Abstract
G-quadruplex secondary structures are four-stranded globular nucleic acid structures form in the specific DNA and RNA G-rich sequences with biological significance such as human telomeres, oncogene-promoter regions, replication initiation sites, and 5′ and 3′-untranslated (UTR) regions. The non-canonical G-quadruplex secondary structures can readily form under physiologically relevant ionic conditions and are considered to be new molecular target for cancer therapeutics. This review discusses the essential progress in our lab related to the structures and functions of biologically relevant DNA G-quadruplexes in human gene promoters and telomeres, and the opportunities presented for the development of G-quadruplex-targeted small- molecule drugs.
Keywords: DNA G-quadruplexes, oncogene promoters, human telomeres, G-quadruplex-targeted small molecules, anticancer drug target
1 Introduction
DNA is widely recognized as a double-helical structure with a crucial role in genetic information storage. However, most recent results from the ENCODE project indicate that only ~3% of human genome is expressed in protein and that RNA and DNA may form non-canonical secondary structures that are functionally important [1]. One such example is the four-stranded G-quadruplex DNA secondary structures, which have been gaining considerable attention for their emerging role in biological systems. First observed in 1910 [2], the G-tetrad structure was not identified until 1962 [3]. DNA G-quadruplexes were first found to form in the single-stranded 3′ overhang of human telomeres [4–6]. More recently, DNA G-quadruplexes were found to form in the proximal promoter regions of human oncogenes to regulate gene transcription [7–10]. Most recently, DNA G-quadruplexes have been associated with replication initiation [11– 13]. In addition, RNA G-quadruplexes have been found to form in mRNA and, recently, in 5′- and 3′-UTRs [14–16].
Many proteins have been identified to interact with G-quadruplex structures; these proteins may bind and stabilize G-quadruplexes or unwind and destabilize G-quadruplexes [17–19]. Very recently, by using a G-quadruplex-specific antibody, G-quadruplex structures have been visualized in human cells at different sites on chromosomes other than telomeres and have been shown to increase in frequency in the presence of a G-quadruplex-interactive compound [20, 21]. Quarfloxin, the first G-quadruplex-interactive drug, has reached Phase II clinical trials for the treatment of cancer [22]. Thus, there is compelling evidence for the existence, biological significance, and potential drug-gability of G-quadruplexes [8, 17]. This review, which is associated with National Conference on Biophysical Chemistry (NCBPC) meeting presentations, will be focused on the progress in our lab related to the structures and functions of biologically relevant G-quadruplexes and the opportunities presented for the development of G-quadruplex-targeted small-molecule drugs.
2 Structural features of G-quadruplexes
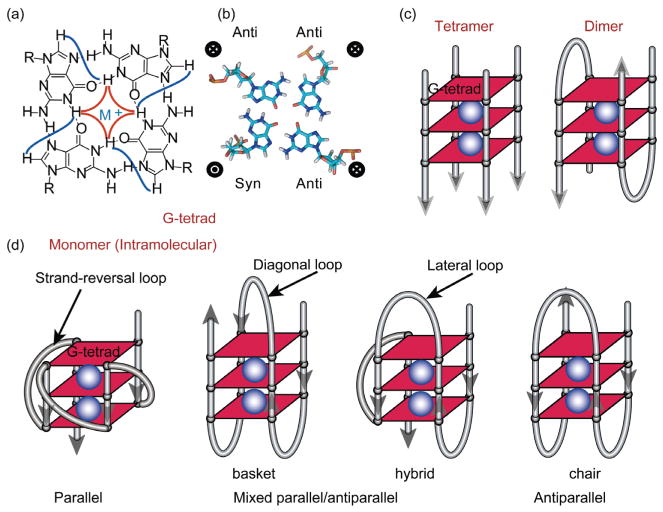
G-quadruplex structures are formed by stacked guanine tetrads (G-tetrads) (Figure 1). Within a G-tetrad, four guanine bases are arranged in a square plane with Hoogsteen hydrogen bonding, instead of the Watson-Crick hydrogen bonding of B-DNA (Figure 1(a, b)). G-quadruplex structures form readily in solution in the presence of either Na+ or K+ [23]. These physiologically relevant monovalent cations are required to stabilize G-quadruplex structures by positioning between the G-tetrad planes in coordination with the O6 atoms of the tetrad guanines (Figure 1). G-quadruplexes can be monomeric or multimeric (e.g., dimeric or tetrameric) (Figure 1(c, d)). Within the G-tetrad, guanine residues may adopt either syn or anti-glycosidic conformation [8] (Figure 1(b)). Adjacent DNA strands in a G-quadruplex can have the same (parallel) or opposite (anti-parallel) orientation (Figure 1(d)); guanines of parallel DNA strands will adopt the same, generally anti, conformation, whereas guanines of anti-parallel strands will adopt opposing conformations [8] (Figure 1(b)).
Figure 1.
(a) Schematic illustration of a G-tetrad, four guanine bases arranged in a square plane with Hoogsteen hydrogen bonding. The H1–H1 and H1–H8 connectivity that are notable in NOESY experiments are shown in red and blue respectively; (b) a G-tetrad structure. Guanines in a G-tetrad may adopt either syn or anti glycosidic conformation; the guanines from parallel G-strands adopt the same glycosidic conformation and the guanines from antiparallel G-strands adopt opposing glycosidic conformations; (c) schematic tetrameric and dimeric G-quadruplexes with three G-tetrads; (d) examples of monomeric (intramolecular) G-quadruplexes with different folding structures and loop conformations. Monovalent cations (K+ or Na+, shown as blue spheres), are required to stabilize G-quadruplexes by coordinating with the O6 atoms of the adjacent G-tetrad planes.
3 Monomeric G-quadruplexes
Most biologically relevant G-quadruplexes are monomeric, or intramolecular, G-quadruplexes [8, 17]. Generally, a monomeric G-quadruplex-forming DNA sequence contains at least four tracts of three or more consecutive guanines, with flanking segments at the two ends and loop residues intervening between the G-tracts. Intramolecular G-quadruplexes are globular structures that form naturally under physiological conditions and exhibit great diversity in their topologies and loop/flanking conformations, which depend on the DNA sequences. Different loop conformations have been observed, such as the strand-reversal/or propeller loop connecting adjacent parallel strands, the lateral/ or edgewise loop connecting adjacent anti-parallel strands, and the diagonal loop connecting anti-parallel strands on opposite sides of a tetrad (Figure 1(d)). A single DNA sequence may adopt different G-quadruplex folding topologies, as in the case of the human telomeric DNA, or it may form multiple structures or loop isomers, as in the case of most G-quadruplex-forming gene promoter sequences. While extensive structural study has elucidated some rules governing intra-molecular G-quadruplex folding, it is not always possible to accurately predict the quadruplex folding in a specific sequence. Thus, structural characterization is a necessity in the determination of G-quadruplex folding. Most intramolecular G-quadruplex structures in the public domain have been determined by NMR spectroscopy in solution.
4 Human telomeric G-quadruplexes
Telomeres are unique DNA-protein complexes that protect the chromosome ends from degradation; the structure and stability of human telomeres are important for cancer, aging, and overall genetic stability [24–26]. Telomerase activity is activated in over 80% of cancer cells to maintain the telomere lengths [27–29]. Human telomeric DNA consists of 5–30 kb tandem repeats of d(TTAGGG)n that end in a single-stranded 3′ overhang that is 35–600 bases long [30–32]. It has been shown to form G-quadruplexes that can be targeted by small molecular compounds to inhibit telomerase activity and disrupt telomere capping in cancer cells [33–35].
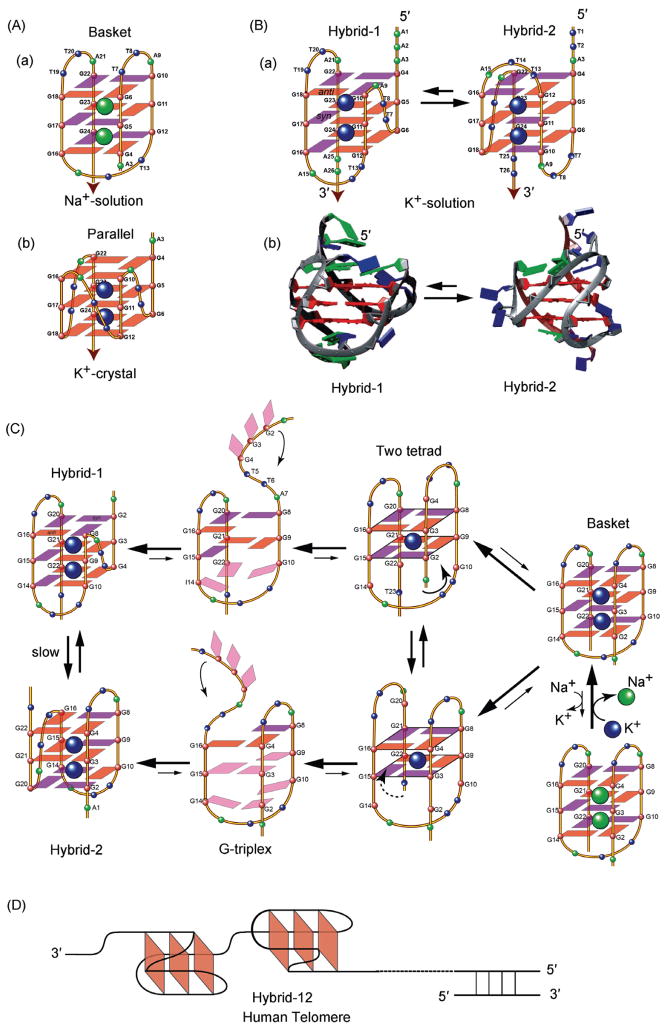
Human telomeric DNA G-quadruplexes have been found to be highly polymorphic in their structures [36]. A 22-mer telomeric DNA was found to form a basket-type structure in Na+ solution [37] (Figure 2(A-a)), and a parallel structure in the crystalline form in the presence of K+ [34] (Figure 2(A-b)). More recently, human telomeric DNA was shown to form two equilibrating hybrid-form G-quadruplexes in K+ solution [38–42] (Figure 2(B)). While both K+ structures adopt hybrid folding (i.e., with three parallel strands and one antiparallel strand connected by two lateral loops and one strand-reversal loop, Figure 2(B-a)) the orders of the loops are different and therefore the molecular structures are distinct, with unique capping and loop conformations (Figure 2(B-b)). Despite the low energy difference between the two equilibrating hybrid structures, interconversion between the two appears to be kinetically slow, suggesting the presence of a high-energy intermediate [40].
Figure 2.
(A) Folding topologies of the basket-type Na+-solution (a) and parallel-stranded K+-crystal (b) intramolecular G-quadruplexes formed by human telomeric sequence wtTel22. Red box = anti guanine, Magenta box = syn guanine; (B) the folding topologies (a) and molecular structures (b) of the two equilibrating hybrid-form human telomeric intramolecular G-quadruplexes, hybrid-1 and hybrid-2, in K+ solution; (C) a strand-reorientation model of the interconversion between different forms of human telomeric G-quadruplexes, likely through the two-tetrad and G-triplex form intermediates; (D) the hybrid structures can effectively form packed multimers in human telomeres.
Based on our NMR structural studies, we proposed a strand-reorientation model with a partially unfolded triplex- type and two-tetrad intermediates [39, 43] (Figure 2(C)). More recently, several studies have provided insights and experimental evidence into the intermediate steps of G-quadruplex folding, including the G-hairpin [44] and the G-triplex [45]. With the 5′ and 3′ ends positioned on opposite sides of the G-quadruplex, the hybrid structures can effectively form packed multimers at the telomere end [39, 40], a conformation that has been confirmed by computer modeling study [46] (Figure 2(D)). All human telomeric G-quadruplexes appear to have small energy differences relative to each other [40, 46]. The structure polymorphism appears to be an intrinsic property of the human telomeric DNA sequence, particularly the TTA loop sequence [36]. As the human telomeric DNA sequence contains the same tandem repeats, the highly conserved telomeric sequence in higher eukaryotes may be naturally selected for its potential to form multiple G-quadruplexes so that different structures may be specifically recognized by different proteins to control biology. This recognition may present a potential opportunity for small molecule targeting.
5 Promoter G-quadruplexes
The potential for G-quadruplex formation in promoter regions is largely concentrated in genes associated with cell growth and proliferation [7]. Computational analyses [47– 51] showed the clustering of G-quadruplex-forming sequences in close proximity (within 1 kb) to the transcriptional start site. These oncogene promoters are typically TATA-less with G-rich regions in their proximal promoters, such as c-MYC [10, 52], VEGF (vascular endothelial growth factor) [53], BCL-2 (B-cell lymphoma 2) [54, 55], and PDGF-R-β [56], as well as HIF-1α (hypoxia-inducible factor 1α) [57], KRAS [58], c-KIT [59, 60], and RET [61]. While telomeric G-quadruplexes form in the single-stranded 3′-overhang of telomeres, promoter G-quadruplexes are formed in regions of double-stranded DNA. Using the c-MYC gene promoter, studies have demonstrated in vivo that active transcription induces negative superhelicity behind the transcriptional machinery [62–64]. This negative superhelicity leads to the intermediate single-stranded DNA, which can spontaneously fold into the more stable G-quadruplex structures [65].
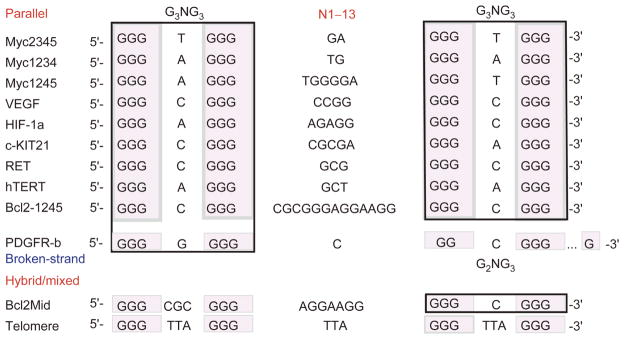
Unlike telomeric DNA, the promoter G-quadruplex- forming sequences are significantly more diverse and often contain more than four tracts of guanine [7, 8, 17]. Consequently, a given sequence can form multiple G-quadruplexes through utilizing varying combinations of G-tracts or different loop isomers through utilizing varying guanines on one G-tract. Three-tetrad G-quadruplexes are most common. A notable feature in these promoter sequences is the presence of the G3NG3 motif, which readily contributes to formation of a robust parallel-stranded structure motif with a single-nucleotide strand-reversal loop (Figure 3). This G3NG3 motif is so widely prevalent in promoter G-quadruplexes that it may have been naturally selected as a basis for promoter G-quadruplex formation [8, 17].
Figure 3.
Comparison of G-quadruplex-forming sequences in selected gene promoters. The human telomeric sequence is also shown. The types of the G-quadruplexes are indicated. The tetrad-guanines are shaded, and the G3NG3 motifs are boxed.
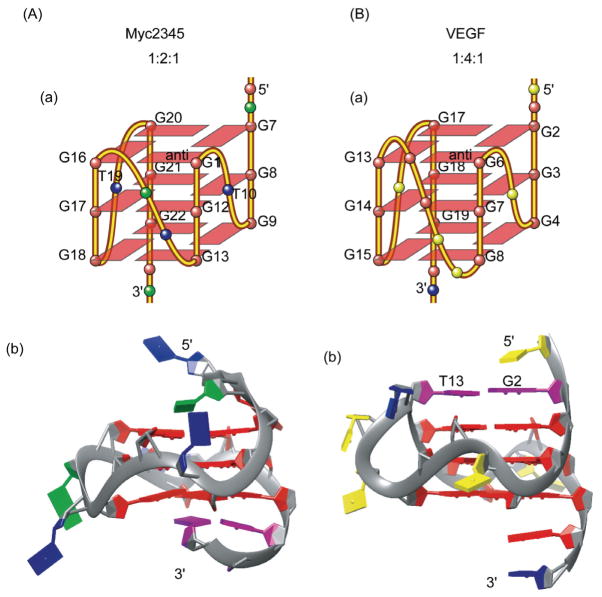
The first example of the G3NG3 structural motif is the major G-quadruplex structure formed in the c-MYC promoter, (1:2:1 are the lengths of the three loops) [66] (Figure 4(A)). We showed that the single-nucleotide loop is highly favored for the robust parallel-stranded structural motif G3NG3 because of the right-handed twist of the adjacent G-strands (Figure 4(A-b)). The G-quadruplex-forming region of the c-MYC promoter is a 27-nucleotide nuclease hypersensitive element (NHE) III1 that has been shown to control about 85% of the c-MYC transcriptional activity [67, 68]. This sequence contains five consecutive runs of guanine. Mutational analysis in conjunction with DMS footprinting and luciferase reporter assays have indicated that the major G-quadruplex formed in K+ solution involves four consecutive 3G-runs that adopt parallel folding (Myc2345), with the major loop isomer being 1:2:1 [10, 69, 70] (Figures 3, 4(A)). While the c-MYC NHE III1 is the first and most extensively studied gene promoter for the G-quadruplex formation, we were very lucky in that it appears to be the simplest, containing only five G-runs with all the G-quadruplexes adopting parallel folding in K+ solution [66, 69–72]. Indeed, the G3NG3 parallel-stranded structural motif was found in all of the c-MYC G-quadruplexes and loop isomers. Structural, thermodynamic and kinetic analysis showed different stabilities associated with loop sizes, loop arrangements, and loop compositions of parallel structures [71, 72].
Figure 4.
The folding topologies (a) and molecular structures (b) of the major G-quadruplexes formed in the human c-MYC gene promoter (A) and human VEGF gene promoter (B) in K+ solution. The capping structures are shown in purple. Guanine = red, adenine = green, thymine = blue, cytosine = yellow.
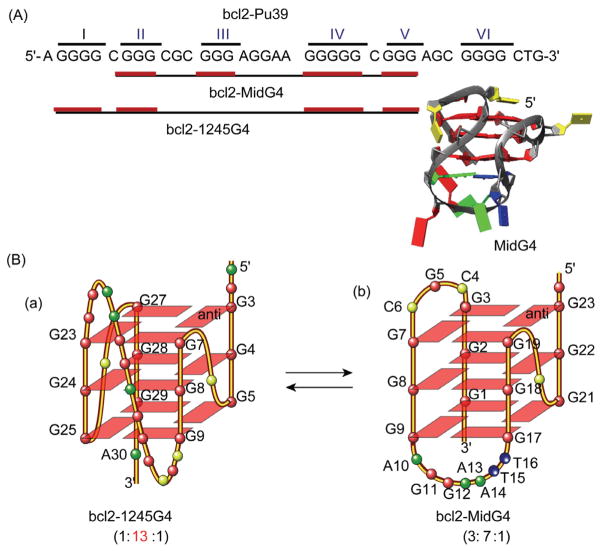
Since the determination of the c-MYC G-quadruplex, parallel-stranded G-quadruplex structures have been found to be common in the human promoter sequences. Notably, most of these parallel structures contain 3 G-tetrads with 1-nt (nt = nucleotide) first and third loops, and a variable-length middle loop (Figure 3). For example, the major G-quadruplex formed in the VEGF promoter (1:4:1) also contains 1-nt first and third loops, but a 4-nt middle loop (Figure 4(B-a)). Interestingly, the 4-nt middle loop of the VEGF G-quadruplex stretches over the 5′ tetrad to form a unique capping structure with the flanking segment [73] (Figure 4(B-b)). In contrast, the 2-nt middle loop of the major MYC-quadruplex (1:2:1) stays in the groove with the capping structures formed solely by the two flanking segments [66] (Figure 4(A-b)). More recently, we found the major G-quadruplex (bcl2-1245G4) formed in the BCL-2 proximal promoter adopts a parallel structure (1:13:1) with a 13-nt middle loop [74] (Figure 5(A, B-a)). This is unexpected because 7 nt has been considered the maximum stable loop length and is used in all available quadruplex-predicting software. It thus appears that, by having two 1-nt loops, a stable parallel G-quadruplex can contain a rather extended middle loop. Therefore, while parallel structures are common to the promoter G-quadruplexes, our studies indicate that each G-quadruplex is likely to adopt unique capping and loop structures by its specific variable middle loop and flanking segments.
Figure 5.
Two interchangeable G-quadruplexes formed in the overlapping region of the BCL-2 gene promoter. (A) The G-quadruplex-forming BCL-2 gene promoter sequence bcl2-Pu39. The regions (black line) and G-runs (red line) for the formation of the two distinct G-quadruplexes, bcl2-MidG4 and bcl2-1245G4, respectively, are shown; (B) the folding topologies of the two interchangeable BCL-2 promoter G-quadruplexes.
Further variation of the 1-nt parallel-strand motif is exemplified by the major G-quadruplex formed in the PDGFR-β (platelet-derived growth-factor receptor β) promoter. Instead of using four runs of three or more consecutive guanines, this 3 G-tetrad quadruplex features a “broken” G-strand of guanines from two different G-runs to form a primarily parallel-stranded structure with three 1-nt strand-reversal loops and one additional lateral loop [75] (Figure 6). The formation of this novel “broken-strand” PDGFR-β G-quadruplex appears to maximize the number of 1-nt loops, again demonstrating the preference for 1-nt loops in the parallel-stranded structural motif, which may more accurately be described as GiNGj. These variations in parallel G-quadruplexes give rise to different overall structure properties, which may be specifically recognized by proteins or small-molecule ligands.
Figure 6.
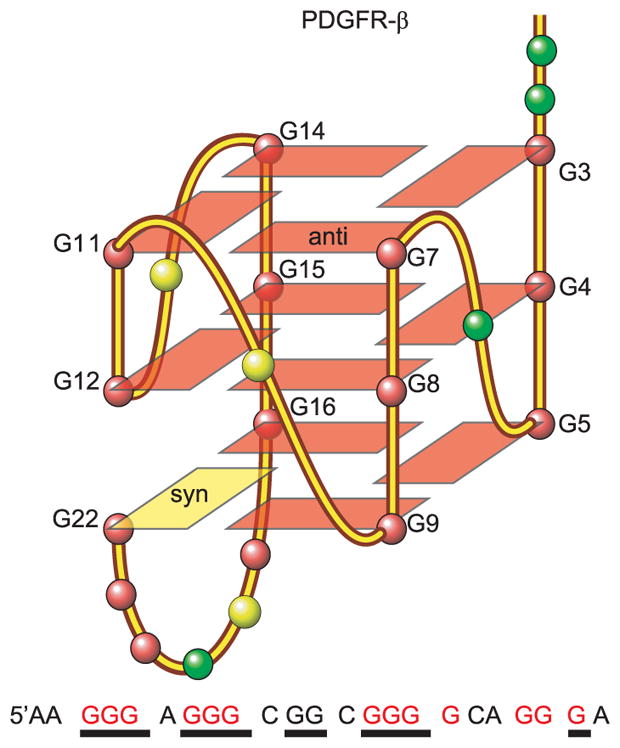
The sequence and the folding topology of the major G-quadruplex formed in the PDGFR-β gene promoter, which forms a novel “broken-strand” structure. The guanines involved in the tetrad formation are underlined, whereas the four runs of three or more consecutive guanines are colored red.
In addition, certain promoter sequences may form multiple stable G-quadruplexes in equilibrium. For example, the BCL2 proximal promoter contains a 39-nucleotide G-rich region with 6 G-runs separated by one or more bases (bcl2-Pu39, Figure 5(A)). This sequence has been shown to form two stable G-quadruplexes, a hybrid-type structure with a G3NG3 motif and two lateral loops [55, 76] (bcl2-MidG4, Figure 5(B-b)) in addition to the 1:13:1 parallel structure which spans 5 G-runs (bcl2-1245G4, Figure 5(B-a)) [74]. The two structures have similar stability. Thermodynamically the parallel 1245G4 structure is slightly more stable than the MidG4; however, the hybrid-type MidG4 structure could be kinetically more favored because it forms on the four consecutive G-runs and thus has shorter loop-lengths. The presence of two distinct interchangeable G-quadruplexes in the overlapping region of the BCL-2 promoter is intriguing, and suggests a novel mechanism for gene transcriptional regulation by different proteins recognizing different G-quadruplex structures. In addition, two interchangeable structures may be recognized by different small molecules for gene modulation.
6 Small-molecule targeting of G-quadruplexes
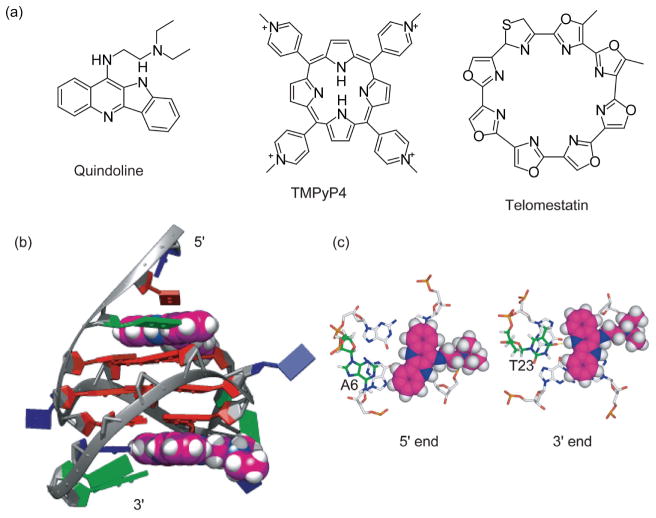
DNA G-quadruplexes, in particular those formed in human telomere and oncogene promoters, have become attractive cancer-specific molecular targets for anticancer therapeutics [17]. Targeting G-quadruplexes in promoters provides a potential means to modulate oncogene transcription [8, 9]. The stabilization of telomeric G-quadruplexes in cancer cells often leads to apoptosis [34, 35]. Elevated interest has been seen in the development of G-quadruplex-interactive small molecules [77]. NMR spectroscopy is a powerful tool for studying small molecule interactions with G-quadruplexes [78]. X-ray crystallography and biophysical methods such as CD, UV, and fluorescence spectroscopies are also commonly utilized in studying quadruplex-ligand interactions [79, 80]. Early dogma on the optimal G-quadruplex-interactive compounds has focused on compounds with large, planar, and symmetric cyclic rings, such as TMPyP4 (tetra- (N-methyl-4-pyridyl)porphyrin) and telomestatin [81, 82] (Figure 7(a)), because they maximize stacking interactions with the external G-tetrad. However, these drugs are found to have low specificity for intramolecular G-quadruplexes [78].
Figure 7.
(a) Structures of G-quadruplex-interactive small molecules; (b) a representative model of the NMR solution structure of the 2:1 complex of quindoline and c-MYC G-quadruplex. The quindoline molecules are shown in space-filling model in magenta. Guanine = red, adenine = green, thymine = blue; (c) axial view of the two drug-induced binding pockets at the 5′-end and the 3′-end.
Our recent NMR solution structure of the 2:1 complex of quindoline and c-MYC G-quadruplex provides some important insights: an asymmetric compound with a smaller stacking moiety and appropriate functional groups is preferred for specific binding to the intramolecular G-quadruplex [83]. Quindoline is a derivative of the natural product cryptolepine, with a crescent-shaped ring system and a diethylamino side chain (Figure 7(a), that has been shown to stabilize the c-MYC promoter G-quadruplex and lower c-MYC protein levels [84]. We have determined the NMR solution structure of a 2:1 complex of quindoline and c-MYC G-quadruplex with one quindoline bound at each end of the quadruplex, the first drug complex structure of a biologically relevant intramolecular promoter quadruplex [83] (Figure 7(b)). This NMR structure shows an unexpected drug-induced repositioning of the flanking sequences at both ends of DNA, namely, the “induced intercalated triad pocket” recognition (Figure 7(b)), in a manner somewhat analogous to a reorganized ligand-induced fit observed in riboswitches [85]. While both 3′ and 5′ complexes show similar overall features, there are identifiable differences, observable only in solution, to emphasize the importance of both stacking and electronic interactions. The 5′ complex involves more hydrophobic and stacking interactions, whereas the 3′ complex at the more hydrophilic 3′-surface involves a H-bonding interaction that is dependent on salt concentration (Figure 7(c)) [83]. The complex structures demonstrate the importance of the shape of the ligand as well as the two flanking bases in determining binding specificity. Unlike the previously recognized paradigm, our results indicate that asymmetric compounds containing a smaller stacking moiety, in particular the crescent-shaped moiety, with appropriate functional groups, are more likely to bind in a specific manner to an intramolecular G-quadruplex. Therefore, the molecular mechanism for G-quadruplex recognition by small molecules may be divided into two steps. The first step involves the induced intercalated triad pocket recognition (by the core of ligand) and second involves the groove/loop interactions (by different functional groups), which could each be exploited in selective recognition of small-molecule drugs.
7 Future directions
Our understanding of G-quadruplexes has expanded from that of a laboratory curiosity to biologically relevant and validated targets for cancer therapeutics. Intramolecular G-quadruplexes form naturally under physiological conditions and are stabilized by monovalent cations present in cells. Depending on their DNA sequence, these structures exhibit great diversity in their structures. Quarfloxin, the first-in-class G-quadruplex-interactive drug, illustrates the potential for quadruplex-targeting drugs to be well tolerated and highly efficacious [22, 86]. We are currently working on structural studies and rational design of different small molecules that bind specifically to intramolecular G-quadruplexes. In addition, proteins have been found to interact with the c-MYC promoter G-quadruplex and be involved in c-MYC gene transcription. Nucleolin specifically binds and stabilizes the c-MYC G-quadruplex and functions as a transcription repressor [87], whereas NM23-H2 binds and unfolds the c-MYC G-quadruplex and functions as a transcription activator [88, 89]. We are studying the interactions of these proteins with the c-MYC G-quadruplex to exploit their cellular functions and the potential for small molecules targeting.
References
- 1.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bang I. Untersuchungen über die guanylsäure. Biochemische Zeitschrif. 1910;26:293–311. [Google Scholar]
- 3.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc Natl Acad Sci USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sundquist WI, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 5.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 6.Henderson E, Hardin CC, Walk SK, Tinoco I, Blackburn EH. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell. 1987;51:899–908. doi: 10.1016/0092-8674(87)90577-0. [DOI] [PubMed] [Google Scholar]
- 7.Qin Y, Hurley LH. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie. 2008;90:1149–1171. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Yang DZ. Sequence, stability, and structure of G-quadruplexes and their interactions with drugs. Curr Protoc Nucleic Acid Chem. 2012;50:17.15.11–17.15.17. doi: 10.1002/0471142700.nc1705s50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat Rev Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao J, Bacolla A, Wang G, Vasquez KM. Non-b DNA structure-induced genetic instability and evolution. Cell Mol Life Sci. 2010;67:43–62. doi: 10.1007/s00018-009-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paeschke K, Capra JA, Zakian VA. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Besnard E, Babled A, Lapasset L, Milhavet O, Parrinello H, Dantec C, Marin JM, Lemaitre JM. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol. 2012;19:837– 844. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- 14.Christiansen J, Kofod M, Nielsen FC. A guanosine quadruplex and two stable hairpins flank a major cleavage site in insulin-like growth factor II MRNA. Nucleic Acids Res. 1994;22:5709–5716. doi: 10.1093/nar/22.25.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bugaut A, Balasubramanian S. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40:4727– 4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beaudoin JD, Perreault JP. Exploring mrna 3′-UTR G-quadruplexes: evidence of roles in both alternative polyadenylation and mrna shortening. Nucleic Acids Res. 2013;41:5898–5911. doi: 10.1093/nar/gkt265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang D, Okamoto K. Structural insights into G-quadruplexes: towards new anticancer drugs. Future Med Chem. 2010;2:619–646. doi: 10.4155/fmc.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fry M. Tetraplex DNA and its interacting proteins. Front Biosci. 2007;12:4336–4351. doi: 10.2741/2391. [DOI] [PubMed] [Google Scholar]
- 19.Oganesian L, Bryan TM. Physiological relevance of telomeric G-quadruplex formation: a potential drug target. Bioessays. 2007;29:155–165. doi: 10.1002/bies.20523. [DOI] [PubMed] [Google Scholar]
- 20.Siddiqui-Jain A, Hurley LH. DNA structure: visualizing the quadruplex. Nat Chem. 2013;5:153–155. doi: 10.1038/nchem.1587. [DOI] [PubMed] [Google Scholar]
- 21.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drygin D, Siddiqui-Jain A, O’Brien S, Schwaebe M, Lin A, Bliesath J, Ho CB, Proffitt C, Trent K, Whitten JP, Lim JKC, von Hoff D, Anderes K, Rice WG. Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Cancer Res. 2009;69:7653–7661. doi: 10.1158/0008-5472.CAN-09-1304. [DOI] [PubMed] [Google Scholar]
- 23.Sen D, Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 24.Blackburn EH. Telomere states and cell fates. Nature. 2000;408:53–56. doi: 10.1038/35040500. [DOI] [PubMed] [Google Scholar]
- 25.van Steensel B, Smogorzewska A, de Lange T. Trf2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 26.Hackett JA, Feldser DM, Greider CW. Telomere dysfunction increases mutation rate and genomic instability. Cell. 2001;106:275– 286. doi: 10.1016/s0092-8674(01)00457-3. [DOI] [PubMed] [Google Scholar]
- 27.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 28.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 29.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 30.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sfeir AJ, Chai WH, Shay JW, Wright WE. Telomere-end processing: the terminal nucleotides of human chromosomes. Molecular Cell. 2005;18:131–138. doi: 10.1016/j.molcel.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 33.Sun D, Thompson B, Cathers BE, Salazar M, Kerwin SM, Trent JO, Jenkins TC, Neidle S, Hurley LH. Inhibition of human telomerase by a G-quadruplex-interactive compound. J Med Chem. 1997;40:2113– 2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 34.Neidle S, Parkinson G. Telomere maintenance as a target for anti-cancer drug discovery. Nat Rev Drug Discov. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- 35.Punchihewa C, Yang DZ. Therapeutic targets and drugs II: G-quadruplex and G-quadruplex inhibitors. In: Hiyama K, editor. Telomeres and Telomerase in Cancer. New York: Humana Press; 2009. [Google Scholar]
- 36.Dai J, Carver M, Yang D. Polymorphism of human telomeric quadruplex structures. Biochimie. 2008;90:1172–1183. doi: 10.1016/j.biochi.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 38.Phan AT, Luu KN, Patel DJ. Different loop arrangements of intra-molecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006;34:5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dai J, Carver M, Punchihewa C, Jones RA, Yang DZ. Structure of the hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35:4927–4940. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dai J, Punchihewa C, Ambrus A, Chen D, Jones RA, Yang DZ. Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation. Nucleic Acids Res. 2007;35:2440–2450. doi: 10.1093/nar/gkm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu Y, Noguchi Y, Sugiyama H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg Med Chem. 2006;14:5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Z, Dai J, Veliath E, Jones RA, Yang DZ. Structure of a two-G-tetrad intramolecular G-quadruplex formed by a variant human telomeric sequence in K+ solution: insights into the interconversion of human telomeric G-quadruplex structures. Nucleic Acids Res. 2010;38:1009–1021. doi: 10.1093/nar/gkp1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rajendran A, Endo M, Hidaka K, Sugiyama H. Direct and single-molecule visualization of the solution-state structures of G-hairpin and G-triplex intermediates. Angew Chem Int Ed. 2014;53:4107– 4112. doi: 10.1002/anie.201308903. [DOI] [PubMed] [Google Scholar]
- 45.Mashimo T, Yagi H, Sannohe Y, Rajendran A, Sugiyama H. Folding pathways of human telomeric type-1 and type-2 G-quadruplex structures. J Am Chem Soc. 2010;132:14910–14918. doi: 10.1021/ja105806u. [DOI] [PubMed] [Google Scholar]
- 46.Petraccone L, Spink C, Trent JO, Garbett NC, Mekmaysy CS, Giancola C, Chaires JB. Structure and stability of higher-order human telomeric quadruplexes. J Am Chem Soc. 2011;133:20951– 20961. doi: 10.1021/ja209192a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verma A, Halder K, Halder R, Yadav VK, Rawal P, Thakur RK, Mohd F, Sharma A, Chowdhury S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J Med Chem. 2008;51:5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- 50.Verma A, Yadav VK, Basundra R, Kumar A, Chowdhury S. Evidence of genome-wide G4 DNA-mediated gene expression in human cancer cells. Nucleic Acids Res. 2009;37:4194–4204. doi: 10.1093/nar/gkn1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rawal P, Kummarasetti VBR, Ravindran J, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brooks TA, Hurley LH. The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nat Rev Cancer. 2009;9:849–861. doi: 10.1038/nrc2733. [DOI] [PubMed] [Google Scholar]
- 53.Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dexheimer TS, Sun D, Hurley LH. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J Am Chem Soc. 2006;128:5404– 5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dai J, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, Yang DZ. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J Am Chem Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qin Y, Fortin JS, Tye D, Gleason-Guzman M, Brooks TA, Hurley LH. Molecular cloning of the human platelet-derived growth factor receptor beta (PDGFR-β) promoter and drug targeting of the G-quadruplex-forming region to repress PDGFR-β expression. Biochemistry. 2010;49:4208–4219. doi: 10.1021/bi100330w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Armond R, Wood S, Sun D, Hurley LH, Ebbinghaus SW. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1alpha promoter. Biochemistry. 2005;44:16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- 58.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J Am Chem Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernando H, Reszka AP, Huppert J, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo K, Pourpak A, Beetz-Rogers K, Gokhale V, Sun D, Hurley LH. Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the ret oncogene. J Am Chem Soc. 2007;129:10220–10228. doi: 10.1021/ja072185g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 63.Kouzine F, Levens D. Supercoil-driven DNA structures regulate genetic transactions. Front Biosci. 2007;12:4409–4423. doi: 10.2741/2398. [DOI] [PubMed] [Google Scholar]
- 64.Kouzine F, Gupta A, Baranello L, Wojtowicz D, Ben-Aissa K, Liu J, Przytycka TM, Levens D. Transcription-dependent dynamic super-coiling is a short-range genomic force. Nat Struct Mol Biol. 2013;20:396–403. doi: 10.1038/nsmb.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun D, Hurley LH. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-MYC promoter: implications for drug targeting and control of gene expression. J Med Chem. 2009;52:2863–2874. doi: 10.1021/jm900055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 67.Berberich SJ, Postel EH. PuF/NM23-H2/NDPK-B transactivates a human c-MYC promoter-cat gene via a functional nuclease hypersensitive element. Oncogene. 1995;10:2343–2347. [PubMed] [Google Scholar]
- 68.Tomonaga T, Levens D. Activating transcription from single stranded DNA. Proc Natl Acad Sci USA. 1996;93:5830–5835. doi: 10.1073/pnas.93.12.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seenisamy J, Rezler EM, Powell TJ, Tye D, Gokhale V, Joshi CS, Siddiqui-Jain A, Hurley LH. The dynamic character of the G-quadruplex element in the c-myc promoter and modification by TMPYP4. J Am Chem Soc. 2004;126:8702–8709. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- 70.Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-MYC promoter. J Am Chem Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hatzakis E, Okamoto K, Yang D. Thermodynamic stability and folding kinetics of the major G-quadruplex and its loop isomers formed in the nuclease hypersensitive element in the human c-MYC promoter: effect of loops and flanking segments on the stability of parallel- stranded intramolecular G-quadruplexes. Biochemistry. 2010;49:9152–9160. doi: 10.1021/bi100946g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mathad RI, Hatzakis E, Dai J, Yang DZ. c-MYC promoter G-quadruplex formed at the 5′-end of nhe iii1 element: insights into biological relevance and parallel-stranded G-quadruplex stability. Nucleic Acids Res. 2011;39:9023–9033. doi: 10.1093/nar/gkr612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Agrawal P, Hatzakis E, Guo K, Carver M, Yang D. Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: insights into loop interactions of the parallel G-quadruplexes. Nucleic Acids Res. 2013;41:10584–10592. doi: 10.1093/nar/gkt784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Agrawal P, Lin C, Mathad RI, Carver M, Yang D. The major G-quadruplex formed in the human BCL-2 proximal promoter adopts a parallel structure with a 13-nt loop in K+ solution. J Am Chem Soc. 2014;136:1750–1753. doi: 10.1021/ja4118945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen Y, Agrawal P, Brown RV, Hatzakis E, Hurley L, Yang D. The major G-quadruplex formed in the human platelet-derived growth factor receptor beta promoter adopts a novel broken-strand structure in K+ solution. J Am Chem Soc. 2012;134:13220–13223. doi: 10.1021/ja305764d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dai J, Chen D, Jones RA, Hurley LH, Yang DZ. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006;34:5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Balasubramanian S, Neidle S. G-quadruplex nucleic acids as therapeutic targets. Curr Opin Chem Biol. 2009;13:345–353. doi: 10.1016/j.cbpa.2009.04.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mathad RI, Yang D. G-quadruplex structures and G-quadruplex-interactive compounds. Methods Mol Biol. 2011;735:77–96. doi: 10.1007/978-1-61779-092-8_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Campbell NH, Parkinson GN. Crystallographic studies of quadruplex nucleic acids. Methods. 2007;43:252–263. doi: 10.1016/j.ymeth.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 80.Vorlickova M, Kejnovska I, Sagi J, Renciuk D, Bednarova K, Motlova J, Kypr J. Circular dichroism and guanine quadruplexes. Methods. 2012;57:64–75. doi: 10.1016/j.ymeth.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 81.Han FXG, Wheelhouse RT, Hurley LH. Interactions of TMPYP4 and TMPYP2 with quadruplex DNA. Structural basis for the differential effects on telomerase inhibition. J Am Chem Soc. 1999;121:3561– 3570. [Google Scholar]
- 82.Shinya K, Wierzba K, Matsuo K, Ohtani T, Yamada Y, Furihata K, Hayakawa Y, Seto H. Telomestatin, a novel telomerase inhibitor from streptomyces anulatus. J Am Chem Soc. 2001;123:1262–1263. doi: 10.1021/ja005780q. [DOI] [PubMed] [Google Scholar]
- 83.Dai J, Carver M, Hurley LH, Yang D. Solution structure of a 2:1 quindoline-c-MYC G-quadruplex: insights into G-quadruplex-interactive small molecule drug design. J Am Chem Soc. 2011;133:17673– 17680. doi: 10.1021/ja205646q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ou TM, Lu YJ, Zhang C, Huang ZS, Wang XD, Tan JH, Chen Y, Ma DL, Wong KY, Tang JCO, Chan ASC, Gu LQ. Stabilization of G-quadruplex DNA and down-regulation of oncogene c-MYC by quindoline derivatives. J Med Chem. 2007;50:1465–1474. doi: 10.1021/jm0610088. [DOI] [PubMed] [Google Scholar]
- 85.Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004;5:451–63. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 86.Brooks TA, Hurley LH. Targeting myc expression through G-quadruplexes. Genes Cancer. 2010;1:641–649. doi: 10.1177/1947601910377493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gonzalez V, Guo K, Hurley L, Sun D. Identification and characterization of nucleolin as a c- MYC G-quadruplex-binding protein. J Biol Chem. 2009;284:23622–23635. doi: 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dexheimer TS, Carey SS, Zuohe S, Gokhale VM, Hu X, Murata LB, Maes EM, Weichsel A, Sun D, Meuillet EJ, Montfort WR, Hurley LH. NM23-H2 may play an indirect role in transcriptional activation of c-myc gene expression but does not cleave the nuclease hypersensitive element III(1) Mol Cancer Ther. 2009;8:1363–1377. doi: 10.1158/1535-7163.MCT-08-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thakur RK, Kumar P, Halder K, Verma A, Kar A, Parent JL, Basundra R, Kumar A, Chowdhury S. Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-myc promoter nuclease hypersensitive element induces c-MYC expression. Nucleic Acids Res. 2009;37:172–183. doi: 10.1093/nar/gkn919. [DOI] [PMC free article] [PubMed] [Google Scholar]