Abstract
Acne vulgaris (AV) affects most adolescents, and of those affected, moderate to severe disease occurs in 20%. Comedones, follicular plugs consisting of desquamated keratinocytes and sebum, are central to its pathogenesis. Despite high heritability in first-degree relatives, AV genetic determinants remain incompletely understood. We therefore employed whole-exome sequencing (WES) in nevus comedonicus (NC), a rare disorder that features comedones and inflammatory acne cysts in localized, linear configurations. WES identified somatic NEK9 mutations, each affecting highly conserved residues within its kinase or RCC1 domains, in affected tissue of three out of three NC-affected subjects. All mutations are gain of function, resulting in increased phosphorylation at Thr210, a hallmark of NEK9 kinase activation. We found that comedo formation in NC is marked by loss of follicular differentiation markers, expansion of keratin-15-positive cells from localization within the bulge to the entire sub-bulge follicle and cyst, and ectopic expression of keratin 10, a marker of interfollicular differentiation not present in normal follicles. These findings suggest that NEK9 mutations in NC disrupt normal follicular differentiation and identify NEK9 as a potential regulator of follicular homeostasis.
Main Text
Acne vulgaris (AV) is a common disorder affecting nearly all adolescents, and it can range in severity from isolated non-inflammatory open and closed comedones to raised inflammatory lesions and pustules.1 The most severe cases demonstrate nodulocystic inflammatory disease, and up to 20% of affected individuals experience permanent scarring.2
Microcomedones, which consist of plugs of desquamated keratinocytes, are thought to be precursors to acne lesions. These plugs trap sebum and keratin within follicles, leading to the formation of dilated plugged follicles known as comedones. The nutrient-rich environment within the comedo can lead to overgrowth of P. acnes and subsequent inflammation.3 Current treatment for AV includes topical or systemic retinoids, antibiotics, and benzoyl peroxide. These agents are thought to prevent comedo formation and inflammation by promoting differentiation of cells within the hair follicle unit, reducing sebum secretion and sebocyte proliferation, and killing P. acnes.4
Genetic investigation of Mendelian acne phenotypes has identified pathways implicated both in the generation of comedones and in subsequent inflammatory responses. Apert syndrome (MIM: 101200), a rare congenital disease that presents with craniosynostosis, syndactyly, and severe acne with comedones and inflammatory cysts, is caused by heterozygous activating mutations in FGFR2 (MIM: 176943).5 Munro acne nevus is a localized form of Apert syndrome that results from somatic FGFR2 mutation and features linear groups of comedones and inflammatory cysts.6 These disorders suggest that FGFR2 signaling might be important both in comedogenesis and inflammation, and mouse models have shown that FGFR2 is necessary for skin and hair follicle homeostasis and regulation of cutaneous inflammation.7, 8
Acne inversa (MIM: 42690), a heritable type of inflammatory acne found in axillary, inguinal, and perianal regions, presents with comedones, inflammatory nodules, and draining sinus tracts.9 A subset of individuals with acne inversa have mutations in PSENEN (MIM: 607632), PSEN1 (MIM: 104311), and NCSTN (MIM: 605254) which act within the γ-secretase pathway, implicating Notch signaling defects in comedogenesis.10 Evidence in mouse models suggests that, as in FGFR2 mutations, γ-secretase haploinsufficiency might promote comedo formation, leading to subsequent inflammation.11
Genome-wide association studies (GWASs) in AV have identified multiple loci associated with severe acne. A GWAS involving Chinese Han individuals found two loci, one near DDB2 (MIM: 600811) and the other within SELL (MIM: 153240). These genes are thought to play a role in anti-microbial activity of sebocytes (DDB2) and mediation of cutaneous inflammation (SELL), respectively.12 A GWAS involving European individuals found three separate loci, containing the genes OVOL1 (MIM: 602313), FST (MIM: 136470), and TGFB2 (MIM: 190220), all active in TGF-β signaling, which might modulate the inflammatory response to comedonal lesions or play a role in tissue remodeling.13, 14
Nevus comedonicus (NC) is a severe, localized form of acne estimated to be present in 1:45,000–1:100,000 births and appears in infancy as a linear shiny patch with development of comedones early in life.15 Some of these comedones can subsequently become inflamed and form acne cysts, suggesting that NC lesions result from a process that most directly leads to comedo development with inflammation occurring as a secondary consequence.1 The linear configuration of NC lesions suggests that NC is a mosaic disorder resulting from acquisition of a somatic mutation during embryonic development.
In cutaneous mosaic disorders due to mutations within keratinocyte precursors, including NC, skin lesions are found in linear configurations known as lines of Blaschko, which represent the dorsoventral migration of epithelial progenitors.16 When multilineage somatic mosaicisim occurs, more widespread involvement is seen, as in the cutaneous-skeletal hypophosphatemic syndrome, which features hypophosphatemic rickets, epidermal and melanocytic nevi, and hamartomas in multiple organs as a result of multilineage somatic activating RAS mutations.17 Extracutaneous manifestations have also been reported in individuals with NC, leading to the diagnosis of nevus comedonicus syndrome, which can feature NC lesions, ipsilateral cataracts, and neurological and skeletal abnormalities.18
To determine the genetic cause of NC, we recruited a cohort of three subjects with NC lesions, and consent to a protocol approved by the Yale University human investigation committee was obtained. All subjects had a history of localized linear lesions featuring intermixed non-inflammatory grouped comedones, scars, and recurrent inflammatory cysts without other intervening skin changes. Subjects provided a skin biopsy from lesional tissue and a peripheral blood sample for analysis.
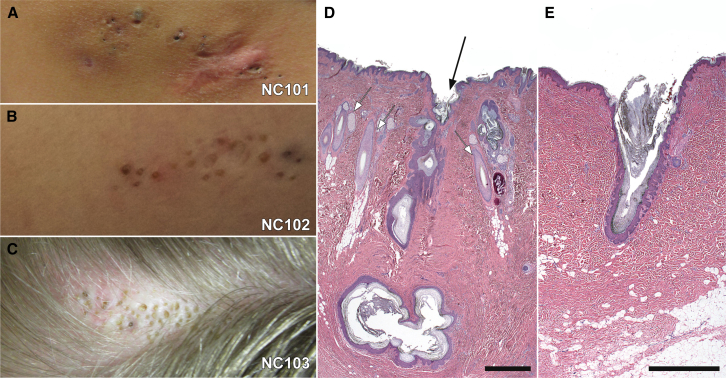
NC101, a 43-year-old woman, initially presented with a 6 cm × 3 cm linear lesion consisting of grouped comedones on the back and a history of inflammatory cysts within the lesion (Figure 1). NC102, a 10-year-old boy, presented at birth with a shiny flat lesion on the right inferior buttock. At the age of 10 months, this lesion began to develop intermittent inflammatory cysts with cribriform scarring, and comedones became apparent. At the time of enrollment, this lesion extended inferiorly to the postero-medial right thigh and was a 10 cm × 2 cm plaque composed of several atrophic cribriform scars and scattered pink comedones that were surmounted by a thin scale crust. Inferior to this plaque was a second, similar appearing one measuring 2 cm × 1 cm (Figure 1). NC103, a 19-year-old woman, presented with a 1 cm × 4 cm lesion on the scalp that appeared at birth and subsequently developed alopecia, comedones, and rare inflammatory cysts (Figure 1).
Figure 1.
Clinical and Histologic Features of Nevus Comedonicus
Three unrelated subjects presented with linear patches of comedones on distinct body sites. NC101, a 43-year-old woman, had a lesion on the back that featured numerous dilated follicular ostia and comedones, with a scar inferiorly at the site of a prior inflammatory lesion (A). NC102, a 10-year-old boy, had a lesion on the thigh that featured numerous comedones and atrophic scars at the sites of prior inflammatory lesions (B). NC103, a 19-year-old woman, had a hairless patch on the scalp. The patch subsequently never grew hair and developed multiple comedones and inflammatory cysts (C). Intervening skin appeared grossly normal in all subjects. Histopathologic examination of affected scalp excision tissue in subject NC103 revealed normal hair follicles (white arrows) adjacent to affected follicles (black arrow), which show dilated follicular ostia, marked acanthosis and papllimatosis of the outer root sheath, and large cystic structures filled with keratin in the deep dermis (D). Examination of tissue excised from the back of subject NC101 shows an isolated follicle with a dilated ostium, acanthosis and papillomatosis of the outer root sheath, and accumulated keratin in place of a hair shaft (E). In both cases, interfollicular epidermis appears to be normal. Scale bars represent 1 mm.
Histopathology of the NC103 lesion revealed follicles with dilated ostia adjacent to normal follicles. These abnormal follicles demonstrated acanthosis and papillomatosis of the outer root sheath. Furthermore, large cystic structures filled with keratin were present in the deep dermis (Figure 1). Examination of tissue from NC101 showed isolated follicles with dilated ostia and acanthosis and papillomatosis of the outer root sheath. Accumulated keratin was present without evident hair shafts (Figure 1). In contrast to the smooth, ordered layer of keratinocytes seen in the outer root sheath of hair follicles, the acanthosis and papillomatosis of cells lining the NC follicle and cyst regions presented a morphology similar to rete ridges in interfollicular epidermis. Although lesional tissue was limited, NC comedonal follicles did not appear to have associated sebaceous lobules. Interfollicular skin in NC lesions was normal on clinical and histologic examination.
These clinical and histologic findings are distinct from Apert syndrome, Munro acne nevus, and acne inversa. The characteristic sinus tracts and double comedones of acne inversa were not observed in any NC lesions. Histologic findings further distinguish Apert syndrome and Munro acne nevus from NC, given that both demonstrate a flat, atrophic cyst wall and numerous associated sebaceous lobules, in contrast to the papillomatosis and acanthosis of the NC follicle and cyst, which also lack sebaceous lobules.19
We isolated DNA from blood and tissue samples by using a phenol-chloroform extraction or DNeasy (QIAGEN), respectively. Using paired samples from each of the three subjects, we performed whole-exome capture (Roche EZ exome V3) followed by high-throughput sequencing (Illumina HiSeq 2500), generating average coverage > 90× for tissue samples and > 85× for blood samples (Table S1). Sequence was aligned to the human genome reference (UCSC Genome Browser, hg19) with the Burrows-Wheeler Aligner (BWA-MEM).20 Reads were then trimmed and PCR duplicates removed with Picard,21 and resultant BAM files were calibrated with GATK.21 To identify somatic single-nucleotide variants (SSNVs) present in tissue samples but absent in the matched blood sample, we employed MuTect22 and a Fisher’s exact test to compare blood and tissue read numbers; SSNVs with a p value greater than 1 × 10−3 were excluded. Remaining SSNVs were annotated with AnnoVar.23 To exclude common and non-damaging variants, we further analyzed nonsynonymous exonic and splice-site SSNVs with a prevalence < 1% in an Exome Aggregation Consortium (ExAC) control dataset (Table S2). For each of the three paired samples, all of these high-confidence SSNVs identified were located within a single gene, NEK9 (MIM: 609798), which encodes a kinase. These occurred at highly conserved residues, and none of the mutations were found in >134,000 alleles in ExAC, Exome Variant Server, and 1000 Genomes control databases. All were confirmed via Sanger sequencing (Figure S2). We found no evidence of loss of heterozygosity or genomic segment amplification or deletion (Figure 2 and Figure S1) in any sample.
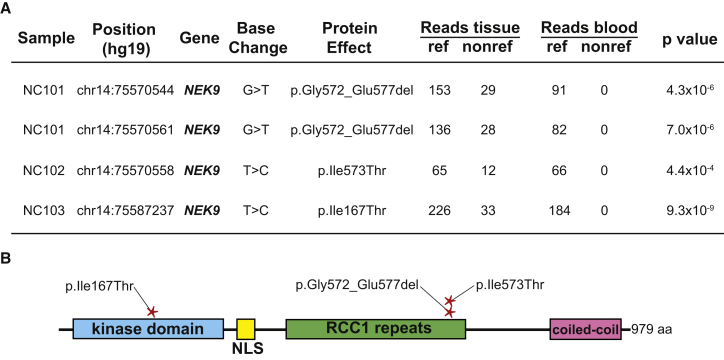
Figure 2.
Somatic NEK9 Mutations Cause Nevus Comedonicus
Whole-exome sequencing of DNA isolated from tissue and blood of individuals NC101, NC102, and NC103 was performed (A). Using MuTect, Perl and Python scripts, we identified tissue-specific SSNVs and indels and ranked them by Fisher’s exact test score. NC101 had two mutations in NEK9 that were in cis on spanning short reads. These mutations led to a loss of six amino acids in the encoded protein, p.Gly572_Glu577del, via abolition of the native exon 14 splice donor and utilization of a cryptic splice donor within the coding region of exon 14 (Figure S3). NC102 had a somatic NEK9 p.Ile573Thr missense variant. NC103 showed a somatic NEK9 p.Ile167Thr missense variant. No other somatic mutations were found in any of the three individuals with a Fisher’s exact test p value < 1 × 10−4, and no non-reference reads were identified in blood DNA in any individual. Mutations were confirmed with Sanger sequencing (Figure S2). NC101’s and NC102’s mutations were localized in the RCC1 domain of NEK9, whereas NC103’s mutation was localized in the kinase domain (B).
To estimate the probability of observing three somatic mutations clustering within a 3 kb coding region (the size of NEK9) by chance, we performed a Monte Carlo simulation, counting the two mutations found in NC101 as a single genetic event. We distributed all three observed somatic mutations in the three samples across a 31 Mb coding region and determined the frequency with which three mutations clustered within a 3 kb segment. In 108 iterations of this simulation, we estimate the probability of observing a mutation in NEK9 in all three samples to be approximately 6 × 10−8.
This analysis revealed two somatic NEK9 mutations in cis in individual NC101: c.1731+1G>T (GenBank: NM_033116.4), which abolishes a native splice donor, at the exon 14 and intron 14 junction and c.1715G>T (p.Gly572Val), which generates a cryptic splice donor site, within exon 14 (Figure 2). To assess mutation consequence, we cloned a segment containing exons 13 to 15 of wild-type and mutant (c.1715G>T and c.1731+1G>T) NEK9 into pcDNA3.1. Resulting constructs were transfected into PLC cells, and we performed RT-PCR 48 hr after transfection, using RNeasy (QIAGEN) for RNA isolation and iScript (BD Biosciences) for cDNA generation. PCR was then used to amplify exons 13 to 15, and products were Sanger sequenced. This assay demonstrated loss of 18 nucleotides in the encoded mRNA (p.Gly572_Glu577del), corresponding to deletion of amino acids 572−577 (Figure S3). All six of these amino acids are highly conserved, demonstrating conservation in 100%, 93%, 84%, 100%, 99%, and 89% of 100 vertebrates, respectively (Figure S4).
Individual NC102 had a single somatic missense mutation (c.1718T>C [p.Ile573Thr]) in exon 14 of NEK9, affecting one of the amino acids that we found deleted in NC101. The isoleucine in this position is conserved in 93% of vertebrates, and no species demonstrates a threonine at this position. The affected amino acids in NC101 and NC102 fall within the regulator of chromosome condensation-like (RCC1) domain of NEK9 (Figure 2). Previous reports have shown that deletion of the NEK9 RCC1 domain causes increased kinase activity via autophosphorylation, suggesting that the observed mutations might abrogate RCC1-mediated suppression of NEK9 kinase activity.24
In individual NC103, we found a somatic mutation (c.500T>C [p.Ile167Thr]) in exon 4 of NEK9, which occurs in the kinase domain of the protein (Figure 2). This isoleucine residue is 100% conserved in vertebrates (Figure S4). There is in vitro evidence that the NEK9 kinase and RCC1 domains bind to each other,24 potentially linking the NC101 and NC102 mutations within the RCC1 domain to the kinase-domain mutation in NC103.
NEK9 is a serine/threonine kinase that functions as an important regulator of cell-cycle and checkpoint control and is a member of a larger family of NEK proteins that share moderate conservation.24 Phosphorylation by PLK1 and CDK1 at Ser29, Thr333, Ser750, and Ser869 are required steps for activation of NEK9 and precede phosphorylation at Thr210, which can also occur auto-catalytically or via PLK1 action.25 Thr210 lies within the activation loop of NEK9, and phosphorylation at this residue is required for kinase activity.25
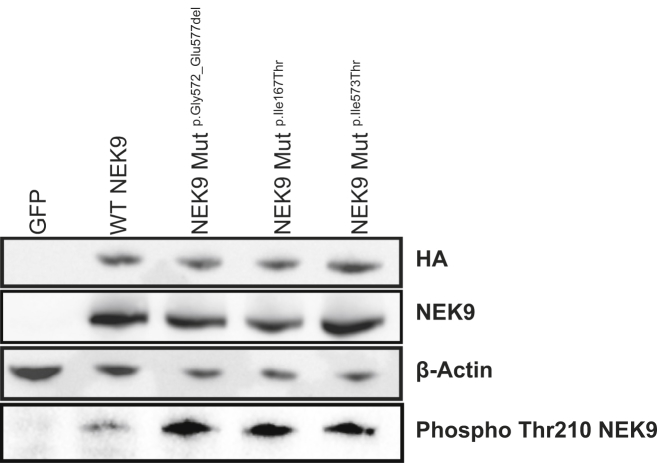
The finding of somatic NEK9 mutations within affected tissue of three NC-affected subjects is consistent with either a gain-of-function or a dominant-negative mechanism. To study the effect of these mutations, we obtained the NEK9 cDNA (Harvard Plasmid) and introduced the observed mutations via QuikChange Site-Directed Mutagenesis (Agilent). An HA tag was introduced at the N terminus via PCR, and tagged wild-type and mutant constructs were transfected into pCAGGS via Gibson assembly (New England Biolabs). We transiently transfected resulting cDNAs of wild-type NEK9 and mutants into HEK293 cells and assayed levels of Thr210-phosphorylated NEK9 by western blotting. We found that, relative to wild-type, all three NC mutations significantly increased NEK9 Thr210 phosphorylation (Figure 3). Given prior observations that Thr210 phosphorylation is necessary for NEK9 kinase activity, and that NEK9 auto-phosphorylation increases kinase activity,25 our data support the hypothesis that NC mutations are gain of function.
Figure 3.
NEK9 Mutations Cause Increased Phosphorylation of NEK9
HEK293 cells were transiently transfected with GFP as a negative control or HA-tagged NEK9 constructs, including those encoding wild-type (WT) NEK9, p.Gly572_Glu577del NEK9, p.Ile167Thr NEK9, and p.Ile573Thr NEK9. 24 hr post-transfection with lipofectamine 2000 (Invitrogen), protein lysates were run on SDS-page gels and transferred to nitrocellulose. Blots were probed with 1:1,000 anti-HA (ab18181; Abcam), 1:1,000 anti-beta-actin (A5316; Sigma), 1:10,000 anti-NEK9 (ab138488; Abcam), and 1:250 anti-phospho-NEK9 (ab63553; Abcam). In contrast to expression of wild-type NEK9, expression of each of the three mutant constructs led to increased phosphorylation of NEK9 despite equivalent protein expression detected by blotting for HA, total NEK9, and actin. This experiment was repeated three times, confirming increased phospho-NEK9 in cells expressing mutant alleles.
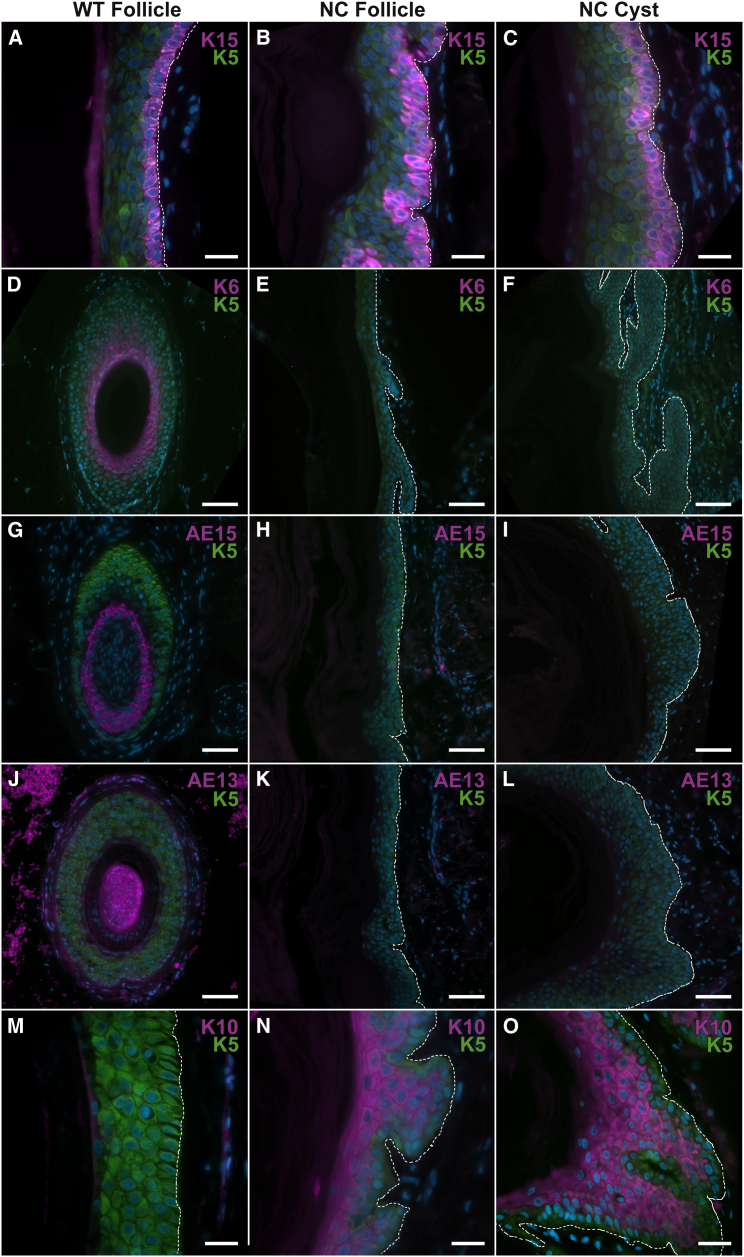
Given the presence of independent somatic mutations in affected tissue of three subjects with NC, including one lesion with comedones replacing hair follicles on the scalp (Figure 1), we sought to investigate localization of NEK9 in hair-bearing skin. Within normal skin, immunolocalization studies showed NEK9 localized to the epidermis and hair follicles. In both interfollicular epidermis and in affected dilated follicles of NC tissue, we found no change in subcellular localization in comparison to normal tissue (Figure S5). To further explore the consequences of NEK9 mutation, we performed immunolocalization studies employing hair follicle and epidermal differentiation markers in NC lesional tissue and a normal control after antigen retrieval (incubation in 10 mM sodium citrate, 0.05% Tween 20 [pH 6.0] at 100°C for 20 min). We found abnormal localization of multiple markers of hair follicle and epidermal differentiation (Figure 4). Expansion of keratin 15 (K15) immunostaining (sc-47697, dilution 1:200; Santa Cruz), a marker of the hair follicle bulge, which is thought to be the location of the hair follicle stem cell pool,26 was found in NC follicles extending from the expected bulge site to dermal cysts (Figure 4 and Figure S6). Noting this result, we further assessed localization of hair follicle differentiation markers, including keratin 6 (K6) (prb-169P, 1:200 dilution; Covance), AE15 (ab58755, 1:100 dilution; Abcam) and AE13 (ab16113, 1:100 dilution; Abcam), which define the companion layer, inner root sheath, and the cuticle and cortex, respectively. K6 was absent within the region of the NC follicular area and cyst (Figure 4). AE13 and AE15 were both absent throughout NC follicles and cysts (Figure 4). Finding an absence of hair follicle differentiation markers in NC, we assessed localization of keratin 10 (K10) (sc-53252, 1:200 dilution; Santa Cruz), a marker of interfollicular epidermal differentiation, which is absent in normal hair follicles. Notably, K10 was ectopically localized suprabasally throughout NC follicles and cysts (Figure 4 and Figure S6). Ki67 (ab15889, 1:300 dilution; Abcam) immunolocalization was performed to assess proliferation, and NC and wild-type skin showed a similar fraction of Ki67-positive basal nuclei, suggesting that comedones are not the direct result of hyperproliferation within follicles or cysts (Figure S7). Presence of the interfollicular epidermis (IFE) marker K10, expansion of the population of cells with K15 immunostaining, and absence of hair-specific markers in NC follicles demonstrates defective differentiation within the NC follicular unit.
Figure 4.
NEK9 Mutation Impairs Follicular Differentiation
In NC103 lesional tissue, follicular differentiation was analyzed by immunofluorescence staining. Keratin 5 staining in all images marks the outer root sheath of the hair follicle. To characterize the stem cell pool in NC, staining for keratin 15 (K15) was performed. K15 localizes to the basal layer of keratinocytes within the bulge in normal hair follicles (A), but in NC was found to strongly localize to the basal layer throughout the follicle (B) and cyst (C). In normal tissue, keratin 6, AE15 and AE13 mark follicular differentiation in the companion layer, inner root sheath, and cuticle and cortex, respectively (D, G, and J). In NC, these were absent in the upper follicular regions (E, H, and K), as well as within cysts (F, I, and L), suggesting a shift in follicular fate to an IFE-like fate within NC tissue. To further assess differentiation of follicular cells, keratin 10 (K10) staining was examined. K10, a marker of suprabasal IFE, which is absent in sub-bulge regions of normal follicles (M), was found within the upper follicular and sub-bulge region of NC (N) and within the cysts of NC tissue (O), in both cases maintaining suprabasal localization. The dermal-epidermal junction is labeled with a dashed white line in each panel. Control tissue was obtained from discarded tips from surgical excision. Scale bar represents 20 μm in (A)–(C) and (M)–(O) and 50 μm in (D)–(L).
Although NEK9 is found in follicle, cyst, and IFE cells, and we have observed via laser capture that somatic mutation is present in both sites (Figure S2), there is no apparent clinical or histologic phenotype in the IFE. Differentiation was assessed via immunolocalization of K10 and filaggrin in the IFE of NC lesional tissue. The localization and immunofluorescence intensity of both markers was identical in normal tissue and in NC IFE (Figure S8).
Mutations in NEK9 paralogs can cause severe phenotypes. Loss-of-function mutations in NEK1 cause short-rib polydactyly syndrome (MIM: 263520),27 and recessive mutations in NEK8 have been implicated in multiple organ dysplasia (MIM: 615415).28 Neither NEK1 nor NEK8 have been shown to interact with NEK9 directly.
NEK6, NEK7, and NEK9 are necessary for proper assembly of the mitotic spindle via interaction with EG5, allowing transition from interphase to mitosis.29 NEK2 separately acts in disassembly of the intercentrosomal linker.30 Activated NEK9 phosphorylates NEK6 and NEK7,24, 25, 31, 32 which act on Eg5 to enable chromosome separation and mitotic spindle formation. NEK9 also functions in replication stress response, activating CHK1,33 and it has been shown to be critical to proliferation in p53-deficient cancer cells, though there is no evidence that NEK9 interacts with p53 directly.34 Notably, NEK9 has been found to be weakly expressed in normal lung parenchyma, but strongly expressed in lung carcinomas of multiple types, regardless of p53 mutation status, suggesting that it might be important in neoplasia.34
Recent reports have shown that recessive germline NEK9 mutations cause skeletal disease without cutaneous phenotypes. Homozygous nonsense mutations in NEK9, c.1489C>T (p.Arg497∗), were found in fetuses with a lethal skeletal dysplasia in two Irish Traveller families.35 In addition, a single affected individual has also been described with a homozygous NEK9 mutation, demonstrating joint contracture and Legg-Calvé-Perthes (MIM: 150600) disease.36 Notably, NC syndrome features skeletal abnormalities, including scoliosis, syndactyly or absence of fingers, and supranumerary digits, suggesting that somatic NEK9 mutation in bone progenitors could account for these findings.37
Overexpression or knockout of NEK9 and its effectors leads to severe cellular dysfunction. NEK9 silencing via RNAi in cell culture prevents effective mitosis, leading to weak or malformed mitotic spindles.25 Further, overexpression of NEK9 in cells is toxic, with the majority of transiently transfected cells unable to successfully progress through a mitotic cycle.24 Similarly severe findings are found with lesions in downstream effectors of NEK9 signaling. Mice with deletion of NEK7 have early mortality due to failure in cytokinesis, and embryonic fibroblasts from NEK7-null animals were found to be polyploid with abnormal cilia morphology.38 Mice overexpressing Eg5 demonstrate increased propensity for tumors and genomic instability with poor chromosome segregation.39
Despite a known role in spindle formation and chromosome separation and in checkpoint regulation, our results further suggest that NEK9 might have additional function in the epidermis. In NC lesional tissue, the ectopic strong basal layer localization of K15 in comedonal follicles and cysts and simultaneous suprabasal localization of K10 suggests a switch in fate of progenitors from hair follicle to an IFE-like lineage. This switch might account for differentiation defects in NC, which bear some similarities to those seen in mouse models featuring cystic hair follicles and IFE-specific localization of differentiation markers in follicular keratinocytes.40, 41, 42
Mice with decreased LEF1 signaling due to deletion of the beta-catenin binding domain of LEF1 develop dermal cysts that resemble those found in NC on histopathologic examination. As in NC, these cysts demonstrate suprabasal K10, but immunolocalization of keratin 6 (K6) is present in cysts and hair follicles, a finding not seen in NC cysts or follicles.42 Similarly, defects in Notch signaling due to loss of mesenchymal RBP-Jκ can lead to K10-positive cyst formation. These lose AE13 and AE15 as seen in NC but retain K6.40 Finally, K15 conditional SOX9 knockout mice also develop K10-positive cysts in hair follicles, though these show normal K6, AE13, and AE15 immunolocalization, unlike NC.41
These mouse models demonstrate that several pathways, involving both epidermal and mesenchymal inputs, can alter hair follicle progenitor fate. In NC, the loss of normal follicular differentiation markers, expansion of strongly K15-positive basal cells, and suprabasal localization of K10 throughout the follicle and cyst suggests that NC demonstrates a similar type of fate switching. This “fate switch” paradigm might partly help explain why NC has no evident IFE pathology; if NEK9 mutation drives cells toward an IFE fate, one might expect that cells intended for such a fate would be less affected or unaffected.
The discovery of somatic NEK9 mutations at highly conserved sites in three unrelated NC cases, with no evidence for additional somatic mutations within the lesions, demonstrates that somatic NEK9 mutation causes NC. NEK9 mutations in NC resulted in increased Thr210 phosphorylation, suggesting gain of function, and affected tissue showed loss of markers of follicular differentiation with concurrent gain of an IFE marker. Although NEK9 localizes to cells within the IFE and hair follicle (Figure S5), is highly conserved across species (Figure S4), and is known to act in pathways regulating cell-cycle and checkpoint control,29 our results suggest that NEK9 might also co-regulate pathways relevant to follicular cell fate. Given the aforementioned mouse models featuring phenotypes similar to NC, it is plausible that NEK9 might function in the Wnt, Notch, or Sox9 signaling pathways. Future experiments addressing these possibilities could reveal a role for NEK9 in hair follicle fate decisions.
Given that comedone formation is critical step in acne pathogenesis and that NEK9 mutations in NC appear to have a follicle-specific effect, it is intriguing to consider whether NEK9 and its effectors might contribute to acne vulgaris pathobiology.
Acknowledgments
We would like to thank Valerie Horsley, Richard Lifton, Young Lim, Haris Mirza, Lynn Boyden, and Peggy Myung for review of the manuscript. We would also like to thank Jing Zhou and Rong Hua Hu for technical assistance. Research was supported by the Doris Duke Charitable Foundation Clinical Scientist Development Award to K.A.C. and a Yale Center for Mendelian Genomics grant (NIH U54 HG006504). J.L.L. is supported by the Medical Scientist Training Program (NIH NIGMS GM007205) at Yale University and is a recipient of a clinical research mentorship award from the Doris Duke Charitable Foundation. J.L.S. is an investigator for Galderma, Ranbaxy, and Activis, a consultant for Galderma, and a Medical Safety Monitor for Seegpharm and Valeant.
Published: May 5, 2016
Footnotes
Supplemental Data include eight figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.03.019.
Accession Numbers
The accession number for the sequencing data reported in this paper is dbGAP: phs000744.
Web Resources
1000 Genomes, http://www.1000genomes.org
ExAC Browser, http://exac.broadinstitute.org/
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Cunliffe W.J., Holland D.B., Jeremy A. Comedone formation: etiology, clinical presentation, and treatment. Clin. Dermatol. 2004;22:367–374. doi: 10.1016/j.clindermatol.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Williams H.C., Dellavalle R.P., Garner S. Acne vulgaris. Lancet. 2012;379:361–372. doi: 10.1016/S0140-6736(11)60321-8. [DOI] [PubMed] [Google Scholar]
- 3.Toyoda M., Morohashi M. Pathogenesis of acne. Med. Electron Microsc. 2001;34:29–40. doi: 10.1007/s007950100002. [DOI] [PubMed] [Google Scholar]
- 4.Zouboulis C.C., Bettoli V. Management of severe acne. Br. J. Dermatol. 2015;172(Suppl 1):27–36. doi: 10.1111/bjd.13639. [DOI] [PubMed] [Google Scholar]
- 5.Wilkie A.O., Slaney S.F., Oldridge M., Poole M.D., Ashworth G.J., Hockley A.D., Hayward R.D., David D.J., Pulleyn L.J., Rutland P. Apert syndrome results from localized mutations of FGFR2 and is allelic with Crouzon syndrome. Nat. Genet. 1995;9:165–172. doi: 10.1038/ng0295-165. [DOI] [PubMed] [Google Scholar]
- 6.Munro C.S., Wilkie A.O. Epidermal mosaicism producing localised acne: somatic mutation in FGFR2. Lancet. 1998;352:704–705. doi: 10.1016/S0140-6736(05)60820-3. [DOI] [PubMed] [Google Scholar]
- 7.Grose R., Fantl V., Werner S., Chioni A.M., Jarosz M., Rudling R., Cross B., Hart I.R., Dickson C. The role of fibroblast growth factor receptor 2b in skin homeostasis and cancer development. EMBO J. 2007;26:1268–1278. doi: 10.1038/sj.emboj.7601583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petiot A., Conti F.J., Grose R., Revest J.M., Hodivala-Dilke K.M., Dickson C. A crucial role for Fgfr2-IIIb signalling in epidermal development and hair follicle patterning. Development. 2003;130:5493–5501. doi: 10.1242/dev.00788. [DOI] [PubMed] [Google Scholar]
- 9.Jansen T., Plewig G. Acne inversa. Int. J. Dermatol. 1998;37:96–100. doi: 10.1046/j.1365-4362.1998.00414.x. [DOI] [PubMed] [Google Scholar]
- 10.Wang B., Yang W., Wen W., Sun J., Su B., Liu B., Ma D., Lv D., Wen Y., Qu T. Gamma-secretase gene mutations in familial acne inversa. Science. 2010;330:1065. doi: 10.1126/science.1196284. [DOI] [PubMed] [Google Scholar]
- 11.Pink A.E., Simpson M.A., Desai N., Trembath R.C., Barker J.N. γ-Secretase mutations in hidradenitis suppurativa: new insights into disease pathogenesis. J. Invest. Dermatol. 2013;133:601–607. doi: 10.1038/jid.2012.372. [DOI] [PubMed] [Google Scholar]
- 12.He L., Wu W.J., Yang J.K., Cheng H., Zuo X.B., Lai W., Gao T.W., Ma C.L., Luo N., Huang J.Q. Two new susceptibility loci 1q24.2 and 11p11.2 confer risk to severe acne. Nat. Commun. 2014;5:2870. doi: 10.1038/ncomms3870. [DOI] [PubMed] [Google Scholar]
- 13.Navarini A.A., Simpson M.A., Weale M., Knight J., Carlavan I., Reiniche P., Burden D.A., Layton A., Bataille V., Allen M., Acne Genetic Study Group Genome-wide association study identifies three novel susceptibility loci for severe Acne vulgaris. Nat. Commun. 2014;5:4020. doi: 10.1038/ncomms5020. [DOI] [PubMed] [Google Scholar]
- 14.Zhang M., Qureshi A.A., Hunter D.J., Han J. A genome-wide association study of severe teenage acne in European Americans. Hum. Genet. 2014;133:259–264. doi: 10.1007/s00439-013-1374-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tchernev G., Ananiev J., Semkova K., Dourmishev L.A., Schönlebe J., Wollina U. Nevus comedonicus: an updated review. Dermatol. Ther. (Heidelb.) 2013;3:33–40. doi: 10.1007/s13555-013-0027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moss C., Larkins S., Stacey M., Blight A., Farndon P.A., Davison E.V. Epidermal mosaicism and Blaschko’s lines. J. Med. Genet. 1993;30:752–755. doi: 10.1136/jmg.30.9.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim Y.H., Ovejero D., Sugarman J.S., Deklotz C.M., Maruri A., Eichenfield L.F., Kelley P.K., Jüppner H., Gottschalk M., Tifft C.J. Multilineage somatic activating mutations in HRAS and NRAS cause mosaic cutaneous and skeletal lesions, elevated FGF23 and hypophosphatemia. Hum. Mol. Genet. 2014;23:397–407. doi: 10.1093/hmg/ddt429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Happle R. The group of epidermal nevus syndromes Part I. Well defined phenotypes. J. Am. Acad. Dermatol. 2010;63:1–22. doi: 10.1016/j.jaad.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 19.Solomon L.M., Fretzin D., Pruzansky S. Pilosebaceous abnormalities in Apert’s syndrome. Arch. Dermatol. 1970;102:381–385. [PubMed] [Google Scholar]
- 20.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013 doi: 10.1002/0471250953.bi1110s43. 11, 11 10 11-11 10 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roig J., Mikhailov A., Belham C., Avruch J. Nercc1, a mammalian NIMA-family kinase, binds the Ran GTPase and regulates mitotic progression. Genes Dev. 2002;16:1640–1658. doi: 10.1101/gad.972202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertran M.T., Sdelci S., Regué L., Avruch J., Caelles C., Roig J. Nek9 is a Plk1-activated kinase that controls early centrosome separation through Nek6/7 and Eg5. EMBO J. 2011;30:2634–2647. doi: 10.1038/emboj.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bose A., Teh M.T., Mackenzie I.C., Waseem A. Keratin k15 as a biomarker of epidermal stem cells. Int. J. Mol. Sci. 2013;14:19385–19398. doi: 10.3390/ijms141019385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thiel C., Kessler K., Giessl A., Dimmler A., Shalev S.A., von der Haar S., Zenker M., Zahnleiter D., Stöss H., Beinder E. NEK1 mutations cause short-rib polydactyly syndrome type majewski. Am. J. Hum. Genet. 2011;88:106–114. doi: 10.1016/j.ajhg.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frank V., Habbig S., Bartram M.P., Eisenberger T., Veenstra-Knol H.E., Decker C., Boorsma R.A., Göbel H., Nürnberg G., Griessmann A. Mutations in NEK8 link multiple organ dysplasia with altered Hippo signalling and increased c-MYC expression. Hum. Mol. Genet. 2013;22:2177–2185. doi: 10.1093/hmg/ddt070. [DOI] [PubMed] [Google Scholar]
- 29.Sdelci S., Bertran M.T., Roig J. Nek9, Nek6, Nek7 and the separation of centrosomes. Cell Cycle. 2011;10:3816–3817. doi: 10.4161/cc.10.22.18226. [DOI] [PubMed] [Google Scholar]
- 30.Bahmanyar S., Kaplan D.D., Deluca J.G., Giddings T.H., Jr., O’Toole E.T., Winey M., Salmon E.D., Casey P.J., Nelson W.J., Barth A.I. beta-Catenin is a Nek2 substrate involved in centrosome separation. Genes Dev. 2008;22:91–105. doi: 10.1101/gad.1596308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roig J., Groen A., Caldwell J., Avruch J. Active Nercc1 protein kinase concentrates at centrosomes early in mitosis and is necessary for proper spindle assembly. Mol. Biol. Cell. 2005;16:4827–4840. doi: 10.1091/mbc.E05-04-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belham C., Roig J., Caldwell J.A., Aoyama Y., Kemp B.E., Comb M., Avruch J. A mitotic cascade of NIMA family kinases. Nercc1/Nek9 activates the Nek6 and Nek7 kinases. J. Biol. Chem. 2003;278:34897–34909. doi: 10.1074/jbc.M303663200. [DOI] [PubMed] [Google Scholar]
- 33.Smith S.C., Petrova A.V., Madden M.Z., Wang H., Pan Y., Warren M.D., Hardy C.W., Liang D., Liu E.A., Robinson M.H. A gemcitabine sensitivity screen identifies a role for NEK9 in the replication stress response. Nucleic Acids Res. 2014;42:11517–11527. doi: 10.1093/nar/gku840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurioka D., Takeshita F., Tsuta K., Sakamoto H., Watanabe S., Matsumoto K., Watanabe M., Nakagama H., Ochiya T., Yokota J. NEK9-dependent proliferation of cancer cells lacking functional p53. Sci. Rep. 2014;4:6111. doi: 10.1038/srep06111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casey J.P., Brennan K., Scheidel N., McGettigan P., Lavin P.T., Carter S., Ennis S., Dorkins H., Ghali N., Blacque O.E. Recessive NEK9 mutation causes a lethal skeletal dysplasia with evidence of cell cycle and ciliary defects. Hum. Mol. Genet. 2016 doi: 10.1093/hmg/ddw054. Published online February 21, 2016. [DOI] [PubMed] [Google Scholar]
- 36.Shaheen R., Patel N., Shamseldin H., Alzahrani F., Al-Yamany R., ALMoisheer A., Ewida N., Anazi S., Alnemer M., Elsheikh M. Accelerating matchmaking of novel dysmorphology syndromes through clinical and genomic characterization of a large cohort. Genet. Med. 2015 doi: 10.1038/gim.2015.147. Published online December 3, 2015. [DOI] [PubMed] [Google Scholar]
- 37.Beck M.H., Dave V.K. Extensive nevus comedonicus. Arch. Dermatol. 1980;116:1048–1050. [PubMed] [Google Scholar]
- 38.Salem H., Rachmin I., Yissachar N., Cohen S., Amiel A., Haffner R., Lavi L., Motro B. Nek7 kinase targeting leads to early mortality, cytokinesis disturbance and polyploidy. Oncogene. 2010;29:4046–4057. doi: 10.1038/onc.2010.162. [DOI] [PubMed] [Google Scholar]
- 39.Castillo A., Morse H.C., 3rd, Godfrey V.L., Naeem R., Justice M.J. Overexpression of Eg5 causes genomic instability and tumor formation in mice. Cancer Res. 2007;67:10138–10147. doi: 10.1158/0008-5472.CAN-07-0326. [DOI] [PubMed] [Google Scholar]
- 40.Hu B., Lefort K., Qiu W., Nguyen B.C., Rajaram R.D., Castillo E., He F., Chen Y., Angel P., Brisken C., Dotto G.P. Control of hair follicle cell fate by underlying mesenchyme through a CSL-Wnt5a-FoxN1 regulatory axis. Genes Dev. 2010;24:1519–1532. doi: 10.1101/gad.1886910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kadaja M., Keyes B.E., Lin M., Pasolli H.A., Genander M., Polak L., Stokes N., Zheng D., Fuchs E. SOX9: a stem cell transcriptional regulator of secreted niche signaling factors. Genes Dev. 2014;28:328–341. doi: 10.1101/gad.233247.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niemann C., Owens D.M., Hülsken J., Birchmeier W., Watt F.M. Expression of DeltaNLef1 in mouse epidermis results in differentiation of hair follicles into squamous epidermal cysts and formation of skin tumours. Development. 2002;129:95–109. doi: 10.1242/dev.129.1.95. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.