− or tBuCO2−) also results in a simplified collection of equilibrating PdII–DAF complexes. Collectively, the results highlight the ability of DAF to equilibrate rapidly among multiple coordination modes, and provide valuable insights into the utility of DAF as a ligand in Pd-catalyzed oxidation reactions.
Graphical abstract
Introduction
The renaissance of Pd-catalyzed aerobic oxidation reactions over the past 10–15 years can be attributed, at least in part, to the identification of ancillary ligands that have enabled new synthetic transformations and support improved catalyst activity, selectivity, and stability.1 The ligands tune catalyst sterics and electronics, and thereby influence elementary steps, such as ligand substitution, β-hydride elimination, and reductive elimination, and provide the basis for catalyst-controlled regio- and stereoselective transformations. Continued identification and/or development of new ligands remains a crucial goal to reduce catalyst loadings, enhance catalyst lifetimes and promote the discovery of novel transformations.
Pd-catalyzed aerobic oxidation reactions require the use of oxidatively stable ligands. Monodentate pyridine derivatives are especially common;2 however, catalysts containing these ligands are often susceptible to decomposition, especially at elevated temperatures.3 Catalyst stability can be enhanced through the use of bidentate ligands, such as 2,2'-bipyridine (bpy) and 1,10-phenanthroline (phen) derivatives. These ligands have been used effectively in Pd-catalyzed aerobic oxidation reactions, although the results are typically observed in polar solvents (e.g., H2O, DMSO, DMF)4 or at high temperatures (120–200 °C),5 which are often needed to enable anionic ligand dissociation from (N~N)PdX2 species to access open coordination at the PdII center. Under mild conditions and/or in non-polar solvents, bidentate ligands such as bpy and phen often strongly inhibit Pd-catalyzed aerobic oxidation reactions.6,7
4,5-Diazafluoren-9-one (DAF) is a rare exception to the observations just noted. In 2010, we reported that DAF is a uniquely active bipyridine-type ligand in Pd-catalyzed allylic acetoxylation of terminal olefins.7 Other bidentate ligands, such as bpy and phen, were found to strongly inhibit the reaction. Subsequent to this discovery, DAF has been used in numerous other Pd-catalyzed reactions, including oxidative C–C and C–O coupling reactions of arenes,8 dehydrogenation of cyclic ketones9 and oxidative Heck reactions.10,11 The unique behavior of DAF, relative to bpy and phen, in several of these reactions is illustrated in Scheme 1.
Scheme 1.
Representative Pd-Catalyzed Aerobic Oxidation Reactions Promoted by DAF.7,8,9a
The beneficial effect of DAF in these reactions could arise from its unique coordination properties relative to more-traditional bidentate nitrogen ligands. Evidence of unusual DAF coordination chemistry are evident from previously reported complexes with first-row transition metals,12 palladium(0),13 and other examples14 in which pentahapto (η5), bridging (μ) and monodentate (κ1), in addition to traditional bidentate, coordination modes have been identified. On the other hand, previously reported DAF/PdII complexes are limited to two examples (Scheme 2),15 both of which exhibit the canonical bidentate coordination mode.
Scheme 2.
Previously Characterized DAF/PdII Coordination Complexes.
In the present study, we show that the DAF/PdII coordination chemistry is much richer and more complex than the structures in Scheme 2 might suggest. Through the use of multinuclear NMR spectroscopy, single crystal X-ray diffraction analysis and density functional theory (DFT) calculations, we have identified and characterized six independent coordination complexes that arise from the combination of DAF and Pd(OAc)2. These complexes include both monomeric and dimeric structures, in which DAF exhibits κ1, κ2, and μ coordination modes. Complementary studies with the structurally similar 9,9-dimethyl-4,5-diazaflourene (Me2DAF) ligand provide insight into the effect of electronic perturbation of the diazafluorene ligand framework. Finally, the synergy between the coordination properties of DAF and the anionic carboxylate ligand is probed by replacing acetate with sterically and electronically different carboxylates, trifluoroacetate (TFA) and pivalate (OPiv). The unique coordination properties of DAF identified herein are compared to other bidenate ligands, and they are proposed to contribute directly with the beneficial, ligand-acceleration effect of DAF in PdII-catalyzed oxidation reactions. This hypothesis is validated in a companion study, in which we compare the mechanistic influence of several different bidentate ligands in Pd-catalyzed aerobic aza-Wacker reactivity and show that the fluxionality and weak coordination of DAF relative to other traditional bidentate ligands underlies its activation of PdII catalysts.16
Results
DAF:Pd(OAc)2 Titration Experiments
1H NMR spectroscopic analysis of a 1:1 mixture of DAF and Pd(OAc)2 in CDCl3 reveals the presence of a complex mixture of species (Figure 1). This spectrum contrasts the spectra obtained from 1:1 mixtures of Pd(OAc)2 and more typical bidentate nitrogen ligands, such as 4,4'-tBu2bpy and 2,9-Me2phen (Figure 1). The latter solutions feature single Pd(κ2-N~N)(OAc)2 species, which have been described previously in the literature.17
Figure 1.
1H NMR spectra of 1:1 solutions of Pd(OAc)2 and three different bidentate nitrogen ligands highlighting the unusual complexity of the DAF/Pd(OAc)2 mixture. [Pd(OAc)2] = 40 mM, [Ligand] = 40 mM, T = −45 °C (top) and 24 °C (middle and bottom), solvent = CDCl3 (0.5 mL); The * designates the CHCl3 solvent peak.
In an effort to begin identifying the species present in the 1:1 DAF:Pd(OAc)2 1H NMR spectrum, we performed a titration experiment in which different quantities of DAF were added to a solution of Pd(OAc)2 (Figure 2A). The most diagnostic region of the 1H NMR spectrum is from 8.6–10.9 ppm, where the ortho protons of the DAF ligand appear. With 0.5 equiv of DAF, a single peak assigned to species A predominates in this region of the spectrum. At 1 equiv of DAF, several pairs of resonances are evident, with the major pair designated as species B. The pairs of resonances are attributed to PdII complexes with unsymmetrically coordinated DAF ligands. Free DAF is also present in this spectrum. When ≥ 2 equiv of DAF are added, only small quantities of A and B are present, and the spectra reveal four new species, three with unsymmetrical DAF ligands, C–E, and one with a symmetrical DAF ligand, F. The growth and/or decay of each of these species were tracked as a function of DAF equivalents (Figure 2C). This plot provided the conditions under which the concentration of individual species could be maximized to facilitate more-thorough structural characterization.
Figure 2.
1H NMR spectra obtained from the titration of DAF with Pd(OAc)2 at −45 °C (A). Description of the nomenclature used to assign resonances in the NMR spectra (B). Speciation plot associated with the six DAF/Pd(OAc)2 complexes identified in the titration experiments (C).
The nomenclature employed for the spectral assignments is presented in Figure 2B. Within a given complex, protons belonging to each DAF ligand are labeled with the lower-case letter of the complex name, a–f. A superscript "0" is used to designate resonances associated with a symmetrical DAF ligand in which both pyridyl rings are coordinated to Pd. For complexes with monodentate (κ1) DAF ligands, a superscript "1" is used to designate the coordinated ring, and a superscript "2" for the uncoordinated ring. Finally, subscripts o, m, and p are used to designate the ortho, meta, and para protons.
Overview of Methods Used to Characterize DAF/Pd(OAc)2 Complexes
The integration of the 1H NMR peaks together with selective 1D TOCSY,18 1D and 2D ROESY19 and 2D 1H-15N HMBC20 experiments provided key insights into the identity of complexes A–F (Scheme 3). The 1D TOCSY experiments enabled assignment of the ortho, meta, and para protons within individual rings of the DAF ligand, even when significant overlap was present among resonances from different complexes. The ROESY experiments revealed through-space interactions between ligands within the Pd coordination sphere, including DAF↔DAF and DAF↔OAc interactions. ROESY experiments were used in place of NOESY experiments at lower temperature because the NOE sign and intensity depend on temperature and molecular size, and use of NOESY led to false-negative results. These data, together with integration of the DAF and OAc resonances, were used to determine the DAF:OAc:Pd stoichiometry for each of the complexes A–F.
Scheme 3.
Application of NMR Techniques for Structure Assignment.
The 1H-15N HMBC experiment provided a means to determine the 15N chemical shift of the DAF nitrogen atoms without isotopic enrichment, and this experiment clearly distinguished between coordinated and uncoordinated nitrogen atoms. Uncoordinated pyridine ligands exhibit 15N chemical shifts at ~300 ppm, whereas PdII-coordinated pyridine ligands appear at ~200 ppm (Scheme 4).21 These differences allowed distinction between monodentate and bidentate DAF coordination modes.
Scheme 4.
Effect of Coordination on 15N Chemical Shift
The NMR spectroscopic data were complemented by X-ray crystallography, which provided solid-state structural information that could be compared to the solution-phase NMR data. Finally, DFT calculations facilitated structural assignment of the closely related structures C–E. Application of these techniques to determine the identity of complexes A–F is elaborated below.
Structural Assignment of A
The first species evident in the ligand titration is complex A. As mentioned previously, A grows to its maximum concentration at 0.5 equiv DAF relative to Pd(OAc)2. The TOCSY experiment correlates a single set of ortho, meta, and para resonances, indicating that the two DAF pyridyl rings are equivalent in A (Figure 3A, Figure S2). The 1H-15N HMBC spectrum reveals a single 15N resonance at 204 ppm, indicating that a0o and a0m are associated with a coordinated pyridyl ligand (Figure 3B). The presence of both 2JNH and 3JNH cross-peaks in the HMBC spectrum complements the results from the TOCSY experiment and confirms that the resonances belong to the same complex.
Figure 3.
1D 1H (A) and 1H-15N HMBC spectra (B) of a solution of Pd(OAc)2 and 0.5 equiv of DAF in CDCl3, focusing on the aromatic spectral region. Cross-peaks for both 2- and 3-bond N–H coupling are observed. [Pd(OAc)2] = 40 mM, [DAF] = 20 mM, T = −45 °C, solvent = CDCl3 (0.5 mL). 1H-15N HMBC: dof = 250 ppm, sw1 = 250 ppm, nt = 16, d1 = 4, ni = 256.
The integrated 1D 1H NMR spectrum of A reveals a DAF:OAc stoichiometry of 1:4, and a 1D ROESY experiment shows through-space interactions between the DAF ligand and two chemically distinct acetate ligands (Scheme 5, Figure S5). Integration of the two acetate resonances reflects three and six protons, corresponding to a DAF:OAc1:OAc2 ratio of 1:1:2. A third acetate peak is evident in the 1D 1H NMR spectrum, which has an integration of three protons. This third acetate resonance does not appear in the ROESY spectra (Figure S5), however, suggesting that it is too distant from DAF to observe an ROE.22
Scheme 5.
Observed DAF-to-OAc ROE Correlations for A.
Crystals suitable for single crystal X-ray analysis were obtained from a 0.5:1 DAF:Pd(OAc)2 mixture in CDCl3, and the structure reveals a Cs-symmetric PdII dimer with a one DAF and two acetate ligands bridging the two Pd atoms (Figure 4). Each of the Pd centers has an additional monodentate (κ1) acetate ligand, which are eclipsed with respect to the Pd–Pd axis. This structure is consistent with the NMR data, and redissolution of the crystals in CDCl3 reproduces the spectrum of A present in the 0.5:1 DAF:Pd(OAc)2 solution.
Figure 4.
X-ray crystal structure of A and summary of NMR spectroscopic data supporting this structure in solution. The crystal structure is drawn with 50% probability ellipsoids and all H atoms are omitted for clarity. See Supporting Information for details.
This structure exhibits a mirror plane perpendicular to the Pd–Pd axis that rationalizes the chemical equivalence of two pyridyl rings of the DAF ligand evident in the 1D 1H NMR spectrum. It also accounts for the 1H-15N HMBC data showing that both DAF pyridyl rings are coordinated to Pd. The μ-OAc and two κ1-OAc ligands adjacent to the DAF ligand are the origin of the observed ROEs. The μ-OAc trans to the DAF ligand is too far from the DAF protons to be detected by the ROESY experiment.23
Structural Assignment of B
Addition of more DAF to the solution of A results in conversion to B. Analysis of the NMR data revealed that B has a DAF ligand with two chemically distinct pyridyl rings, b1 and b2 (Figure 5A). A Pd-bound pyridyl ring (b1) is clearly evident from the 15N resonance at 189 ppm, and an unbound ring (b2) is associated with the resonance at 305 ppm (Figure 5B).
Figure 5.
1D 1H (A) and 1H-15N HMBC spectra (B) of a solution of Pd(OAc)2 and 1 equiv of DAF in CDCl3, focusing on the aromatic spectral region. The resonances appearing at <220 ppm correspond to coordinated nitrogen atoms of DAF ligands, while those appearing at >300 ppm correspond to unbound nitrogen atoms. [Pd(OAc)2] = 40 mM, [DAF] = 40 mM, T = −45 °C, solvent = CDCl3 (0.5 mL). 1H-15N HMBC: dof = 250 ppm, sw1 = 250 ppm, nt = 40, d1 = 6 s, ni = 256.
The 2D ROESY spectrum reveals two acetate cross-peaks, indicating that B contains chemically inequivalent acetate ligands (Figures S6 and S7). Furthermore, ROEs are observed from b1 and b2 to both of these acetates, demonstrating that b1 and b2 are present in the same complex (i.e., B = b1 + b2) (Scheme 6). These conclusions are supported by 1D ROESY experiments in which ROEs were observed between each of the acetate resonances and the b1o and b2o ring resonances (Figure S8). Integration of the DAF and OAc resonances in the 1D 1H NMR spectrum reveals a DAF:OAc stoichiometry of 1:2.
Scheme 6.
Observed DAF to OAc ROE Correlations for B.
Crystals suitable for single crystal X-ray analysis were obtained from a 1.5:1 DAF:Pd(OAc)2 mixture in CDCl3, and the structure reveals a C2-symmetric PdII dimer with two acetate ligands bridging the Pd atoms (Figure 6). Each Pd center is also coordinated by a κ1-DAF and a κ1-OAc ligand.
Figure 6.
X-ray crystal structure of B and summary of NMR spectroscopic data supporting this structure in solution. The crystal structure is drawn with 50% probability ellipsoids and all H atoms are omitted for clarity. See Supporting Information for details.
This structure has a C2 axis perpendicular to the Pd–Pd axis that bisects the two bridging acetate ligands, rationalizing the chemical equivalence of the two κ1-DAF ligands and the pairs of μ- and κ1-OAc ligands evident in the 1D 1H NMR spectrum. This structure also explains the 1H-15N HMBC data showing both bound and unbound DAF pyridyl rings. The ROESY data showing through-space interactions between DAF and two chemically distinct acetate ligands are also readily rationalized by this structure.
Structural Assignment of C, D, and E
Addition of more DAF results in the conversion of species A and B into a mixture of four new species C–F. At a DAF:Pd(OAc)2 ratio of 6:1, seven resonances associated with the ortho protons of the DAF ligand are present in the 1H NMR spectrum, together with four distinct OAc peaks (Figures 7A, S9 and S11). Six of the seven DAF resonances correspond to three pairs of peaks with 1:1 integration, and they are assigned to species C, D, and E. The remaining resonance is assigned to F and will be discussed separately below. The concentrations of C, D, and E exhibit a squared dependence on the DAF concentration, suggesting that these structures exhibit a 2:1 DAF:Pd stoichiometry (cf. Figure 2C). The 1H-15N HMBC spectrum reveals six cross-peaks for C, D, and E (Figure 7B), with c1, d1, and e1 corresponding to Pd-bound DAF pyridyl rings (δ 203–207 ppm) and c2, d2, and e2 corresponding to unbound rings (δ 304–310 ppm). These data indicate that each of the complexes C, D, and E contains DAF ligands bound in a κ1 coordination mode.
Figure 7.
1D 1H (A) and 1H-15N HMBC spectra (B) of a solution of Pd(OAc)2 and 6 equiv of DAF in CDCl3, focusing on the aromatic spectral region. The resonances appearing at <220 ppm correspond to coordinated nitrogen atoms of DAF ligands, while those appearing at >300 ppm correspond to unbound nitrogen atoms. [Pd(OAc)2] = 40 mM, [DAF] = 240 mM, T = −45 °C, solvent = CDCl3 (0.5 mL). 1H-15N HMBC: dof = 250 ppm, sw1 = 250 ppm, nt = 32, d1 = 4 s, ni = 256.
1D ROESY spectra for each complex reveal through-space interactions between each of the two (inequivalent) DAF ortho protons and a single acetate resonance (Scheme 7A and Figures S9 and S10). Furthermore, integrations of the DAF and OAc resonances reveal a DAF:OAc ratio of 1:1 for each complex. The 1D ROESY experiments also reveal through-space interactions between the chemically inequivalent DAF ortho protons in C and E (i.e., from c1o ↔ c2o and e1o ↔ e2o), but analogous ROE correlations are not present in D. The distance between the ortho protons within an individual DAF ligand (d ~ 7.0 Å) is too large to show an ROE; however, these signals can arise from interactions between two separate DAF ligands within the same complex. Computational modeling confirms that two DAF ligands with an anti relationship have ortho protons with sufficient proximity to exhibit an ROE correlation (Scheme 7B and Figures S9 and S10). No ROE correlation is expected for two DAF ligands with a syn relationship.
Scheme 7.
Observed DAF↔DAF and DAF↔OAc ROE Correlations for C–E.
Four possible isomeric structures, 1–4, satisfy the constraints of the NMR spectroscopic data of species C, D, and E (Table 1) and correspond to Pd(κ1-DAF)2(OAc)2 complexes that are distinguished by cis vs. trans and syn vs. anti relationships between two κ1 DAF ligands in the Pd coordination sphere. The viability of these structures is supported by X-ray crystallographic characterization of one of these complexes (Figure 8). The X-ray quality crystals were obtained from a 6:1 DAF:Pd(OAc)2 mixture in CDCl3.
Table 1.
Assignments of C–E from NMR Spectroscopic and DFT Results.
Figure 8.
X-ray crystal structure of trans/anti-Pd(κ1-DAF)2(OAc)2 2, species E. The crystal structure is drawn with 50% probability ellipsoids and all H atoms are omitted for clarity. See Supporting Information for details.
The relative energies of structures 1–4 were assessed by performing DFT computations.24 Structures 1–3 are within 1.6 kcal/mol of each other, with 1 having the lowest energy. Structure 4, however, is significantly higher in energy (ΔG = +6.0 kcal/mol relative to 1) and would have a population that would not be detected by NMR spectroscopy (Table 1). The higher energy of this complex may be rationalized by the steric clash between the cis/syn relationship between the two DAF ligands. These data suggest the three species detected in solution consist of a mixture of structures 1, 2 and 3.
ROESY experiments reveal chemical exchange between D and E (Scheme 8A and Figure S9). The exchange process converts one of the DAF ortho protons in D (d1o) into an equal mixture of the two ortho protons in E (e1o and e2o). The temperature dependence of this exchange process was used to estimate an activation energy of ΔG‡ = 13 ± 0.5 kcal/mol (Figure S12–13).25 These observations may be rationalized by an intramolecular exchange process that interconverts the trans/syn and trans/anti bis-DAF complexes 1 and 2 (cf. Table 1). A mechanism for this process was identified by DFT methods, and the calculated barrier is 11 kcal/mol, which is in good agreement with the experimental value.
Scheme 8.
Chemical Exchange Process between D and E.
Together, the ROESY and DFT computational data enable structural assignment of complexes C, D, and E. The DAF↔DAF ROEs observed for C and E (cf. Scheme 7) require an anti relationship between the DAF ligands and thereby limit these species to structures 2 and 3 (cf. Table 1). The chemical exchange between D and E permits assignment of complex E to the trans/anti structure 2 and complex D to the trans/syn structure 1. Structure 4 is not believed to be among the observed structures on the basis of the DFT energy and, therefore, complex C is assigned to the cis/anti structure 3. These assignments are supported by comparison of the 1H NMR chemical shifts of the acetate -CH3 groups for these three complexes with the general trends observed in chemical shifts of cis- and trans-acetate ligands.26
Structural Assignment of F
Complex F appears together with C, D, and E as the DAF concentration increases. Unlike C–E, however, its concentration increases with a linear, rather than squared, dependence on DAF concentration, suggesting that it has a 1:1 DAF:Pd(OAc)2 stoichiometry (cf. Figure 2C). The 1D 1H NMR spectrum for F reveals a symmetrical DAF ligand, and 1H-13C HSQC, variable temperature and 1D TOCSY data show that the resonances at 8.66 ppm corresponds to both ortho and para protons (fo/p in Figure 7A; see also Figures S17–19). 1D ROESY data reveal a through-space interaction between the ortho proton of DAF and an acetate ligand, and integration of the acetate peak reveals a DAF:OAc stoichiometry of 1:2 (Figure S11). Finally, the 1H-15N HMBC spectrum (cf. Figure 7B) exhibits only a single cross-peak for F at 213.3 ppm, showing that the symmetrical pyridyl rings of the DAF ligand are coordinated to Pd. The monomeric structure in Scheme 9 is consistent with all of these data.
Scheme 9.
Structure of F Derived from NMR Spectroscopic Methods.
Characterization of Me2DAF/Pd(OAc)2 and DAF/Pd(O2CR) (R = CF3, tBu) Complexes
Several variants of the DAF/Pd(OAc)2 catalyst system have been used in catalytic aerobic oxidation reactions. For example, as shown in Scheme 1, DAF has been replaced with 9,9-dimethyl-4,5-diazafluorene (Me2DAF) and acetate has been replaced with trifluoroacetate (TFA) or pivalate (OPiv) (cf. Scheme 1).8,9 In an effort to assess the potential influence of these catalyst modifications, we investigated Me2DAF/Pd(OAc)2, DAF/Pd(TFA)2, and DAF/Pd(OPiv)2 in a manner analogous to DAF/Pd(OAc)2.
Me2DAF/Pd(OAc)2
Titration of Me2DAF into a solution of Pd(OAc)2 reveals the formation of only two species G and H, and only H is observed when >1 equiv of Me2DAF is present, relative to Pd(OAc)2 (Figure 9). The simplicity of this mixture contrasts with the complex speciation observed with DAF/Pd(OAc)2 (cf. Figure 2). Full NMR spectroscopic analysis of these species was performed (see Supporting Information, Figures S20–S25). TOCSY data reveal that the Me2DAF ligand in both G and H have equivalent pyridyl rings, and 1H-15N HMBC data reveal a single 15N resonance for each complex, with chemical shifts in a region associated with bound N atoms: 199.4 ppm (G) and 209.7 ppm (H). For G, an ROE correlation was observed between the DAF ligand and two chemically distinct acetates, designated OAc1 and OAc2, and another between OAc2 and a third acetate ligand, OAc3. Integration of the 1D 1H NMR spectrum of G reveals a DAF:OAc stoichiometry of 1:4, with a DAF:OAc1:OAc2:OAc3 ratio of 1:1:2:1. In contrast, the acetate ligands in H are equivalent and the DAF:OAc stoichiometry is 1:2.
Figure 9.
1H NMR spectra obtained from the titration of Me2DAF with 40 mM Pd(OAc)2 at 24 °C (A). Speciation plot associated with the two Me2DAF/Pd(OAc)2 complexes identified in the titration experiments (B).
Crystals of G and H suitable for single crystal X-ray analysis were obtained from chloroform solutions of 0.5:1 and 1:1 Me2DAF:Pd(OAc)2, respectively, and the resulting structures are consistent with the solution NMR data (Figure 10). G is a dimeric species with a bridging Me2DAF ligand and two bridging and two κ1 acetate ligands, matching the DAF/Pd(OAc)2 structure A (cf. Figure 4). H is a monomeric species with the Me2DAF ligand in a traditional κ2 bidentate coordination mode, analogous to the DAF/Pd(OAc)2 structure F.
Figure 10.
X-ray crystal structures and summary of NMR spectroscopic data supporting of Pd(μ-Me2DAF)(μ-OAc)2(κ1-OAc)2 dimer G (left) and Pd(κ2-Me2DAF)(κ1-OAc)2 monomer H (right). The crystal structures are drawn with 50% probability ellipsoids and all H atoms are omitted for clarity. See Supporting Information for details.
DAF/Pd(TFA)2
Pd(TFA)2 is insoluble CDCl3, but it dissolves in a 1:1 ratio of CDCl3 and trifluoroacetic acid (TFAH). Titration of DAF into a solution of Pd(TFA)2 in this solvent mixture reveals the sequential appearance of three species, I, J, and K (Figure 11), each of which was extensively characterized by NMR spectroscopic methods (see Supporting Information, Figures S28–S32). Complex I maximizes at 0.5:1 DAF:Pd(TFA)2 stoichiometry, but coexists with J. Complex J is essentially the only species present at a 1:1 DAF:Pd(TFA)2 stoichiometry, while it begins to disappear in favor of K at higher DAF concentrations. It was possible to obtain X-ray crystal structures of I and J (Figure 12), and the solid-state structures are consistent with the solution NMR data. Complex I is the dimeric species Pd2(μ-DAF)(μ-TFA)2(κ1-TFA)2, which resembles structures A and G above (cf. Figures 4 and 10), while J is the monomeric structure Pd(κ2-DAF)(κ1-TFA)2, which resembles structures F and H above (Scheme 9 and Figure 10). Complex K only forms at high DAF concentrations, and it was not possible to obtain crystals of this complex. NMR data for this complex reveals dynamic behavior arising from rapid exchange between uncoordinated DAF and coordinated to J and K. The parabolic growth of K with increasing [DAF] and NMR integrations are consistent with a 2:1 DAF:Pd(TFA)2 stoichiometry, and all of the data point toward this species being a mononuclear complex with two κ1-DAF ligands, Pd(κ1-DAF)2(TFA)2, similar to species C, D and E (cf. Table 1).
Figure 11.
1H NMR spectra obtained from the titration of DAF with 20 mM Pd(TFA)2 in 1:1 TFAH:CDC13 at 24 °C (A). Speciation plot associated with the DAF/Pd(TFA)2 complexes identified in the titration experiments (B).
Figure 12.
X-ray crystal structures and summary of NMR spectroscopic data for Pd(μ-DAF)(μ-TFA)2(κ1-TFA)2 dimer I (left) and Pd(κ1-TFA)2 J (right). The crystal structures are drawn with 50% probability ellipsoids and all H atoms are omitted for clarity. See Supporting Information for details.
DAF/Pd(OPiv)2
Titration of DAF into a solution of Pd(OPiv)2 reveals a single major DAF-ligated Pd species L at all DAF:Pd(OPiv)2 stoichiometries (Figure 13). Only minor (<10%) contributions from other DAF-ligated species are evident. The NMR spectroscopic data show that the DAF and OPiv ligands are present in 1:2 stoichiometry and that the two pyridyl groups of DAF are equivalent and coordinated to Pd (see Supporting Information, Figures S33 and S34). An X-ray crystal structure of L reveals that this complex is a C2-symmetric dimer in which two trans DAF ligands bridge two PdII centers, each of which also has two κ1-OPiv ligands (Figure 14).27 An approximately 45° twist is evident between the N–Pd–N vectors of the two PdII centers, when the dimer is viewed down the Pd···Pd axis. This structural feature differs from the DAF/Pd(OAc)2 dimers A and B, for which the two PdII square planes have an eclipsed relationship (i.e., θ = 0°). Upon redissolving the crystals of L into CDCl3, identical spectral data are observed. It seems likely that the symmetrical monomeric structure, Pd(κ2-DAF)(OAc)2, is disfavored by the steric bulk of the pivalate groups.
Figure 13.
1H NMR spectra obtained from the titration of DAF with 40 mM Pd(OPiv)2 at 24 °C (A). Speciation plot associated with the two DAF/Pd(OPiv)2 complexes identified in the titration experiments (B).
Figure 14.
X-ray crystal structure and summary of NMR spectroscopic data supporting Pd2(μ-DAF)2(κ1-OPiv)4 dimer L. The crystal structures are drawn with 50% probability ellipsoids and all H atoms are omitted for clarity. See Supporting Information for details.
Discussion
Origin of Diverse Coordination Geometries with DAF
The results presented here highlight the structural diversity of DAF coordination to Pd-carboxylate salts. The results show that DAF exhibits bridging (μ), κ1- and κ2-binding modes when coordinated to PdII centers bearing carboxylate anionic ligands, and most of the structures deviate from the classical κ2-bidentate coordination mode typically associated with bipyridyl-type ligands. This rich diversity is evident in mixtures of DAF and Pd(OAc)2. Under conditions with 1:1 DAF:Pd(OAc)2 stoichiometry (a typical stoichiometry for catalytic reactions), six different structures are present in significant quantity (A–F, cf. Figure 2). The complexes are highly fluxional, but static structures suitable for NMR spectroscopic characterization were accessible at – 45 °C. Upon replacing DAF with the more electron-rich ligand Me2DAF or the acetate ligands with more electron-deficient trifluoroacetate ligands, equilibrium mixtures are still observed, but a smaller number of species are present (cf. Figures 9 and 11). The corresponding complexes are also less fluxional than the DAF/Pd(OAc)2 species, allowing NMR spectroscopic studies to be performed at room temperature. As expected, increasing the DAF concentration promotes the formation of monomeric structures, including complexes bearing a single κ2-DAF and/or two κ1-DAF ligands. Only the sterically encumbered Pd(OPiv)2 species favors a single DAF complex at all DAF concentrations and, in this case, favors a unique dimeric Pd complex with two bridging DAF ligands.
This unique DAF/Pd-carboxylate coordination chemistry may be rationalized by the structural distortion of the DAF ligand relative to traditional bpy-type ligands. Two structural parameters that facilitate comparison of these ligands are the angle α, defined by the C2–N and C2'–N bond vectors of the two pyridyl rings, and the N···N distance between the two pyridyl nitrogen atoms (Scheme 10). Introduction of a one-atom linker between the 3- and 3'-positions of 2,2'-bipyridine expands α from 53° in bpy to 73° in the unligated bpy and DAF ligands, and similarly increases the N···N distance from 2.66 Å to 3.06 Å, respectively.28 The parameters for Me2DAF (73°, 3.07 Å) are nearly identical to those for DAF.29 The α angle for unligated bpy is similar to that observed in bpy-ligated complexes of PdII. For example, α = 49° and 54° in Pd(bpy)(OAc)2 and Pd(2,9-Me2phen)(OAc)2, respectively.17a,c In contrast, the κ2-DAF complexes of PdII require significant contraction of their α angle relative to unligated DAF: α = 61° and 60° in complexes H and J, respectively. The α angle expands considerably when DAF serves as a bridging ligand: α = 82–84° in complexes A, G, I, and L, while virtually no distortion is evident in the structures bearing κ1-DAF ligands: α = 76°, complexes B, C, D, E (Scheme 10). Commensurate changes are evident in the N···N bond distances (Scheme 10). In short, unligated DAF exhibits structural features intermediate between the κ2 (bidentate) and μ (bridging) coordinated DAF ligands, thereby enabling it to access both of these coordination modes. In addition, DAF can access the κ1 coordination mode, which relieves the modest strain present in the other two modes. The small α value for unligated bpy makes it well suited for coordination to PdII as a bidentate (κ2) ligand, but poorly suited to serve as a bridging (μ) ligand. Rare examples of Pd2(μ-bpy) complexes exist,30 but they accommodate this coordination via a significant dihedral angle between the two pyridyl rings of the bpy ligands (53° and 59° respectively).
Scheme 10.
Structural Comparison of 2,2'-Bipyridine, DAF and Corresponding PdII Complexes.
Relevance to Aerobic Oxidation Catalysis
The unique coordination chemistry of DAF has important implications for Pd-catalyzed aerobic oxidation reactions. The simplified mechanism of these catalytic reactions consists of two half-reactions: substrate oxidation by PdII and catalyst reoxidation by O2 (Scheme 11). The turnover-limiting step in most (if not all) of these reactions is associated with the PdII-mediated substrate oxidation half-reaction and corresponds to one of various fundamental steps, such as ligand substitution (i.e., substrate binding), alkene insertion (e.g., into a Pd–C or Pd–N bond), β-hydride elimination or C–H activation. Ligand steric and electronic properties influence the rate of the PdII-mediated reaction steps, and one or more ligands typically must dissociate from PdII to provide coordination sites for substrate reactivity. Steps associated with the catalyst reoxidation half-reaction are typically fast, by comparison, but ancillary ligands still play an important role.31 Ligands not only promote the reaction of O2 with Pd0, but they also can minimize catalyst aggregation into inactive metallic Pd ("Pd-black").32
Scheme 11.
Representative Catalytic Cycle for Pd-Catalyzed Aerobic Oxidations
Neutral monodentate ligands, such as pyridine, triethylamine (NEt3) and DMSO, have been widely used in Pd-catalyzed aerobic oxidation reactions,1 and typical catalyst systems employ a 2:1 L:PdII stoichiometry. Mechanistic studies have revealed that faster initial rates may be obtained with a 1:1 L:PdII stoichiometry, which enhances substrate accessibility to the PdII coordination sphere.33 The use of ≥ 2:1 L:PdII stoichiometry in reported catalyst systems represents the optimal balance between catalyst activity and stability. At L:PdII ratios < 2:1, the catalyst often decomposes before the reaction reaches full conversion.
Bidentate ligands are appealing because they enhance catalyst stability, but they often inhibit catalytic turnover. The chelate effect disfavors neutral ligand dissociation and, therefore, anionic ligand dissociation is required to enable substrate access to the PdII coordination sphere. Formation of a charged complex has a strong solvent dependence, as illustrated by a recent study of amidopalladation of alkenes with a (bpy)PdII(amidate)Cl complex (Scheme 12).34 Alkene insertion does not proceed in toluene, while it proceeds readily in DMSO at room temperature. The reaction was shown to proceed via pre-equilibrium dissociation of chloride, which is strongly inhibited in a non-polar solvent.
Scheme 12.
Solvent Effect on X-Type Ligand Dissociation
The structures of DAF/Pd(OAc)2 complexes presented herein show that DAF exhibits properties of both mono- and bidentate ligands. And, the empirical success of DAF in Pd-catalyzed aerobic oxidation reactions suggests that DAF can take advantage of the beneficial features of both ligand types. The κ1 coordination mode, evident in structures B-E, provides access to open coordination sites at Pd11 without requiring dissociation of an anionic ligand. This feature should stabilize intermediates and/or transition states associated with key substrate oxidation steps, such as C-H activation, alkene insertion or (3-hydride elimination (Scheme 13).8,10 On the other hand, the bidentate character of DAF evident in F, H, J, Pd(DAF)(r|3-allyl)+ 6b and Pd(DAF)Cl215 provides a means to enhance catalyst stability upon formation of Pd° in the catalytic cycle.
Scheme 13.
Proposed Intermediates and Transition States in DAF-Promoted Catalysis.
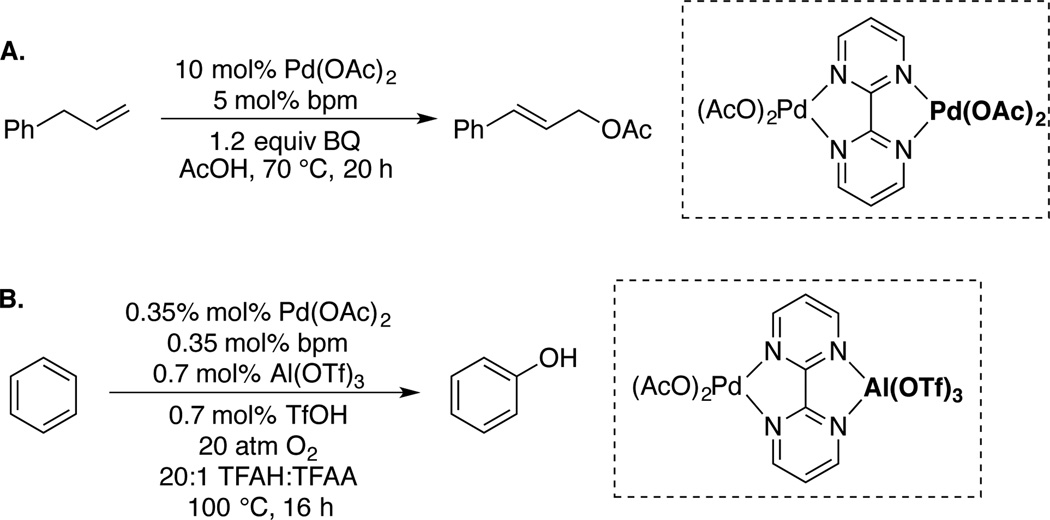
The principles discussed here have implications for other Pd-catalyzed oxidation reactions. Of particular note are the allylic and aromatic C–H oxidation reactions that employ 2,2'-bipyrimidine (bpm)-PdII catalysts, reported recently by Bercaw/Labinger35a and Yin,35b respectively (Scheme 14). These reactions are promoted by coordination of the bpm ligand to a second metal, PdII or AlIII. This coordination could lead to a structural distortion similar to that has been characterized for DAF in the present study.36,37 Further investigation of this concept in catalytic reactions is the focus of ongoing investigation.
Scheme 14.
Pd(bpm)(OAc)2-Catalyzed C–H Oxidation Reactions Promoted by Ligand Coordination to a Second Metal.
Conclusion
The rich coordination chemistry of 4,5-diazafluoren-9-one (DAF)-Pd(OAc)2 has been characterized by diverse NMR spectroscopic methods, X-ray crystallography and DFT calculations. In particular, 1H-15N HMBC and ROESY experiments played a critical role in determining the coordination mode of the DAF ligand in solution. The data show that DAF is able to adopt bridging (μ), chelating (κ2) and monodentate (κ1) coordination modes in monomeric and dimeric PdII complexes. Six different DAF/Pd(OAc)2 species have been characterized and compared to analogous Me2DAF/Pd(OAc)2, DAF/Pd(TFA)2, DAF/Pd(OPiv)2 and DAF/Pd(O2CC6F5)2 species. The equilibrium population changes systematically as a function of [DAF], with monomeric structures favored at higher [DAF]. The hemilabile character of DAF provides a compelling rationale for the beneficial reactivity of DAF, relative to other bidentate ligands, in Pd-catalyzed aerobic oxidation reactions, and it provides a valuable foundation for future studies in this field.
Supplementary Material
Acknowledgments
We are grateful for financial support from the NIH (R01 GM67173). Computational resources are supported in part by the NSF (CHE-0840494). NMR instrumentation was partially funded by the NIH (S10 RR13866-01).
Footnotes
ASSOCIATED CONTENT
Supporting Information. Experimental and computational details, NMR spectra, exchange measurements and crystallography reports are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.For reviews, see: Stahl SS. Angew. Chem. Int. Ed. 2004;43:3400–3420. doi: 10.1002/anie.200300630. Gligorich KM, Sigman MS. Chem. Commun. 2009;26:3854–3867. doi: 10.1039/b902868d.
- 2.For representative examples, see the following: Nishimura T, Onoue T, Ohe K, Uemura S. J. Org. Chem. 1999;64:6750–6755. doi: 10.1021/jo9906734. Fix SR, Brice JL, Stahl SS. Angew. Chem. Int. Ed. 2002;41:164–166. doi: 10.1002/1521-3773(20020104)41:1<164::aid-anie164>3.0.co;2-b. Ferreira EM, Stoltz BM. J. Am. Chem. Soc. 2003;125:9578–9579. doi: 10.1021/ja035054y. Iwasawa T, Tokunaga M, Obora Y, Tsuji Y. J. Am. Chem. Soc. 2004;126:6554–6555. doi: 10.1021/ja031936l. Zhang Y-H, Shi B-F, Yu J-Q. J. Am. Chem. Soc. 2009;131:5072–5074. doi: 10.1021/ja900327e. Izawa Y, Stahl SS. Adv. Synth. Catal. 2010;352:3223–3229. doi: 10.1002/adsc.201000771. Izawa Y, Pun D, Stahl SS. Science. 2011;333:209–213. doi: 10.1126/science.1204183. John LC, Gunay A, Wood AJ, Emmert MH. Tetrahedron. 2013;69:5758–5764.
- 3.(a) Steinhoff BA, Guzei IA, Stahl SS. J. Am. Chem. Soc. 2004;126:11268–11278. doi: 10.1021/ja049962m. [DOI] [PubMed] [Google Scholar]; (b) Komano T, Iwasawa T, Tokunaga M, Obora Y, Tsuji Y. Org. Lett. 2005;7:4677–4679. doi: 10.1021/ol051845o. [DOI] [PubMed] [Google Scholar]; (c) Kubota A, Emmert MH, Sanford MS. Org. Lett. 2012;14:1760–1763. doi: 10.1021/ol300281p. [DOI] [PubMed] [Google Scholar]
- 4.(a) ten Brink G-J, Arends IWCE, Papadogianakis G, Sheldon RA. Appl. Catal., A. 2000:435–442. [Google Scholar]; (b) Hu J, Gu Y, Guan Z, Li J, Mo W, Li T, Li G. Chem. Sus. Chem. 2011;4:1767–1772. doi: 10.1002/cssc.201100337. [DOI] [PubMed] [Google Scholar]; (c) Ye M, Gao G-L, Yu J-Q. J. Am. Chem. Soc. 2011;133:6964–6967. doi: 10.1021/ja2021075. [DOI] [PubMed] [Google Scholar]; (d) Ben-Yahia A, Naas M, Kazzouli SE, Essassi EM, Guillaumet G. Eur. J. Org. Chem. 2012;36:7075–7081. [Google Scholar]
- 5.Ref 4b and 4c. See also: Shiotani A, Itatani H, Inagaki T. J. Mol. Catal. 1986;34:57–66. Gasperini M, Ragaini F, Cenini S, Gallo E, Fantauzzi S. Applied Organomet. Chem. 2007;21:782–787.
- 6.Ref 2g and the following: Andappan MMS, Nilsson P, Larhed M. Chem. Commun. 2004;2:218–219. doi: 10.1039/b311492a. Zheng CW, Wang D, Stahl SS. J. Am. Chem. Soc. 2012;134:16496–16499. doi: 10.1021/ja307371w.
- 7.Campbell AN, White PB, Guzei IA, Stahl SS. J. Am. Chem. Soc. 2010;132:15116–15119. doi: 10.1021/ja105829t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Campbell AN, Meyer EB, Stahl SS. Chem. Commun. 2011;47:10257–10259. doi: 10.1039/c1cc13632a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Xiao B, Gong T-J, Liu Z-J, Liu J-H, Luo D-F, Xu J, Liu L. J. Am. Chem. Soc. 2011;133:9250–9253. doi: 10.1021/ja203335u. [DOI] [PubMed] [Google Scholar]
- 9.(a) Gao W, He Z, Qian Y, Zhao J, Huang Y. Chem. Sci. 2012;3:883–886. [Google Scholar]; (b) Diao T, Wadzinski TJ, Stahl SS. Chem. Sci. 2012;3:887–891. doi: 10.1039/C1SC00724F. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Buter J, Moezelaar R, Minnaard AJ. Org. Biomol. Chem. 2014;12:5883–5890. doi: 10.1039/c4ob01085j. [DOI] [PubMed] [Google Scholar]
- 10.(a) Piotrowicz M, Zakrzewski J. Organometallics. 2013;32:5709–5712. [Google Scholar]; (b) Piotrowicz M, Zakrzewski J, Métivier R, Brosseau A, Makal A, Woźniak K. J. Org. Chem. 2015;80:2573–2581. doi: 10.1021/jo502619k. [DOI] [PubMed] [Google Scholar]; (c) Vasseur A, Laugel C, Harakat D, Muzart J, Le Bras J. Eur. J. Org. Chem. 2015;5:944–948. [Google Scholar]
- 11.For other applications of DAF in Pd-catalyzed oxidation reactions, see: Sharma A, Hartwig JF. J. Am. Chem. Soc. 2013;135:17983–17989. doi: 10.1021/ja409995w. Malik HA, Taylor BLH, Kerrigan JR, Grob JE, Houk KN, Du Bois J, Hamann LG, Patterson AW. Chem. Sci. 2014;5:2353–2361. doi: 10.1039/C3SC53414F.
- 12.(a) Li B, Li B, Zhu X, Zhang Y. Inorg. Chem. Commun. 2003;6:1304–1306. [Google Scholar]; (b) Siemeling U, Scheppelmann I, Neumann B, Stammler H-G, Schoeller WW. Organometallics. 2004;23:626–628. [Google Scholar]; (c) Yang H-J, Kou H-Z, Gao F, Cui A-L, Wang R-J. Acta Crystallogr., Sec. E: Struct. Rep. Online. 2004;60:m611–m613. [Google Scholar]; (d) Zhang R-L, Zhao J-S, Yang S-Y, Ng SW. Acta Crystallogr., Sec. E: Struct. Rep. Online. 2004;60:m262–m263. [Google Scholar]; (e) Tian A, Han Z, Peng J, Ying J, Sha J, Dong B, Zhai J, Liu H. Inorg. Chim. Acta. 2008;361:1332–1338. [Google Scholar]; (f) Feng X-L, Zhang Y-P. Acta Crystallogr., Sec. E: Struct. Rep. Online. 2011;67:m1171. doi: 10.1107/S1600536811029060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein RA, Witte P, van Belzen R, Fraanje J, Goubitz K, Numan M, Schenk H, Ernsting JM, Elsevier CJ. Eur. J. Inorg. Chem. 1998;3:319–330. [Google Scholar]
- 14.For numerous other examples, see the following "perspective" article and references cited therein: Annibale VT, Song D. Dalton Trans. 2016;45:32–49. doi: 10.1039/c5dt03665h.
- 15.See ref. 7 and the following: Xu Z-G, Liu H-Y, Zhan Q-G, Chen J, Xu M-J. Acta Crystallogr., Sec. E. 2009;65:m1166. doi: 10.1107/S1600536809035272.
- 16.White PB, Jaworski JN, Zhu GH, Stahl SS. submitted for publication. [Google Scholar]
- 17.(a) Milani B, Alessio E, Mestroni G, Sommazzi A, Garbassi F, Zangrando E, Bresciani-Pahor N, Randaccio L. J. Chem. Soc,. Dalton Trans. 1994;13:1903–1911. [Google Scholar]; (b) Ragaini F, Gasperini M, Cenini S, Arnera L, Caselli A, Macchi P, Casati N. Chem. Eur. J. 2009;15:8064–8077. doi: 10.1002/chem.200801882. [DOI] [PubMed] [Google Scholar]; (c) Bercaw JE, Day MW, Golisz SR, Hazari N, Henling LM, Labinger JA, Schofer SJ, Virgil S. Organometallics. 2009;28:5017–5024. [Google Scholar]; (d) Ye Y, Ball ND, Kampf JW, Sanford MS. J. Am. Chem. Soc. 2010;132:14682–14687. doi: 10.1021/ja107780w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.TOCSY = Total Correlation Spectroscopy. This technique uses scalar coupling to transfer magnetization from one nuclei to another. The extent of transfer depends on the mixing time, with longer mixing times unveiling more of the coupling network, and will proceed until a quaternary center or non-protonated heteroatom is encountered.
- 19.ROESY = Rotating-frame Nuclear Overhauser Effect Spectroscopy. This technique is similar to NOESY in that it detects through-space interactions between protons. The ROESY experiment locks the selected magnetization along a specific direction in the transverse plane (rotating frame) by a high-power spinlock rather than along the z-axis (NOESY). The sign of an ROE is always positive, whereas the sign of an NOE is dependent on the tumbling frequency of the molecule relative to the static magnetic field. At the temperatures used in the experiments in this study, NOE data cannot unambiguously distinguish between through-space interactions and chemical exchange. See the Supporting Information for more details.
- 20.HMBC = Heteronuclear Multiple-Bond Correlation spectroscopy is a long-range scalar coupling experiment that involves polarization transfer from 1H to X-nuclei that are typically 2 or 3 bonds away. It is often used to resolve the chemical shift of insensitive nuclei in structure determination studies.
- 21.This upfield shift in the 15N resonance when PdII is coordinated is due to the occupation of the lone-pair in bonding. The 15N chemical shift is also sensitive to the oxidation state of the metal, the coordination geometry (axial vs equatorial) and the ligand trans to the nitrogen See: Mason J. Chem. Rev. 1981;81:205–227. Pazderski L, Szłyk E, Sitkowski J, Kamieński B, Kozerski L, Toušek J, Marek R. Magn. Reson. Chem. 2006;44:163–170. doi: 10.1002/mrc.1740.
- 22.ROESY experiments aimed at observing an ROE from OAc1 or OAc2 to OAc3 are complicated by chemical exchange that occurs among the acetate resonances (see Figure S5).
- 23.The chemical shifts of the acetate -CH3 groups seem to follow some general trends that are helpful to guide structural assignment (cf. Table S1). For example, we find that the cis acetates in Pd(κ2-N-N)(OAc)2 complexes typically have -CH3 1H resonances >2.1 ppm, while the trans acetates in PdL2(OAc)2 complexes typically have 1H resonances <1.9 ppm. The bridging acetates that are present in the Pd dimers observed in this study have chemical shifts fall into a chemical shift region between the cis- and trans-κ1-acetates (1.9–2.1 ppm).
- 24.See Supporting Information for full computational details. Optimization and frequencies calculations were performed with rB3LYP/6-31+G* for all non-Pd atoms and the Stuttgart 1997 basis set/ECP was used for Pd. PCM solvation calculations were performed with CHCl3 solvent parameters and at a higher basis set (6–311+G**) for all non-Pd atoms.
- 25.The experimental barrier was calculated from observing the lineshape of the acetate peaks for D and E variable-temperature NMR experiments (Figure S11–S12).
- 26.The chemical shift of the acetate -CH3 groups of C at 1.85 ppm is significantly more downfield than the acetate resonances associated with D (1.66 ppm) and E (1.52 ppm). These relative chemical shifts align with the general trends that we observe for chemical shifts of cis- versus trans-acetates ligands in other complexes (cf. ref. 23).
- 27.A similar structure was obtained with pentafluorobenzoate ligands. See Supporting Information for X-ray crystallographic data for this species (Structure M, S52).
- 28.(a) 1,10-Phenanthroline. Nelyubina YV, Korlyukov AA, Lyssenko KA. Mendeleev Commun. 2014;24:286–289. [Google Scholar]; (b) 4,4’-Di-tert-butyl-2,2’-bipyridine. Amarante TR, Figueiredo S, Lopes AD, Gonçalves IS, Paz FAA. Acta Cryst. 2009;E65:o2047. doi: 10.1107/S1600536809029109. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) 2,2'-bipyridine. Kavitha SJ, Panchanatheswaran K, Low JK, Glidewell C. Acta Crystallogr., Sect. C: Cryst. Struct. Commun. 2005;61:o473. doi: 10.1107/S0108270105018810. [DOI] [PubMed] [Google Scholar]; (d) DAF. Fun H-K, Sivakumar K, Zhu D-R, You X-Z. Acta Cryst. 1995;C51:2076–2078. [Google Scholar]
- 29.See Supporting Information for crystal structure data.
- 30.(a) Klein A, Lepski R. Z. Anorg. Allg. Chem. 2009;635:878–884. [Google Scholar]; (b) Fornies J, Navarro R, Sicilia V, Tomas M. Inorg. Chem. 1993;32(17):3675–3681. [Google Scholar]
- 31.(a) Wilke G, Schott H, Heimbach P. Angew. Chem. Int. Ed. 1967;6:92–93. [Google Scholar]; (b) Konnick MM, Guzei IA, Stahl SS. J. Am. Chem. Soc. 2004;126:10212–10213. doi: 10.1021/ja046884u. [DOI] [PubMed] [Google Scholar]; (c) Konnick MM, Stahl SS. J. Am. Chem. Soc. 2008;130:5753–5762. doi: 10.1021/ja7112504. [DOI] [PubMed] [Google Scholar]; (d) Konnick MM, Decharin N, Popp BV, Stahl SS. Chem. Sci. 2011;2:326–330. [Google Scholar]
- 32.Steinhoff BA, Stahl SS. J. Am. Chem. Soc. 2006;128:4348–4355. doi: 10.1021/ja057914b. [DOI] [PubMed] [Google Scholar]
- 33.See refs. 2f, 3c and the following for examples: Steinhoff BA, Stahl SS. Org. Lett. 2002;4:4179–4181. doi: 10.1021/ol026988e. Schultz MJ, Park CC, Sigman MS. Chem. Commun. 2002:3034–3035. doi: 10.1039/b209344h. Ye XA, Liu GS, Popp BV, Stahl SS. J. Org. Chem. 2011;76:1031–1044. doi: 10.1021/jo102338a.
- 34.White PB, Stahl SS. J. Am. Chem. Soc. 2011;133:18594–18597. doi: 10.1021/ja208560h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.(a) Lin B-L, Labinger JA, Bercaw JE. Can. J. Chem. 2009;87:264–271. [Google Scholar]; (b) Guo HJ, Chen Z, Mei F, Zhu D, Xiong H, Yin G. Chem.-Asian J. 2013;8:888–891. doi: 10.1002/asia.201300003. [DOI] [PubMed] [Google Scholar]
- 36.We thank Dr. Jay A. Labinger (Caltech) for drawing our attention to this relationship between DAF/Pd(OAc)2 and the bpm-[Pd(OAc)2]2 catalyst system.
- 37.For related observations made recently in the study of stoichiometric reactions of Pd(bpym)Ar2, see: Liberman-Martin AL, Levine DS, Liu W, Bergman RG, Tilley TD. Organometallics. 2016 T. D.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.