How sensory neurons coordinate organismal aging in response to changes in environmental signals remains elusive. Artan et al. show that a subset of sensory neurons shortens C. elegans’ life span by differentially regulating the expression of a specific insulin-like peptide (ILP), INS-6. INS-6 in turn relays the longevity signals to nonneuronal tissues by decreasing the activity of the transcription factor DAF-16/FOXO.
Keywords: sensory neurons, aging, C. elegans, insulin-like peptide, DAF-16/FOXO
Abstract
Environmental fluctuations influence organismal aging by affecting various regulatory systems. One such system involves sensory neurons, which affect life span in many species. However, how sensory neurons coordinate organismal aging in response to changes in environmental signals remains elusive. Here, we found that a subset of sensory neurons shortens Caenorhabditis elegans’ life span by differentially regulating the expression of a specific insulin-like peptide (ILP), INS-6. Notably, treatment with food-derived cues or optogenetic activation of sensory neurons significantly increases ins-6 expression and decreases life span. INS-6 in turn relays the longevity signals to nonneuronal tissues by decreasing the activity of the transcription factor DAF-16/FOXO. Together, our study delineates a mechanism through which environmental sensory cues regulate aging rates by modulating the activities of specific sensory neurons and ILPs.
It is vital for the survival of an organism to properly perceive and respond to environmental fluctuations, such as changes in chemical cues and ambient temperatures. These environmental changes are perceived by sensory neurons to elicit appropriate physiological responses for the optimal survival of animals. Sensory neurons also affect long-term physiological processes, such as development and metabolism (Bargmann 2006; Allen et al. 2015). In addition, the sensory systems of Caenorhabditis elegans, Drosophila, and mice influence life span (Pletcher 2009; Jeong et al. 2012; Riera et al. 2014), although the molecular mechanisms involved are poorly understood.
Gustatory and olfactory neurons, which perceive chemical signals, are some of the sensory neurons that have been shown to affect life span (Alcedo and Kenyon 2004; Libert et al. 2007; Poon et al. 2010; Ostojic et al. 2014; Waterson et al. 2014). In C. elegans, chemosensory neuronal signaling is initiated by the binding of chemical ligands to sensory G protein-coupled receptors (GPCRs), which in turn activates G protein signaling cascades to transduce the signals that increase cyclic GMP (cGMP) levels (Bargmann 2006). The cGMP binds to cyclic nucleotide-gated channels composed of TAX-2 and TAX-4 subunits and triggers cation influx (Coburn and Bargmann 1996; Komatsu et al. 1996). The role of chemosensory neurons in life span regulation has been established through inhibition of ciliated sensory neurons in C. elegans (Apfeld and Kenyon 1999; Alcedo and Kenyon 2004).
Chemosensory neurons appear to regulate the life span of C. elegans via modulation of insulin/IGF-1 signaling (IIS) components, including insulin/IGF-1 receptor homolog DAF-2 and the FOXO transcription factor DAF-16 (Apfeld and Kenyon 1999; Alcedo and Kenyon 2004). Nevertheless, the identities of actual environmental cues generating a longevity response and endocrine signals transmitting these longevity signals that regulate IIS remain elusive. Some of the insulin-like peptides (ILPs), the predicted ligands of the DAF-2 receptor, are reasonable candidates that could transmit the longevity-influencing signals from the sensory neurons to the IIS. The C. elegans genome contains 40 genes encoding putative ILPs, and many of these are expressed in sensory neurons (Pierce et al. 2001; Cornils et al. 2011; Chen et al. 2013; Ritter et al. 2013). Hence, at least some of the ILPs may modulate longevity through IIS acting in the sensory neurons.
In this study, we found that the sensory TAX-2/TAX-4 cation channel modulates the expression of specific sets of ILPs. Two of these ILPs are ins-6 and daf-28, which we show mediate the longevity effects of the sensory mutations by decreasing the activity of DAF-16/FOXO in a tissue-nonautonomous manner. We also found that food cues act as environmental life span-regulating signals that induce ins-6 in a subset of chemosensory neurons. Moreover, optogenetic activation of specific sensory neurons increases the expression of ins-6 and reduces DAF-16/FOXO activity, which decreases life span in food-deprived conditions. Thus, specific ILPs act as systemic neuroendocrine factors that mediate the sensory regulation of C. elegans longevity upon changes in food conditions. Our finding provides mechanistic insights regarding how environmental cues alter organismal longevity by fine-tuning the IIS via neuroendocrine peptides.
Results
Sensory tax-2/tax-4 mutations extend C. elegans’ life span by activating DAF-16/FOXO in multiple tissues
We sought to identify the mechanisms through which sensory neurons mediate the longevity effects of different sensory cues. Specifically, we focused our study on tax-2 and tax-4, which together encode a cyclic nucleotide-gated ion channel required for sensory function (Coburn and Bargmann 1996; Komatsu et al. 1996). Mutations in tax-2 and/or tax-4 shortened life span at 25°C (high temperature) (Lee and Kenyon 2009) but increased life span at 15°C (low temperature) (Fig. 1A; Supplemental Fig. S1A,B). Interestingly, while the short life span of tax-2 mutants at 25°C is due to impaired sterol hormone signaling (Lee and Kenyon 2009), the tax-2 mutation still extended the life span of the sterol hormone receptor daf-12-null mutants at 15°C (Supplemental Fig. S1C). This suggests that the extended longevity of these mutants at lower temperatures is independent of sterol signaling. In contrast, the long life phenotype of tax-2 mutants at 15°C requires daf-16/FOXO (Fig. 1A), a downstream effector of the IIS pathway (Lin et al. 1997; Ogg et al. 1997). As would be predicted, loss-of-function mutations in this ion channel also increased the expression of known transcriptional targets of daf-16/FOXO (Fig. 1C–E; Supplemental Fig. S2). Consistent with the idea that tax-2 affects life span through IIS, we observed that the tax-2 mutation did not further increase the 15°C long life phenotype of insulin/IGF-1 receptor daf-2 mutants (Fig. 1B). Our data are similar to previous findings at 20°C (Apfeld and Kenyon 1999) but unlike findings at 25°C (Lee and Kenyon 2009), which suggests that sensory function regulates longevity via distinct signaling pathways under different environments.
Figure 1.
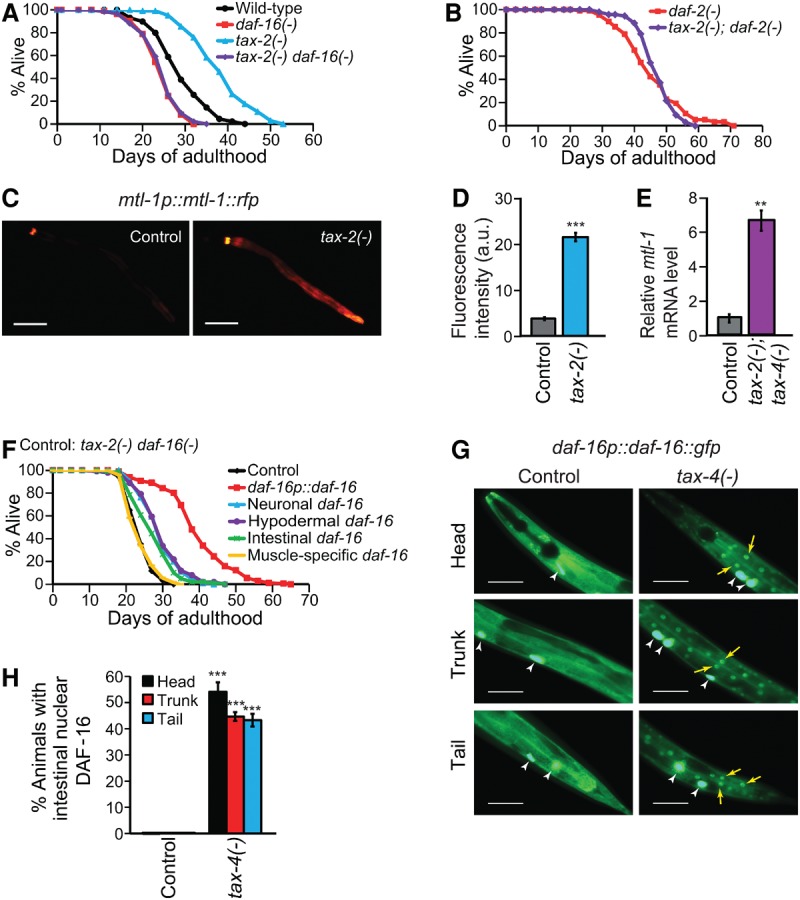
Sensory tax-2/tax-4 mutations extend life span via activating DAF-16/FOXO in a tissue-nonautonomous manner. (A,B) The effects of daf-16(mu86) [daf-16(−)] (A) and daf-2(e1370) [daf-2(−)] (B) on the longevity of tax-2(p671) [tax-2(−)] animals at 15°C. (C,D) Fluorescence images of a FOXO reporter, mtl-1p::mtl-1::rfp, in wild-type (control) or tax-2(−) animals (C) and the quantification (D) (three independent trials, ≥90 total worms). (a.u.) Arbitrary unit. Bars, 50 μm. (E) The levels of mtl-1 mRNA in wild-type and tax-2(p671); tax-4(p678) [tax-2(−); tax-4(−)] animals measured by quantitative RT–PCR (qRT–PCR). n = 3. (F) The life span of tax-2(−) daf-16(−) animals expressing daf-16::gfp from its own (daf-16p::daf-16::gfp), a neuronal, a hypodermal, an intestinal, or a muscle-specific promoter. (G,H) The effects of tax-4(p678) [tax-4(−)] on the subcellular localization of DAF-16::GFP in the intestinal cells (G) ([yellow arrows] nuclear DAF-16::GFP; [white arrowheads] coinjection marker ofm-1p::gfp) and the quantification in the head, trunk, and tail regions (H) (at least three independent trials, >60 total worms). Error bars represent SEM. (***) P < 0.001; (**) P < 0.01, two-tailed Student's t-test. See Supplemental Table S1 for additional repeats and statistical analysis for the life span data shown in this figure.
Interestingly, the tax-2-dependent modulation of IIS in affecting low-temperature life span (Fig. 1A,B) is just as specific as the tax-2-dependent modulation of sterol signaling in affecting high-temperature life span (Lee and Kenyon 2009). At 15°C, the tax-2 mutation lengthened the life span of animals carrying mutations in heat-shock transcription factor 1 (hsf-1) (Hsu et al. 2003; Morley and Morimoto 2004), the dietary restriction mimetic eat-2 (Lakowski and Hekimi 1998), or the mitochondrial protein gene isp-1 (Supplemental Fig. S1D–F; Feng et al. 2001).
Since tax-2 is expressed only in neurons (Coburn and Bargmann 1996), we next asked whether the long life phenotype of tax-2 mutants was sufficiently explained by increased DAF-16/FOXO activity in neurons. Surprisingly, the expression of a functional DAF-16::GFP fusion protein in neurons only partly rescued the short life span of tax-2(−) daf-16(−) double mutants (Fig. 1F). This partial restoration of longevity was similar to nonneuronal daf-16::gfp expression in the hypodermis or intestine but not in the muscle (Fig. 1F). In contrast, a daf-16 promoter-driven DAF-16::GFP (daf-16p::daf-16) fully rescued the life span phenotype of the double mutants (Fig. 1F). These results suggest that DAF-16/FOXO activity in several tissues contributes to the longevity of tax-2 mutants. This is also consistent with the induction of DAF-16/FOXO target genes in the intestine and hypodermis by tax-2 and tax-4 mutations (Fig. 1C; Supplemental Fig. S2A,C,G,I). Thus, a neuroendocrine signal appears to link tax-2 and tax-4 activity in neurons to DAF-16/FOXO activity in other tissues.
We then asked how tax-2/tax-4 mutations increase DAF-16/FOXO activity. Unlike wild type, which has diffused intracellular DAF-16::GFP, long-lived daf-2 insulin/IGF-1 receptor mutants or sensory daf-10 mutants displayed enhanced nuclear translocation of DAF-16/FOXO (Lin et al. 2001). Likewise, long-lived tax-4(−) mutants at 15°C displayed enhanced nuclear DAF-16::GFP (Fig. 1G,H). This suggests that the TAX-2/TAX-4 ion channel regulates the nucleocytoplasmic shuttling of nonneuronal DAF-16/FOXO in a tissue-nonautonomous manner.
Sensory neurons regulate longevity by modulating the expression of specific ILPs
How, then, does sensory function nonautonomously regulate longevity? The ILPs are reasonable candidates mediating this nonautonomous regulation, since many ILPs are expressed in sensory neurons (Pierce et al. 2001; Li et al. 2003; Cornils et al. 2011; Chen et al. 2013; Ritter et al. 2013). Because the transcriptional expression of multiple ILPs can be coordinately regulated (Chen et al. 2013; Ritter et al. 2013; Fernandes de Abreu et al. 2014), we determined whether the tax-2(−); tax-4(−) mutations affected the steady-state mRNA levels of the ILPs. Among the 33 ILP genes that we examined by quantitative RT–PCR (qRT–PCR), tax-2(−); tax-4(−) mutations significantly increased or decreased the expression of 10 ILP genes (Fig. 2A,B). We also confirmed that the expression of two ILPs, INS-6 and DAF-28, was decreased by the tax-4 mutation by using fluorescence reporter transgenic animals (Fig. 2C,D). Thus, the sensory TAX-2/TAX-4 ion channel dynamically regulates the mRNA levels of many, but not all, ILPs.
Figure 2.
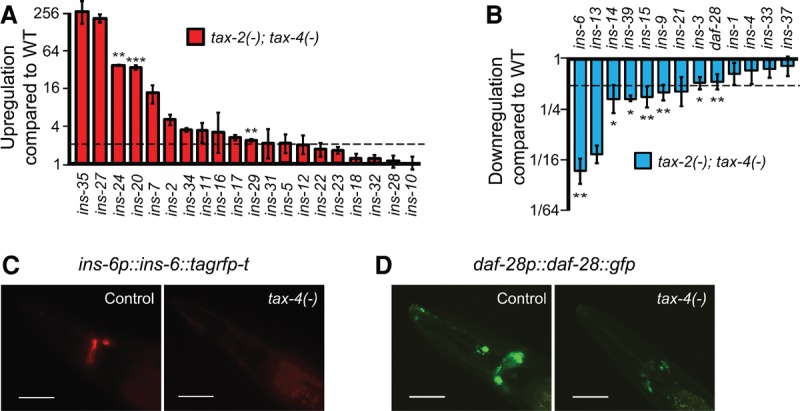
Sensory tax-2/tax-4 mutations regulate the expression of various ILPs. (A,B) The effects of tax-2(−); tax-4(−) on mRNA levels of 33 ILP genes, determined by qRT–PCR. (Dotted line) Twofold increase or decrease. The graph was drawn in log2 scale. Note that although the mean expression levels of several ILPs, including ins-35, ins-27, ins-7, and ins-13, were highly altered in tax-2(−); tax-4(−) mutants, the changes in expression were not statistically significant. Error bars represent SEM. (***) P < 0.001; (**) P < 0.01; (*) P < 0.05, two-tailed Student's t-test. (C,D) The expression of an RFP-fused INS-6 (ins-6p::ins-6::tagrfp-t) (C) or GFP-fused DAF-28 (daf-28p::daf-28::gfp) (D) was decreased by mutations in tax-4 compared with that in wild-type animals (control). Bar, 50 μm. Please note that the expression of daf-28p::gfp (daf-28 promoter-driven gfp transgene) was shown to be higher in tax-4(−) mutants than in wild-type animals (Li et al. 2003). This difference may have originated from the fact that daf-28p::gfp is a transcriptional fusion transgene and that they measured the GFP expression at the L3 stage, whereas we did so at the young adult stage.
Next, we analyzed the functional significance of this differential expression of ILPs in the longevity of sensory mutants. Previous studies and our current data showed that individual loss-of-function mutations in ILP genes displayed either weak or no life span phenotypes, perhaps due to functional redundancy (Fig. 3A,B; Supplemental Fig. S3; Cornils et al. 2011; Ritter et al. 2013; Fernandes de Abreu et al. 2014). We therefore overexpressed several ILPs that were down-regulated in sensory tax-2 and tax-4 mutants. We specifically focused on ins-6 and daf-28, which not only are expressed in ASI and ASJ sensory neurons that coexpress tax-2 and tax-4 (Coburn and Bargmann 1996; Komatsu et al. 1996; Li et al. 2003; Cornils et al. 2011) but their individual mutations also significantly increased life span (Fig. 3B; Supplemental Fig. S3K,L). Importantly, overexpression of ins-6 or daf-28 under their endogenous promoters suppressed the long life span of tax-2(−) and tax-4(−) mutants without shortening the life span of the wild type (Fig. 3C,D; Supplemental Fig. S4A–F). ins-6 or daf-28 overexpression also significantly decreased the nuclear localization of DAF-16::GFP in nonneuronal tissues (Fig. 3E,F; Supplemental Fig. S4G–J) and the induction of daf-16 target genes in tax-4 mutants (Fig. 3G,H; Supplemental Fig. S4K–N). These data suggest that sensory function regulates ins-6 and daf-28 expression and the activity of DAF-16/FOXO to modulate longevity.
Figure 3.
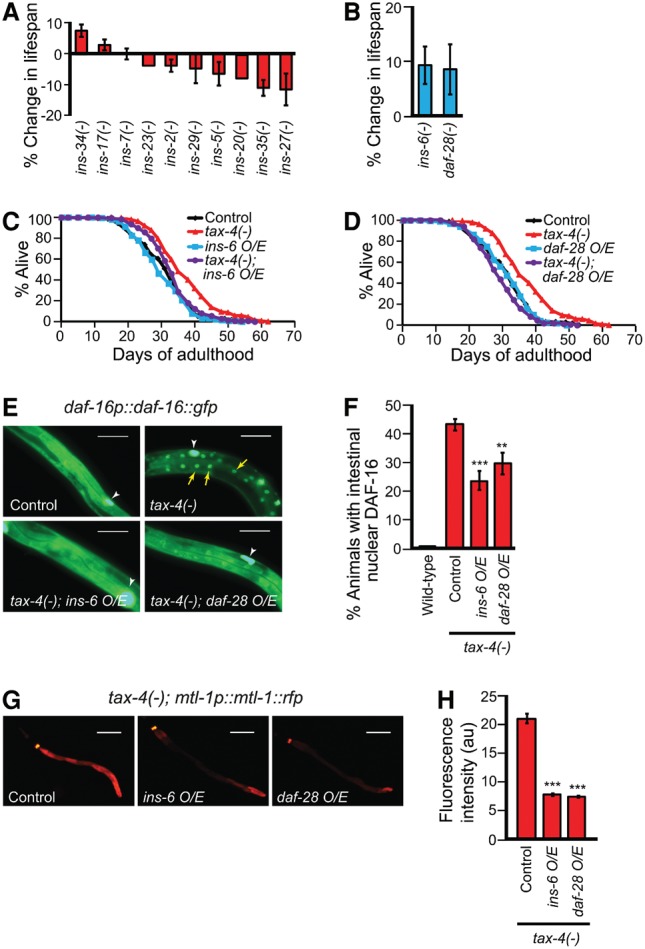
Overexpression of ins-6 or daf-28 decreases longevity and DAF-16/FOXO activity in sensory tax-4 mutants. (A,B) The life span changes by mutations in selected ILPs whose gene expression was increased (A) or decreased (B) in tax-2(−); tax-4(−) animals. See Supplemental Table S2 for additional repeats and statistical analysis for the life span data shown in this figure. (C,D) Suppression of tax-4(−) longevity by overexpression (O/E) of ins-6 (C) or daf-28 (D). Note that transgenic animals that express low copies of the ins-6 transgene also suppressed the long life span of tax-4 mutants (Supplemental Table S2). (E,F) The effect of ins-6 or daf-28 overexpression on the nuclear localization of DAF-16::GFP in tax-4(−) worms (E) ([yellow arrows] nuclear DAF-16::GFP; [white arrowheads] coinjection marker ofm-1p::gfp) and the quantification (F). n ≥ 6, >100 total worms per data set. Bar, 50 μm. (G,H) The effect of ins-6 or daf-28 overexpression on the induction of mtl-1p::mtl-1::rfp in tax-4(−) animals (G) and the quantification (H) (n = 3, >100 total worms). Error bars represent SEM. (***) P < 0.001; (**) P < 0.01, two-tailed Student's t-test.
Food-derived cues perceived by sensory neurons decrease life span by inducing ins-6 and down-regulating DAF-16/FOXO
To identify the actual sensory cues that modulate ILP expression and DAF-16/FOXO activity, we tested two candidates: changes in food signals and temperatures. We found that ins-6 and daf-28 mRNA levels were decreased by Escherichia coli food deprivation but are hardly affected by temperature changes (Fig. 4A,B; Supplemental Fig. S5A,B). To characterize the nature of longevity-influencing food components, we treated animals with cell-free E. coli supernatants (Supplemental Fig. S5C), which contain potential food-derived odorants and tastants for C. elegans (Bargmann 2006). We found that the exposure of food-deprived animals to E. coli supernatants was sufficient to increase ins-6 and daf-28 mRNA levels (Fig. 4A,B). E. coli supernatant supplementation also partly reversed the effects of food deprivation on the DAF-16/FOXO nuclear translocation (Fig. 4C,D) and the induction of several DAF-16/FOXO target genes (Fig. 4E,F; Supplemental Fig. S5E,F). As previously reported (Smith et al. 2008), E. coli supernatant treatment partly suppressed the longevity caused by food deprivation (Fig. 4G). These data suggest that food cues can increase the expression of ins-6 and daf-28, which in turn decrease DAF-16/FOXO activity and life span.
Figure 4.
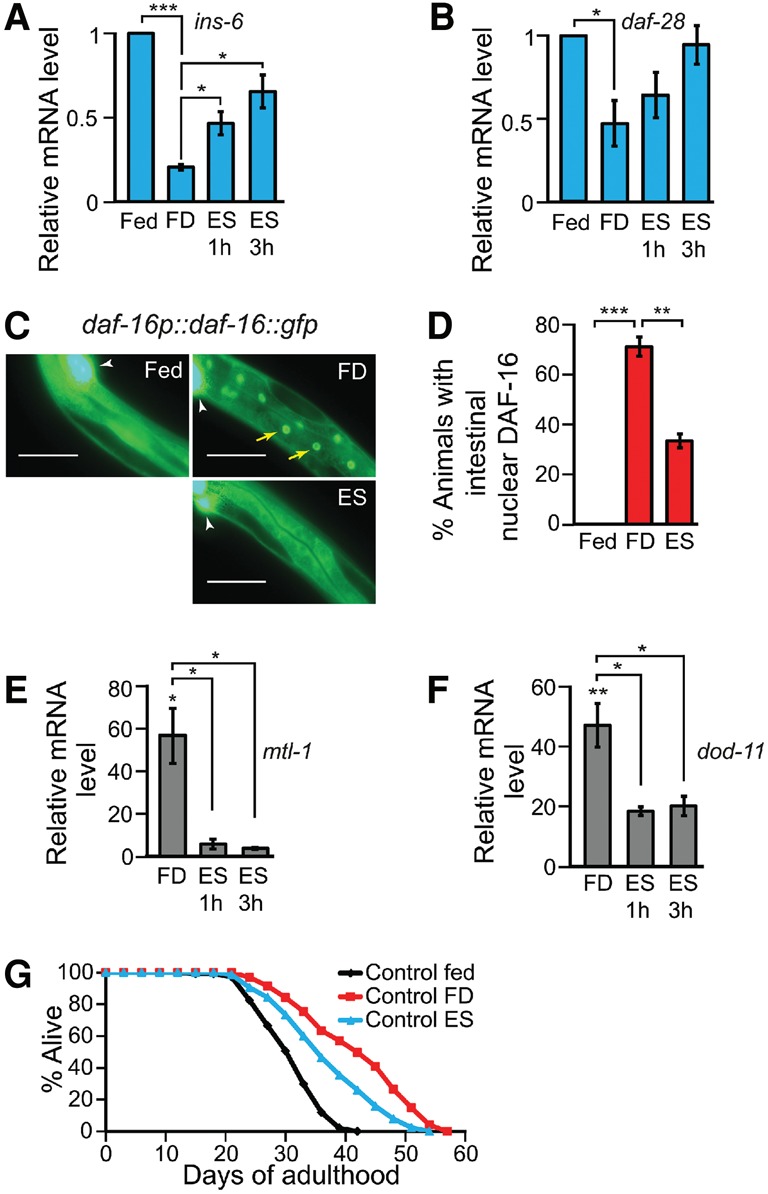
Sensory food cues regulate ILP expression and DAF-16/FOXO activity. (A,B) qRT–PCR analysis of ins-6 (A) and daf-28 (B) expression in fed, food-deprived (FD), or E. coli supernatant (ES)-supplemented animals. n = 3. (C,D) The effects of food deprivation or supplementation of E. coli supernatants on DAF-16/FOXO nuclear localization (C) ([yellow arrows] nuclear DAF-16::GFP; [white arrowhead] coinjection marker ofm-1p::gfp) and the quantification (D). n = 3, >30 total worms. Bar, 50 μm. (E,F) The mRNA levels of mtl-1 (E) or dod-11 (F) in food-deprived or E. coli supernatant-treated animals compared with those in fed animals using qRT–PCR. n = 3. Error bars represent SEM. (***) P < 0.001; (**) P < 0.01; (*) P < 0.05, two-tailed Student's t-test. (G) The effects of E. coli supernatant treatment on the life span of food-deprived worms. See Supplemental Table S3 for additional repeats and statistical analysis for the life span data.
Several lines of evidence indicate that the ILP ins-6 mediates the effects of food-derived cues on DAF-16/FOXO signaling and life span, acting in or downstream from sensory neurons. We found that E. coli supernatants did not affect the nucleocytoplasmic shuttling of DAF-16/FOXO and the long life span of sensory-defective tax-4 mutants that are food-deprived (Fig. 5A,B). This suggests that E. coli supernatants indeed decrease FOXO activity and life span through intact sensory neurons. Overexpression of ins-6 or daf-28 decreased the nuclear localization of DAF-16/FOXO in wild-type animals upon food deprivation while not further reducing it upon treatment with food-derived cues, suggesting that food-derived cues and these ILPs act in the same pathway (Fig. 5C,D). We also found that overexpression of ins-6 or daf-28 was sufficient to diminish the nuclear localization of DAF-16/FOXO in both food-deprived and E. coli supernatant-treated tax-4 mutants (Fig. 5E,F). These data are consistent with the idea that INS-6 and DAF-28 mediate the effects of food-derived cues on decreasing DAF-16/FOXO activity by acting in or downstream from sensory neurons. We found that ins-6 overexpression shortened the life span of food-deprived animals (Fig. 5G), although daf-28 overexpression did not (Fig. 5H). Thus, food-derived cues perceived by sensory neurons appear to decrease life span by increasing the expression of ILPs, such as ins-6, which subsequently down-regulates DAF-16/FOXO.
Figure 5.
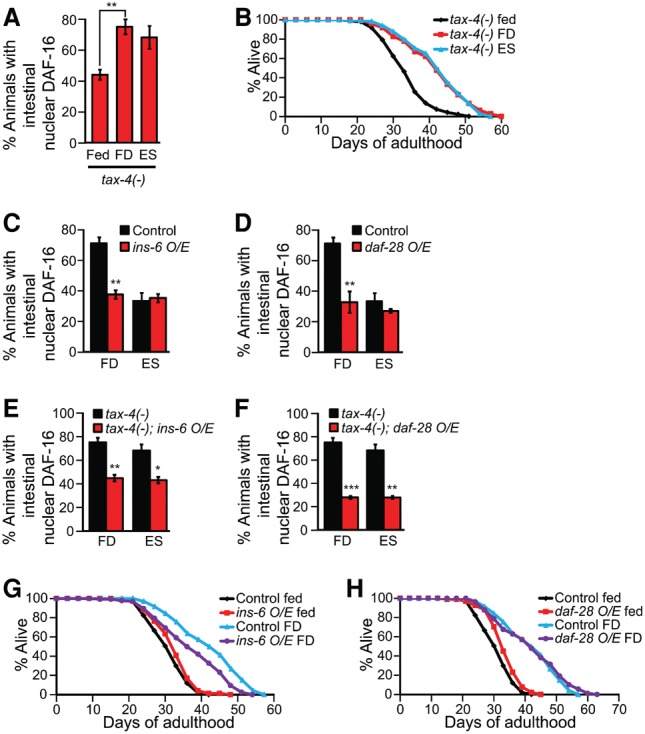
ins-6 acts via sensory neurons to mediate the effects of food cues on DAF-16/FOXO activity and life span. (A) The effect of E. coli supernatant (ES) supplementation on DAF-16 nuclear localization in food-deprived (FD) tax-4(−) mutants. n = 3, >30 total worms. (B) The life span of fed, food-deprived, and E. coli supernatant-supplemented food-deprived tax-4(−) worms. See Supplemental Table S3 for additional repeats and statistical analysis for the life span data shown in this figure. (C,D) The effects of ins-6 (C) or daf-28 (D) overexpression (O/E) on the localization of DAF-16::GFP in food-deprived or E. coli supernatant-supplemented wild-type worms. n = 3, >30 total worms. (E,F) The effects of ins-6 (E) or daf-28 (F) overexpression on the localization of DAF-16::GFP in food-deprived or E. coli supernatant-supplemented tax-4(−) worms. n = 3, >30 total worms. (G,H) The effects of ins-6 (G) and daf-28 (H) overexpression on the life span of food-deprived worms. Error bars represent SEM. (***) P < 0.001; (**) P < 0.01; (*) P < 0.05, two-tailed Student's t-test.
INS-6 acts in sensory ASI and ASJ neurons to decrease DAF-16/FOXO activity and longevity upon changes in food-derived environmental cues
We next asked whether the longevity regulatory roles of ins-6 and daf-28 can be generalized to other sensory mutants (Supplemental Fig. S6A–E). Overexpression of ins-6 significantly suppressed the long life span of the sensory intraflagellar transport-defective osm-5(−) (Perkins et al. 1986) and TRPV ion channel-deficient ocr-2(−) (Tobin et al. 2002) mutant animals, whereas daf-28 overexpression did not (Fig. 6A–D). Thus, ins-6, rather than daf-28, appears to be a more general longevity ILP signal from sensory neurons, leading us to focus further on ins-6.
Figure 6.
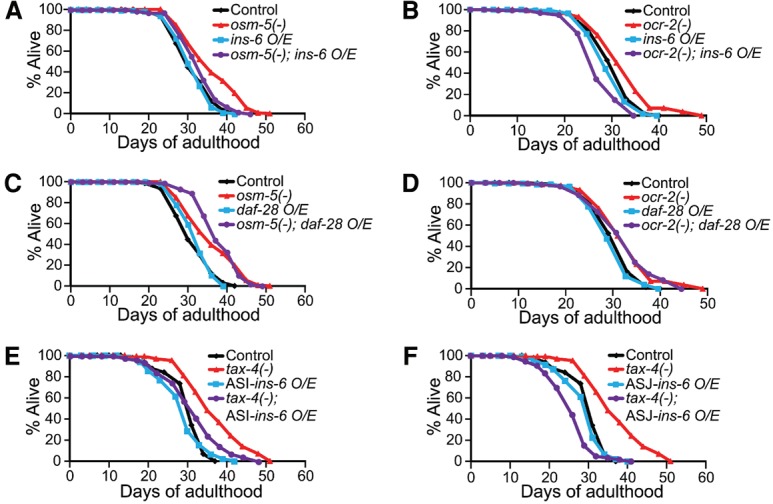
ins-6 mediates the longevity of sensory mutants acting in ASI and ASJ neurons. (A,B) Overexpression (O/E) of ins-6 suppressed the long life span of osm-5(p813) [osm-5(−)] (A) and ocr-2(ak47) [ocr-2(−)] (B) mutant worms. (C,D) Overexpression of daf-28 did not suppress the long life span of osm-5(−) (C) or ocr-2(−) (D) mutants. See Supplemental Table S4 for additional repeats and statistical analysis for the life span data shown in this figure. (E,F) The effects of ASI-specific (E) or ASJ-specific (F) ins-6 expression on the life span of wild-type (control) and tax-4(−) animals. Note that ASI- or ASJ-specific expression of high or low copy of the ins-6 transgene had similar effects on life span (Supplemental Table S4).
ins-6 is expressed in specific sensory neurons, ASI and ASJ, and has neuron-specific functions in development and behavior (Cornils et al. 2011; Chen et al. 2013). We tested whether ins-6 from ASI or ASJ neurons decreased the longevity of sensory mutants. We found that ins-6 acted from either neuron to shorten life span (Fig. 6E,F).
We then asked whether the activation of ASI or ASJ in food-deprived conditions was sufficient to shorten life span via ILP signaling by expressing the blue light-gated ion channel channelrhodopsin 2 (ChR2) from either neuron. The activation of ASI or ASJ by blue light triggered the gradual translocation of intestinal DAF-16::GFP from the nucleus to the cytosol in food-deprived animals only in the presence of all-trans retinal (ATR), a cofactor of ChR2 (Fig. 7A–D; Supplemental Fig. S7A,B; Supplemental videos 1–4). The ChR2-dependent activation of either ASI or ASJ neuron also increased ins-6 expression and decreased DAF-16/FOXO target gene expression (Fig. 7E–H). Importantly, the optogenetic activation of ASI or ASJ by blue light specifically shortened the life span of food-deprived worms (Fig. 7I,J; Supplemental Fig. S7C–F). Altogether, these data suggest that the activation of ASI or ASJ sensory neurons induces INS-6, which signals to distal tissues to down-regulate DAF-16/FOXO and decreases organismal life span.
Figure 7.
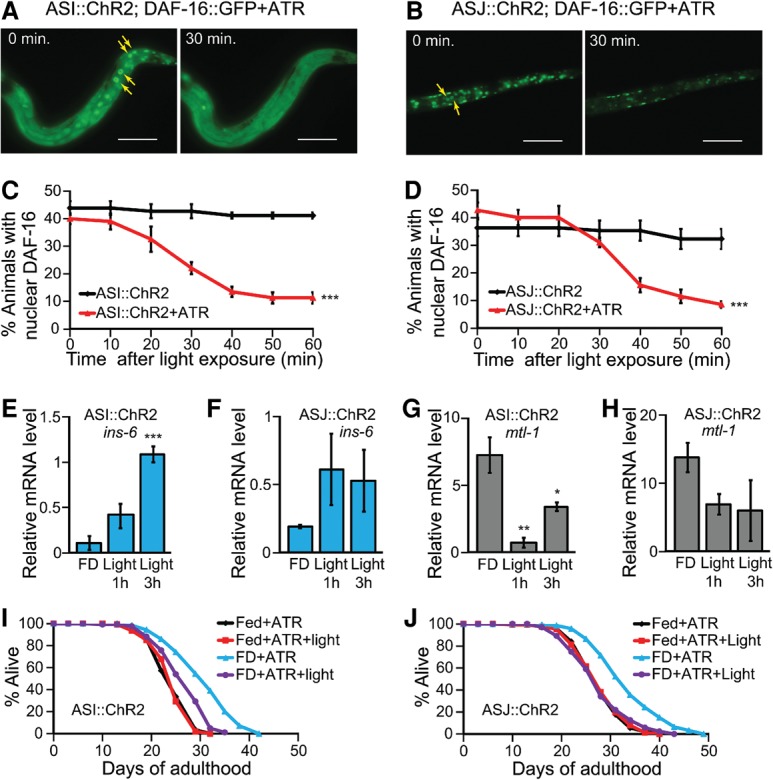
Optogenetic activation of ASI or ASJ neurons modulate sensory neuron-regulated longevity via INS-6 and DAF-16/FOXO. (A,B) The effects of optogenetic activation of ASI (A) or ASJ (B) neurons by using ChR2 on DAF-16::GFP localization in food-deprived (FD) animals. (Yellow arrow) Nuclear DAF-16::GFP. Bar, 50 μm. (C,D) Quantification of the A and B by performing time-course experiments. n = 3, >90 total worms. (***) P < 0.001, calculated using two-way ANOVA test. (E,F) Changes in the mRNA levels of ins-6 upon optogenetic activation of ASI (E) (n = 4) or ASJ (F) (n = 3) neurons in food-deprived animals. Error bars represent SEM. (***) P < 0.001; (**) P < 0.01; (*) P < 0.05, two-tailed Student's t-test. (G,H) Changes in the mRNA levels of mtl-1 upon optogenetic activation of ASI (G) (n = 4) or ASJ (H) (n = 3) neurons in food-deprived animals. (I,J) Life span of fed or food-deprived animals upon repetitive optogenetic activation of ASI (I) or ASJ (J) neurons with ATR. We found that treatment with either ATR or blue light alone did not affect life span (Supplemental Fig. S7C–F). See Supplemental Table S5 for additional repeats and statistical analysis for the life span data.
Discussion
Although the life span regulatory role of C. elegans sensory neurons has been known for more than a decade (Apfeld and Kenyon 1999; Alcedo and Kenyon 2004), much of the molecular mechanisms remained elusive. In the present study, we addressed how sensory neurons process environmental changes and relay the signals to distal tissues to modulate the life span of C. elegans. Our current report provides functional evidence showing that the ILP INS-6 acts as a neuroendocrine signal that regulates the longevity conferred by sensory cues via IIS. The inhibition of sensory neurons by deprivation of food cues decreases IIS through the down-regulation of INS-6, which could act as an agonist of DAF-2. We also showed that food-derived cues inhibit the nuclear localization of the downstream effector DAF-16/FOXO. Our study suggests that sensory neurons respond to the presence or absence of specific food-derived cues to modulate IIS and life span.
Sensory neurons will mediate many mechanisms, ranging from food level to food quality (Libert et al. 2007; Maier et al. 2010). Since we know that ILPs can have different activities in different physiological processes and have distinct neuronal expression patterns (for review, see Allen et al. 2015 and references therein), certain ILPs may mediate the DR effects on life span, whereas other ILPs may mediate the non-DR effects on life span. However, it remains unclear how all of these ILPs will affect the activities of the IIS pathway. Some of them might be agonists of the pathway, whereas others may antagonize or have no effect on the pathway. Since the expression of different ILPs is regulated by distinct environmental cues (Li et al. 2003; Cornils et al. 2011; Ritter et al. 2013), the variable effects of IIS under different dietary regimens (Greer and Brunet 2009; Mair et al. 2009) could be explained by the nature of the ILPs that are present versus those that are absent under these conditions.
Cell-nonautonomous regulation of longevity through IIS has previously been demonstrated in C. elegans (Apfeld and Kenyon 1999; Wolkow et al. 2000; Libina et al. 2003; Zhang et al. 2013). Loss of a functional daf-2 gene in specific subsets of cells is sufficient to extend C. elegans life span (Apfeld and Kenyon 1998). Conversely, neuronal expression of daf-2 can rescue the long life phenotype of daf-2 mutants (Wolkow et al. 2000). At the same time, DAF-16/FOXO can act in multiple tissues (i.e., neurons, intestine, and hypodermis) to promote the longevity of daf-2 mutants (Libina et al. 2003; Zhang et al. 2013). Here, we show that expression of daf-16 in these same tissues contributes to the longevity of sensory mutants, which corroborates the life span-regulating role of DAF-16/FOXO in different tissue types. Furthermore, our study delineates a complete signaling pathway by specifying upstream environmental cues, sensory neurons, and neuroendocrine ILPs, which regulate downstream IIS components in various tissues that regulate longevity.
Many genes encoding IIS components that influence longevity also play roles in other physiological processes. For example, the ILPs INS-6 and DAF-28 regulate the animal's cold-shock response in a synergistic manner (Ohta et al. 2014). INS-6 and DAF-28 are also negative regulators of the developmental program known as dauer (Li et al. 2003; Cornils et al. 2011; Fernandes de Abreu et al. 2014), which is an alternative, hibernation-like larval stage that occurs under conditions of stress (Hu 2007). daf-28 plays a more primary role in inhibiting dauer formation, whereas ins-6 is more important in preventing dauer maintenance (Cornils et al. 2011). Consistently, ins-6 and daf-28 also appear to have distinct effects on aging. First, daf-28::gfp expression is increased during aging (Li et al. 2003), whereas ins-6::gfp is decreased (Ritter et al. 2013). Second, we show that the roles of ins-6 and daf-28 in the longevity of different sensory mutants diverge, as ins-6 appears to be a more general life span regulator than daf-28 (See Fig. 6). Thus, these two ILPs can have nonredundant and specific functions in development and adult life span. However, as we functionally tested only two of the potential insulin/IGF-1 receptor DAF-2 agonists, INS-6 and DAF-28, overexpression of some other ILPs may also decrease the activity of DAF-16. It will be interesting to determine the role of other ILPs in the life span regulation of sensory mutants in future studies.
An intriguing feature of our study is that the tax-2 and tax-4 sensory mutations decrease or increase the expression of specific sets of ILPs in different ways (Fig. 2). Recently, the C. elegans ILP family has been shown to consist of an ILP-to-ILP regulatory network (Fernandes de Abreu et al. 2014). Hence, changes in the expression of one ILP may lead to changes in the expression of other ILPs. This is illustrated by the up-regulation of ins-35 when ins-6 expression is down (Fig. 2A,B), which would be expected from the network organization that has been previously described (Fernandes de Abreu et al. 2014). However, some of the observed changes in ILP expression at a low temperature, 15°C (this study), do not necessarily fit all of the predictions that could arise from the ILP network that has been delineated at a higher temperature, 20°C (Fernandes de Abreu et al. 2014). This could mean that the state of the ILP network is dynamic and changes under different environments and thus is subject to sensory regulation.
Some sensory neurons only affect life span under certain environments (Maier et al. 2010), since these neurons, which detect a given set of environmental cues, would presumably only affect longevity based on the presence of these cues. However, a neuron can also have opposing activities within a given physiological process. The modulation of such a neuron's activities (also known as neuromodulation) will depend on changes in the neuron's local environment as well as the animal's global environment (Strand 1999; Marder 2012). For example, the sensory neuron ASI inhibits longevity in well-fed animals (Alcedo and Kenyon 2004) but promotes longevity in diet-restricted animals (Bishop and Guarente 2007), which suggests that food-derived cues will change ASI's local environment. In this study, we show that a second sensory neuron, ASJ, has opposing effects on life span and that its longevity effects again depend on environmental context. ASJ lengthens life span at 20°C (Alcedo and Kenyon 2004) but shortens life span at the lower temperature of 15°C through the ILP INS-6 (this study). Together, these studies show that the neuronal effects on life span will involve different types of environmental cues (e.g., food-derived cues and temperature) (for review, see Allen et al. 2015), which could change a neuron's local environment and thus its consequent effects on life span.
The present work provides direct evidence for the role of ILPs in the regulation of longevity in C. elegans sensory mutants. Although the direct role of ILPs, insulin, or IGF in the regulation of life span by sensory neurons in Drosophila or mammals remains unknown, several intriguing clues are present. For example, in Drosophila, the ablation of ILP-expressing cells increases life span (Broughton et al. 2005), and the activation of c-Jun N-terminal kinase specifically in ILP-producing neurons results in the reduction of ILP production and life span extension (Wang et al. 2005). Similarly, long-lived Drosophila taste mutants have altered expression of certain ILPs (e.g., an increase in head expression of dilp6) (Ostojic et al. 2014), which has been shown earlier to promote longevity through up-regulation of the IIS effector dFOXO (Bai et al. 2012). Together, these reports support the idea that neuronal ILPs act as mediators of life span regulation in flies.
In mammals, blood glucose levels alter the secretion of insulin. Interestingly, the perception of food increases blood insulin levels in humans (Sjostrom et al. 1980), suggesting that chemosensation also directly influences insulin signaling in mammals. In addition, a recent study in mice illustrates that pain perception alters longevity through the regulated secretion of pancreatic insulin (Riera et al. 2014). Thus, other sensory modalities may have a regulatory function in mammalian life span by altering IIS through modulation of ILPs, such as insulin, IGF1/2, or the relaxin subfamily (Sherwood 2004).
Materials and methods
Strains
Strains used in this study are described in the Supplemental Material.
Preparation of E. coli OP50 cell-free supernatant
Cell-free E. coli OP50 supernatant was obtained as described (Smith et al. 2008) with some modifications. Briefly, E. coli OP50 was grown overnight at 37°C and centrifuged at 4000 rpm for 5 min, and the supernatant was filtered with a 0.2-μm pore size filter.
Life span analysis
Life span assays were conducted as described previously (Lee et al. 2010; Kim et al. 2014), starting at day 1 of adulthood. OASIS (Online Application for Survival Analysis, http://sbi.postech.ac.kr/oasis) was used for statistical analysis of the data (Yang et al. 2011), and P-values were calculated using the log rank (Mantel-Cox) method. Strains were grown for at least two generations at 15°C before being used for life span assays at 15°C. Prefertile young adult worms were transferred to plates containing 5 µM 5-fluoro-2′-deoxyuridine (FUdR) (Sigma-Aldrich) to prevent their progeny from developing unless stated otherwise. Recent studies suggest that FUdR confers stress resistance by activating a heat-shock response (Brunquell et al. 2014) and increases life span under mild osmotic stress conditions (Anderson et al. 2016). We noticed that these studies used relatively high concentrations of FUdR (at least 25 µM). In contrast, we used 5 µM FUdR for most of our life span assays. In addition, we assayed non-FUdR-treated animals to confirm the life span phenotypes of key sensory and ILP mutants as well as ILP transgenic animals. For life span assays with food-derived cues, prefertile young adult worms were transferred to streptomycin-resistant E. coli OP50-seeded plates. Three days later, the worms were transferred to new OP50-seeded plates. At the sixth day, worms were transferred to OP50-seeded plates for setting up fed groups or to empty agar plates for food-deprived groups. Experimental groups were treated with 100 μL of filtered supernatant of overnight E. coli culture, which was grown in Luria broth (LB) medium supplemented with streptomycin every 3 d, whereas control groups were treated with 100 μL of LB–streptomycin medium only. For life span assays using optogenetics, the worms were grown on OP50-seeded plates in the presence or absence of ATR (Sigma-Aldrich) for at least two generations at 15°C before conducting life span analysis. ATR-treated animals were always maintained on ATR-treated plates (both OP50-seeded and empty unseeded plates). ATR was dissolved in ethanol at a concentration of 100 mM. The E. coli OP50 culture on the NGM (nematode growth medium) plates or unseeded plates were supplemented with 50 μL of 10 mM ATR diluted in M9 buffer to a final concentration of 100 μM. Control plates were treated with 50 μL of M9 buffer with 10% ethanol. Prefertile young adult worms were transferred to OP50-seeded plates and, 3 d later, transferred to new OP50-seeded plates. At the sixth day, fed groups were transferred to OP50-seeded plates, and food-deprived animals were transferred to empty agar plates. Light-exposed groups were subjected to blue light (∼470 nm) every day for 2 min. For all the life span experiments, at least five plates were used per condition unless some plates were discarded due to bacterial or fungal contamination. For the majority of the experiments, 25 animals per plate were used for a total of 150 animals per trial. Most of the life span assays were performed at least twice per condition by at least two independent researchers. We noticed that the censoring rate is higher for life span assays that are conducted in experiments at 15°C, and therefore, on average, 38% of the animals were censored. Animals that ruptured, bagged, or crawled off the plates were censored but included in the life span analysis as censored worms.
Microscopy and quantification of fluorescence
Fluorescence imaging was conducted as reported previously (Lee et al. 2015). The images of transgenic animals were captured using an AxioCam (Zeiss Corporation) attached to a HRc Zeiss Axioscope A.1 (Zeiss Corporation). Quantification of GFP intensity was carried out using ImageJ (National Institutes of Health, http://rsbweb.nih.gov/ij). The normalization of the mean fluorescence intensity was achieved by subtracting mean fluorescence intensity of pictures of nontransgenic control worms taken with the same exposure time. Levamisole (2 mM) was used for immobilization of the worms on 2% agarose pads before taking pictures. P-values were calculated using the unpaired Student's t-test (two-tailed). For the experiments measuring the localization of DAF-16::GFP, gravid adults were synchronized for 12 h, and the progeny were transferred to empty plates at L2/L3 larval stages after washing three times with M9 buffer. After 24 h of food deprivation, the worms were scored for nuclear localization of DAF-16::GFP. For food cue experiments, the nuclear localization of DAF-16::GFP was scored after 100 μL of E. coli supernatant (overnight bacterial culture in LB–streptomycin medium) or control (LB–streptomycin medium only) treatment. For optogenetic activation of sensory neurons, animals were food-deprived for 24 h in the presence or absence of ATR and subjected to blue light (∼470 nm) for ChR2 excitation for 2 min. Changes in the intestinal nuclear localization of DAF-16::GFP were recorded every 1 min for time-lapse experiments and every 10 min for quantification during a period of 1 h. The images used in Figure 7, A and B, are the same images used in the videos. We created a video with the images that were taken every minute during the time span of 30 min. Although the quantitation of fluorescence in transgenic animals was not scored blindly, many of the experiments were replicated independently in several trials by different researchers. For example, the quantitation of fluorescence of transgenic animals expressing DAF-16 target genes in a tax-2(p671) mutant background as well as DAF-16 subcellular localization and DAF-16 target gene expression in a tax-4(p678) mutant background was performed by independent researchers; the results were very similar. In addition, optogenetic analysis of the changes in DAF-16 subcellular localization was performed and reproduced independently by two researchers at two different institutions.
Quantitative real-time PCR analysis
Synchronized L4 animals grown at 15°C were used for RNA extraction and subsequent experiments. Extraction, purification, and reverse transcription of RNA were performed as described (Seo et al. 2015) with some modifications. qPCR from the cDNA was carried out using a 7300 real-time PCR system (Applied Biosystems) and analyzed by the comparative Ct method described in the manufacturer's manual. The average values of the mRNA levels of nhr-23 and/or ama-1 genes were used for normalization. The average of at least two technical repeats was applied for each biological data point. For the qRT–PCR analysis of sensory food cue experiments, wild-type worms at L2/L3 stages were food-deprived for 24 h, treated with E. coli supernatants (bacteria were grown overnight in LB–streptomycin medium) or LB–streptomycin control medium, and harvested 1 or 3 h after treatment. For the qRT–PCR analysis using ChR2-expressing transgenic animals, worms at L2/L3 stages were food-deprived for 24 h in the presence of ATR and subjected to blue light (∼470 nm) for ChR2 excitation for 2 min. Subsequently, the animals were harvested 1 or 3 h after light exposure. Please see the Supplemental Material for primer sequences.
Generation of transgenic constructs
To clone ins-6, the promoter and coding region of ins-6 (∼2.1 kb) was PCR-amplified from C. elegans genomic DNA using 5′-TGCAGGTCGACTCTAGATGTTGCTCCACTGATTGCAGCT-3′ and 5′-CCCTTAGACACCATCGCTGGACAAGCAGATCTTATG-3′ primers and inserted into the pPD95.75 vector (a gift from Dr. Andrew Fire), which was previously modified and contained tagrfp-t and unc-54 3′ untranslated region (UTR), digested with XbaI and KpnI. To clone trx-1p::ChR2::gfp and str-3p::ChR2::gfp expression vectors, ChR2 cDNA was PCR-amplified from the pAAV-TRE-hChR2(H134R)-EYFP vector (a gift from Dr. Joung-Hun Kim) using 5′-GCAGGTCGACTCTAGAGCCACCATGGACTATGGCGG-3′ and 5′-TCATTTTTTCTACCGGTACCGCTGGCACGGCTCCGGCCTCGGC-3′ primers and inserted into the pPD95.75 vector, which contained gfp and unc-54 3′ UTR, digested with XbaI and KpnI. trx-1 (∼1 kb) and str-3 (∼3.1 kb) promoters were PCR-amplified from C. elegans genomic DNA using 5′-GAAATGAAATAAGCTTAGAGAATGGATACCTGATCATTC-3′ and 5′-CCATGGTGGCTCTAGATCTGTTACCATATCAGCAAGCTC-3′, and 5′-GAAATGAAATAAGCTTGCTGGTGAAGATTTGTTCAAGGA-3′ and 5′-CCATGGTGGCTCTAGAGTTCCTTTTGAAATTGAGGCAGT-3′ primers, respectively. Amplified trx-1 and str-3 promoters were inserted into the upstream ChR2 cDNA by using HindIII and XbaI sites in the pPD95.75 vector. All cloning reactions were performed using In-Fusion cloning (Clontech) by following the manufacturer's instructions.
Generation of transgenic animals
ins-6p::ins-6::tagrfp-t, trx-1p::ChR2::gfp, or str-3p::ChR2::gfp expression vectors (25 ng/μL) were coinjected with an injection marker, myo-3p::rfp (75 ng/μL), into the gonad of day 1 adult wild-type animals. The rol-6D vector was used as a coinjection marker for the ins-6p::ins-6::tagrfp-t (25 ng/μL) expression vector.
Dye filling assay
Dye filling assay was performed as previously described (Gaglia et al. 2012). Well-fed worms were washed three times with M9 buffer and centrifuged at 2000 rpm to spin down the worms. The worms were then resuspended in 1 mL of M9 buffer and 5 μL of 2 mg/mL DiI stock solution. The worms were kept on a slow shaker for 3 h and then washed three times with M9 buffer and transferred to agar pads for visualization under a HRc Zeiss Axioscope A.1 (Zeiss Corporation).
Supplementary Material
Acknowledgments
We thank Dr. Yun Zhang, Dr. Shohei Mitani, Dr. Cynthia Kenyon, and Dr. Joung-Hun Kim for providing several strains and DNA constructs. We are also grateful for Dr. Cynthia Kenyon for allowing S.-J.V.L. to use some preliminary data generated in her laboratory at University of California at San Francisco. We also thank all Lee laboratory members for helpful discussions. This work was supported by a grant funded by the Ministry of Education, Science, and Technology (NRF-2012R1A4A1028200) through the National Research Foundation (NRF) of Korea and an academic research grant for aging support project of the Seokchun Daewoong Foundation to S.-J.V.L.; DGIST R&D Program of the Ministry of Science, ICT, and Future Planning (16-BD-06) to K.K.; and the National Institutes of Health/National Institute of General Medical Sciences (R01GM108962) to J.A. D.-E.J is supported by the Fostering Core Leaders of the Future Basic Science Program (NRF-2013H1A8A1003751).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.279448.116.
References
- Alcedo J, Kenyon C. 2004. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron 41: 45–55. [DOI] [PubMed] [Google Scholar]
- Allen EN, Ren J, Zhang Y, Alcedo J. 2015. Sensory systems: their impact on C. elegans survival. Neuroscience 296: 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson EN, Corkins ME, Li JC, Singh K, Parsons S, Tucey TM, Sorkac A, Huang H, Dimitriadi M, Sinclair DA, et al. 2016. C. elegans lifespan extension by osmotic stress requires FUdR, base excision repair, FOXO, and sirtuins. Mech Ageing Dev 154: 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfeld J, Kenyon C. 1998. Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell 95: 199–210. [DOI] [PubMed] [Google Scholar]
- Apfeld J, Kenyon C. 1999. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature 402: 804–809. [DOI] [PubMed] [Google Scholar]
- Bai H, Kang P, Tatar M. 2012. Drosophila insulin-like peptide-6 (dilp6) expression from fat body extends lifespan and represses secretion of Drosophila insulin-like peptide-2 from the brain. Aging Cell 11: 978–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI. 2006. Chemosensation in C. elegans. WormBook (ed. The C. elegans Research Community), WormBook, 10.1895/wormbook.1.123.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Bishop NA, Guarente L. 2007. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature 447: 545–549. [DOI] [PubMed] [Google Scholar]
- Broughton SJ, Piper MD, Ikeya T, Bass TM, Jacobson J, Driege Y, Martinez P, Hafen E, Withers DJ, Leevers SJ, et al. 2005. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci 102: 3105–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunquell J, Bowers P, Westerheide SD. 2014. Fluorodeoxyuridine enhances the heat shock response and decreases polyglutamine aggregation in an HSF-1-dependent manner in Caenorhabditis elegans. Mech Ageing Dev 141–142: 1–4. [DOI] [PubMed] [Google Scholar]
- Chen Z, Hendricks M, Cornils A, Maier W, Alcedo J, Zhang Y. 2013. Two insulin-like peptides antagonistically regulate aversive olfactory learning in C. elegans. Neuron 77: 572–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn CM, Bargmann CI. 1996. A putative cyclic nucleotide-gated channel is required for sensory development and function in C. elegans. Neuron 17: 695–706. [DOI] [PubMed] [Google Scholar]
- Cornils A, Gloeck M, Chen Z, Zhang Y, Alcedo J. 2011. Specific insulin-like peptides encode sensory information to regulate distinct developmental processes. Development 138: 1183–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bussiere F, Hekimi S. 2001. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev Cell 1: 633–644. [DOI] [PubMed] [Google Scholar]
- Fernandes de Abreu DA, Caballero A, Fardel P, Stroustrup N, Chen Z, Lee K, Keyes WD, Nash ZM, Lopez-Moyado IF, Vaggi F, et al. 2014. An insulin-to-insulin regulatory network orchestrates phenotypic specificity in development and physiology. PLoS Genet 10: e1004225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglia MM, Jeong DE, Ryu EA, Lee D, Kenyon C, Lee SJ. 2012. Genes that act downstream of sensory neurons to influence longevity, dauer formation, and pathogen responses in Caenorhabditis elegans. PLoS Genet 8: e1003133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Brunet A. 2009. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell 8: 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. 2003. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300: 1142–1145. [DOI] [PubMed] [Google Scholar]
- Hu PJ. 2007. Dauer. WormBook (ed. The C. elegans Research Community), WormBook, 10.1895/wormbook.1.144.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong DE, Artan M, Seo K, Lee SJ. 2012. Regulation of lifespan by chemosensory and thermosensory systems: findings in invertebrates and their implications in mammalian aging. Front Genet 3: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YI, Bandyopadhyay J, Cho I, Lee J, Park DH, Cho JH. 2014. Nucleolar GTPase NOG-1 regulates development, fat storage, and longevity through insulin/IGF signaling in C. elegans. Mol Cells 37: 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu H, Mori I, Rhee JS, Akaike N, Ohshima Y. 1996. Mutations in a cyclic nucleotide-gated channel lead to abnormal thermosensation and chemosensation in C. elegans. Neuron 17: 707–718. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. 1998. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci 95: 13091–13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Kenyon C. 2009. Regulation of the longevity response to temperature by thermosensory neurons in Caenorhabditis elegans. Curr Biol 19: 715–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Hwang AB, Kenyon C. 2010. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr Biol 20: 2131–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Jeong DE, Son HG, Yamaoka Y, Kim H, Seo K, Khan AA, Roh TY, Moon DW, Lee Y, et al. 2015. SREBP and MDT-15 protect C. elegans from glucose-induced accelerated aging by preventing accumulation of saturated fat. Genes Dev 29: 2490–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Kennedy SG, Ruvkun G. 2003. daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev 17: 844–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libert S, Zwiener J, Chu X, Vanvoorhies W, Roman G, Pletcher SD. 2007. Regulation of Drosophila life span by olfaction and food-derived odors. Science 315: 1133–1137. [DOI] [PubMed] [Google Scholar]
- Libina N, Berman JR, Kenyon C. 2003. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 115: 489–502. [DOI] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. 1997. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278: 1319–1322. [DOI] [PubMed] [Google Scholar]
- Lin K, Hsin H, Libina N, Kenyon C. 2001. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet 28: 139–145. [DOI] [PubMed] [Google Scholar]
- Maier W, Adilov B, Regenass M, Alcedo J. 2010. A neuromedin U receptor acts with the sensory system to modulate food type-dependent effects on C. elegans lifespan. PLoS Biol 8: e1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W, Panowski SH, Shaw RJ, Dillin A. 2009. Optimizing dietary restriction for genetic epistasis analysis and gene discovery in C. elegans. PLoS One 4: e4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder E. 2012. Neuromodulation of neuronal circuits: back to the future. Neuron 76: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley JF, Morimoto RI. 2004. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell 15: 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. 1997. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389: 994–999. [DOI] [PubMed] [Google Scholar]
- Ohta A, Ujisawa T, Sonoda S, Kuhara A. 2014. Light and pheromone-sensing neurons regulates cold habituation through insulin signalling in Caenorhabditis elegans. Nat Commun 5: 4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostojic I, Boll W, Waterson MJ, Chan T, Chandra R, Pletcher SD, Alcedo J. 2014. Positive and negative gustatory inputs affect Drosophila lifespan partly in parallel to dFOXO signaling. Proc Natl Acad Sci 111: 8143–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. 1986. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol 117: 456–487. [DOI] [PubMed] [Google Scholar]
- Pierce SB, Costa M, Wisotzkey R, Devadhar S, Homburger SA, Buchman AR, Ferguson KC, Heller J, Platt DM, Pasquinelli AA, et al. 2001. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev 15: 672–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher SD. 2009. The modulation of lifespan by perceptual systems. Ann N Y Acad Sci 1170: 693–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon PC, Kuo TH, Linford NJ, Roman G, Pletcher SD. 2010. Carbon dioxide sensing modulates lifespan and physiology in Drosophila. PLoS Biol 8: e1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera CE, Huising MO, Follett P, Leblanc M, Halloran J, Van Andel R, de Magalhaes Filho CD, Merkwirth C, Dillin A. 2014. TRPV1 pain receptors regulate longevity and metabolism by neuropeptide signaling. Cell 157: 1023–1036. [DOI] [PubMed] [Google Scholar]
- Ritter AD, Shen Y, Fuxman Bass J, Jeyaraj S, Deplancke B, Mukhopadhyay A, Xu J, Driscoll M, Tissenbaum HA, Walhout AJ. 2013. Complex expression dynamics and robustness in C. elegans insulin networks. Genome Res 23: 954–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Seo K, Hwang W, Koo HJ, Hahm JH, Yang JS, Han SK, Hwang D, Kim S, Jang SK, et al. 2015. RNA helicase HEL-1 promotes longevity by specifically activating DAF-16/FOXO transcription factor signaling in Caenorhabditis elegans. Proc Natl Acad Sci 112: E4246–E4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood OD. 2004. Relaxin's physiological roles and other diverse actions. Endocr Rev 25: 205–234. [DOI] [PubMed] [Google Scholar]
- Sjostrom L, Garellick G, Krotkiewski M, Luyckx A. 1980. Peripheral insulin in response to the sight and smell of food. Metabolism 29: 901–909. [DOI] [PubMed] [Google Scholar]
- Smith ED, Kaeberlein TL, Lydum BT, Sager J, Welton KL, Kennedy BK, Kaeberlein M. 2008. Age- and calorie-independent life span extension from dietary restriction by bacterial deprivation in Caenorhabditis elegans. BMC Dev Biol 8: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand FL. 1999. Neuropeptides—regulators of physiological processes (ed. Stevens CK). The MIT Press, Cambridge, MA. [Google Scholar]
- Tobin D, Madsen D, Kahn-Kirby A, Peckol E, Moulder G, Barstead R, Maricq A, Bargmann C. 2002. Combinatorial expression of TRPV channel proteins defines their sensory functions and subcellular localization in C. elegans neurons. Neuron 35: 307–318. [DOI] [PubMed] [Google Scholar]
- Wang MC, Bohmann D, Jasper H. 2005. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell 121: 115–125. [DOI] [PubMed] [Google Scholar]
- Waterson MJ, Chung BY, Harvanek ZM, Ostojic I, Alcedo J, Pletcher SD. 2014. Water sensor ppk28 modulates Drosophila lifespan and physiology through AKH signaling. Proc Natl Acad Sci 111: 8137–8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkow CA, Kimura KD, Lee MS, Ruvkun G. 2000. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science 290: 147–150. [DOI] [PubMed] [Google Scholar]
- Yang JS, Nam HJ, Seo M, Han SK, Choi Y, Nam HG, Lee SJ, Kim S. 2011. OASIS: online application for the survival analysis of lifespan assays performed in aging research. PLoS One 6: e23525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Judy M, Lee SJ, Kenyon C. 2013. Direct and indirect gene regulation by a life-extending FOXO protein in C. elegans: roles for GATA factors and lipid gene regulators. Cell Metab 17: 85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


