Abstract
Although exome sequencing data are generated primarily to detect single-nucleotide variants and indels, they can also be used to identify a subset of genomic rearrangements whose breakpoints are located in or near exons. Using >4,600 tumor and normal pairs across 15 cancer types, we identified over 9,000 high confidence somatic rearrangements, including a large number of gene fusions. We find that the 5′ fusion partners of functional fusions are often housekeeping genes, whereas the 3′ fusion partners are enriched in tyrosine kinases. We establish the oncogenic potential of ROR1-DNAJC6 and CEP85L-ROS1 fusions by showing that they can promote cell proliferation in vitro and tumor formation in vivo. Furthermore, we found that ∼4% of the samples have massively rearranged chromosomes, many of which are associated with upregulation of oncogenes such as ERBB2 and TERT. Although the sensitivity of detecting structural alterations from exomes is considerably lower than that from whole genomes, this approach will be fruitful for the multitude of exomes that have been and will be generated, both in cancer and in other diseases.
Introduction
Genomic profiling of tumors with high-throughput sequencing technologies has provided an unprecedented opportunity for in-depth studies of genome rearrangements. Whole-genome sequencing (WGS) data are now routinely used for detection of a wide range of rearrangements with base-pair resolution of breakpoints, including those breakpoints in non-coding regions. These events are typically identified on the basis of read depth,1 discordant paired-end reads,2 split-read (reads spanning the breakpoint) alignment,3 genome assembly,4 local assembly,5 or by a combination of these methods.6 RNA-seq data can be used to interrogate gene fusions when the fusion is expressed at a sufficiently high amount.
Whole-exome sequencing (WES) data are generated to detect single-nucleotide variants (SNVs) and small indels. An enormous number of exomes have been generated by researchers around the world: the latest release from the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project7 includes 6,500 samples; the Exome Aggregation Consortium (ExAC), an international collaboration to collect exome data, has more than 60,000 exomes in its current release. Despite the decreasing cost of WGS, WES data will continue to be generated because many somatic variants occur at low variant allelic frequency, and the necessary high-depth (e.g., >100–500×) sequencing is affordable only with a capture-based approach given current technologies. An important question, therefore, is whether genomic rearrangements can also be detected in exomes. If that were possible, we would be able to identify a large number of rearrangements with datasets that were generated for other purposes.
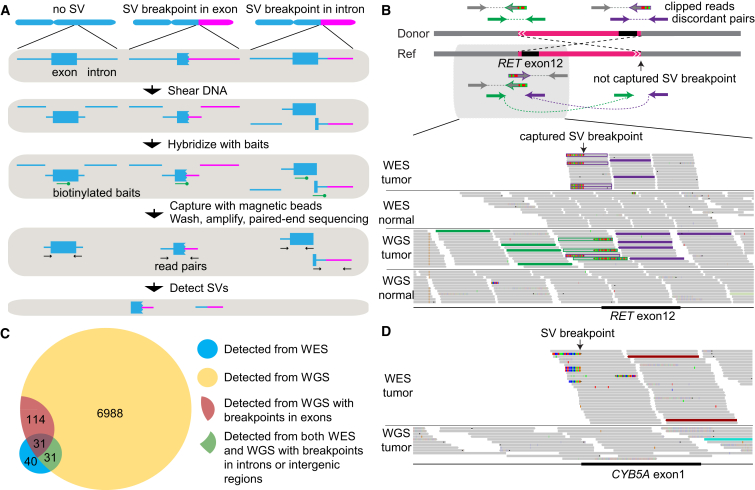
Here, we describe an approach to identify structural variations (SVs) from WES data. In a typical WES protocol, genomic DNA is sheared into fragments (∼150–250 bp), and those containing exons are enriched by hybridization with shorter biotinylated probes (∼50–100 nucleotides long). These probes are usually densely tiled across exons, extending just past the exon-intron boundaries. Thus, when the breakpoint of an SV occurs in or near the targeted region, the DNA fragment that contains the breakpoint can be captured if there is sufficient overlap between a probe and the DNA on either side of the breakpoint (Figure 1A). The sensitivity of detection from WES is clearly much lower than that from WGS, given that just a subset of rearrangements with breakpoints in or near exons can be detected and the fragment capture process introduces inefficiencies. However, with the large number of available exomes and the higher coverage than WGS, we demonstrate that re-analyzing existing large-scale WES data for genomic rearrangements can yield valuable findings.
Figure 1.
Detecting Somatic SVs from WES Data
(A) Workflow showing how DNA fragments are captured and sequenced when SV breakpoints occur in exons and near exon-intron boundaries.
(B) A true somatic CCDC6-RET fusion resulting from a balanced inversion (chr10:61,655,977–43,611,997) in a thyroid cancer (TCGA-FK-A3SE) is detected by both WES and WGS. The scheme of the inversion is shown on the top (not to scale). The Integrated Genome Viewer screen shot for the captured breakpoint is shown on the bottom. Green and purple read pairs represent discordant pairs from two different breakpoints; one breakpoint is captured by WES and the other is not. The gray reads are concordant read pairs. The half-gray and half-striped reads with green or purple outlines are partially aligned (clipped) reads spanning the breakpoints.
(C) A Venn diagram showing the overlap between somatic SVs called from WES and WGS data.
(D) A true somatic deletion (chr18:71,930,713–71,958,983) in a lung adenocarcinoma (TCGA-91-6840) is detected by WES but not by WGS and is validated by PCR. The coverage in WES is >100× and there are six discordant read pairs (two displayed), whereas the coverage of WGS in the same region is 30× and no discordant read pair is present. The red reads are discordant read pairs supporting the somatic deletion.
We applied our proposed method to survey somatic SVs in 4,609 samples across 15 tumor types from The Cancer Genome Atlas (TCGA). We focus on somatic variants here, but the approach we describe applies to detection of both germline and somatic rearrangements. We chose the TCGA data because they are high-quality, multi-dimensional data from a large number of samples, including cases that have undergone both WES and WGS. The availability of these two data types for the same samples allows us to characterize the sensitivity and specificity of exome-based SV detection. Although exome-based fusion detection has been recently used to identify recurrent NAB2-STAT6 (MIM: 602381 and 601512) fusion in solitary fibrous tumors,8 our study expands this approach to a much larger scale to discover additional cancer-driving gene fusions and characterize their features. Our results demonstrate the association of oncogene upregulation with massive rearrangements. We also report experimental validation that two of the candidate fusions we identified are cancer drivers, including the report of an activating genetic event related to ROR1 (MIM: 602336).
Material and Methods
TCGA Sample Acquisition and WES
The details of data production were described in a previous publication.9 The procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national). Tumor samples were obtained from the TCGA network with appropriate consent from the relevant institutional review board. Tumors were resected, flash-frozen, and shipped to a centralized processing center (Biospecimen Core Resource) for additional pathologic review and extraction of nucleic acids. The three genome sequencing centers (Baylor Human Genome Sequencing Center, Broad Institute, and The Genome Institute at Washington University) collectively sequenced the exomes from tumor tissues and matched normal tissues (mostly blood samples). Exome capturing procedures differ among sequencing centers and evolve over time. The details can be found in individual TCGA marker papers. Sequencing reads were aligned to the reference genome with the Burrows-Wheeler Aligner,10 and quality control was performed. A single BAM file that includes reads, calibrated quantities, and alignments to the genome was generated for each sample.
Data Access
All primary sequence files can be downloaded by registered users from CGHub. Clinical data are available through the TCGA Data Portal. All coordinates are based on the hg19 human reference genome, downloaded from the UCSC Genome Browser.
Detecting Somatic Genome Rearrangements in WES Data
Somatic genome rearrangements were called by Meerkat, a software package we developed.6 In brief, all discordant read pairs (reads that do not form a proper pair with expected orientations and distance between the reads) are first identified from the BAM files. Then, discordant read pairs supporting the same breakpoint are merged into clusters, which are used to call SV candidates. Reads spanning SV breakpoints (clipped reads and unmapped reads) are mapped back to the SV candidates (split-read mapping). Breakpoints are refined to the basepair resolution once split-read supports are identified. Variants are filtered by a large database of germline variants obtained by merging all matched normal BAM files from different tumor types together. The final somatic variants must have discordant read-pair support and split-read support totaling at least six reads and/or read pairs, with at least three discordant read-pair support. We have previously used these criteria to identify somatic SVs from WGS samples and have demonstrated that such a workflow offers great sensitivity and specificity. Samples with >100 somatic SVs were discarded from further analysis. Additional filters were applied to obtain high-confidence somatic rearrangements: at least four supporting discordant read pairs were required for each somatic event, and the size of an intra-chromosomal event could not be less than 20 kb. For comparison with WGS results, if the somatic rearrangement detected from WES data and the one detected from WGS data were the same type of event on the same chromosome(s) and the breakpoints differed by less than 50 bp, they were considered to be the same event. In most cases, the breakpoints predicted from WES and WGS were exactly the same. PCR primers were designed by Primer3.11
Detecting Activating Gene Fusions
RNA was extracted, prepared into Illumina TruSeq mRNA libraries, and sequenced by an Illumina sequencing platform with a target of 60 million read pairs per tumor (48 bp paired-end reads) and subjected to quality control. RNA reads were aligned to the reference genome with Mapsplice.12 Gene expression was quantified for the transcript models (TCGA GAF2.1) with RSEM13 and normalized within sample to a fixed upper quartile of total reads. RNA-seq results (normalized gene-level expression and exon-level expression) were downloaded from the Genome Data Analysis Center at the Broad Institute. RNA data were available only for tumor tissues because TCGA collected blood (rather than adjacent normal tissues, which are generally unavailable) as the matched normal control for the majority of the cases. Therefore, to normalize exonic expression, we computed a Z score for each exon on the basis of its expression across all samples in that tumor type. Gene Ontology (GO) term enrichment analyses were performed with DAVID.14 All 5′ and 3′ fusion partners were entered into DAVID as a gene list to identify over-represented GO categories, and the functional annotation clustering of GO terms was performed. The p value was calculated by one-tail Fisher’s exact test.
Analysis of Massive Rearrangements
A binomial model was used to identify the samples in which the number of somatic rearrangement breakpoints observed on one chromosome significantly exceeded the expected number, given the total number of somatic rearrangement breakpoints in one sample (the likelihood of observing at least n breakpoints on one chromosome given the total N breakpoints in that sample, with the probability p being the mappable coding-sequence (CDS) size for the chromosome divided by the mappable CDS size for the whole genome). Bonferroni correction was used to adjust for multiple testing. The mappability of the reference genome was downloaded from UCSC Genome browser and was used to normalize the chromosome size.
Statistical Analysis
All statistical analyses were conducted in R package (v.2.14.1). A p value of 0.01 was used for statistical significance.
Fusion Gene Cloning
Constructs of CEP85L-ROS1(C9;R36) (MIM: 165020), GOPC (MIM: 606845)-ROS1(G7;R35), and EML4 (MIM: 607442)-ALK (MIM: 105590) gene fusions were synthesized by CosmogeneTech and then transferred into pLenti6.3/V5-DEST (Life Technologies) and pLenti6.3-EF1α lentiviral vectors. ROR1-DNAJC6 (MIM: 608375) fusion fragments were cloned from cDNA prepared from U87MG cells with overlapping ends, fused ROR1-DNAJC6 was then generated by overlap-extension PCR, and the resulting fusion gene was then transferred into the pLenti6.3/V5-DEST vector. Expression of the ROR1-DNAJC6 fusion gene was confirmed via RT-PCR and western blots with the following primer sets: forward, ROR1, 5′-GTGATGAAGATGGGACTGTGAA-3′; reverse, DNAJC6, 5′-CTAGAAGATGTGTCTTTGAGGGTGT-3′.
Ba/F3 Cell Viability and Inhibitor Assays
The Ba/F3 cell line was maintained in RPMI 1640 medium with 5% fetal bovine serum and 2.5 ng/ml recombinant mouse IL-3. CEP85L-ROS1, BCR (MIM: 151410) -ABL ([MIM: 189980] positive control), and GFP (negative control) were transduced into Ba/F3 cells. At 72 hr post-transduction, cells were re-suspended in medium without IL-3. Cell viability was determined with Cell Titer-Glo (Promega) at 7 days after IL-3 depletion. Ba/F3 cells stably expressing CEP85L-ROS1 (no IL-3 medium) and parental Ba/F3 cells (IL-3 medium) were seeded in 96-well plates in quadruplicates at 1,000 cells per well. For the dose-dependent inhibitor assay, cells were treated with dimethyl sulfoxide (DMSO) or crizotinib (5 nM to 0.5 μM) and cell viability was determined with Cell Titer-Glo (Promega). Cell survival was normalized to non-treated (DMSO control treated) cells. IC50, which is the concentration of an inhibitor causing 50% inhibition of cell survival normalized to non-treated cells, was calculated from a sigmoidal curve. The response of CEP85L-ROS1-expressing cells (without IL3) to crizotinib was compared to parental cells without treatment of crizotinib as control. Two independent experiments were performed.
Western Blot
Whole-cell and mouse tumor tissue lysates were prepared with radioimmunoprecipitation assay (50 mM Tris-HCl, 150 mM NaCl, 1% NP-40, and 0.25% sodium deoxycholate) plus protease inhibitors cocktail (GenDepot). Cell and tissue lysates were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes. The blots were probed with antibodies for ROS1, phosphorylated, and total STAT3 (MIM: 102582), AKT and ERK (Cell Signaling Technology), and ROR1 (Abcam) were then detected with chemiluminescent substrate (EMD Millipore). All western blot images are representative of at least three independent experiments.
In Vitro Cell Proliferation and Transforming Assays
NIH 3T3 cells were obtained from the Korean Cell Line Bank, and BEAS-2B cells (ATCC CRL-9609) were obtained from the American Type Culture Collection (Manassas, VA). They were expanded in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 mg/ml streptomycin. NIH 3T3 cells and BEAS-2B cells were transduced with LacZ (negative control), CEP85L-ROS1, GOPC-ROS1 (positive control), ROR1-DNAJC6, and EML4-ALK (positive control). Then, stable cell lines were selected with blasticidin. Cell proliferation was determined by a EZ-Cytox cell viability assay kit (Daeil Lab Service). The transforming activity was assessed by transformed foci formation in Matrigel. NIH 3T3 stable cells expressing CEP85L-ROS1, GOPC-ROS1, and EML4-ALK, and BEAS-2B stable cells expressing ROR1-DNAJC6 and EML4-ALK were seeded in Matrigel (BD Sciences; 10,000 cells per well), on which medium with 10% FBS was overlaid. The images of transformed foci were taken after culturing for 7 or 14 days.
Anchorage Independent Growth Assay
MCF-10A cells were cultured as described previously15 and transduced with CEP85L-ROS1, PIK3CAH1047R (positive control), and GFP (negative control). Soft agar assays were performed in six-well plates in triplicate. First, bottom layers were prepared at 0.8% Noble agar (Affymetrix) with complete MCF-10A growth medium. After solidification, 10,000 cells were mixed with 0.45% agar in complete growth medium and laid on top of the bottom layer. 2 mL of medium was added in each well after 3 days, and the medium was refreshed every 3 days. For NIH 3T3 and BEAS-2B cells expressing LacZ, CEP85L-ROS1, GOPC-ROS1, ROR1-DNAJC6, and EML4-ALK in 0.35% agar (BD Sciences), 20,000 cells were seeded on top of 0.5% agar in each well. Cells were cultured for 14 or 21 days, colonies were stained with 0.05% crystal violet, and images were taken by phase-contrast microscope (Olympus CKX41) and analyzed by i-Solution Lite image analysis software, and cells were counted in ten randomly selected fields.
Xenograft Tumor Formation Assay
All animal experiments were approved by the institutional review board of Samsung Medical Center. 5 × 106 cells were re-suspended in 1:1 PBS and Matrigel (BD Biosciences) and then subcutaneously injected into the right dorsal flank of six-week-old male nude mice (Orient Bio). Mice were monitored three times weekly until reaching maximal tumor size (approximately 2 cm × 2 cm). Mice were then sacrificed and photographed on day 23 after injection, and tumors were collected for further analysis.
Results
Detecting Somatic SVs in WES
In a standard WES protocol (Figure 1A), probes are designed to capture coding exons. The enriched exonic regions are subsequently amplified and subjected to paired-end sequencing. Due to the capturing and amplification steps, the coverage of resulting sequencing data is uneven across the genome. SV detection tools using read-depth information will suffer from this uneven sequencing coverage, whereas tools that depend on discordant read pairs and split reads to detect genomic rearrangements can be used in WES data as long as the breakpoints are captured and sequenced. We first tested the efficacy of detecting somatic SVs using discordant read pairs and split reads but not read depth. We selected 120 TCGA samples that had both WES and WGS data (Table S1) for initial analysis, with the assumption that somatic SVs called on both platforms are true positives (example in Figure 1B). We did not define the truth set purely on the basis of WGS data because some SVs are missed and some SV calls are artifacts even in WGS.
A major challenge in reliably identifying somatic SVs in WES data is to remove a large number of artifacts arising from chimeric molecules in the library preparation. This requires designing data processing steps to remove WES-specific artifacts. When we applied the Meerkat algorithm we originally developed6 for WGS to WES data, we found a small subset of the samples containing a large number (>100) of somatic SVs, with the majority of SVs not found in the matched WGS (Figure S1A; examples shown in Figures S1B and S1C). WES-specific artifacts were distinguishable by their even distribution across all chromosomes, enrichment of small tandem duplications, and no homology at the breakpoints (Figures S1D–S1F). These samples therefore failed our quality control steps and were discarded from further analysis. For the remaining comparisons, we also removed two WGS cases whose normal data had poor quality (Figure S2).
We designed additional computational filters (see Material and Methods) to remove such artifacts in the remaining samples by testing different combinations of thresholds and comparing the resulting set against WGS calls. This filtration resulted in high-confidence somatic calls from WES data with a substantial reduction in the number of WES-specific calls (Figure S3A). Overall, 61% of the WES calls were shared by WGS (Figure 1C). Many calls found in WGS are missed by WES; out of 145 SVs detected from WGS data with breakpoints in exons (excluding UTRs), 21% (31/145) were recovered from WES data. This low rate is mainly due to the insufficient number of supporting read pairs (Figure S3B) in addition to the uneven read coverage in the targeted regions in WES (Figure S3C). The allele fractions of somatic SVs detected in WES are generally smaller than those in WGS data (Figure S3D). We suspect that the exon capture efficiency is lower for the chimeric DNA molecules that contain the breakpoints, resulting in lower coverage and hence not enough supporting reads for detecting SVs. Conversely, it is important to note that ∼39% of the WES calls were not found in WGS. At least a few of these are true positives that were detected by the higher sequencing coverage in WES data than in WGS (Figure 1D and Figure S4). The concordance between WES and WGS calls depends on the quality of the libraries and may vary among datasets.
To test the accuracy of our calls, we performed PCR on all high-confidence somatic SVs called from WES data for which we could obtain the DNA. We found that 78% (21/27) were validated (Table S2). Overall, these results suggest that, despite its modest sensitivity, WES-based SV analysis is likely to yield additional SV candidates that are biologically meaningful.
A Catalog of Gene Fusions and the Properties of Driver Fusions
We analyzed WES data for 4,859 cancer samples across 15 tumor types from TCGA (Table 1). A total of 9,171 high-confidence somatic SVs were detected from 4,609 samples (Table S3) after excluding 250 samples because of low quality. The breast cancers (MIM: 114480) have the highest number of somatic SVs, whereas the kidney cancers (both clear cell [MIM: 144700] and papillary cell [MIM: 605074] carcinomas) have the fewest, consistent with our previous findings6 (Table 1). The genes with somatic rearrangements are expressed significantly higher (∼2-fold increase) than the ones without any rearrangements (Figure S5). Although a previous study16 associated somatic SV breakpoints with expression, the SV and expression data came from different sets of samples. Here, we used a large number of samples that have each undergone both WES and RNA-seq for a more direct comparison.
Table 1.
Summary of Somatic SVs in 15 Tumor Types
| Tumor Type | Abbreviation | Sample Size | Bad Samples | Good Samples | Total SVs | Average SVs per Sample | Massively Rearranged |
|---|---|---|---|---|---|---|---|
| Urothelial bladder cancer | BLCA | 185 | 3 | 182 | 370 | 2.03 | 6 |
| Breast cancer | BRCA | 781 | 93 | 688 | 3123 | 4.54 | 65 |
| Glioblastoma multiforme | GBM | 318 | 63 | 255 | 626 | 2.45 | 24 |
| Head and neck squamous cell carcinoma | HNSC | 377 | 0 | 377 | 413 | 1.10 | 4 |
| Clear cell kidney carcinoma | KIRC | 322 | 13 | 309 | 191 | 0.62 | 4 |
| Papillary kidney carcinoma | KIRP | 147 | 0 | 147 | 80 | 0.54 | 4 |
| Lower grade glioma | LGG | 272 | 0 | 272 | 218 | 0.80 | 6 |
| Liver hepatocellular carcinoma | LIHC | 98 | 0 | 98 | 350 | 3.57 | 2 |
| Lung adenocarcinoma | LUAD | 485 | 27 | 458 | 791 | 1.73 | 12 |
| Lung squamous cell carcinoma | LUSC | 460 | 23 | 437 | 837 | 1.92 | 9 |
| Prostate adenocarcinoma | PRAD | 235 | 1 | 234 | 331 | 1.41 | 6 |
| Cutaneous melanoma | SKCM | 311 | 1 | 310 | 577 | 1.86 | 24 |
| Stomach adenocarcinoma | STAD | 234 | 0 | 234 | 570 | 2.44 | 11 |
| Papillary thyroid carcinoma | THCA | 485 | 2 | 483 | 342 | 0.71 | 0 |
| Uterine corpus endometrial carcinoma | UCEC | 149 | 24 | 125 | 352 | 2.82 | 1 |
| Total | – | 4,859 | 250 | 4,609 | 9,171 | 1.99 | 178 |
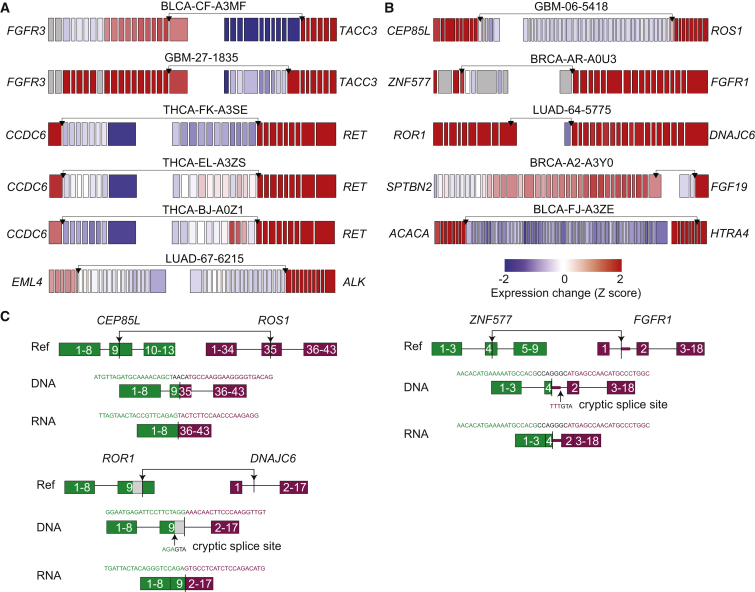
Our exome-based SV calling identified many biologically important variants. Some SVs disrupted tumor suppressors, such as TP53 (MIM: 191170), CDKN2A (MIM: 600160), and PTEN (MIM: 601728) (Table S4). Many SVs were known driver fusions (examples in Figure 2A). For example, we detected four RET (MIM: 164761) fusions (three CCDC6 [MIM: 601985]-RET fusions and one FKBP15-RET fusion) in thyroid carcinomas, an EML4-ALK fusion in lung adenocarcinoma, and five FGFR3 (MIM: 134934)-TACC3 (MIM: 605303) fusions in three cancer types (glioblastoma [GBM], bladder cancer [MIM: 109800], and renal papillary cancer). FGFR3-TACC3 was originally described in GBM, with 3 out of 97 tumors examined carrying this fusion.17 This was an important discovery because this subset of individuals could potentially benefit from targeted FGFR kinase inhibition. We had also found the same fusion in about 3% of the bladder cancer samples, based on analysis of WGS data, as we reported recently in the TCGA consortium paper.18 Our analysis of the exome data reveals that FGFR3-TACC3 also occurs in papillary kidney carcinoma. We also detected two prostate adenocarcinoma (MIM: 176807) cases with TMPRSS2 (MIM: 602060)-ERG (MIM: 165080) fusions. As expected, the frequencies of these known drivers are much lower than the previously reported numbers due to limited sensitivity. However, we were able to discover a wide range of variants as a result of the large sample size.
Figure 2.
Activating Gene Fusions Detected
(A) Exon-specific expression profiles for known cancer-driving fusions.
(B) Exon-specific expression profiles for additional activating fusions. Black arrows in (A) and (B) denote fusion breakpoints. Each box represents an exon. The expression of each exon was normalized to its average expression across all individuals of the same tumor type. A gray box indicates that the exon is not expressed in more than 70% of the samples.
(C) Examples of fusion breakpoints at the DNA and RNA level for CEP85L-ROS1, ZNF577-FGFR1, and ROR1-DNAJC6. Green and purple boxes denote exons of 5′ and 3′ fusion partners, respectively. Breakpoint junction sequences are shown above the fusions, with letters in black denoting non-reference sequences. The thick purple line in FGFR1 denotes exonized intronic sequence. The gray box in ROR1 denotes the part of the exon being spliced out.
Distinguishing drivers (alterations that increase the fitness of cells) from passengers (neutral alterations) is challenging for any type of genetic alteration. For SNVs and copy-number variants, computational methods (e.g., MutSigCV19 and GISTIC,20 respectively) aim to assess the statistical significance of the observed mutation frequencies by using a background model. Recurrence is the most obvious factor in estimating the likelihood of fusion being a driver; however, understanding the molecular characteristics of driver fusion is critical, given that some driver fusions have very low frequency, many studies have small sample sizes, or, as in the case here, detection sensitivity might be low. Furthermore, recurrent events can also result from frequent breaks of certain genomic regions such as fragile sites and might not drive cancer. We previously observed that most of the known driver fusions are activating fusions and that the 3′ fusion partners are almost always upregulated, typically with expression change at the fusion breakpoints21, 22 (Figure 2A). To identify activating gene fusions, we thus propose three criteria: (1) the gene fusion must maintain the same transcription orientation; (2) the fused 3′ partner must be upregulated; and (3) a significant expression change must be observed at or near the fusion breakpoints in at least one of the two source regions (e.g., red versus blue exons on the two sides of the TACC3 breakpoint in the FGFR3-TACC3 fusion in Figure 2A). There are driver fusions that do not have an upregulated 3′ partner, but these are hard to identify unless they recur across many samples. Expression change at the breakpoint was also used to identify fusion candidates from expression array data, followed by 5′ rapid amplification of cDNA ends to search for the fusion partners.23, 24, 25 Using the three criteria above, we uncovered a total of 150 activating fusions (Table S5). Five activating fusions (CEP85L-ROS1, ZNF577-FGFR1 [MIM: 136350], ROR1-DNAJC6, SPTBN2 [MIM: 604985]-FGF19 [MIM: 603891], ACACA [MIM: 200350]-HTRA4 [MIM: 610700]) are shown in Figure 2B as examples. We note that these activating fusions are candidate driver fusions, but the criteria we used are not sufficient to define them as cancer drivers. In vitro and in vivo experiments are needed to definitively address their role in tumorigenesis (see Functional Validation of Fusion Genes In Vitro and In Vivo).
Not surprisingly, GO analysis of the activating fusions revealed that the 3′ fusion partners are enriched for protein tyrosine kinases (p = 1.7E-4) (Table 2) as previously observed.26, 27, 28 The protein tyrosine kinases RET, ALK, and ROS1 are known oncogenes and often form fusions with various partners in lung (MIM: 211980), thyroid, and colorectal cancers (MIM: 114500)22, 29, 30, 31, 32, 33 (e.g., for RET: CCDC6, FKBP15, TBL1XR1 [MIM: 608628], AKAP13 [MIM: 604686], KIF5B [MIM: 602809]; for ALK: EML4, STRN [MIM: 614765], GTF2IRD1 [MIM: 604318], MROH2B, C2orf44 [MIM: 616234]; for ROS1: SLC34A2 [MIM: 604217], CD74 [MIM: 142790], SDC4 [MIM: 600017], EZR [MIM: 123900], LRIG3 [MIM: 608870]). Some of the kinase fusions detected from WES were known previously. For instance, NFASC (MIM: 609145)-NTRK1 ([MIM: 191315] neurotrophic tyrosine receptor kinase type 1) was found in two TCGA GBM samples via RNA-seq data and validated as a cancer driver.34 Other fusions identified here were not reported previously: for example, INSRR (MIM: 147671), an insulin receptor-related receptor, is paralogous to many oncogenes such as ROS1, NTRK1, and ALK, but has never been described as a fusion partner in cancer even though it is involved in the AKT and MAPK signaling pathways and its expression has been correlated with a favorable prognosis in neuroblastoma.35 The fusion GON4L (MIM: 610393)-INSRR found in low-grade glioma activates the protein kinase domain of INSRR, suggesting that it is likely to be a driver fusion.
Table 2.
Activating Fusions with 3′ Tyrosine Protein Kinases
| ID | Chr A | Breakpoint A | Gene A | Chr B | Breakpoint B | Gene B | Discord Pair | Split Read | Homology |
|---|---|---|---|---|---|---|---|---|---|
| THCA-FK-A3SE | 10 | 61655977 | CCDC6 | 10 | 43611997 | RET | 13 | 17 | 3 |
| THCA-EL-A3ZS | 10 | 61659539 | CCDC6 | 10 | 43611930 | RET | 4 | 4 | 0 |
| THCA-BJ-A0ZJ | 10 | 61626050 | CCDC6 | 10 | 43611953 | RET | 13 | 5 | 1 |
| THCA-ET-A3DQ | 9 | 115932783 | FKBP15 | 10 | 43610457 | RET | 5 | 2 | −3 |
| LUAD-67-6215 | 2 | 42491894 | EML4 | 2 | 29447037 | ALK | 6 | 5 | 2 |
| THCA-EM-A4FR | 5 | 41038833 | MROH2B | 2 | 29481156 | ALK | 7 | 7 | 3 |
| GBM-06-5418 | 6 | 118801608 | CEP85L | 6 | 117642526 | ROS1 | 55 | 46 | −4 |
| BRCA-AR-A0U3 | 19 | 52383621 | ZNF577 | 8 | 38317439 | FGFR1 | 104 | 29 | −7 |
| BRCA-AR-A0TT | 19 | 16243092 | RAB8A | 19 | 4115139 | MAP2K2 | 45 | 32 | 0 |
| GBM-06-5411 | 1 | 204951828 | NFASC | 1 | 156844167 | NTRK1 | 534 | 398 | 2 |
| LGG-E1-5319 | 1 | 155784108 | GON4L | 1 | 156813488 | INSRR | 29 | 24 | 1 |
Genes on the left denoted by “Gene A” are 5′ fusion partners, and genes on the right denoted by “Gene B” are 3′ fusion partners.
We also found that the 5′ fusion partners of activating fusions are often housekeeping genes, such as those related to the cytoskeleton (p = 7.4E-5) and biosynthesis (p = 2.8E-3) (Table S6). For example, CCDC6, FKBP15, and EML4 are cytoskeleton proteins that fuse to RET and ALK. Furthermore, both the 5′ fusion partners (p = 8.5E-3) and the 3′ fusion partners (p = 4.9E-3) of the activating fusions are enriched in chromatin regulators (Tables S7 and S8). Many of the chromatin regulator fusions occur in the breast cancer samples. USP21 (ubiquitin specific protease 21 [MIM: 604729]), which deubiquitinates histone H2A and removes the transcriptional repression tag, is upregulated in 33% of the breast cancer samples.36 KDM2A (MIM: 605657), a histone demethylase that maintains heterochromatin and genome stability, and C11orf30 (MIM: 608574), a protein-coding gene that can repress transcription and might play a central role in the DNA-repair function of BRCA2 (MIM: 600185), are upregulated in 17% and 11% of the breast cancer samples, respectively.36 The chromatin regulators are upregulated upon fusions and might alter expressions of many other genes and play important roles in tumor progression.
Given the functional categories enriched in the fusion partners, we propose a general model of driver fusions in cancer. The 3′ partners are often oncogenes, which can promote cell growth and proliferation but are typically not expressed in differentiated cells. The 5′ partners are enriched in housekeeping genes, which are expressed in normal cells but whose production is controlled by various mechanisms, including negative feedback loops. Upon fusion, the active housekeeping gene in cancer cells turns on its oncogenic partner. However, because no housekeeping protein is produced, the housekeeping genes remain on. As a result, both the 5′ and 3′ fusion partners are upregulated. In the case of TMPRSS2-ERG in prostate cancers (the predominant recurrent aberration in that tumor type), TMPRSS2 is activated by the androgen receptor and serves as a housekeeping gene in the prostate tissue. The 3′ fusion partners are different ETS family oncogenes (e.g., ERG, ETV1 [MIM: 600541], ETV4 [MIM: 600711], and ETV5 [MIM: 601600])23 that are activated by TMPRSS2.
With sequencing data available from both DNA and RNA, it is also possible to interrogate how the fusion genes are spliced. Three cases are shown in Figure 2C: (1) The CEP85L-ROS1 fusion occurs between exon 9 of CEP85L and exon 35 of ROS1. The breakpoints at the DNA level are out of frame; however, upon alternative splicing (the fusion exon 9-35 being spliced out), the fusion is in frame at the RNA level. (2) The ZNF577-FGFR1 fusion is between exon 4 of ZNF577 and intron 1 of FGFR1. A small portion of the FGFR1 intron becomes part of an exon through a cryptic splice site, and the resulting transcript is in frame. (3) The ROR1-DNAJC6 fusion is between exon 9 of ROR1 and intron 1 of DNAJC6. After fusion, part of the ROR1 exon 9 is spliced out through a cryptic splice site along with the intron 1 of DNAJC6, resulting in an in-frame transcript. These examples illustrate how alternative splicing and/or cryptic splice sites can be used after gene-fusion events to produce in-frame transcripts even if the fusions are out of frame at the DNA level. Therefore, prediction of functional consequences for gene fusions on the basis of the DNA sequence must account for these mechanisms.
Functional Validation of Fusion Genes In Vitro and In Vivo
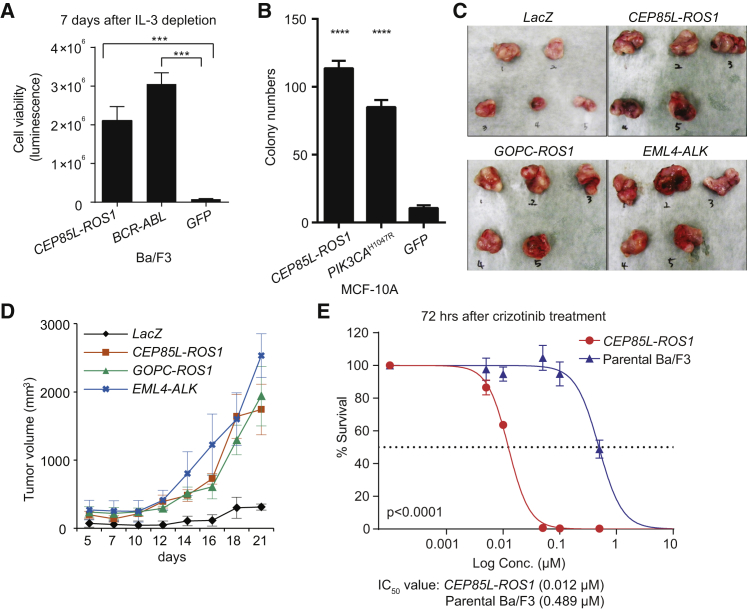
We performed extensive in vitro and in vivo validation for two fusions. Various fusions involving the ROS1 receptor tyrosine kinase have been identified previously, primarily in non-small cell lung cancer (NSCLC),33 and they are known to induce cell foci formation and anchorage-independent growth.37, 38 The CEP85L-ROS1 fusion in particular was reported in angiosarcoma and epithelioid hemangioendothelioma,25 and we found it in GBM in our analysis. However, its function in tumorigenesis has not yet been established. To test the oncogenic potential of this fusion, we utilized Ba/F3, a murine pro-B cell line that depends on interleukin-3 (IL-3) for survival and proliferation. Ba/F3’s dependence on IL-3 is readily transferred to expressed oncogenes, thus representing a sensitive assay to quantitate oncogenic activity of fusion genes after Ba/F3 transduction and IL-3 removal from growth medium.39, 40, 41 Introduction of the CEP85L-ROS1 fusion gene into Ba/F3 cells revealed a robust, >100-fold increase (p < 0.0001) in survival after IL3 removal in comparison to GFP-expressing control cells (Figure 3A). Notably, the growth-promoting activity exhibited by CEP85L-ROS1 was similar to that of BCR–ABL1, whose oncogenic activity has been well characterized.42 Next, we delivered CEP85L-ROS1 fusion into MCF-10A human breast epithelial cells43 which are widely used in anchorage-independent growth assays to assess the transforming activity of oncogenes.44 As shown in Figure 3B, expression of CEP85L-ROS1 in MCF-10A cells significantly increased colony formation (11-fold, p < 0.0001), as did the oncogenic PIK3CAH1047R control.45 We also found that CEP85L-ROS1 expression in NIH 3T3 murine fibroblasts induced their anchorage independent growth and cellular proliferation in vitro (Figures S6A and S6B) and tumor-forming activity in vivo (Figures 3C and 3D). Immunoblot analysis showed elevated phosphorylation of ERK1/2 (T202/Y204) in all three cell lines (Ba/F3, MCF-10A, and NIH 3T3; Figures S6C–S6E), which suggested that the MAPK signaling pathway was activated. We tested the effectiveness of this fusion as a drug target. Crizotinib is a small molecular protein kinase inhibitor for ALK and ROS1. It is approved for use in NSCLC cases with ALK fusion, and it has shown great anti-tumor activity in clinical trials targeting advanced NSCLC with a ROS1 rearrangement.46 We observed a marked inhibitory activity of crizotinib on CEP85L-ROS1-transformed Ba/F3 cells in comparison to parental cells (CEP85L-ROS1 IC50 = 0.012 μM; parental IC50 = 0.489 μM) as shown in Figure 3E. Our results show that individuals harboring a ROS1 fusion in tumor types other than NSCLC might also benefit from the ROS1 inhibitor.
Figure 3.
Functional Validation of CEP85L-ROS1
(A) CEP85L-ROS1 expression relieves Ba/F3 cells from dependency on IL-3.
(B) Anchorage-independent colony formation assays for CEP85L-ROS1 in MCF-10A cells (mean colony count from ten random areas).
(C and D) The transforming potential of the CEP85L-ROS1 fusion in vivo. The tumor volume was calculated with the modified ellipsoidal formula (volume = 1/2 [length × width2]) and the greatest longitudinal diameter (length) and the greatest transverse diameter (width) were used. Mice were sacrificed and photographed on day 23.
(E) Compared to parental cells (IC50 = 0.489 μM), CEP85L-ROS1-transformed Ba/F3 cells are significantly more sensitive (log-rank test) to crizotinib (IC50 = 0.012 μM). Error bars indicate SD.
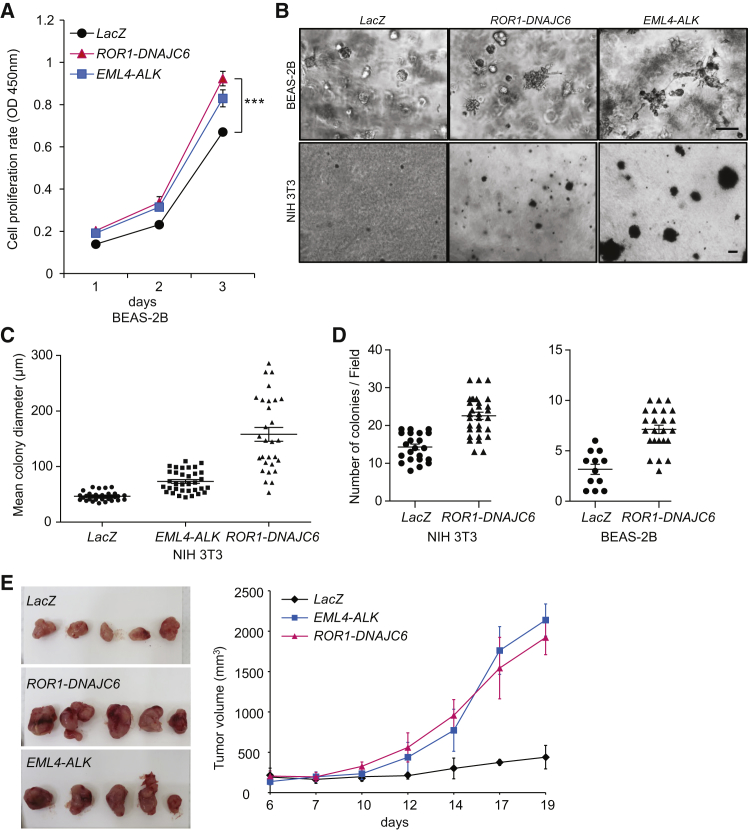
Our second candidate fusion for experimental validation was ROR1-DNAJC6 in lung adenocarcinoma. ROR1 is a receptor tyrosine kinase that modulates neurite growth in the CNS and might interact with the Wnt signaling pathway.47 It has not yet been reported as a cancer-driving fusion partner. Our experiments showed that the ROR1-DNAJC6 fusion can promote in vitro cell proliferation in BEAS-2B cells (non-cancerous human bronchial epithelium; Figures 4A and S7). It can also induce anchorage-independent cell growth (Figures 4B–4D) in both BEAS-2B and NIH 3T3 cells, and promote in vivo tumor formation in mice (Figure 4E) as well. Interestingly, the receptor tyrosine kinase ROR1 is the 5′ partner in this fusion, in contrast to most other fusions in which protein tyrosine kinases are activated as 3′ fusion partners. Another example with a protein tyrosine kinase on the 5′ side is the FGFR3-TACC3 fusion,17 in which FGFR3 loses its 3′ UTR and escapes from silencing to promote cellular growth.
Figure 4.
Functional Validation of ROR1-DNAJC6
(A) Growth rate of cells expressing ROR1-DNAJC6 fusion protein in BEAS-2B cells.
(B) BEAS-2B cells cultured in Matrigel after 7 days and NIH 3T3 cells cultured in soft agar after 14 days expressing ROR1-DNAJC6 fusion protein. Scale bar, 50 μm.
(C and D) Anchorage independent growth in soft agar. BEAS-2B or NIH 3T3 cells transformed with ROR-DNAJC6 were cultured in soft agar for 21 days.
(E) The transforming potential of ROR1-DNAJC6 fusion in vivo.
Our results showing the oncogenic potential of these two fusions demonstrate that previously unknown cancer-driving fusions can be detected from WES data, including some that are potential drug targets.
Massive Rearrangements
A small percentage of cancers might have one or more chromosomes massively rearranged, often with copy numbers oscillating between two or three states (chromothripsis),48, 49 segments amplified to many copies (chromoanasynthesis),6, 50 or chains of rearrangements (chromoplexy).51 These rearrangements have been proposed to form through shattering and rejoining of DNA fragments by non-homologous end joining,48 pulverization of chromosomes in the micronuclei,52 and template switching during DNA replication.6, 50 When we searched for chromosomes with statistically significant enrichment of SV breakpoints compared to the rest of the genome by using WES data (taking into account the gene densities on different chromosomes), we found a total of 196 chromosomes in 178 samples (3.8% of 4,609 samples; Table 1, Figure 5A, and Table S9). Our statistical threshold was based on the binomial test with a cutoff of p = 0.01 after the Bonferroni correction (see Material and Methods); given this stringent threshold, the number of samples we report with massively rearranged chromosomes is likely to be an underestimation.
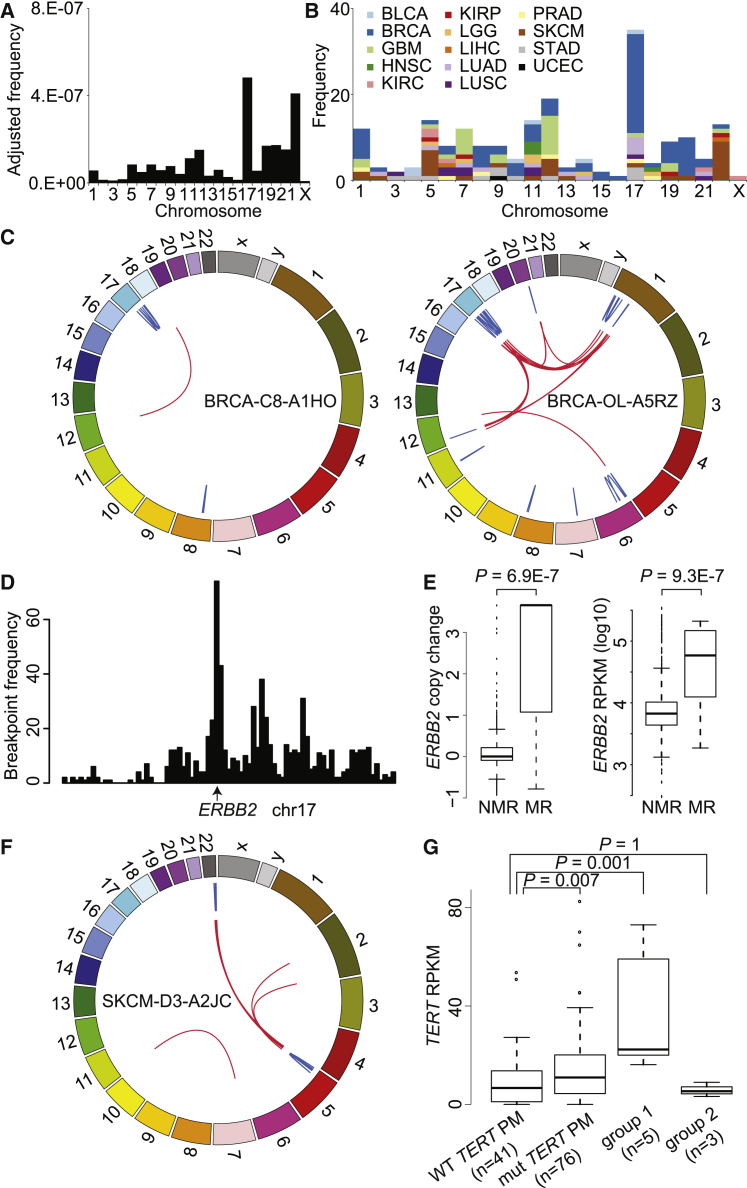
Figure 5.
Massive Rearrangements Are Often Associated with Upregulation of Oncogenes
(A) The frequencies of massively rearranged chromosomes normalized by the uniquely mappable size of CDS in each chromosome.
(B) The frequencies of massively rearranged chromosomes colored by tumor type.
(C) Examples of two breast cancers with massively rearranged chr17. Blue and red lines denote intra-chromosomal and inter-chromosomal rearrangements, respectively.
(D) The breakpoint distribution of massively rearranged chr17 of breast cancers with the peak at ERBB2.
(E) Association (Wilcoxon one-side rank test) of massive rearrangements with copy change and expression of ERBB2 in breast cancers. NMR, not massively rearranged; MR, massively rearranged. Error bars indicate SD.
(F) An example of massively rearranged chr22 that involves chr5 in melanoma.
(G) Association (Wilcoxon one-side rank test) of massively rearranged chr22 with TERT expression. Group 1 includes melanomas with massively rearranged chr22 that involves chr5. Group 2 includes melanomas with massively rearranged chr22 that does not involve chr5 with wild-type TERT promoter. Error bars indicate SD.
The frequency of massive rearrangements was highly variable across chromosomes (Figure 5A), with up to an ∼100-fold difference in the normalized frequencies (e.g., chr17 versus chrX). The highest frequencies were found in chromosomes 17 and 22, consistent with a previous study53 that found amplification breakpoints to be most frequent on chromosome 17. Different chromosomes were enriched for the SV clusters from different tumor types (Figure 5B). Chromosomes 7 and 12 are enriched for rearrangements in GBMs, and chromosome 22 is enriched for melanomas (MIM: 155600). On chromosome 17, 23 out of 35 occurrences are in breast cancers (examples in Figure 5C), and their breakpoints are highly abundant at the ERBB2 (MIM: 164870) locus (Figure 5D). Significantly higher copy numbers and expression at the ERBB2 locus suggest that the massively rearranged chromosome 17 is associated with upregulation of oncogene ERBB2 (Figure 5E). Those breast cancers with any massively rearranged chromosome, as well as those with massively rearranged chromosome 17 among the HER2+ subtype, have poorer prognosis with marginal statistical significance (p = 0.06 and 0.08, respectively; Figure S8).
There are co-occurrence patterns among the chromosomes that have massive rearrangements. For example, of the nine melanomas with chromosome 22 rearrangements, seven involve other chromosomes, including five involving chromosome 5 (Figures 5F and S9). Conversely, there are three melanomas with massively rearranged chromosome 5, and all of them co-occur with massively rearranged chromosome 22 (Table S9). In melanoma cases, it is known that ∼70% have TERT ([MIM: 187270] on chromosome 5) upregulated by promoter mutations.54, 55 We found that the individuals with massively rearranged chromosome 22 have significantly higher expression of TERT when chromosome 5 is also involved (Figure 5G). In GBM, CDK4 (MIM: 123829) is often amplified and expressed at a significantly higher amount in individuals with massively rearranged chromosome 12 (Figure S10A). On the other hand, the expression of EGFR (MIM: 131550) is not significantly different in individuals with massively rearranged chromosome 7 because the ones without massively rearranged chromosome 7 also have EGFR amplifications (Figure S10B). This is consistent with our previous study6 showing that most (14 out of 16) GBM samples have EGFR amplified and that some of the amplifications are achieved through very complex rearrangements. These results suggest that massive rearrangements are often associated with upregulation of oncogenes, which provides selective advantage to the cells, and these rearrangements are thus maintained in the genome.
Discussion
Here, we report the somatic genome rearrangements detected in the WES data for nearly 5,000 human cancer samples. WES data present challenges for SV identification, with ligation artifacts formed during exome capture and/or DNA amplification steps often manifesting as small tandem duplications. Many of the samples we excluded on the basis of quality were whole-genome amplified (WGA) samples, but other WGA samples did not suffer from the same problem. Although it is not possible to determine whether a specific tandem duplication is a true or artifactual one, their genome-wide distribution is strongly indicative of the sample data quality. The large number of samples with both WES and WGS data allowed us to set proper filtering thresholds.
SV identification based on WES data has much lower sensitivity than that based on WGS data. Therefore, it is not sensible to generate WES data to profile SVs or to replace WGS with WES. Our goal here was to re-analyze existing WES data, given that the number of samples with WES data is larger than that with WGS by an order of magnitude. In TCGA, for example, almost all of the samples were profiled by WES, whereas about 10% of the cases were profiled by WGS.
The number of exomes sequenced will continue to grow, especially as we search for somatic mutations with low variant allelic frequency. For instance, some somatic driver SNVs in cancer have been shown to occur in <5% of the cells. In neuroscience, there is now a great deal of interest in identifying somatic mutations in the brain to potentially explain neurological diseases such as epilepsy and developmental brain malformations.56 For such variants, high-coverage WES will be the preferred platform for most investigators until WGS at very high coverage becomes more affordable. Identification of even a fraction of the SVs in these datasets will be valuable. As we showed in one example (Figure 1D), somatic SV with low variant allele frequency cannot be detected by WGS as a result of its much lower coverage than WES. Importantly, the framework we described here is also applicable to germline rearrangements, and the number of germline exomes from individuals with a variety of disease phenotypes as well as from healthy individuals is already enormous.
As another application of exome-based SV analysis, we investigated massive rearrangements in our cohort and found that WES data can capture the presence of these events and their association with other factors. Because these events are rare (∼4% of the cases), their enrichment in specific chromosomes or tumor types, as well as their correlations with copy number and gene expression, became apparent with a large sample size (hundreds of samples per tumor type). Our finding that massive rearrangements are often associated with oncogene upregulation would not have been possible from WGS data. Copy-number profiles from microarray have been used to detect chromothripsis events on the basis of oscillating copy numbers on one or more chromosomes,48 including in our own work.57 However, inter-chromosomal events cannot be detected from array profiles, and the association between chromosome 22 massive rearrangements and upregulation of TERT could only be detected with WES data. Overall, our study of somatic genome rearrangements utilizing WES data provides insights into how gene fusions drive cancer and demonstrates the utility of re-analyzing existing data.
Acknowledgments
The results published here are in whole or in part based upon data generated by The Cancer Genome Atlas (TCGA) project established by the National Cancer Institute and the National Human Genome Research Institute. Information about the TCGA and the investigators and institutions who constitute the TCGA research network can be found at http://cancergenome.nih.gov. This work was supported by the Harvard Ludwig Center (P.J.P.), NIH grant U24CA144025 (R.K. and P.J.P.), National Research Foundation grants NRF-2013M3C8A1078501, NRF-2013R1A2A2A01068922 (Y.L.C.) and 2015R1C1A1A02037066 (M.L.) funded by the Korean Ministry of Science, Information and Communications Technology & Future Planning, Cancer Prevention and Research Institute of Texas grants RP140102 (H.L.) and RP120046 (K.L.S), and NIH grant U01CA168394 (K.L.S). This work made use of the Bionimbus Protected Data Cloud.
Published: May 5, 2016
Footnotes
Supplemental Data include ten figures and nine tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.03.017.
Contributor Information
Yoon-La Choi, Email: ylachoi@skku.edu.
Peter J. Park, Email: peter_park@harvard.edu.
Web Resources
Bionimbus, http://bionimbus.opensciencedatacloud.org
CGhub, https://cghub.ucsc.edu/
Genome Data Analysis Center (GDAC), http://gdac.broadinstitute.org
Meerkat software, http://compbio.med.harvard.edu/Meerkat
OMIM, http://www.omim.org/
The Cancer Genome Atlas Data Portal, https://tcga-data.nci.nih.gov/tcga/
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Abyzov A., Urban A.E., Snyder M., Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen K., Wallis J.W., McLellan M.D., Larson D.E., Kalicki J.M., Pohl C.S., McGrath S.D., Wendl M.C., Zhang Q., Locke D.P. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye K., Schulz M.H., Long Q., Apweiler R., Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iqbal Z., Caccamo M., Turner I., Flicek P., McVean G. De novo assembly and genotyping of variants using colored de Bruijn graphs. Nat. Genet. 2012;44:226–232. doi: 10.1038/ng.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J., Mullighan C.G., Easton J., Roberts S., Heatley S.L., Ma J., Rusch M.C., Chen K., Harris C.C., Ding L. CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nat. Methods. 2011;8:652–654. doi: 10.1038/nmeth.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang L., Luquette L.J., Gehlenborg N., Xi R., Haseley P.S., Hsieh C.H., Zhang C., Ren X., Protopopov A., Chin L. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell. 2013;153:919–929. doi: 10.1016/j.cell.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu W., O’Connor T.D., Jun G., Kang H.M., Abecasis G., Leal S.M., Gabriel S., Rieder M.J., Altshuler D., Shendure J., NHLBI Exome Sequencing Project Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chmielecki J., Crago A.M., Rosenberg M., O’Connor R., Walker S.R., Ambrogio L., Auclair D., McKenna A., Heinrich M.C., Frank D.A., Meyerson M. Whole-exome sequencing identifies a recurrent NAB2-STAT6 fusion in solitary fibrous tumors. Nat. Genet. 2013;45:131–132. doi: 10.1038/ng.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kandoth C., McLellan M.D., Vandin F., Ye K., Niu B., Lu C., Xie M., Zhang Q., McMichael J.F., Wyczalkowski M.A. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Misener S., Krawetz S.A., editors. Bioinformatics Methods And Protocols. Springer; 1999. pp. 365–386. [Google Scholar]
- 12.Wang K., Singh D., Zeng Z., Coleman S.J., Huang Y., Savich G.L., He X., Mieczkowski P., Grimm S.A., Perou C.M. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38 doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 15.Debnath J., Muthuswamy S.K., Brugge J.S. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 16.Drier Y., Lawrence M.S., Carter S.L., Stewart C., Gabriel S.B., Lander E.S., Meyerson M., Beroukhim R., Getz G. Somatic rearrangements across cancer reveal classes of samples with distinct patterns of DNA breakage and rearrangement-induced hypermutability. Genome Res. 2013;23:228–235. doi: 10.1101/gr.141382.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh D., Chan J.M., Zoppoli P., Niola F., Sullivan R., Castano A., Liu E.M., Reichel J., Porrati P., Pellegatta S. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–1235. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas Research Network Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawrence M.S., Stojanov P., Polak P., Kryukov G.V., Cibulskis K., Sivachenko A., Carter S.L., Stewart C., Mermel C.H., Roberts S.A. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beroukhim R., Getz G., Nghiemphu L., Barretina J., Hsueh T., Linhart D., Vivanco I., Lee J.C., Huang J.H., Alexander S. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc. Natl. Acad. Sci. USA. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research Network Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomlins S.A., Rhodes D.R., Perner S., Dhanasekaran S.M., Mehra R., Sun X.-W., Varambally S., Cao X., Tchinda J., Kuefer R. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 24.Wang L., Motoi T., Khanin R., Olshen A., Mertens F., Bridge J., Dal Cin P., Antonescu C.R., Singer S., Hameed M. Identification of a novel, recurrent HEY1-NCOA2 fusion in mesenchymal chondrosarcoma based on a genome-wide screen of exon-level expression data. Genes Chromosomes Cancer. 2012;51:127–139. doi: 10.1002/gcc.20937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giacomini C.P., Sun S., Varma S., Shain A.H., Giacomini M.M., Balagtas J., Sweeney R.T., Lai E., Del Vecchio C.A., Forster A.D. Breakpoint analysis of transcriptional and genomic profiles uncovers novel gene fusions spanning multiple human cancer types. PLoS Genet. 2013;9:e1003464. doi: 10.1371/journal.pgen.1003464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitelman F., Johansson B., Mertens F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 27.Yoshihara K., Wang Q., Torres-Garcia W., Zheng S., Vegesna R., Kim H., Verhaak R.G. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34:4845–4854. doi: 10.1038/onc.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stransky N., Cerami E., Schalm S., Kim J.L., Lengauer C. The landscape of kinase fusions in cancer. Nat. Commun. 2014;5:4846. doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipson D., Capelletti M., Yelensky R., Otto G., Parker A., Jarosz M., Curran J.A., Balasubramanian S., Bloom T., Brennan K.W. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat. Med. 2012;18:382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soda M., Choi Y.L., Enomoto M., Takada S., Yamashita Y., Ishikawa S., Fujiwara S., Watanabe H., Kurashina K., Hatanaka H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 31.Kohno T., Ichikawa H., Totoki Y., Yasuda K., Hiramoto M., Nammo T., Sakamoto H., Tsuta K., Furuta K., Shimada Y. KIF5B-RET fusions in lung adenocarcinoma. Nat. Med. 2012;18:375–377. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ju Y.S., Lee W.-C., Shin J.-Y., Lee S., Bleazard T., Won J.-K., Kim Y.T., Kim J.-I., Kang J.-H., Seo J.-S. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res. 2012;22:436–445. doi: 10.1101/gr.133645.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeuchi K., Soda M., Togashi Y., Suzuki R., Sakata S., Hatano S., Asaka R., Hamanaka W., Ninomiya H., Uehara H. RET, ROS1 and ALK fusions in lung cancer. Nat. Med. 2012;18:378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 34.Kim J., Lee Y., Cho H.-J., Lee Y.-E., An J., Cho G.-H., Ko Y.-H., Joo K.M., Nam D.-H. NTRK1 fusion in glioblastoma multiforme. PLoS ONE. 2014;9:e91940. doi: 10.1371/journal.pone.0091940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weber A., Huesken C., Bergmann E., Kiess W., Christiansen N.M., Christiansen H. Coexpression of insulin receptor-related receptor and insulin-like growth factor 1 receptor correlates with enhanced apoptosis and dedifferentiation in human neuroblastomas. Clin. Cancer Res. 2003;9:5683–5692. [PubMed] [Google Scholar]
- 36.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davies K.D., Doebele R.C. Molecular pathways: ROS1 fusion proteins in cancer. Clin. Cancer Res. 2013;19:4040–4045. doi: 10.1158/1078-0432.CCR-12-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaw A.T., Hsu P.P., Awad M.M., Engelman J.A. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat. Rev. Cancer. 2013;13:772–787. doi: 10.1038/nrc3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warmuth M., Kim S., Gu X.J., Xia G., Adrián F. Ba/F3 cells and their use in kinase drug discovery. Curr. Opin. Oncol. 2007;19:55–60. doi: 10.1097/CCO.0b013e328011a25f. [DOI] [PubMed] [Google Scholar]
- 40.Liang H., Cheung L.W., Li J., Ju Z., Yu S., Stemke-Hale K., Dogruluk T., Lu Y., Liu X., Gu C. Whole-exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer. Genome Res. 2012;22:2120–2129. doi: 10.1101/gr.137596.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grubbs E.G., Ng P.K., Bui J., Busaidy N.L., Chen K., Lee J.E., Lu X., Lu H., Meric-Bernstam F., Mills G.B. RET fusion as a novel driver of medullary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2015;100:788–793. doi: 10.1210/jc.2014-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daley G.Q., Van Etten R.A., Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- 43.Soule H.D., Maloney T.M., Wolman S.R., Peterson W.D., Jr., Brenz R., McGrath C.M., Russo J., Pauley R.J., Jones R.F., Brooks S.C. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- 44.Shin S.-I., Freedman V.H., Risser R., Pollack R. Tumorigenicity of virus-transformed cells in nude mice is correlated specifically with anchorage independent growth in vitro. Proc. Natl. Acad. Sci. USA. 1975;72:4435–4439. doi: 10.1073/pnas.72.11.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Isakoff S.J., Engelman J.A., Irie H.Y., Luo J., Brachmann S.M., Pearline R.V., Cantley L.C., Brugge J.S. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992–11000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 46.Shaw A.T., Ou S.-H.I., Bang Y.-J., Camidge D.R., Solomon B.J., Salgia R., Riely G.J., Varella-Garcia M., Shapiro G.I., Costa D.B. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N. Engl. J. Med. 2014;371:1963–1971. doi: 10.1056/NEJMoa1406766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Green J.L., Kuntz S.G., Sternberg P.W. Ror receptor tyrosine kinases: orphans no more. Trends Cell Biol. 2008;18:536–544. doi: 10.1016/j.tcb.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stephens P.J., Greenman C.D., Fu B., Yang F., Bignell G.R., Mudie L.J., Pleasance E.D., Lau K.W., Beare D., Stebbings L.A. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maher C.A., Wilson R.K. Chromothripsis and human disease: piecing together the shattering process. Cell. 2012;148:29–32. doi: 10.1016/j.cell.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu P., Erez A., Nagamani S.C., Dhar S.U., Kołodziejska K.E., Dharmadhikari A.V., Cooper M.L., Wiszniewska J., Zhang F., Withers M.A. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baca S.C., Prandi D., Lawrence M.S., Mosquera J.M., Romanel A., Drier Y., Park K., Kitabayashi N., MacDonald T.Y., Ghandi M. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crasta K., Ganem N.J., Dagher R., Lantermann A.B., Ivanova E.V., Pan Y., Nezi L., Protopopov A., Chowdhury D., Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zack T.I., Schumacher S.E., Carter S.L., Cherniack A.D., Saksena G., Tabak B., Lawrence M.S., Zhsng C.Z., Wala J., Mermel C.H. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang F.W., Hodis E., Xu M.J., Kryukov G.V., Chin L., Garraway L.A. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Horn S., Figl A., Rachakonda P.S., Fischer C., Sucker A., Gast A., Kadel S., Moll I., Nagore E., Hemminki K. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 56.Poduri A., Evrony G.D., Cai X., Walsh C.A. Somatic mutation, genomic variation, and neurological disease. Science. 2013;341:1237758. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim T.-M., Xi R., Luquette L.J., Park R.W., Johnson M.D., Park P.J. Functional genomic analysis of chromosomal aberrations in a compendium of 8000 cancer genomes. Genome Res. 2013;23:217–227. doi: 10.1101/gr.140301.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.