Abstract
Peritoneal dialysis (PD) is the most readily feasible home-dialysis method for renal replacement therapy. However, repeated use of PD can lead to induction of mesothelial/epithelial–mesenchymal transition (MMT/EMT) and fibrosis, eventually leading to ultrafiltration failure and discontinuation of PD. MicroRNA-129-5p (miR-129-5p) is believed to be a potent downstream inhibitor of TGF-β1 in renal fibrosis, but the effect of miR-129-5p on MMT/EMT relevant to PD is unknown. In this study, as determined by microRNA array analysis and confirmed by northern blot analysis and real-time PCR, we demonstrate that miRNA-129-5p is decreased in mesothelial cells isolated from effluent of patients having PD for more than 6 months extending to several years compared with those who have undergone PD for less than 6 months. The decreased expression of miR-129-5p was accompanied with alterations in EMT-related genes and the expression of respective proteins in vivo. In addition, in in vitro studies we noted that the expression of E-cadherin and claudin-1 were significantly reduced with increased cell migration in HMrSV5, a human peritoneal mesothelial cell line (HPMC), treated with TGF-β1, whereas expression of vimentin, fibronectin and transcription factors SIP1 and SOX4 increased significantly, as assessed by real-time PCR, western blot analysis and immunofluorescence microscopy. Furthermore, alteration in EMT-related genes and proteins were reversed by overexpression of miR-129-5p. No effect was observed in cells treated with miR-negative control. Meanwhile, inhibition of SIP1 and SOX4 with their respective siRNA also could decrease the expression of EMT-related genes and protein levels in HPMCs induced with TGF-β1. Finally, we demonstrate that SIP1 can inhibit the promoter activity of E-cadherin while enhancing the promoter activity of vimentin. We also observed that miR-129-5p could directly target the 3′UTR of SIP1 and SOX4 genes, and repressed their post-transcriptional activities. These data suggest that there is a novel TGF-β1/miR-129-5p/SIP-1 or SOX4 pathway that has a significant role in MMT and fibrosis in the setting of PD.
Peritoneal dialysis (PD) is an alternative treatment for end-stage renal disease that uses the peritoneal membrane (PM) as a permeable barrier for the exchange of nocuous substances and water.1,2 During PD, long-term exposure to bioincompatible PD solutions and episodes of infection cause damage to the PM, which then can lead to denudation of the mesothelial cells (MCs), submesothelial fibrosis and angiogenesis, eventually leading to ultrafiltration failure and discontinuation of PD.2 Emerging evidence has indicated phenotypic conversion of MCs to mesenchymal cells. The mesothelial-to-mesenchymal transition (MMT) is an initial and reversible event leading to structural and functional peritoneal lining deterioration.3,4 Recent studies indicate that the MMT is a complex process, which is characterized by a loss of epithelial phenotype and acquisition of myofibroblast-like properties,5,6 and the cellular and molecular mechanisms and factors that participate in this process are not fully delineated. TGF-β1 is a potent inducer of epithelial-to-mesenchymal transition (EMT) in several tissues and organs.7–9 TGF-β1 binds to its receptor and triggers Smad/non-Smad signaling pathways, which results in a profound molecular reprogramming, including the downregulation of the intracellular adhesion molecule E-cadherin and the upregulation of mesenchymal-associated molecules, such as, vimentin, fibronectin (FN), etc.7,8 TGF-β1 is known to have a pathogenetic role in the MMT in states of PD dysfunction.1,10–12 However, the mechanism(s) by which TGF-β1 induces MMT in PD dysfunction remains to be clearly defined.
MicroRNAs (miRNAs) are small 18–25-nucleotide non-coding RNAs that are believed to be involved in the modulation of gene expression by inducing mRNA degradation or repressing mRNA translation,13 and thus have a globalized role in a wide variety of biological functions, such as cellular proliferation, apoptosis, tumorigenesis, and fibrosis in various target organs.13–15 Recent studies indicate that miRNAs may contribute to the process of EMT in PD dysfunction10,16,17 as well as in development of fibrosis.18–20 For example, miR-30 and miR-589 have been reported to be associated with the MMT process and peritoneal fibrosis in certain models of PD10,16, suggesting that miRNA may h ave a critical role in the pathogenesis of TGF-β-induced fibrosis in PD.
To identify other miRNAs involved in the MMT/EMT of peritoneal MCs (PMCs), we employed miRNA array analyses of the PMCs from the effluent of PD patients. The results indicate that some miRNAs, including miR-129-5p, were downregulated in patients undergoing PD for more than 6 months when compared with those undergoing PD therapy for less than 6 months. We further demonstrate that miR-129-5p expression is notably decreased, which closely correlated with the development of MMT/EMT and peritoneal fibrosis in the PD patients. We also observed that overexpression of miR-129-5p in human PMCs (HPMCs) reversed the TGF-β1-induced MMT/EMT phenotypic transformation cells and the expression of SIP1, a member of the zinc-finger E-box-binding (ZEB) homeobox factor family, as well as the sex-determining region Y-related high-mobility group box-4 gene (SOX4). In addition, we noted that miR-129-5p regulate promoter activity of E-cadherin and vimentin in HPMCs treated with TGF-β1. Finally, we report that miR-129-5p could directly target the 3′ untranslated region (UTR) regions of SIP1 and SOX4 genes, and modulate their translational efficiency. From these data, we suggest that miR-129-5p could serve as a potential therapeutic target for the amelioration of peritoneal fibrosis in PD.
MATERIALS AND METHODS
Peritoneal Equilibrium Test (PET)
PET measurements were carried out as previously reported.21,22 Briefly, a 2.5% glucose exchange was performed overnight using a PD patient’s standard dwell volume. The abdomen cavity was drained for over 20 min, following which 2.5% dextrose dialysate (2 l) was instilled into the peritoneal cavity. A 10-ml sample was removed at 4 h to determine the urea, creatinine, and glucose levels. PM transport function was designated as high, high average, low average or low transporter, as classified previously.21,22
Human PMCs (HPMCs)
Sixteen patients were enrolled for the continuous ambulatory PD (CAPD), including eight patients who received CAPD for ‘more’ than 6 months (from 6 to 50 months) and were designated as the PD >6 months group, and eight patients who received CAPD for ‘less’ than 6 months and were designated as the PD-start group. The study was approved by the Institutional Review Board and Ethics Committee of The Second Xiangya Hospital, Central South University, China. All of the participants had signed the informed consent form. HPMCs were harvested from the effluents of PD patients before the first episode of peritonitis occurred. The HPMCs were isolated using low-speed (1500 r.p.m.) centrifugation, washed with DMEM, cultured in DMEM containing l-glutamine supplemented with 15% FBS, 0.5 μg/ml insulin, 5 μg/ml transferrin, and 1 μg/ml hydrocortisone in humidified air with 5% CO2 at 37 °C. Nonadherent cells were removed the next day with two brief washes with DMEM, and the adherent cells were incubated in fresh culture medium. The cells were allowed to reach confluence in 7–10 days, and then they were split and seeded onto two to three cultured flasks. After achieving 80% confluency, the cells were used for various experiments.
miRNA Microarray Analyses
Total RNA was extracted from HPMCs using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) as per the manufacturer’s instruction, and miRNA microarray was performed at Kangcheng BioCorp (Shanghai, China). Briefly, Total RNA was purified and hybridized onto the miRCURY LNA Array (v.11.0), which contains probes of over 1700 unique miRNAs based on the miRBase version 11.0 release of hsa, mmu & rno array (online at http://microrna.sanger.ac.uk). The intensity values of the microarray data were subjected to GenePix pro V6.0 for subtraction of background values and normalization, followed by mean summarization. A list of miRNA candidates was generated based on their differential expression between the two populations of RNA from PD patients (<6 months PD vs >6months).
Culture of MC Line
HMrSV5 (a human peritoneal mesothelial cell line) was obtained from Dr Pierre Ronco (Tenon Hospital, Paris).23,24 Cells were maintained in DMEM/F12 medium (Invitrogen) containing 10% fetal bovine serum, and incubated at 37 °C in a humidified incubator with 5% CO2. After achieving 80% confluency, the cells were cultured in serum-free medium in the presence or absence of 5 ng/ml of TGF-β1 (R&D Systems, Minneapolis, MN). In various combinations, the cells were transfected with pre-miR-129-5p or miRNA-negative control (miR-neg-ctrl) (Life Technologies, CA, USA), or SIP1-siRNA and SOX4-siRNA (Santa Cruz Biotechnology, TX, USA), or with pBK-CMV-hNHERF2 plasmid (Entrez Gene: SIP1, Plasmid #47801 SIP1), a mammalian cell expression plasmid (Addgene, Cambridge, MA, USA), or pWPXL-Sox4 plasmid (Entrez Gene: SOX4, Plasmid #36984) from Addgene. For transfection, the Lipofectamine 2000 kit was used following the manufacturer’s instructions (Invitrogen). The cells were then used for various expression studies and assays.
TaqMan Reverse Transcription-PCR (RT-PCR) Studies
For measurement of miR-129-5p expression, a RT reaction was performed using the TaqMan MicroRNA Reverse Transcription Kit. Three μl of cDNA products were used for the measurement of miR-129-5p by employing the TaqMan MicroRNA Assays-miR-129-5p kit (Life Technologies) as per the manufacturer’s instructions. For mRNA expression of various genes, total RNA (500 ng) was used for RT and the cDNA was synthesized as described.25 The mRNA expression of SIP1, SOX4, E-cadherin, claudin-1, vimentin, and FN was detected by TaqMan gene expression assays using Real-time PCR system 7300 (Applied Biosystems, Life Technologies).26–29 The expression of miR-129-5p and of various genes were normalized to β-actin and U6snRNA. The PCR primers used were as follows: E-cadherin sense: 5′-GTCACTGACACCAACGATAATCCT-3′ and antisense: 5′-TTTCAGTGTGGTGATTACGACGTTA-3′, vimentin sense: 5′-TTGAACGCAAAGTGGAATC-3′ and antisense: 5′-AGGTCAGGCTTGGAAACA-3′, claudin-1 sense: 5′-CCAGTCAATGCCAGGTACGAAT-3′ and antisense: 5′-TTGGTGTTGGGTAAGAGGTTGTT-3′, FN sense: 5′-TGGAGGAAGCCGAGGTTT-3′ and antisense: 5′-CAGCGGTTTGCGATGGTA-3′, β-actin sense: 5′-CCTTCCTGGGCATGGAGTC-3′ and antisense: 5′-GAGGAGCAATGATCTTGATCTTC-3′, SOX4 sense: 5′-GTGAGCGAGATGATCTCGGG-3′ and antisense: 5′-CAGGTTGGAGATGCTGGACTC-3′ and SIP1 sense: 5′-CCCTTCTGCGACATAAATACA-3′ and antisense: 5′-TGTGATTCATGTGCTGCGAGT-3′.
Northern Blot Analysis of miRNAs
To confirm the miRNA-129-5p expression in cells from PD patients, northern blotting procedures were performed, as described previously.30,31 Briefly, 50 μg of low-molecular weight RNA was subjected to denaturing 10% polyacrylamide gel electrophoresis, transferred to a PVDF membrane (Amersham, NJ, USA), and probed. The probes used were γ-32 P-ATP (PerkinElmer Life Sciences, Akron, OH, USA) end labeled of locked nucleic acid (LNA)-modified miR-129-5p oligonucleotide from Exiqon (Woburn, MA, USA). The northern blot analysis of miRNAs was carried out following their quantification using National Institutes of Health Image J version 1.42q software, as described previously.31
Western Blot Analyses
Protein expression was assessed by western blot analysis. Protein (50 μg) samples were resolved by 10% SDS–PAGE and transferred to PVDF membranes, then probed with various primary antibodies against E-cadherin, claudin-1, and vimentin (Santa Cruz Biotechnology); FN (Sigma-Aldrich, St Louis, MO, USA); SIP1 and SOX4 (Abcam Trading Company, Shanghai, China). After incubating with an appropriate secondary antibody, the protein expression was detected by western blot analysis, as described previously.27–29 β-Actin (Santa Cruz Biotechnology) was used as an internal control.
Immunofluorescence Studies
HPMCs were grown on coverslips, washed twice with PBS, fixed in 4% paraformaldehyde for 20 min, and permeabilized using 0.1% Triton X-100. The antibodies used were as follows: anti-E-cadherin (1:100 dilution), anti-vimentin (1:100) from Santa Cruz Biotechnology; anti-SIP1 (1:100), and anti-SOX4 (1:100) from Abcam. The antibodies were diluted in a blocking buffer and cells were incubated with them overnight at 4 °C, followed by incubation with FITC-labeled secondary antibody (Santa Cruz Biotechnology) for 2 h at 22 °C. They were then examined using a UV fluorescence microscope. The intensity of fluorescence was analyzed by image analysis software (Path, QC, Logene Biological Medical Engineering) as described in (ref. 31).
Cell Migration Studies
For cell migration assays, trypsinized HPMCs were seeded onto 12-well plates. A wound was created by scratching the cell culture with a pipette tip. The status of the wound was monitored for several hours after treating the cells with TGF-β1 with or without transfection of pre-miR-129-5p. The wound width in the cell culture was measured at the termination of the experiments, as described in previous publications.32,33
Promoter Activity Assay
Promoter activity assays were carried out using a luciferase system. A E-cadherin-luciferase reporter was constructed by cloning the human E-cad promoter fragment spanning −420 to +32 from the transcription start site upstream of the luciferase segment in the pGL3 vector (Promega, Madison, WI, USA), as described,34 and designated as pGL3-E-cad vector. Briefly, the promoter fragment was first cloned into PCR II vector by PCR using oligonucleotide sense primer: 5′-AGAACCGTGCAGGTCCCATAA-3′ and antisense primer: 3′-AACTGACTTCCGCAAGCTCAC-5′ and the genomic DNA from HPMCs that served as the template. Following cloning, its sequence was confirmed to ensure that there were no mutations. The promoter fragment was then subcloned into pGL3 vector (Promega). To construct the human vimentin promoter luciferase reporter plasmid (VimPro), a 1.5-kb fragment of human vimentin 5′-upstream region was cloned in the EcoRI–BamHI site of the firefly luciferase reporter plasmid pGL3 (Promega).35 Briefly, the promoter (−1416 to +85 bp) of human vimentin 5′-upstream region was cloned into PCR II vector and amplified using the sense primer: 5′-AATTCCTTTTGAAAACAGAACCTATCATT-3′ and the antisense primer: 5′-GAAGAGGGAAGAGGGGGGTGGGTGTGGGT-3′. Following confirmation of the sequence, the fragment then was subcloned at the EcoRI–BamHI site of the firefly luciferase reporter plasmid pGL3 as described previously.35,36 For the luciferase assay, pGL3-E-cad and vimPro constructs were then transiently transfected into HPMCs in a 24-well plate that were serum starved for 18 h prior to treatment with TGF-β1, or co-transfected with SIP plasmid or pre-miR-129-5p siRNA. After 24 h, luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega) following the manufacturer’s instructions. The luciferase activities were subsequently normalized with β-galactosidase activity where cells were co-transfected with pSVb-galactosidase vector (pSVb) (Promega) as in (ref. 36).
Luciferase Reporter Assay
For construction of the 3′UTR of SIP1 and SOX4 reporter plasmids, SIP1 and SOX4 3′UTR segments were amplified by PCR. They were then subcloned into pGL3 luciferase vector (Promega), as described previously,28,29 and the plasmids were designated as pGL3-SIP1-wt or pGL3-SOX4-wt. Mutations in SIP1 and SOX4 3′UTR were then created using the pGL3-SIP1-wt or pGL3-SOX4-wt as PCR templates while replacing four nucleotides in the miR-129-5p-binding site using a QuikChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA, USA). These constructs were designated as pGL3-SIP1-mut or pGL3-SOX4-mut, as previously reported.28,29 For luciferase reporter analysis, HMrSV5 cells were transfected with the reporter constructs as indicated above pre-miR-129-5p with or without TGF-β1 treatment. Following 48 h of transfection, luciferase activity was measured using a Dual-Glo Luciferase Reporter Assay Kit (Promega) as described previously.28,29
Statistical Analysis
Data are expressed as the mean ± s.e.m. One-way ANOVA was performed to assess the statistical analysis. P-values <0.05 were considered statistically significant.
RESULTS
Mir-129-5p was Downregulated in the PMCs from the Effluent of PD Patients, Which Negatively Correlated with the MMT Transformation and the Expression of SIP1 and SOX4
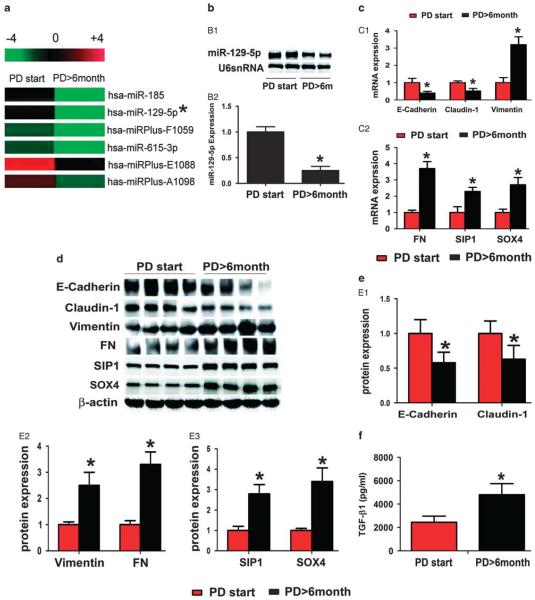
Average duration of dialysis in the PD-start group was 3.90 ± 1.79 months and 19.33 ± 11.82 months for the PD>6 months group (P<0.01). There are no significant differences in other clinical characteristics, including gender, age, BMI, renal failure cause, and creatinine ratio (dialysis to plasma), at 4 h in a peritoneal equilibrium function test between PD-start and PD>6 months groups (Table 1). By miRNA array analysis, expression of 91 miRNAs was noted to be dysregulated in the cultured PMCs from the effluent of PD patients. Among them included 33 miRs upregulated and 58 miRs downregulated in the PMCs from the PD patients having dialyses over 6 months compared with the PD-start group (Tables 2 and 3). The miR-129-5p was found to be downregulated nearly ninefold in the PMCs from the effluent of patients having PD >6 months (Figure 1a). The differential expression of miR-129-5p in PMCs from the effluent of PD patients was further validated by northern blot analyses and TaqMan real-time PCR (Figure 1b B1 and B2).
Table 1.
Clinical characteristics of patients in each group
| PD-start group |
PD>6 month group |
P-value | |
|---|---|---|---|
| Total number of patients (n) | 8 | 8 | |
| Males (n) | 4 | 4 | |
| Females (n) | 4 | 4 | >0.05 |
| Age (years) | 55.30 ± 8.90 | 56.30 ± 5.82 | |
| BMI (kg/m2) | 23.50 ± 3.55 | 22.92 ± 3.24 | >0.05 |
| Renal failure cause | |||
| Chronic glomerulonephritis (n) | 5 | 4 | |
| Diabetic nephropathy (n) | 1 | 2 | |
| Hypertensive nephropathy (n) | 2 | 2 | |
| Dialysis duration (months) | 3.90 ± 1.79 | 19.33 ± 11.82 | <0.01 |
| Peritoneal equilibrium test | |||
| D/PCr(4 h) | 0.69 ± 0.15 | 0.71 ± 0.25 | >0.05 |
| High (n) | 1 | 2 | |
| High average (n) | 3 | 2 | |
| Low average (n) | 4 | 4 | |
| Low (n) | 0 | 0 |
Table 2.
Upregulated microRNAs in PMCs from effluents of PD patients
| miR ID | Fold change | miR ID | Fold change |
|---|---|---|---|
| hsa-miR-146b-5p | 1.511596907 | hsa-miR-181a | 1.976029581 |
| hsa-miRPlus-E1066 | 2.51784185 | hsa-miRPlus-E1225 | 1.547911548 |
| hsa-miR-152 | 2.515094014 | hsa-miR-768-3p | 1.994051822 |
| hsa-let-7i | 1.753991528 | hsa-miR-23a | 1.720172866 |
| hsa-miR-26a | 1.796862942 | hsa-miRPlus-E1168 | 3.579545455 |
| hsa-miR-30e | 1.609776609 | hsa-miRPlus-E1234 | 2.415971126 |
| hsa-miR-923 | 2.919836761 | hsa-miR-193a-3p | 1.786047914 |
| hsa-miR-142-5p | 6.46627566 | hsa-miR-381 | 1.571759968 |
| hsa-miR-29c | 1.684491979 | hsa-miR-191 | 1.581974901 |
| hsa-miR-195 | 3.265361795 | hsa-miR-19b | 1.598804421 |
| hsa-miR-30a | 1.506479992 | hsa-miR-101 | 2.121212121 |
| hsa-miR-148b | 2.021994135 | hsa-miRPlus-C1115 | 2.40412813 |
| hsa-miR-142-3p | 3.854761537 | hsa-miRPlus-E1038 | 1.575815077 |
| hsa-miR-374a | 1.616047609 | hsa-miR-335 | 1.622201559 |
| hsa-miR-574-3p | 1.72550213 | hsa-miR-19a | 1.549112676 |
| hsa-miR-584 | 1.590380022 | hsa-miR-874 | 2.192191535 |
| hsa-miRPlus-E1141 | 1.855575049 |
Table 3.
Downregulated microRNAs in PMCs from effluents of PD patients
| miR ID | Fold change | miR ID | Fold change |
|---|---|---|---|
| hsa-miR-638 | 0.12995774 | hsa-miRPlus-E1013 | 0.557412989 |
| hsa-miR-887 | 0.344830655 | hsa-miRPlus-E1013 | 0.557412989 |
| hsa-miRPlus-D1116 | 0.276039331 | hsa-miRPlus-A1056 | 0.305602716 |
| hsa-miRPlus-E1065 | 0.432457926 | hsa-miR-668 | 0.459077579 |
| hsa-miRPlus-E1063 | 0.108676902 | hsa-miR-18b | 0.667155425 |
| hsa-miR-585 | 0.008474804 | hsa-miR-302c | 0.37485656 |
| hsa-miRPlus-D1036 | 0.116578778 | hsa-miRPlus-E1271 | 0.661862752 |
| hsa-miRPlus-E1108 | 0.349294355 | hsa-miRPlus-F1187 | 0.362256339 |
| hsa-miR-665 | 0.386159319 | hsa-miR-340 | 0.617657776 |
| hsa-miRPlus-E1088 | 0.327549335 | hsa-miR-519d | 0.488210082 |
| hsa-miRPlus-F1159 | 0.23318974 | hsa-miR-424 | 0.539301273 |
| hsa-miRPlus-F1059 | 0.35227894 | hsa-miRPlus-E1045 | 0.085989973 |
| hsa-miR-1264 | 0.653599292 | hsa-miRPlus-F1231 | 0.417888563 |
| hsa-miRPlus-F1065 | 0.084296912 | hsa-miR-615-3p | 0.342130987 |
| hsa-miR-1290 | 0.643846245 | hsa-miR-203 | 0.489369501 |
| hsa-miRPlus-F1029 | 0.500866436 | hsa-miR-642 | 0.587843242 |
| hsa-miRPlus-E1035 | 0.340330298 | hsa-miRPlus-E1202 | 0.351906158 |
| hsa-miR-130b | 0.620431567 | hsa-miR-1285 | 0.559024359 |
| hsa-miR-654-3p | 0.549258936 | hsa-miR-185 | 0.099529014 |
| hsa-miR-197 | 0.595691616 | hsa-miR-129-5p | 0.11153682 |
| hsa-miR-939 | 0.460133901 | hsa-miR-589 | 0.576422287 |
| hsa-miR-1274a | 0.581932299 | hsa-miR-888 | 0.41211142 |
| hsa-miRPlus-E1247 | 0.184014674 | hsa-miRPlus-E1170 | 0.556634372 |
| hsa-miRPlus-F1063 | 0.609677419 | hsa-miRPlus-E1024 | 0.652822229 |
| ebv-miR-BHRF1-2 | 0.015710096 | hsa-miR-15b | 0.562997796 |
| hsa-miR-138-1 | 0.628589773 | hsa-miR-634 | 0.482134589 |
| hsa-miRPlus-A1098 | 0.122377622 | hsa-miR-602 | 0.474924201 |
| ebv-miR-BART2-3p | 0.364387259 | hsa-miR-202 | 0.017106549 |
| hsa-miRPlus-E1253 | 0.634025019 | hsa-miRPlus-B1114 | 0.622996658 |
Figure 1.
miR-129-5p, epithelial–mesenchymal transition-related genes, SIP1, SOX4, and TGF-β1 expression in the mesothelial cells isolated from effluent of patients with peritoneal dialysis (PD). (a) MicroRNA (miRNA) expression profiling in the mesothelial cells isolated from effluent of patients with PD, as detected by miRNA array analyses, a decreased expression of miR-129-5p was seen (asterisks). (b, B1) Northern blot analysis showed that miR-129-5p expression decreased in the PD patients (PD<6 months) as compared with that of the control group (PD<6months). There was no change in U6snRNA expression, which served as an internal control. (B2) The bar graph represents the summary of the messenger RNA (mRNA) expression of miR-129-5p from northern blot analysis. (c) The bar graph represents the summary of the mRNA expression of E-cadherin, claudin-1, vimentin (C1), fibronectin, SIP1, and SOX4 (C2) in the cells by real-time PCR. (d) Expression profiling of their protein levels as assessed by western blot analysis. (e, E1–E3) The bar graphs represent the density of relative bands depicted by western blot analyses. (f) TGF-β1 expression in the effluent of PD patients by an enzyme-linked immunosorbent assay. Values are the mean ± s.e.m., n = 4, *P<0.01 vs control (PD<6 months).
At the same time, the expression of MMT/EMT-related genes and proteins was also altered. As shown in Figure 1c C1, C2, 1d and 1e E1, E2, the mRNA and protein expression of E-cadherin significantly decreased in the PMCs from the effluent of PD>6 months patients, as compared with that from PD-start patients, whereas that of mesenchymal markers, such as vimentin and FN expression, increased. A negative correlation between miR-129-5p expression and that of vimentin and FN was seen, suggesting that miR-129-5p may be involved in the MMT process during PD.
Next, we investigated the expression of SIP1 and SOX4 transcription factors, as they may be the predicted targets of miRNA-129-5p by analysis of the 3′-UTR sequence using biological analysis software (www.microrna.org). These factors have been reported to have an important role in the EMT process. As expected, both mRNA and protein expression of SIP1 and SOX4 were increased in the PMCs from the effluent of the PD<6 months group (Figure 1e E3), which positively correlated with the MMT transformation, and correlated with the expression of miR-129-5p. Moreover, an upregulated level of TGF-β1 was also observed in effluent of the PD<6 months group, as assessed by ELISA methods (Figure 1f).
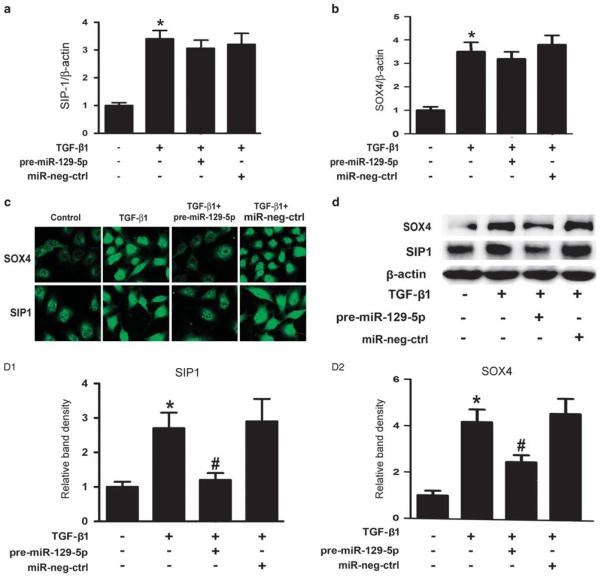
TGF-β1 Suppressed the Levels of miR-129-5p While Enhancing that of the SIP1 and SOX4 in HPMCs
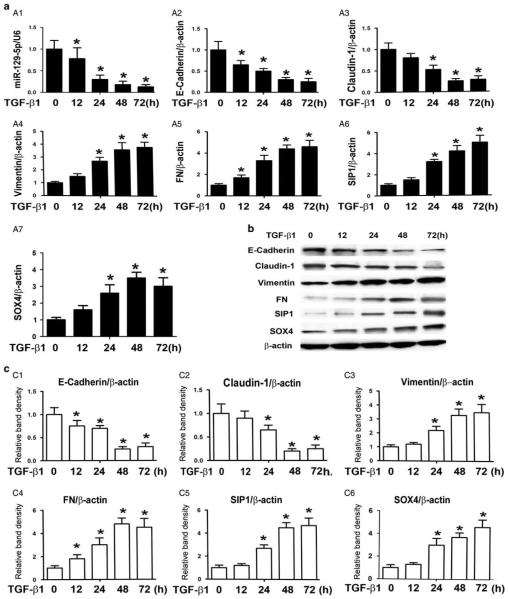
Real-time PCR showed that the expression of miR-129-5p decreased in HPMCs incubated with 5 ng/ml TGF-β1 for 12 h (Figure 2a A1), and it was further suppressed in a time-dependent manner. The change in the expression paralleled to the decreased expression of E-cadherin (Figure 2a A2) and claudin-1 mRNA (Figure 2a A3). The expression of vimentin (Figure 2a A4) and FN (Figure 2a A5) increased in a time-dependent manner in HPMCs following TGF-β1 treatment. Expression of both SIP 1 (Figure 2a A6) and SOX4 mRNA (Figure 2a A7) also significantly increased in the HPMCs treated with TGF-β1, and peaked at 48–72 h. Similar results were also seen in their protein expression levels (Figure 2b and c).
Figure 2.
Expression of epithelial–mesenchymal transition-related genes, SOX4, and SIP1 in human peritoneal mesothelial cell lines (HPMCs) induced by TGF-β1. (a) The bar graph represents the summary of the results by real-time PCR. (A1) An overt decrease in messenger RNA expression of miRNA-129-5p was observed in HPMCs, a human peritoneal mesothelial cell line, following treatment with TGF-β1. Real-time PCR showed that E-cadherin (A2) and Claudin-1(A3) expressions were significantly decreased in a time-dependent manner in HPMCs treated with TGF-β1, whereas expression of vimentin (A4) and fibronectin (A5), SIP1 (A6) and SOX4 (A7) were upregulated. (b) Western blot assays showed the expression of E-cadherin, claudin, vimentin, fibronectin (FN), SIP1, and SOX4 proteins in HPMCs induced with TGF-β1. (c, C1–C6) The bar graphs representing the western blot relative band densities from Figure 7b. Values are the mean ± s.e.m., n = 3, *P<0.01 vs control.
Overexpression of miR-129-5p Blocked MMT Transformation in HPMCs Treated with TGF-β1
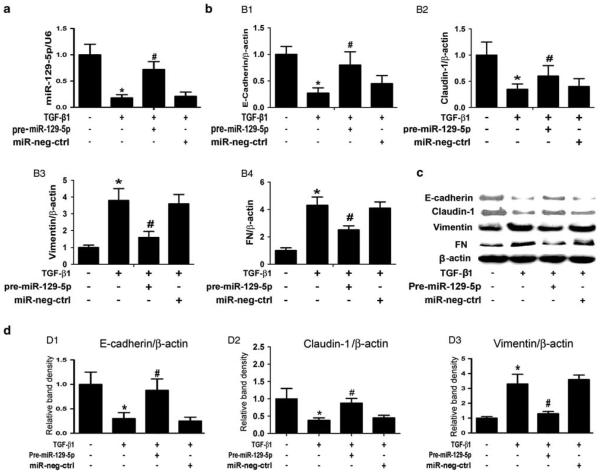
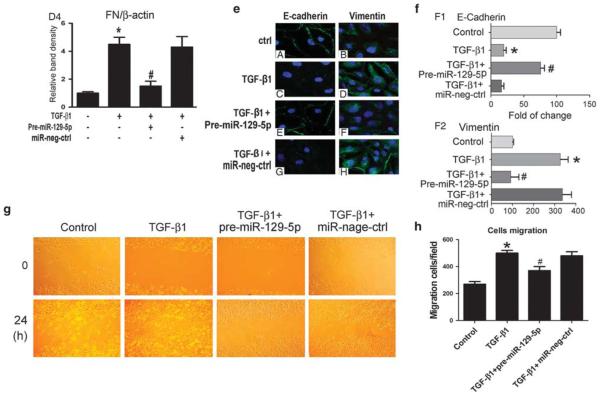
To verify the precise role of miR-129-5p, expression of E-cadherin and vimentin was induced with TGF-β1. The pre-miR-129-5p was transfected into HPMCs and then exposed to TGF-β1. The results showed that overexpression of miR-129-5p significantly reversed the TGF-β1-reduced miRNA-129-5p expression, and there was no change seen in that treated with miR-negative control (Figure 3a). In addition, the inhibitory effect of TGF-β1 on the mRNA expression of the E-cadherin (Figure 3b B1) and claudin-1 (Figure 3b B2) was also blocked partially in cells transfected with pre-miR-129-5p, whereas opposite results were observed for vimentin (Figure 3b B3) and FN expression (Figure 3b B4). Similar results were seen in their protein expression by western blot analyses (Figure 3c and d). The immunofluorescence staining of E-cadherin was significantly decreased in the membranes of HPMCs stimulated with TGF-β1, whereas the signal intensity was restored in cells transfected with pre-miR-129-5p (Figure 3e, left panels and Figure 3f F1). On the other hand, over-expression of pre-miR-129-5p significantly inhibited TGF-β1- induced vimentin expression (Figure 3e, right panels and Figure 3f F2). There were no differences in cells transfected with miR-negative control compared with the control group. Since cell migration has been described to be modulated by the EMT process, the studies were performed to further understand the effect of miR-129-5p on cell migration in HMPCs. We observed that the overexpression of miR-129-59 significantly dampened the TGF-β1-induced HMPC migration, as shown in Figure 3g and h. In other words, treatment of cells with TGF-β1 leads to increased HMPC migration, whereas miR-129-59 treatment decreases cell migration. These results indicated that miR-129-5p partially blocks the effect of TGF-β1 during the MMT process in HPMCs.
Figure 3.
Overexpression of pre-miR-129-5p reverses epithelial–mesenchymal transition-related gene and protein expression in human peritoneal mesothelial cell lines (HPMCs) following treatment with TGF-β1. (a) By real-time PCR, a significantly reduced expression of miR-129-5p with time dependency was observed in HPMCs induced by TGF-β1 (5 ng/ml) for 0–72 h, as compared with control, whereas the effect was abolished in cells transfected with pre-miRNA-129-5p. No effect was seen in cells treated with miR-negative control. The measured transcript level was normalized to U6snRNA expression. (b) Overexpression of miR-129-5p in cells transfected with pre-miR-129-5p reversed the mesenger RNA (mRNA) expression of E-cadherin (B1) and claudin-1 (B2) in TGF-β1-induced HPMCs, whereas it partially blocked vimentin (B3) and fibronectin (FN) mRNA (B4) expression, as assessed by real-time PCR analysis. (c) Western blot analysis showed that the protein expression of E-cadherin, claudin-1, vimentin, and FN in TGF-β1-stimulated HPMCs with or without transfection with pre-miR-129-5p. The bar graphs represent expression of the above proteins relative to β-actin (d, D1–D4). (e) Immunofluorescence microscopy showed downregulated E-cadherin and upregulated vimentin expression in HPMCs subjected to TGF-β1, which was normalized with transfection with pre-miR-129-5p, and no effect was seen in transfection of miR-neg control. Magnification, × 400. (f) The bar graph represents the density of immunofluorescence intensity of E-cadherin (F1) and vimentin (F2). Values are the mean ± s.e.m., n = 3, *P>0.01 vs control, #P>0.01 vs TGF-β1. (g) Representative fields of cells showing migration after scratching the HPMC culture and following TGF-β1 treatment with or without pre-miR-129-5p transfection. Magnification, × 0100. (h) Quantitative analyses of cell migration in the scratch area. *P<0.01 vs control, #P<0.01 vs TGF-β1.
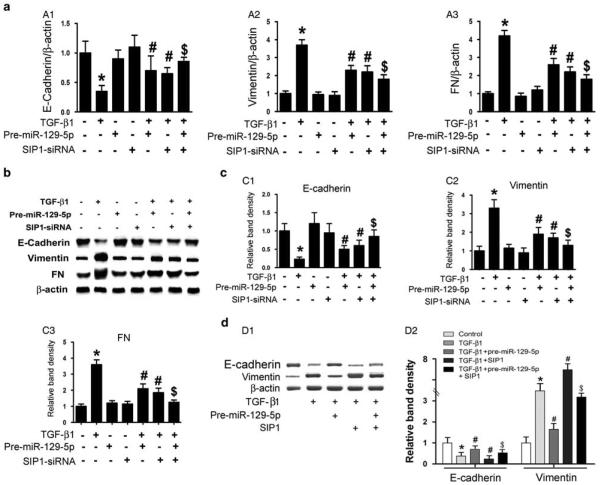
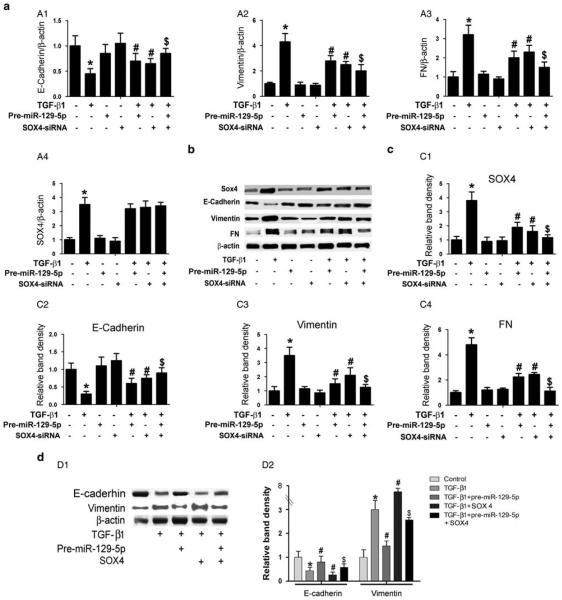
Knockdown of SIP1 or SOX4 Ameliorated TGF-β1-Induced MMT/EMT Changes in HPMCs
To clarify the precise mechanism by which miR-129-5p modulates TGF-β1-induced MMT/EMT mRNA and protein expression, SIP1-siRNA and SOX4-siRNA were used to knock down the endogenous expression of SIP1 or SOX4 in HPMCs in vitro. As shown in Figure 4, significantly downregulated mRNA and protein expression of E-cadherin in HPMCs induced by TGF-β1 was observed, as demonstrated by real-time PCR and immunoblotting (Figure 4a A1 line 2 vs line 1; Figure 4b upper panels, Figure 4c C1). The expression was restored partially by transfection of pre-miR-129-5p or SIP1-siRNA (lines 5 and 6 vs line 2). In contrast to E-cadherin, the mRNA and protein expression of both vimentin and FN were upregulated in HPMCs stimulated by TGF-β1 (Figure 4a A2, line 2 vs line 1, Figure 4b middle and Figure 4c C2), and the expression was inhibited by transfection with pre-miR-129-5p or SIP1-siRNA (lines 5 and 6 vs line 2). This effect was enhanced by co-treatment of pre-miR-129-5p and SIP1-siRNA (line 7 vs lines 5 and 6). By western blot analysis, a significantly decreased expression of E-cadherin protein in HPMCs treated with TGF-β1 or transfected with SIP1 plasmid was seen, and it was reversed by co-treatment with pre-miR-129-5p (Figure 4d D1 upper panel, line 2 and line 4 vs line 1, and Figure 4d D2, left panel). An opposite result was observed for vimentin protein expression (Figure 4d D1 middle panel, and Figure 4d D2, right panel). It is interesting to note that the above changes were also seen in cells transfected with SOX4-siRNA under same conditions (Figure 5a–d). These data suggested that SIP1 and SOX4 probably exert their effects downstream of miR-129-5p and are thus involved in the modulation of MMT-related genes and protein expression in HPMCs induced by TGF-β1.
Figure 4.
SIP1-siRNA reverses the altered expression of epithelial–mesenchymal transition-related genes in human peritoneal mesothelial cell lines (HPMCs) induced by TGF-β1. (a) By real-time PCR, a decreased messenger RNA (mRNA) expression of E-cadherin was seen in HPMCs subjected to TGF-β1 treatment (5 ng/ml) for 48 h, whereas it was reversed by transfection with pre-miR-129-5p or SIP1-siRNA. Inhibition of SIP1 expression further enhanced the reversal effect of pre-miR-129-5p in TGF-β1-reduced E-cadherin mRNA expression (A1). In addition, compared with E-cadherin expression, an opposite effect was observed for vimentin and fibronectin (FN) mRNA expression (A2, A3). (b) Western blot analysis showed similar results that SIP1-siRNA reversed the altered expression of E-cadherin, vimentin, and FN in HPMCs subjected to TGF-β1 treatment. The bar graphs represent the densitometric measurements of the relative band density observed by western blot analyses (c, C1–C3). (d, D1) Western blot analysis revealed that overexpression of pre-miR-129-5p partially blocked the altered protein expression of E-cadherin and vimentin in HPMCs treated with TGF-β1 or TGF-β1 +SIP1. (D2) The bar graphs represent the densitometry measurements of relative band density of autoradiograms of D1. Values are the mean ± s.e.m., n = 3, *P<0.01 vs control, P<0.01 vs TGF-β1, P<0.01 vs $P<0.01 vs TGF-β1+pre-miR-129-5p.
Figure 5.
SOX4-siRNA reverses the altered expression of E-cadherin, vimentin, and fibronectin (FN) in human peritoneal mesothelial cell lines (HPMCs) induced by TGF-β1. (a, A1) Real-time PCR showed SOX4-siRNA reversed the TGF-β1-reduced messenger RNA (mRNA) expression of E-cadherin in HPMCs, whereas this effect was partially blocked in cells transfected with pre-miR-129-5p. In contrast, opposite results were seen for the expression of vimentin and FN mRNA (A2, A3). Furthermore, real-time PCR also revealed that TGF-β1 significantly increased SOX4 mRNA expression, whereas there was no change following treatment with pre-miR-129-5p (A4). (b) Western blot analyses revealed comparable results for their protein expression compared with mRNA expression. (c) The bar graphs represent the western blot relative band densities for the SOX4 (C1), E-cadherin(C2), vimentin (C3), and FN (C4) compared with β-actin. (d, D1) Western blot analyses show that overexpression of pre-miR-129-5p normalizes the altered expression of E-cadherin and vimentin in HPMCs treated with TGF-β1 or TGF-β1+SOX4. (D2) The bar graphs represent the protein expression relative band densities of E-cadherin and vimentin compared with β-actin. Values are the mean ± s.e.m., n = 3, *P<0.01 vs control, #P<0.01 vs TGF-β1, P<0.01 vs $P<0.01 vs TGF-β1+pre- miR-129-5p.
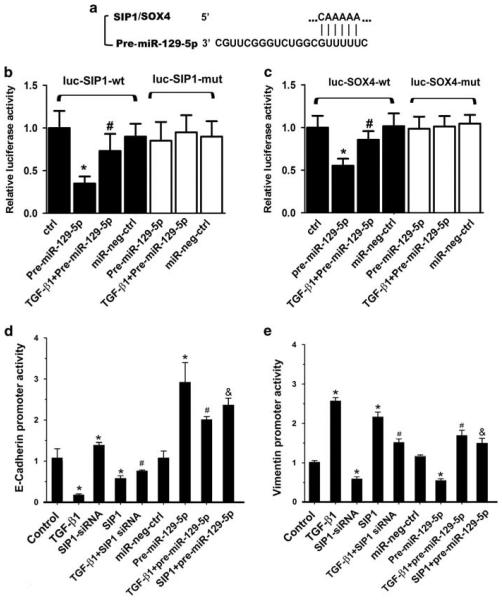
MiR-129-5p Inhibits Protein Expression of SIP1 and SOX4 by Downregulation of their 3′UTR Activity and Modulation of the Promoter Activity of E-cadherin and Vimentin
By real-time PCR, a significantly increased mRNA expression of SIP1 and SOX4 was seen in HPMCs induced by TGF-β1 (Figure 6a and b, line 2 vs line 1), whereas no significant differences were seen between TGF-β1 and TGF-β1+miR- -129-5p groups (line 3 vs line 2). However, immunostaining with anti-SIP1 or anti-SOX4 antibodies showed that the SIP1 and SOX4 expression in HPMCs increased, which was mainly reflected by a significant increase in the nucleus following treatment with TGF-β1. This was significantly dampened by the treatment with pre-miR-129-5p (Figure 6c), whereas treatment with the miR-negative control had no effect. Furthermore, immunoblotting revealed an increased SOX4 and SIP1 protein expression in HPMCs treated with TGF-β1 (Figure 6d, line 2 vs line 1), whereas it was partially blocked by transfection with pre-miR-129-5p (line 3 vs line 2). This indicated that miR-129-5p repressed the translation of SIP1 and SOX4, but did not cause degradation of their mRNA.
Figure 6.
Effect of miR-129-5p on the expression of SIP1 and SOX4 in human peritoneal mesothelial cell lines (HPMCs) treated with TGF-β1. (a and b) By real-time PCR, a significant increase in the SIP1 and SOX4 messenger RNA expression in HPMCs subjected to TGF-β1 treatment was seen, whereas it was not significantly different between the TGF-β1-treated group or group co-treated with TGF-β1+miR-129-5p. (c) Immunofluorescence microscopy revealed an increased SIP1 and SOX4 expression in the nuclei of HPMCs treated with TGF-β1, which was normalized with transfection with pre-miR-129-5p. Magnification, × 400. (d) Similar results were observed by western blot assays. (D1, D2) The bar graphs represent the expression of SIP1 and SOX4 protein band densities relative to β-actin. Values are the mean ± s.e.m., n = 3, *P<0.01 vs control, #P<0.01 vs TGF-β1.
To confirm whether miR-129-5p directly targets the 3′UTR of SIP1 or SOX4 gene (5′-CAAAAA-3′), a 1048-bp fragment of the 3′UTR of SIP1 and a 786-bp fragment of the 3′UTR of SOX4 were cloned into PGL3 luciferase reporter vector. These segments included a predicated binding site for miR-129-5p (Figure 7a). Furthermore, the mutant plasmids with a mutated binding site of miR-129-5p in the 3′UTR of SIP1 and SOX4 were generated and used in the promoter activity analysis. As shown in Figure 7b and c, overexpression of pre-miR-129-5p in HPMCs notably decreased the luciferase activity of 3′UTR in the wild-type reporter of SIP1 and SOX4 by 36 and 53%, respectively. However, the activity was not significantly different in cells transfected with the mutant reporter plasmids of SIP1 or SOX4. In addition, there was no effect on the 3′UTR activity following treatment with the miR-negative control.
Figure 7.
miR-129-5p is predicted to directly target SIP1/SOX4 genes and modulates the promoter activity of both E-cadherin and vimentin in HPMCs. (a) Location of the predicted miR-129-5p target site in SIP1 and SOX4 3′UTR, as assessed by biological analysis (www. microrna.org). (b) A decreased luciferase activity of SIP1-wt in HPMCs treated with pre-miRNA-129-5p while the decreased activity was partially restored with co-treatment of TGF-β. There was no effect in cells transfected with miR-neg control. (c) Similar results were seen for SOX4 activity. Luciferase activities were normalized to β-gal activities. Results were obtained from three independent experiments. Data are shown as mean ± s.e.m. *P<0.01 vs control, #P<0.01 vs pre-miR-129-5p. (d and e) The bar graphs represent luciferase activity in HPMC following various treatments. Opposite effects were observed for the E-cadherin and vimentin promoters. Luciferase activity was normalized with β-galactosidase activity, and it is depicted as mean value ± s.e.m. from triplicate measurements. Data are shown as mean ± s.e.m. *P<0.01 vs control, #P<0.01 vs TGF-β1, &P<0.05 vs SIP1.
The promoters of E-cadherin and vimentin contain binding sites for the transcription factor, SIP1.34,36 We therefore tested the E-cadherin promoter activity in HPMCs using a PGL3 luciferase reporter system. As show in Figure 7d, a significantly decreased luciferase activity was observed, which basically represented E-cadherin promoter activity in HPMCs treated with TGF-β1 or overexpression of SIP1 (Figure 7d, line 2 and line 4 vs line 1). This effect was blocked partially with the pre-treatment of pre-miR-129-5p (line 8 vs line 2, line 9 vs line 4). Interestingly, opposite results were observed in assays for the vimentin promoter activity (Figure 7e). These results suggest that miR-129-5p modulates E-cadherin and vimentin expression by targeting the 3′UTR region of SIP1 and SOX4 genes or by modulating the promoter activity of E-cadherin and vimentin by the TGF-β1/ SIP1 pathway.
DISCUSSION
Recent studies have demonstrated that TGF-β1-induced MMT has a central role in the pathogenesis of peritoneal fibrosis in PD.1,16 However, the precise mechanism(s) by which TGF-β1 induces EMT/MMT process in PD still remains to be clearly defined. In the present study, we identified that the expression of miR-129-5p is decreased both in PD patients and in the HPMCs cell line treated with TGF-β1, and it negatively correlated with MMT and peritoneal fibrosis. Overexpression of miR-129-5p ameliorated TGF-β1-induced MMT, and additionally inhibited cell migration following wound scratch in HPMCs culture. We also investigated the role SIP1 and SOX4, two well-known EMT-associated transcription factors that can directly bind to the promoter E-box site of E-cadherin and vimentin, in the process of MMT in patients undergoing PD. We observed that the inhibition of SIP1 and SOX4, using their respective siRNA, could dampen EMT by rescuing the altered promoter activity of both the E-cadherin and vimentin in HPMCs following TGF-β1 treatment. Our results also indicate that miR-129-5p could directly target the 3′UTR of SIP1 and SOX4 genes, and repressed their post-translational activities. These data suggest that there is a new pathway of TGF-β1/ miR-129-5p/SIP1 or SOX4 that may have a significant role in the process of MMT and fibrosis in PD.
An emerging body of evidence suggests that miRNAs are dysregulated and they mediate the TGF-β-induced EMT process in various neoplastic processes.37–40 With respect to peritoneal fibrosis, we reported that miRNA589 is decreased and it negatively regulates EMT in cultured human PMCs treated with TGF-β1 (ref. 10), and likewise, Zhou Q et al. reported that another miRNA, miR30a, ameliorates TGF-β1-induced EMT and peritoneal fibrosis by targeting snail1. This suggests that miRNAs may have a key role in peritoneal fibrosis of PD. To comprehensively study the role of miRNAs, their expression profile in PD state was carried out along with in vitro studies, using PMCs, where the process of MMT was investigated following TGF-β1 treatment. As shown in Figure 1 and Tables 2 and 3, 33 miRNAs were upregulated and 58 miRNAs were downregulated in patients having PD over 6 months compared with the patients of the PD-start group. Among those downregulated, the expression of miR-129-5p was noted to be significantly decreased. This change in the expression was confirmed by northern blot analyses and real-time PCR in effluents of patients with PD >6 months, as well as in HPMCs treated with TGF-β1 (Figure 2). Recent studies have shown that downregulation of miR-129-5p expression was mediated by blocking the IL-17A receptor in fibroblasts following the TGF-β1 treatment,20 suggesting that decreased miR-129-5p in PD may be modulated by the TGF-β1/IL-17A pathway. The miR-129-5p is one of the mature forms of miR-129-1 and miR-129-2, and previous studies have shown that miR-129, especially the miR-129-5p, has a vital role in various neoplastic processes where it serves as a tumor suppressor.13,18,19,41 Interestingly, it is also known that the expression of miR-129-5p is down-regulated in states of fibrosis, such as, systemic sclerosis where it modulates the collagen I expression20 and also under circumstances where invasion/migration of cells is affected, such as, in thyroid carcinoma cells.42 This would suggest that miR-129-5p has a potential role in regulating the EMT process and fibrosis. However, whether miR-129-5p is involved in peritoneal fibrosis during PD is unknown. Thus, we selected the miRNA-129-5p for the investigation in this study among various miRNAs that were detected during expression profiling.
In this study, we discovered that expression of miR-129-5p is significantly downregulated in both MCs isolated from the effluent of PD or HPMCs cells treated with TGF-β1. This change was associated with decreased expression of E-cadherin and claudin-1 and increased expression of vimentin and FN (Figures 1 and 2). On the other hand, overexpression of miR-129-5p markedly reversed the effect of TGF-β1-induced expression of EMT-related genes and protein expression (Figure 3), suggesting that miR-129-5p acts as a negative modulator during the process of MMT that is induced by TGF-β1 in PD.
There are many transcription factors and pathways involved in the TGF-β1-induced EMT process, such as ZEB1,43 SIP1,44 snail,45 slug,46 SOX4,47 Jagged/Notch,48 and the Wnt3a/β-catenin wnt/β-cat pathway.49 To delineate the mechanism by which miR-129-5p modulates TGF-β1-induced MMT gene expression in PD, we focused to investigate the status of SIP1 and SOX4. The SIP1, also known as ZEB2, is thought to be a transcription factor that acts as a repressor by binding to two widely separated clusters of c2H2-type zinc fingers and paired CAGGTA/G E-box-like promoter elements of E-cadherin and vimentin genes. Conceivably, SIP1 could also induce the TGF-β-stimulated EMT process by suppressing the expression of many genes characteristic of epithelia27,50,51 and also through targeting miR-132, miR-205, and miR-200 to ultimately regulate the EMT process.50,52–54 Moreover, SIP1 is also known to coordinate miR-200a-3p/141-3p to induce renal mesangial cells to undergo EMT43,55 while it can also regulate E-cadherin expression utilizing miR-192/215.56
SOX4 modulates transcription responses to TGF-β1, Wnt signaling, Notch pathway, and various miRNA processing.57–60 SOX4 is a downstream target of TGF-β1 (ref. 61) and have been shown to mediate the TGF-β-induced EMT process in mammary cells.47 Moreover, many studies suggest that SOX4 is a downstream of miR-129-5p47,61–63 and the SOX4 3′UTR is one of the targets of miR-129-5p.18,19,42,64 Interestingly, Sand (Sp100, AIRE-1, NucP41/75, DEAF-1) epigenetic repression of miRNA-129 leads to overexpression of SOX4 in certain neoplastic processes.63 Furthermore, biological software analyses indicate that miRNA-129-5p target genes include SIP1 and SOX4, and not others like snail1 and snail3, etc (www.microrna org; www.targetscan.org). This information suggests that SIP1 and SOX4 may have a critical role, unlike the other downstream molecules, such as ZEB1 and snail1, etc. In this scenario, they would conceivably participate in the TGF-β1-induced EMT process and fibrosis by interacting with miRNA-129-5p in states of PD. In view of this, we investigated the effect of SIP1 and SOX4 in TGF-β1-induced EMT modulated by miR-129-5p.
Our findings indicate that expression of SIP1 significantly increased in HPMCs both in patients undergoing PD patients and cells treated with TGF-β1. This upregulation tightly correlated with the expression of EMT-related genes and protein, and negatively correlated with the expression of miR-129-5p (Figures 1 and 2). In addition, we observed that inhibiting SIP1 expression with its siRNA blocked MMT induced by TGF-β1 in vitro (Figure 4). In contrast, over-expression of miR-129-5p decreased the mRNA and protein expression of SIP1 (Figure 6). These data suggest that SIP1 may serve as a critical modulator of miR-129-5p in the MMT process during PD. We also observed that expression of SOX4 increased in PMCs isolated from PD patients and HPMCs cell lines stimulated by TGF-β1, and it negatively correlated with the expression of miR-129-5p and MMT changes (Figures 1 and 2). Similar to SIP1, SOX4-siRNA also blocked the MMT induced by TGF-β1 in vitro (Figure 5). Overexpression of miR-129-5p decreased the protein expression of SOX4 (Figure 6), suggesting that both SIP1 and SOX4 can serve as the key downstream molecules utilizing different pathways but ultimately converging into TGF-β1/miR-129-5p pathway that modulated EMT/MMT process in PD.
This study also demonstrates that TGF-β1 and SIP1 independently confer a repression of E-cadherin expression by inhibiting its promoter activity, as assessed by luciferase activity analysis (Figure 7a). In contrast to E-cadherin, an increased promoter activity of vimentin was observed (Figure 7b), and these perturbations in the activities were reversed by the treatment with pre-miR-129-59. Incidentally, it has been reported that miR-129 directly targets SOX4 as indicated by the 3′UTR analysis.62,63 Likewise, we noted that miR-129-5p directly interacts with the 3hUTR of SIP1 and SOX4, and overexpression of pre-miR-129-5p decreased the luciferase activity of the 3′UTR wild-type reporter of SIP1 and SOX4. However, no effect was observed in the mutant of 3′ UTR reporter of SIP1 or SOX4 (Figure 7c and d), suggesting that miR-129-5p exerts a protective role in MMT in PD, which may be partly through the downstream SIP1 and SOX4 transcription factors.
In summary, we suggest that miR-129-5p is a critical molecule in protection of MCs undergoing MMT transformation induced by TGF-β1 during of PD. Our findings also suggest that miR-129-5p exerts its protective effect perhaps through direct targeting of SIP1 and SOX4. Finally, it is anticipated that these findings may provide new insights into the role of miR-129-5p in the MMT and peritoneal fibrosis in PD, and yield an opportunity for developing novel therapeutic interventions.
ACKNOWLEDGMENTS
This work was supported by the Natural Sciences Foundation of China (81100541, 81370832, 81270812, 81470960 and 81300600), the Doctoral Fund of Ministry of Education of China (20110162110012), Furong Scholars Fund from Hunan Province Education Department and the NIH Grant (DK60635).
Footnotes
DISCLOSURE/CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Loureiro J, Gonzalez-Mateo G, Jimenez-Heffernan J, et al. Are the mesothelial-to-mesenchymal transition, sclerotic peritonitis syndromes, and encapsulating peritoneal sclerosis part of the same process? Int J Nephrol. 2013;2013:263285. doi: 10.1155/2013/263285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perez-Lozano ML, Sandoval P, Rynne-Vidal A, et al. Functional relevance of the switch of VEGF receptors/co-receptors during peritoneal dialysis-induced mesothelial to mesenchymal transition. PLoS One. 2013;8:e60776. doi: 10.1371/journal.pone.0060776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Del Peso G, Jimenez-Heffernan JA, Bajo MA, et al. Epithelial-to-mesenchymal transition of mesothelial cells is an early event during peritoneal dialysis and is associated with high peritoneal transport. Kidney Int Suppl. 2008;108:S26–S33. doi: 10.1038/sj.ki.5002598. [DOI] [PubMed] [Google Scholar]
- 4.Mizutani M, Ito Y, Mizuno M, et al. Connective tissue growth factor (CTGF/CCN2) is increased in peritoneal dialysis patients with high peritoneal solute transport rate. Am J Physiol Renal Physiol. 2010;298:F721–F733. doi: 10.1152/ajprenal.00368.2009. [DOI] [PubMed] [Google Scholar]
- 5.Aroeira LS, Aguilera A, Sanchez-Tomero JA, et al. Epithelial to mesenchymal transition and peritoneal membrane failure in peritoneal dialysis patients: pathologic significance and potential therapeutic interventions. J Am Soc Nephrol. 2007;18:2004–2013. doi: 10.1681/ASN.2006111292. [DOI] [PubMed] [Google Scholar]
- 6.Margetts PJ, Bonniaud P. Basic mechanisms and clinical implications of peritoneal fibrosis. Perit Dial Int. 2003;23:530–541. [PubMed] [Google Scholar]
- 7.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 8.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desmouliere A, Geinoz A, Gabbiani F, et al. Transforming growth factor-beta 1 induces alpha-smooth muscle actin expression in granulation tissue myofibroblasts and in quiescent and growing cultured fibroblasts. J Cell Biol. 1993;122:103–111. doi: 10.1083/jcb.122.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang K, Zhang H, Zhou X, et al. miRNA589 regulates epithelial-mesenchymal transition in human peritoneal mesothelial cells. J Biomed Biotechnol. 2012;2012:673096. doi: 10.1155/2012/673096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Q, Mao H, Nie J, et al. Transforming growth factor {beta}1 induces epithelial-mesenchymal transition by activating the JNK-Smad3 pathway in rat peritoneal mesothelial cells. Perit Dial Int. 2008;28:S88–S95. [PubMed] [Google Scholar]
- 12.Jang YH, Shin HS, Sun Choi H, et al. Effects of dexamethasone on the TGF-beta1-induced epithelial-to-mesenchymal transition in human peritoneal mesothelial cells. Lab Invest. 2013;93:194–206. doi: 10.1038/labinvest.2012.166. [DOI] [PubMed] [Google Scholar]
- 13.Yu X, Luo L, Wu Y, et al. Gastric juice miR-129 as a potential biomarker for screening gastric cancer. Med Oncol. 2013;30:365. doi: 10.1007/s12032-012-0365-y. [DOI] [PubMed] [Google Scholar]
- 14.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 15.Jiang X, Tsitsiou E, Herrick SE, et al. MicroRNAs and the regulation of fibrosis. FEBS J. 2010;277:2015–2021. doi: 10.1111/j.1742-4658.2010.07632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Q, Yang M, Lan H, et al. miR-30a negatively regulates TGF-beta1-induced epithelial-mesenchymal transition and peritoneal fibrosis by targeting Snai1. Am J Pathol. 2013;183:808–819. doi: 10.1016/j.ajpath.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 17.Yu JW, Duan WJ, Huang XR, et al. MicroRNA-29b inhibits peritoneal fibrosis in a mouse model of peritoneal dialysis. Lab Invest. 2014;94:978–990. doi: 10.1038/labinvest.2014.91. [DOI] [PubMed] [Google Scholar]
- 18.Yu X, Song H, Xia T, et al. Growth inhibitory effects of three miR-129 family members on gastric cancer. Gene. 2013;532:87–93. doi: 10.1016/j.gene.2013.09.048. [DOI] [PubMed] [Google Scholar]
- 19.Li M, Tian L, Wang L, et al. Down-regulation of miR-129-5p inhibits growth and induces apoptosis in laryngeal squamous cell carcinoma by targeting APC. PLoS One. 2013:8–e77829. doi: 10.1371/journal.pone.0077829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakashima T, Jinnin M, Yamane K, et al. Impaired IL-17 signaling pathway contributes to the increased collagen expression in scleroderma fibroblasts. J Immunol. 2012;188:3573–3583. doi: 10.4049/jimmunol.1100591. [DOI] [PubMed] [Google Scholar]
- 21.Al-Malki HA, Fawzy AA, Rashid AH, et al. Standard continuous ambulatory peritoneal dialysis therapy provides similar initial T-Kt/V regardless of the patient's peritoneal membrane transporter category: single-center experience. Saudi J Kidney Dis Transpl. 2014;25:788–792. doi: 10.4103/1319-2442.135004. [DOI] [PubMed] [Google Scholar]
- 22.Vazquez-Rangel A, Soto V, Escalona M, et al. Spironolactone to prevent peritoneal fibrosis in peritoneal dialysis patients: a randomized controlled trial. Am J Kidney Dis. 2014;63:1072–1074. doi: 10.1053/j.ajkd.2014.01.426. [DOI] [PubMed] [Google Scholar]
- 23.Rougier JP, Guia S, Hagege J, et al. PAI-1 secretion and matrix deposition in human peritoneal mesothelial cell cultures: transcriptional regulation by TGF-beta 1. Kidney Int. 1998;54:87–98. doi: 10.1046/j.1523-1755.1998.00955.x. [DOI] [PubMed] [Google Scholar]
- 24.Rougier JP, Moullier P, Piedagnel R, et al. Hyperosmolality suppresses but TGF beta 1 increases MMP9 in human peritoneal mesothelial cells. Kidney Int. 1997;51:337–347. doi: 10.1038/ki.1997.42. [DOI] [PubMed] [Google Scholar]
- 25.Lin S, Sahai A, Chugh SS, et al. High glucose stimulates synthesis of fibronectin via a novel protein kinase C, Rap1b, and B-Raf signaling pathway. J Biol Chem. 2002;277:41725–41735. doi: 10.1074/jbc.M203957200. [DOI] [PubMed] [Google Scholar]
- 26.Zhang D, Sun L, Xian W, et al. Low-dose paclitaxel ameliorates renal fibrosis in rat UUO model by inhibition of TGF-beta/Smad activity. Lab Invest. 2010;90:436–447. doi: 10.1038/labinvest.2009.149. [DOI] [PubMed] [Google Scholar]
- 27.Lindley LE, Briegel KJ. Molecular characterization of TGFbeta-induced epithelial-mesenchymal transition in normal finite lifespan human mammary epithelial cells. Biochem Biophys Res Commun. 2010;399:659–664. doi: 10.1016/j.bbrc.2010.07.138. [DOI] [PubMed] [Google Scholar]
- 28.Kato M, Zhang J, Wang M, et al. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci USA. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang M, Li Y, Liu W, et al. miR-129-2 suppresses proliferation and migration of esophageal carcinoma cells through downregulation of SOX4 expression. Int J Mol Med. 2013;32:51–58. doi: 10.3892/ijmm.2013.1384. [DOI] [PubMed] [Google Scholar]
- 30.Long J, Wang Y, Wang W, et al. MicroRNA-29c is a signature microRNA under high glucose conditions that targets Sprouty homolog 1, and its in vivo knockdown prevents progression of diabetic nephropathy. J Biol Chem. 2011;286:11837–11848. doi: 10.1074/jbc.M110.194969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun L, Zhang D, Liu F, et al. Low-dose paclitaxel ameliorates fibrosis in the remnant kidney model by down-regulating miR-192. J Pathol. 2011;225:364–377. doi: 10.1002/path.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu L, Ding GF, He C, et al. MicroRNA-424 is down-regulated in hepatocellular carcinoma and suppresses cell migration and invasion through c-Myb. PLoS One. 2014;9:e91661. doi: 10.1371/journal.pone.0091661. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Li F, Li S, Cheng T. TGF-beta1 promotes osteosarcoma cell migration and invasion through the miR-143-versican pathway. Cell Physiol Biochem. 2014;34:2169–2179. doi: 10.1159/000369660. [DOI] [PubMed] [Google Scholar]
- 34.Miyoshi A, Kitajima Y, Sumi K, et al. Snail and SIP1 increase cancer invasion by upregulating MMP family in hepatocellular carcinoma cells. Br J Cancer. 2004;90:1265–1273. doi: 10.1038/sj.bjc.6601685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rittling SR, Baserga R. Functional analysis and growth factor regulation of the human vimentin promoter. Mol Cell Biol. 1987;7:3908–3915. doi: 10.1128/mcb.7.11.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bindels S, Mestdagt M, Vandewalle C, et al. Regulation of vimentin by SIP1 in human epithelial breast tumor cells. Oncogene. 2006;25:4975–4985. doi: 10.1038/sj.onc.1209511. [DOI] [PubMed] [Google Scholar]
- 37.Jiang Z, Yin J, Fu W, et al. MiRNA 17 family regulates cisplatin-resistant and metastasis by targeting TGFbetaR2 in NSCLC. PLoS One. 2014;9:e94639. doi: 10.1371/journal.pone.0094639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kitamura K, Seike M, Okano T, et al. MiR-134/487b/655 cluster regulates TGF-beta-induced epithelial-mesenchymal transition and drug resistance to gefitinib by targeting MAGI2 in lung adenocarcinoma cells. Mol Cancer Ther. 2014;13:444–453. doi: 10.1158/1535-7163.MCT-13-0448. [DOI] [PubMed] [Google Scholar]
- 39.Sun Y, Shen S, Liu X, et al. MiR-429 inhibits cells growth and invasion and regulates EMT-related marker genes by targeting Onecut2 in colorectal carcinoma. Mol Cell Biochem. 2014;390:19–30. doi: 10.1007/s11010-013-1950-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor MA, Davuluri G, Parvani JG, et al. Upregulated WAVE3 expression is essential for TGF-beta-mediated EMT and metastasis of triple-negative breast cancer cells. Breast Cancer Res Treat. 2013;142:341–353. doi: 10.1007/s10549-013-2753-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brest P, Lassalle S, Hofman V, et al. MiR-129-5p is required for histone deacetylase inhibitor-induced cell death in thyroid cancer cells. Endocr Relat Cancer. 2011;18:711–719. doi: 10.1530/ERC-10-0257. [DOI] [PubMed] [Google Scholar]
- 42.Duan L, Hao X, Liu Z, et al. MiR-129-5p is down-regulated and involved in the growth, apoptosis and migration of medullary thyroid carcinoma cells through targeting RET. FEBS Lett. 2014;588:1644–1651. doi: 10.1016/j.febslet.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 43.Gao J, Zhu Y, Nilsson M, et al. TGF-beta isoforms induce EMT independent migration of ovarian cancer cells. Cancer Cell Int. 2014;14:72. doi: 10.1186/s12935-014-0072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 45.Chen XH, Liu ZC, Zhang G, et al. TGF-beta and EGF induced HLA-I downregulation is associated with epithelial-mesenchymal transition (EMT) through upregulation of snail in prostate cancer cells. Mol Immunol. 2015;65:34–42. doi: 10.1016/j.molimm.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Shi J, Chai K, et al. The Role of Snail in EMT and tumorigenesis. Curr Cancer Drug Targets. 2013;13:963–972. doi: 10.2174/15680096113136660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vervoort SJ, Lourenco AR, van Boxtel R, et al. SOX4 mediates TGF-beta-induced expression of mesenchymal markers during mammary cell epithelial to mesenchymal transition. PLoS One. 2013;8:e53238. doi: 10.1371/journal.pone.0053238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen X, Xiao W, Liu X, et al. Blockade of Jagged/Notch pathway abrogates transforming growth factor beta2-induced epithelial-mesenchymal transition in human retinal pigment epithelium cells. Curr Mol Med. 2014;14:523–534. doi: 10.2174/1566524014666140331230411. [DOI] [PubMed] [Google Scholar]
- 49.Zou W, Zou Y, Zhao Z, et al. Nicotine-induced epithelial-mesenchymal transition via Wnt/beta-catenin signaling in human airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2013;304:L199–L209. doi: 10.1152/ajplung.00094.2012. [DOI] [PubMed] [Google Scholar]
- 50.Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop–a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11:670–677. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vandewalle C, Van Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci. 2009;66:773–787. doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.You J, Li Y, Fang N, et al. MiR-132 suppresses the migration and invasion of lung cancer cells via targeting the EMT regulator ZEB2. PLoS One. 2014;9:e91827. doi: 10.1371/journal.pone.0091827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee JY, Park MK, Park JH, et al. Loss of the polycomb protein Mel-18 enhances the epithelial-mesenchymal transition by ZEB1 and ZEB2 expression through the downregulation of miR-205 in breast cancer. Oncogene. 2014;33:1325–1335. doi: 10.1038/onc.2013.53. [DOI] [PubMed] [Google Scholar]
- 54.Kim T, Veronese A, Pichiorri F, et al. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J Exp Med. 2011;208:875–883. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei J, Zhang Y, Luo Y, et al. Aldose reductase regulates miR-200a-3p/ 141-3p to coordinate Keap1-Nrf2, Tgfbeta1/2, and Zeb1/2 signaling in renal mesangial cells and the renal cortex of diabetic mice. Free Radic Biol Med. 2014;67:91–102. doi: 10.1016/j.freeradbiomed.2013.10.811. [DOI] [PubMed] [Google Scholar]
- 56.Wang B, Herman-Edelstein M, Koh P, et al. E-cadherin expression is regulated by miR-192/215 by a mechanism that is independent of the profibrotic effects of transforming growth factor-beta. Diabetes. 2010;59:1794–1802. doi: 10.2337/db09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Medina PP, Castillo SD, Blanco S, et al. The SRY-HMG box gene, SOX4, is a target of gene amplification at chromosome 6p in lung cancer. Hum Mol Genet. 2009;18:1343–1352. doi: 10.1093/hmg/ddp034. [DOI] [PubMed] [Google Scholar]
- 58.Tavazoie SF, Alarcon C, Oskarsson T, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sinner D, Kordich JJ, Spence JR, et al. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol Cell Biol. 2007;27:7802–7815. doi: 10.1128/MCB.02179-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scharer CD, McCabe CD, Ali-Seyed M, et al. Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells. Cancer Res. 2009;69:709–717. doi: 10.1158/0008-5472.CAN-08-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuwahara M, Yamashita M, Shinoda K, et al. The transcription factor Sox4 is a downstream target of signaling by the cytokine TGF-beta and suppresses T(H)2 differentiation. Nat Immunol. 2012;13:778–786. doi: 10.1038/ni.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen R, Pan S, Qi S, et al. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 in gastric cancer. Biochem Biophys Res Commun. 2010;394:1047–1052. doi: 10.1016/j.bbrc.2010.03.121. [DOI] [PubMed] [Google Scholar]
- 63.Huang YW, Liu JC, Deatherage DE, et al. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69:9038–9046. doi: 10.1158/0008-5472.CAN-09-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang J, Li S, Yan Q, et al. Interferon-beta induced microRNA-129-5p down-regulates HPV-18 E6 and E7 viral gene expression by targeting SP1 in cervical cancer cells. PLoS One. 2013;8:e81366. doi: 10.1371/journal.pone.0081366. [DOI] [PMC free article] [PubMed] [Google Scholar]