Supplemental Digital Content is available in the text
Abstract
DNA (cytosine-5)-methyltransferase 3 alpha (DNMT3A) mutations were widely believed to be independently associated with inferior prognosis in acute myeloid leukemia (AML) patients. As dominant missense alterations in DNMT3A mutations, R882 mutations cause the focal hypomethylation phenotype. However, there remains debate on the influence of R882 mutations on AML prognosis. Thus, this meta-analysis aimed at further illustrating the prognostic power of DNMT3A R882 mutations in AML patients.
Eligible studies were identified from 5 databases containing PubMed, Embase, Web of Science, Clinical Trials, and the Cochrane Library (up to October 25, 2015). Effects (hazard ratios [HRs] with 95% confidence interval [CI]) of relapse-free survival (RFS) and overall survival (OS) were pooled to estimate the prognostic power of mutant DNMT3A R882 in overall patients and subgroups of AML patients.
Eight competent studies with 4474 AML patients including 694 with DNMT3A R882 mutations were included. AML patients with DNMT3A R882 mutations showed significant shorter RFS (HR = 1.40, 95% CI = 1.24–1.59, P < 0.001) and OS (HR = 1.47, 95% CI = 1.17–1.86, P = 0.001) in the overall population. DNMT3A R882 mutations predicted worse RFS and OS among the subgroups of patients under age 60 (RFS: HR = 1.44, 95% CI = 1.25–1.66, P < 0.001; OS: HR = 1.48, 95% CI = 1.15–1.90, P = 0.002), over age 60 (RFS: HR = 2.03, 95% CI = 1.40–2.93, P < 0.001; OS: HR = 1.85, 95% CI = 1.36–2.53, P < 0.001), cytogenetically normal (CN)-AML (RFS: HR = 1.52, 95% CI = 1.26–1.83, P < 0.001; OS: HR = 1.67, 95% CI = 1.16–2.41, P = 0.006), and non-CN-AML (RFS: HR = 1.96, 95% CI = 1.20–3.21, P = 0.006; OS: HR = 2.51, 95% CI = 1.52–4.15, P = 0.0038).
DNMT3A R882 mutations possessed significant unfavorable prognostic influence on RFS and OS in AML patients.
INTRODUCTION
Acute myeloid leukemia (AML) is a clinical and biological heterogeneous clonal stem cell disorder characterized by clonal and aggressive expansion of myeloid progenitor cells or “blast” cells in bone marrow.1,2 AML usually presents with a broad spectrum of prognosis-related cytogenetic abnormities, genetic mutations, and aberrant expression of genes.3,4 Currently, AML is healed in 35% to 40% among younger patients with age <60, and 5% to 15% among older patients with age ≥60.5 The huge molecular heterogeneity of AML has become growingly distinct over the past 15 years, despite the cytogenetic heterogeneity of the disease has been realized for over 30 years.5 The prognostic significance of this biological heterogeneity is well-accepted, but there remains a need to identify better and more precise predictors of disease outcome.
Recently, genetic mutations and epigenetic alterations have been identified in the bone-marrow leukemogenesis and are reported to be associated with AML outcomes.6,7 Previous studies have suggested that internal tandem duplication in fms-related tyrosine kinase 3 (FLT3-ITD), mutations in nucleophosmin (NPM1), and CCAAT/enhancer binding protein alpha (CEBPA) can be used to stratify risk among patients with normal karyotype.8 Later reports have identified novel prognosis-related mutations in AML patients, which include mutational isocitrate dehydrogenase 2 (IDH2), additional sex combs like 1 (ASXL1), and DNA (cytosine-5)-methyltransferase 3 alpha (DNMT3A).9 DNMT3A is responsible for de novo methylation of genome DNA during mammalian development, and DNMT3A alterations are thought to play important roles in etiology of various diseases including AML.10DNMT3A is one of the most frequently mutated genes in AML patients, being found mutated in approximately 20% of the patients.11,12DNMT3A somatic mutation was first identified by whole-genome sequencing in an AML patient with normal karyotype,13 which was associated with worse clinical outcomes.14,15 Overall, mountains of studies have declared that DNMT3A could be a prognostic indicator in AML patients.
With the announcement of the Precision Medicine Initiative in USA, it is urgent to find out the function of more and finer biomarkers, thus to generate knowledge applicable to the whole range of health and disease.16,17 And AML is no exception. In AML patients with DNMT3A mutations, about 60% patients exhibit heterozygous mutations at Arginine 882 (R882), which results in loss-of-function effect and disruption of normal methylation function.18–20 Four R882 mutations included R882C (arginine → cysteine), R882H (arginine → histidine), R882S (arginine → serine), and R882P (arginine → phenylalanine) are reported.14,21 Therefore, DNMT3A mutations are usually classified as R882 mutations and non-R882 mutations.22 However, there existed an inconsistent opinion on whether DNMT3A R882 mutations have the potential to predict AML prognosis. For example, Renneville and colleagues reported that patients with R882 mutations showed shorter RFS and OS in cytogenetically normal (CN)-AML,23 while some studies showed negative findings on OS time.24,25 So, this meta-analysis was aimed at systematically elaborating the prognostic values of DNMT3A R882 mutations in AML patients, in order to guide precisely clinical decision-making even to improve the prognosis of the patients.
MATERIALS AND METHODS
Literature Search
Literature search was conducted in PubMed, Embase, Web of Science, ClinicalTrials, and the Cochrane Library with the following search terms: “AML,” “acute myeloid leukemia,” “Leukemia, Myeloid, Acute,” “acute myelogenous leukemia,” “acute myelocytic leukemia,” AND “DNMT3A,” “DNA methyltransferase 3 alpha,” “DNA methyltransferase 3A,” “DNA (cytosine-5-)-methyltransferase 3 alpha,” “DNA (cytosine-5)-methyltransferase 3A,” AND “R882,” “Arginine 882,” “Arg-882,” “Arg 882,” “882.”
Study Selection
No related review protocol has been existed or registered. Studies were included when they fulfilled all criteria as follows. Published in English before October 25, 2015; original articles as cohort studies; focused on prognostic effect of DNMT3A containing R882 mutations on AML patients; offered data on overall survival (OS) and/or relapse-free survival (RFS). Exclusion criteria: pediatric AML; meta-analysis, letters, comments, case reports and reviews; duplicate publications. And repetitive literature was managed and removed by Endnote X4.
Data Extraction and Quality Assessment
Two researchers independently went over all the articles that were satisfied with the inclusion criteria, and the discrepancies between reviewers were resolved via discussion. Information including first author, year of publication, study region, sample size, sex distribution, median age, the French-American-British (FAB) subtype and cytogenetic features from each eligible study was extracted. Furthermore, the corresponding hazard ratios (HRs) with 95% confidence interval (95% CI) for RFS and OS were calculated from COX multivariable models, or from analysis of original data in supplemental information via COX models, or from corresponding Kaplan–Meier (K-M) curves by the methods.26,27
The methodological quality of included literatures was evaluated through the Newcastle-Ottawa-Scale (NOS).28 The NOS consisted of 3 dimensions (selection, comparability, and exposure or outcome), which assigned, respectively, 4, 2, and 3 points for the 3 dimensions with a total maximum of 9 scores. On the basis of the NOS, the quality of these studies was classified into 3 types: high qualities (7–9 scores), intermediate qualities (4–6 scores), and low qualities (1–3 scores).28,29
Statistical Analysis
Meta-analysis was carried out with the software of Review Manager (RevMan) (version 5.3.5; the Nordic Cochrane Centre, Copenhagen, Denmark), while meta-regression analysis was performed with STATA software (version 12.0; College Station, TX). Prognostic role of DNMT3A R882 mutations on RFS and OS were assessed by estimation of the pooled HRs and their respective 95% CI with the inverse variance method in total population and subgroups. Statistical heterogeneity was assessed by using the Chi-squared test (the significance of heterogeneity was artificially expressed as P′-value to distinguish from the significance of outcomes) and I2 statistics. When there was no significant heterogeneity (P-value > 0.1 and I2 < 50%), the pooled HRs were assessed by fixed-effect model. Otherwise, random-effect model was applied to enhance the stability of the meta-analysis. Subgroup analysis and meta-regression analysis were implemented to probe the potential sources of heterogeneity. Sensitivity analysis was conducted to test the robustness of incorporative HRs for RFS and OS of AML patients. Publication bias was evaluated by Begg funnel plot and Egger test. This study was written following the PRISMA guidelines. As a meta-analysis study, ethical approval of this study is not required.
RESULTS
Search Results
A total of 50 publications were identified by the systematic literature search, of which 1 review was excluded and 13 duplicates were removed, resulting in 36 publications. Also, 28 articles were removed in view of relevance, design, and suitable outcome data through the title, abstract, and full-text screening regarding the aforementioned inclusion criteria (Figure 1). Ultimately, 8 publications were included in the meta-analysis.
FIGURE 1.
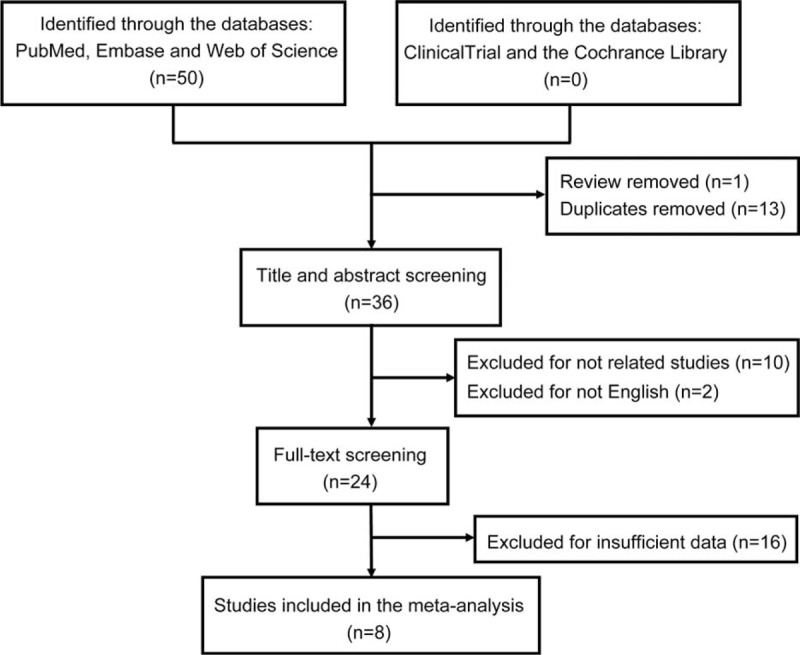
Flow chart of the procedure for the literature search.
Characteristics and Bias Risk of the Included Studies
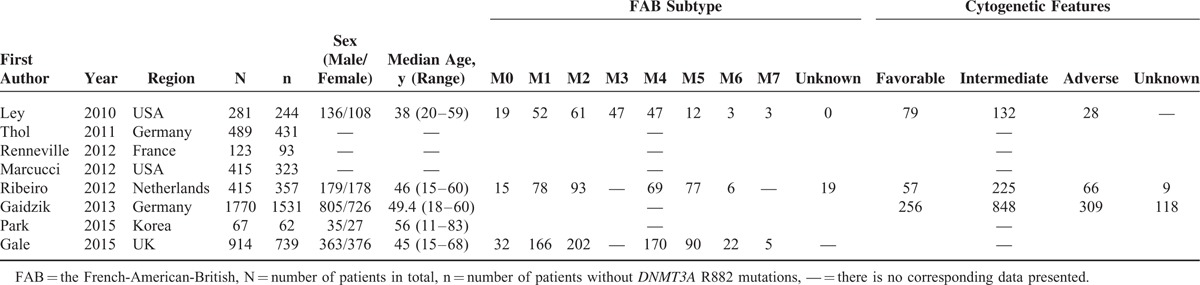
Eight studies containing a total of 4474 subjects (694 with DNMT3A R882 mutations) were included in the meta-analysis. The principal features of these subjects with or without DNMT3A R882 mutations are displayed in Tables 1 and 2, respectively, and the accessional characteristics are shown in Tables S1 and S2, respectively. Meanwhile, sample size of the studies ranged from 67 to 1770 patients. Of these studies, 5 studies originated from Europe, 1 from Asia, and 2 from USA. The frequency of DNMT3A R882 mutations ranged from 7.46% to 24.39%.
TABLE 1.
Clinical and Laboratory Characteristics of AML Patients With DNMT3A R882 Mutations From the 8 Included Studies
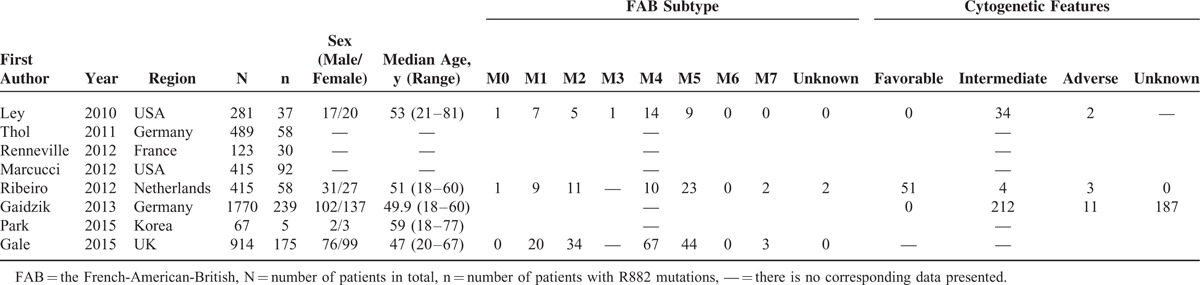
TABLE 2.
Clinical and Laboratory Characteristics of AML Patients Without DNMT3A R882 Mutations From the 8 Included Studies
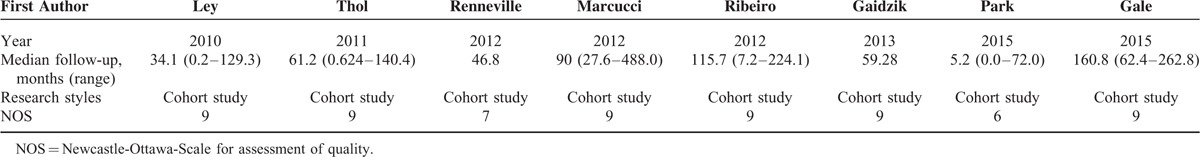
Risk of bias (see quality assessment in the part of methods) was evaluated based on 9 assessment items of NOS. The qualities of 7 studies (87.5%) were regarded as high, and the rest 1 study (12.5%) was treated as moderate. Relevant details are presented in Table 3.
TABLE 3.
Quality Assessment of 8 Included Studies
Prognostic Power of DNMT3A R882 Mutations in Total Population
-
RFS
Data were extracted from 6 studies, totaling 3915 AML patients, containing 631 patients with DNMT3A R882 mutations and 3284 without R882 mutations. As presented in Figure 2, results showed no distinct heterogeneity (P′ = 0.21, I2 = 30 %). With a fixed-effect model, a significant shorter RFS was observed in AML patients with DNMT3A R882 mutations compared with those without R882 mutations in total population (HR = 1.40, 95% CI = 1.24–1.59, P < 0.001).
-
OS
Data were derived from 8 studies, totaling 4474 AML patients, containing 694 patients with DNMT3A R882 mutations and 3780 without R882 mutations. With a random-effect model, AML patients with the DNMT3A R882 mutations presented an evident shorter OS time than those without R882 mutations in total population (HR = 1.47, 95% CI = 1.17–1.86, P = 0.001, Figure 3). These results suggested that DNMT3A R882 mutations could predict inferior clinical outcomes in AML patients.
FIGURE 2.
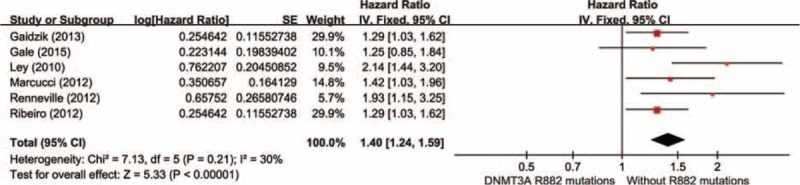
Forest plots of the HRs with 95% CI for RFS in overall AML patients. The size of the blocks or diamonds represents the weight for the fixed-effect model in the meta-analysis. HR >1 indicates that the presence of DNMT3A R882 mutations is associated with a shorter relapse-free survival (RFS).
FIGURE 3.
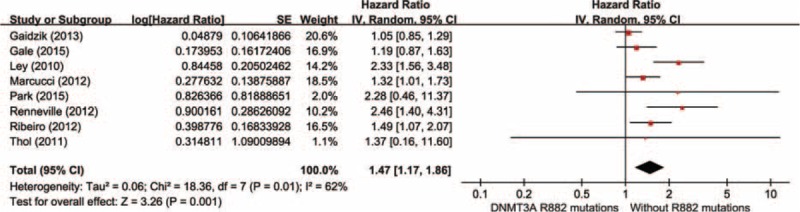
Forest plots of the HRs with 95% CI for OS in overall AML patients. The size of the blocks or diamonds represents the weight for the random-effect model in the meta-analysis. HR >1 indicates that the presence of DNMT3A R882 mutations is associated with a shorter overall survival (OS).
Prognostic Power of DNMT3A R882 Mutations in Different Subgroups
Prognostic Power of DNMT3A R882 Mutations Stratified by Age (<60 and ≥60)
Age is the most important factor influencing the prognosis of AML, and adults with age older than 60 have a shorter OS than adults with age younger than 60.30,31 Meanwhile, based on the NCCN Clinical Practice Guidelines in Acute myeloid leukemia (available free of charge on the NCCN web site: http://www.nccn.org/ and in the NCCN Guidelines for Acute Myeloid Leukemia), we divided AML patients into different subgroups containing different age (<60 and ≥60) to further validate the prognostic ability of DNMT3A R882 mutations. The incorporative results of HRs for RFS and OS among AML patients of different age (<60 and ≥60) were presented in fixed- and random-effect model, respectively (Tables 4 and 5). Also, the matching forest plots are shown in Figures S1 and S2. With a fixed-effect model, significant shorter RFS or/and OS were observed in AML patients with the DNMT3A R882 mutations in comparison with those without R882 mutations in both subgroups of age < 60 (RFS: HR = 1.44, 95% CI = 1.25–1.66, P < 0.001) and age ≥60 (RFS: HR = 2.03, 95% CI = 1.40–2.93, P < 0.001; OS: HR = 1.85, 95% CI = 1.36–2.53, P < 0.001). With a random-effect model, significant shorter OS was observed in AML patients with the DNMT3A R882 mutations in comparison with those without R882 mutations in subgroup of age < 60 (OS: HR = 1.48, 95% CI = 1.15–1.90, P = 0.002). Meanwhile, similar results were also observed in the other model. This suggested that the DNMT3A R882 mutations could predict shorter RFS and OS in AML patients regardless of patient age.
TABLE 4.
Outcomes of Subgroups Analysis in Fixed-Effect Models
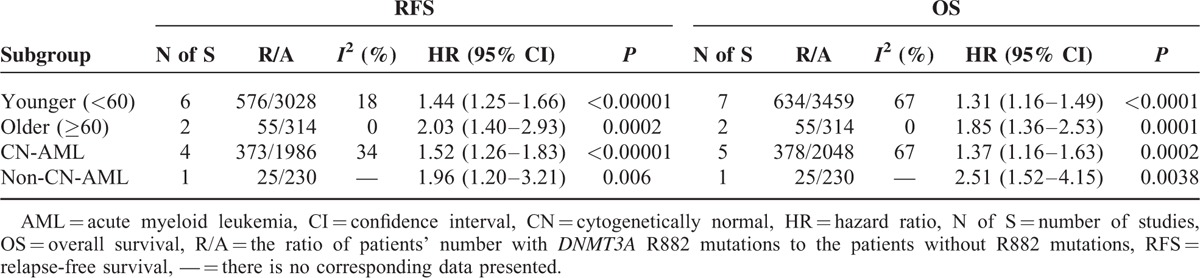
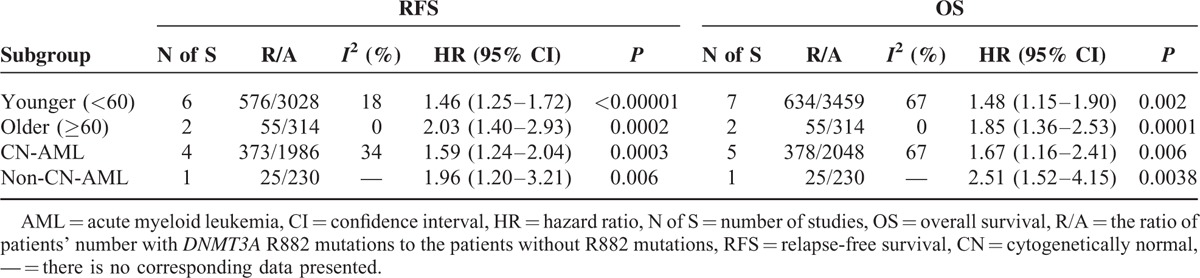
TABLE 5.
Outcomes of Subgroup Analysis in Random-Effect Models
Prognostic Power of DNMT3A R882 Mutations in the Population of CN-AML and Non-CN-AML
Cytogenetic influenced dramatically the clinical outcome of AML patients, so subgroup analysis was also carried out in CN-AML and non-CN-AML patients, respectively. Tables 4 and 5 show the pooled results of HRs for RFS and OS among AML patients in subgroups of CN-AML and non-CN-AML in fixed- and random-effect model, respectively. Also, the matching forest plots in subgroups of CN-AML are presented in Figure S3. With a fixed-effect model, a significant shorter RFS was observed in AML patients with the DNMT3A R882 mutations compared with those without R882 mutations in subgroup of CN-AML (HR = 1.52, 95% CI = 1.26–1.83, P < 0.001). With a random-effect model, a significant shorter OS was observed in AML patients with the DNMT3A R882 mutations compared with those without R882 mutations in subgroup of CN-AML (HR = 1.67, 95% CI = 1.16–2.41, P = 0.006). Similarly, the consistent results were seen in the other effect model.
RFS and OS in non-CN-AML patients were analyzed by using the original data of Ley study. As presented in Figure 4, remarkable shorter RFS and OS was shown in AML patients with the DNMT3A R882 mutations than those without R882 mutations in the non-CN-AML patients (RFS: HR = 1.96, 95% CI = 1.20–3.21, P = 0.006; OS: HR = 2.51, 95% CI = 1.52–4.15, P = 0.0038). In brief, DNMT3A R882 mutations may act as a poor prognostic indicator in both CN-AML and non-CN-AML patients.
FIGURE 4.
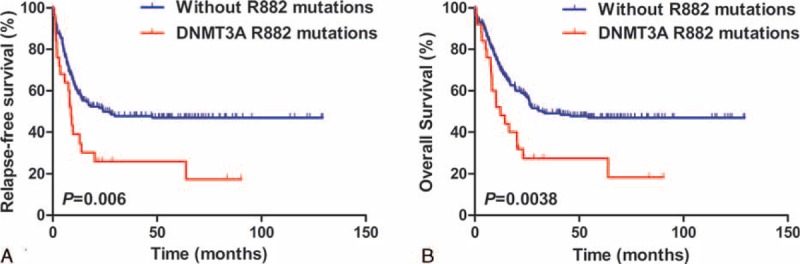
Kaplan–Meier estimates of RFS and OS in the non-CN-AML patients. The relapse-free survival [RFS] (A) and overall survival [OS] (B) of DNMT3A R882 mutations were shown in noncytogenetically normal (CN)-AML patients, including 25 with DNMT3A R882 mutations and 205 without R882 mutations (n = 230). The median survival of RFS: DNMT3A R882 mutations vs without R882 mutations = 8.8 vs 24.2, P = 0.006; and the median survival of OS: DNMT3A R882 mutations vs without R882 mutations = 12.3 vs 32.5, P = 0.0038.
Comparison of Prognostic Power Between DNMT3A R882 Mutations and Non-R882 Mutations
-
RFS
Data were derived from 3 researches summing up 465 AML patients with DNMT3A mutations, including 306 DNMT3A R882 mutations and 159 DNMT3A non-R882 mutations. As shown in Figure 5, the results showed no visible heterogeneity (P′ = 0.47, I2 = 0%). With a fixed-effect model, there was no obvious difference in RFS time between the group of DNMT3A R882 mutations and non-R882 mutations (HR = 1.23, 95% CI = 1.00–1.52, P = 0.05).
-
OS
Data were extracted from 4 studies, totaling 954 AML patients with DNMT3A mutations, including 364 DNMT3A R882 mutations and 590 DNMT3A non-R882 mutations. With a random-effect model, there was no distinct difference in OS time between the group of DNMT3A R882 mutations and non-R882 mutations (Figure 6; HR = 0.95, 95% CI = 0.59–1.54, P = 0.84). These results suggested that DNMT3A R882 mutations may not differentiate the prognosis of AML patients at least on OS with other DNMT3A mutations.
FIGURE 5.
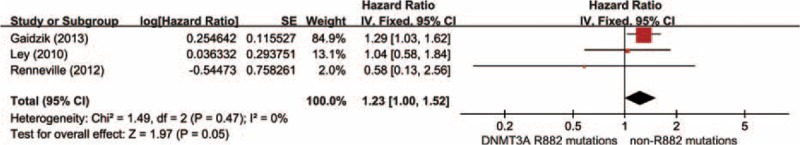
Forest plots of the HRs with 95% CI for RFS in AML patients with DNMT3A mutations. The size of the blocks or diamonds represents the weight for the fixed-effect model in the meta-analysis. HR >1 indicates that the presence of DNMT3A R882 mutations is associated with a shorter relapse-free survival (RFS).
FIGURE 6.
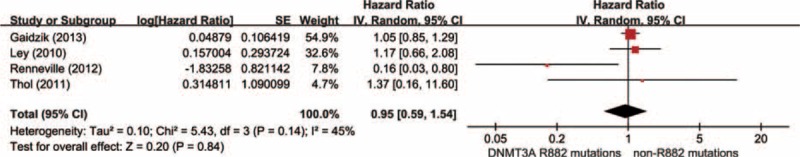
Forest plots of the HRs with 95% CI for overall survival in AML patients with DNMT3A mutations. The size of the blocks or diamonds represents the weight for the random-effect model in meta-analysis.
Meta-Regression, Publication Bias, and Sensitive Analysis
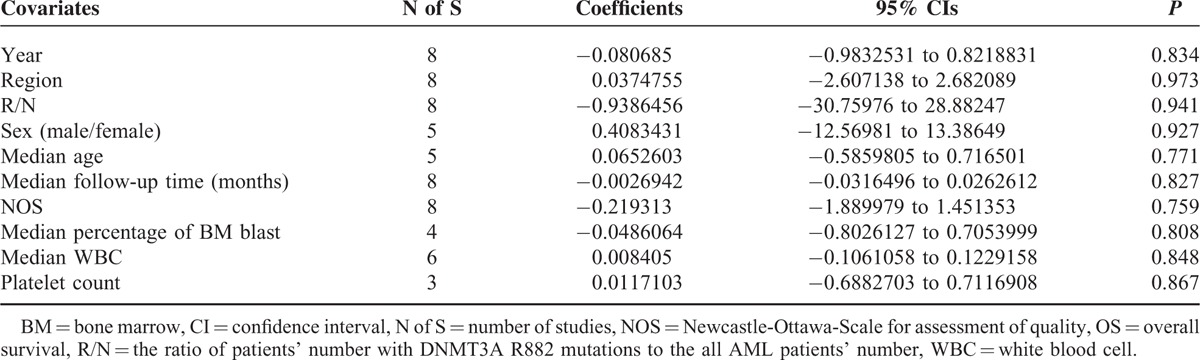
Meta-regression analysis was analyzed by using the software Stata 12.0. Fixed-effect meta-regression analysis was applied in RFS study due to its relatively low heterogeneity (P′ = 0.21 > 0.1 and I2 = 30% < 50%), while random-effect model was applied in OS study. Fixed-effects meta-regression analysis showed that none of the following covariates affected the prognostic values of R882 mutations on RFS in AML patients (Table 6 and Figure S4): publication year (coefficient = −0.0939653, P = 0.835), region (coefficient = −0.1422338, P = 0.923), R/N (the ratio of patients’ number with DNMT3A R882 mutations to the all AML patients’ number, coefficient = 0.3002279, P = 0.986), sex (the ratio of males’ number to females’ number, coefficient = 0.3981209, P = 0.936), median age (coefficient = 0.0685541, P = 0.866), median follow-up time (months, coefficient = −0.0023704, P = 0.873), NOS (Newcastle-Ottawa-Scale for assessment of quality, coefficient = −0.1587112, P = 0.884), median percentage of BM blast (coefficient = −0.0224812, P = 0.926), and median WBC (coefficient = 0.0005318, P = 0.991). Furthermore, the random-effects meta-regression analysis showed that none of the following covariates affected the prognostic values of R882 mutations on OS in AML patients (Table 7 and Figure S5): publication year (coefficient = −0.080685, P = 0.834), region (coefficient = 0.0374755, P = 0.973), R/N (coefficient = −0.9386456, P = 0.941), sex (coefficient = 0.4083431, P = 0.927), median age (coefficient = 0.0652603, P = 0.771), median follow-up time (months, coefficient = −0.0026942, P = 0.827), NOS (coefficient = −0.219313, P = 0.759), median percentage of BM blast (coefficient = −0.0486064, P = 0.808), median WBC (coefficient = 0.008405, P = 0.848), and platelet count (coefficient = 0.0117103, P = 0.867).
TABLE 6.
Fixed-Effects Meta-Regression Analysis for RFS Studies
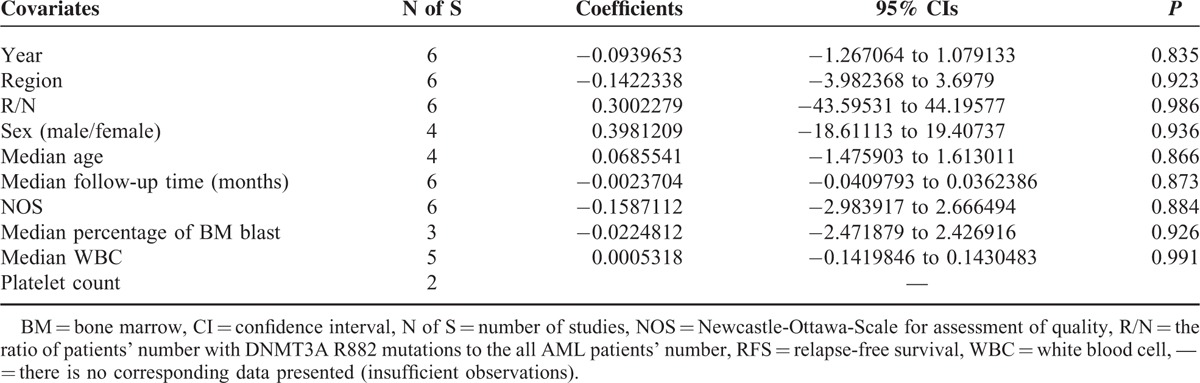
TABLE 7.
Random-Effects Meta-Regression Analysis for OS Studies
Publication bias was analyzed by using RevMan 5.3.5 software. The funnel plot of RFS outcomes showed that the points were evenly distributed, and most of the points were within 95% CI. This may indicate no obvious publication bias in RFS analysis, and thus, the corresponding results of the study were credible. Although the shape of these funnel plot in OS studies did not amount to gross asymmetry except for the OS outcome in the population of age ≥60, these results merit consideration. In addition, the z-value of 4.54 or something like that and a corresponding 2-tailed P-value of <0.001 on OS between the group of DNMT3A R882 mutations and without R882 mutations in total patients is also worthy of our attention. The results of funnel plot are shown in Supplemental Materials (Figure S6).
Furthermore, sensitivity tests were conducted during the process of the meta-analysis. Exclusion of any single study did not alter dramatically the over-all findings (Tables S3–S5).
DISCUSSION
DNA methylation is a key mechanism of epigenetic regulation in eukaryotes. As one of the enzymatically active mammalian DNA methyltransferases (DNMTs), DNMT3A can regulate gene expression and maintain cellular homeostasis by mediating the de novo methylation of DNA.32 Recently, with the ever-accelerated development of cancer genome sequencing, DNMT3A was exposed as one of the most frequently mutated genes, raising questions concerning the prominent part of DNMT3A mutations in AML patients.14,33 As dominant missense alterations in DNMT3A mutations, R882 mutations cause directly the focal hypomethylation phenotype.20 In addition, DNMT3A R882 mutations are frequent in AML, but rare in other hematological diseases,34 suggesting that DNMT3A R882 mutations own the potential to act as an independent prognostic marker in AML. This systematic meta-analysis showed that mutant DNMT3A R882 was associated with poor prognosis in AML patients.
In this meta-analysis, 8 studies containing a total of 4474 AML patients were included, which included 694 AML patients with DNMT3A R882 mutations and 3780 AML patients without R882 mutations. And we found that AML patients with the DNMT3A R882 mutations presented significant shorter RFS and OS than those without R882 mutations in overall AML patients. Although there was a considerable but acceptable heterogeneity in those of OS study except for the population over age 60, the outcomes still deserve being considered. And various possible reasons contributed to the production of heterogeneity. First of all, the constituent ratio of patients’ age and cytogenetic abnormalities were diverse in each study. For example, 7 studies just or almost included patients under age 60,14,15,23,24,35–37 and 4 studies merely included CN-AML patients.23–25,35 Secondly, in consideration of less numbers of AML patients with R882 mutations in few included studies, we cannot further reckon the RFS and OS of AML patients when they were included. For example, only 5 AML patients with R882 mutations was appeared in K-M curves of RFS and OS in Park study, which can not accurately even not roughly calculate the values of HRs and 95% CI. They are likely to a source of heterogeneity. Thirdly, there was a lot of between-study heterogeneity on some other hands, such as the time of follow-up, region origin of patients among the 8 studies. At last, general information of individual patient just like Ley study did was not available for other studies, which also led to the heterogeneity of our analysis.
AML is a clonal disorder of hemopoietic stem cells.38 The survival of AML is influenced by factors such as age, cytogenetics, somatic mutations, etc.39 Age is the most vital prognostic factor for AML patients, and adults with age older than 60 have a shorter OS in comparison with adults with age younger than 60.30,31 Meanwhile, cytogenetic normality or abnormality influenced clinical outcome of AML dramatically.40 For these reasons, we stratified AML patients into different subgroups including age (<60 and ≥60) and cytogenetics (CN-AML and non-CN-AML) to further validate the prognostic effect of mutant DNMT3A R882 in AML patients. Our findings showed shorter RFS and OS in AML patients with DNMT3A R882 mutations compared with those without R882 mutations in subgroups of age <60, age ≥60, CN-AML, and non-CN-AML, respectively. These results indicated that DNMT3A R882 mutations may act as a poor prognostic indicator in AML patients, which is independent of the age and cytogenetics. While, a relatively considerable heterogeneity remained in OS study, except for the subgroup of age ≥60. This may result from the small number of literatures included in the OS study of age ≥60 AML patients. Or else, it suggested that the DNMT3A R882 mutations may be particularly appropriate for predicting the clinical outcome OS in the AML population of age ≥60.
Presently, there is still a controversy on prognostic effect of R882 mutations compared with DNMT3A non-R882 mutations (DNMT3A mutations affecting other codons). Ley et al14 suggested no difference in the OS between the 2 groups. Meanwhile, Marcucci et al35 reported that DNMT3A R882 mutations had no prognostic value in younger patients whereas were independently associated with worse outcome in older patients. Yet, Gaidzik et al's24 findings showed unfavorable for DNMT3A R882 mutations on RFS while favorable for non-R882 mutations on OS in a cohort study. While in our meta-analysis, we observed no difference in either RFS or OS between AML patients with the DNMT3A R882 mutations and those with non-R882 mutations in patients positive for DNMT3A mutations. What merit our concern is that the P-value coincidently was 0.05, and this makes it essential do more studies with larger sample size to validate the OS significance of DNMT3A non-R882 mutations in AML patients. Our results were in lines with the studies of Ley and Marcucci, while were partly inconsistent with Gaidzik's study. The inconsistence was possibly due to the differences in biometrical analysis, such as selection bias and variances in model building. In Gaidzik's study, a potential selection bias may exist because of the high percentage of patients was selected for the analysis in relation to the whole study populations with 90%, while low percentage of patients was selected for the analysis in relation to the whole study populations with 6% in Marcucci study35 and 18% in Ley study.14 Anyway, DNMT3A R882 mutations could predict shorter RFS and OS in total AML population or in patients with DNMT3A mutations, especially for RFS.
Three main limitations should be considered in our meta-analysis. First, there may be language bias, because the included studies were totally published in English. Second, selectional reporting was existed in some studies, such as the incorporated HRs for RFS were displayed in relative fewer studies than those for OS, and certain subgroups, like the older and non-CN-AML patients, were not analyzed in most of the studies, which leaded to the unavailability of useful information. Third, if aforementioned authors could offer complete patient data, like Ley, our paper would have been quite more flawless.
In conclusion, our meta-analysis presented definitively an independent inferior prognostic effect of mutant DNMT3A R882 on the RFS and OS in AML patients. This was true also for AML patients in subgroups of age <60, age ≥60, CN-AML, and non-CN-AML. These results of meta-analysis may provide an insight for the prognostic prediction of AML patients, as well as infuse a drop into the ocean of precision prediction. Further studies with larger sample size and open individual data of patients are needed to validate the prognostic significance of mutant DNMT3A R882 in AML patients.
Supplementary Material
Footnotes
Abbreviations: 95% CI = 95% confidence interval, AML = acute myeloid leukemia, ASXL1 = additional sex combs like 1, CEBPA = CCAAT/enhancer binding protein (C/EBP) alpha, CN = cytogenetically normal, DNMT3A = DNA (cytosine-5)-methyltransferase 3 alpha, FAB = the French-American-British, FLT3-ITD = internal tandem duplication in fms-related tyrosine kinase 3, HR = hazard ratio, IDH2 = isocitrate dehydrogenase 2, K-M = Kaplan–Meier, NOS = Newcastle-Ottawa-Scale, NPM1 = nucleophosmin, OS = overall survival, RFS = relapse-free survival.
Author contributions: XPC, GCL, and XQY conceived and designed the meta-analysis. XQY and LP collected the references and drafted the manuscript. XPC, GCL, WJZ, and BYJ revised critically the manuscript. All authors read and approved the final manuscript.
This work was supported by Chinese National Science Foundation (81422052), Special topic of the major subject of national science and technology (2012ZX09509–107), Hunan Provincial Natural Science Foundation of China (13JJ1010), and the Fundamental Research Funds for the Central Universities of Central South University (2015zzts096). Furthermore, we thank Doctor Timothy J. Ley and his colleagues for the original data in supplemental information, which provided great convenience for our meta-analysis especially in subgroups analysis.
X-QY and LP contributed equally to this work.
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
REFERENCES
- 1.Kato T, Sakata-Yanagimoto M, Nishikii H, et al. Hes1 suppresses acute myeloid leukemia development through FLT3 repression. Leukemia 2015; 29:576–585. [DOI] [PubMed] [Google Scholar]
- 2.Ignatz-Hoover JJ, Wang H, Moreton SA, et al. The role of TLR8 signaling in acute myeloid leukemia differentiation. Leukemia 2015; 29:918–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mrozek K, Marcucci G, Paschka P, et al. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: are we ready for a prognostically prioritized molecular classification? Blood 2007; 109:431–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niederwieser C, Kohlschmidt J, Volinia S, et al. Prognostic and biologic significance of DNMT3B expression in older patients with cytogenetically normal primary acute myeloid leukemia. Leukemia 2015; 29:567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med 2015; 373:1136–1152. [DOI] [PubMed] [Google Scholar]
- 6.Yan P, Frankhouser D, Murphy M, et al. Genome-wide methylation profiling in decitabine-treated patients with acute myeloid leukemia. Blood 2012; 120:2466–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcucci G, Yan P, Maharry K, et al. Epigenetics meets genetics in acute myeloid leukemia: clinical impact of a novel seven-gene score. J Clin Oncol 2014; 32:548–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlenk RF, Dohner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med 2008; 358:1909–1918. [DOI] [PubMed] [Google Scholar]
- 9.Patel JP, Gonen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 2012; 366:1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen BF, Chan WY. The de novo DNA methyltransferase DNMT3A in development and cancer. Epigenetics 2014; 9:669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan XJ, Xu J, Gu ZH, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet 2011; 43:309–315. [DOI] [PubMed] [Google Scholar]
- 12.Hahn CN, Ross DM, Feng J, et al. A tale of two siblings: two cases of AML arising from a single pre-leukemic DNMT3A mutant clone. Leukemia 2015; 29:2101–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang DC, Liu HC, Yang CP, et al. Cooperating gene mutations in childhood acute myeloid leukemia with special reference on mutations of ASXL1, TET2, IDH1, IDH2, and DNMT3A. Blood 2013; 121:2988–2995. [DOI] [PubMed] [Google Scholar]
- 14.Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med 2010; 363:2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thol F, Damm F, Ludeking A, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J Clin Oncol 2011; 29:2889–2896. [DOI] [PubMed] [Google Scholar]
- 16.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 2015; 372:793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chun M. Precision medicine initiative in the offing. Cancer Discov 2015; 5:456. [DOI] [PubMed] [Google Scholar]
- 18.Mayle A, Yang L, Rodriguez B, et al. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood 2015; 125:629–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim SJ, Zhao H, Hardikar S, et al. A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood 2013; 122:4086–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russler-Germain DA, Spencer DH, Young MA, et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell 2014; 25:442–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin J, Yao DM, Qian J, et al. Recurrent DNMT3A R882 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS One 2011; 6:e26906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Im AP, Sehgal AR, Carroll MP, et al. DNMT3A and IDH mutations in acute myeloid leukemia and other myeloid malignancies: associations with prognosis and potential treatment strategies. Leukemia 2014; 28:1774–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Renneville A, Boissel N, Nibourel O, et al. Prognostic significance of DNA methyltransferase 3A mutations in cytogenetically normal acute myeloid leukemia: a study by the Acute Leukemia French Association. Leukemia 2012; 26:1247–1254. [DOI] [PubMed] [Google Scholar]
- 24.Gaidzik VI, Schlenk RF, Paschka P, et al. Clinical impact of DNMT3A mutations in younger adult patients with acute myeloid leukemia: results of the AML Study Group (AMLSG). Blood 2013; 121:4769–4777. [DOI] [PubMed] [Google Scholar]
- 25.Park SH, Choi JC, Kim SY, et al. Incidence and prognostic impact of DNMT3A mutations in Korean normal karyotype acute myeloid leukemia patients. Biomed Res Int 2015; 2015:723682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007; 8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med 1998; 17:2815–2834. [DOI] [PubMed] [Google Scholar]
- 28.Wells GA, Shea B, O’Connell D, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Ottawa Hospital Research Institute, 2013, Available at: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp. Accessed 2013 Jun 23 [Google Scholar]
- 29.Tie R, Zhang T, Fu H, et al. Association between DNMT3A mutations and prognosis of adults with de novo acute myeloid leukemia: a systematic review and meta-analysis. PLoS One 2014; 9:e93353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Estey E, Dohner H. Acute myeloid leukaemia. Lancet 2006; 368:1894–1907. [DOI] [PubMed] [Google Scholar]
- 31.Gangatharan SA, Grove CS, P’ng S, et al. Acute myeloid leukaemia in Western Australia 1991–2005: a retrospective population-based study of 898 patients regarding epidemiology, cytogenetics, treatment and outcome. Intern Med J 2013; 43:903–911. [DOI] [PubMed] [Google Scholar]
- 32.Deplus R, Denis H, Putmans P, et al. Citrullination of DNMT3A by PADI4 regulates its stability and controls DNA methylation. Nucleic Acids Res 2014; 42:8285–8296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang L, Rau R, Goodell MA. DNMT3A in haematological malignancies. Nat Rev Cancer 2015; 15:152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Zhu B. Acute myeloid leukemia with DNMT3A mutations. Leuk Lymphoma 2014; 55:2002–2012. [DOI] [PubMed] [Google Scholar]
- 35.Marcucci G, Metzeler KH, Schwind S, et al. Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. J Clin Oncol 2012; 30:742–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gale RE, Lamb K, Allen C, et al. Simpson's paradox and the impact of different DNMT3A mutations on outcome in younger adults with acute myeloid leukemia. J Clin Oncol 2015; 33:2072–2083. [DOI] [PubMed] [Google Scholar]
- 37.Ribeiro AF, Pratcorona M, Erpelinck-Verschueren C, et al. Mutant DNMT3A: a marker of poor prognosis in acute myeloid leukemia. Blood 2012; 119:5824–5831. [DOI] [PubMed] [Google Scholar]
- 38.Ferrara F, Schiffer CA. Acute myeloid leukaemia in adults. Lancet 2013; 381:484–495. [DOI] [PubMed] [Google Scholar]
- 39.Singh H, Asali S, Werner LL, et al. Outcome of older adults with cytogenetically normal AML (CN-AML) and FLT3 mutations. Leuk Res 2011; 35:1611–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Byrd JC, Mrozek K, Dodge RK, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood 2002; 100:4325–4336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


