Abstract
Polyketide synthases (PKSs) of Aspergillus species are multidomain and multifunctional megaenzymes that play an important role in the synthesis of diverse polyketide compounds. Putative PKS protein sequences from Aspergillus species representing medically, agriculturally, and industrially important Aspergillus species were chosen and screened for in silico studies. Six candidate Aspergillus species, Aspergillus fumigatus Af293, Aspergillus flavus NRRL3357, Aspergillus niger CBS 513.88, Aspergillus terreus NIH2624, Aspergillus oryzae RIB40, and Aspergillus clavatus NRRL1, were selected to study the PKS phylogeny. Full-length PKS proteins and only ketosynthase (KS) domain sequence were retrieved for independent phylogenetic analysis from the aforementioned species, and phylogenetic analysis was performed with characterized fungal PKS. This resulted into grouping of Aspergilli PKSs into nonreducing (NR), partially reducing (PR), and highly reducing (HR) PKS enzymes. Eight distinct clades with unique domain arrangements were classified based on homology with functionally characterized PKS enzymes. Conserved motif signatures corresponding to each type of PKS were observed. Three proteins from Protein Data Bank corresponding to NR, PR, and HR type of PKS (XP_002384329.1, XP_753141.2, and XP_001402408.2, respectively) were selected for mapping of conserved motifs on three-dimensional structures of KS domain. Structural variations were found at the active sites on modeled NR, PR, and HR enzymes of Aspergillus. It was observed that the number of iteration cycles was dependent on the size of the cavity in the active site of the PKS enzyme correlating with a type with reducing or NR products, such as pigment, 6MSA, and lovastatin. The current study reports the grouping and classification of PKS proteins of Aspergilli for possible exploration of novel polyketides based on sequence homology; this information can be useful for selection of PKS for polyketide exploration and specific detection of Aspergilli.
Keywords: Aspergillus, polyketide synthases, phylogeny, polyketide, ketosynthase
Introduction
Aspergillus species have recently gained great attention in view of their impact on humans and agriculture and due to the production of bioactive secondary metabolites (SMs). Sequencing of Aspergillus species genome led to the identification of more than 200 SM gene clusters with the potential to produce SMs, which still need to be explored. Many of these clusters include polyketide synthases (PKSs) as the principal enzyme.1 The SM compounds presently identified from Aspergillus species under studied culture conditions are only handful, and various research groups are exploring SM compounds using different approaches. Web-based online tools, such as SMURF: genomic mapping of fungal SM clusters, have been developed and are extremely useful as they give the annotation of gene clusters for sequenced fungal genomes.2 Considering the wealth of information provided by whole-genome sequencing and the presence of schematically arranged gene clusters in Aspergilli, researchers are encouraged to explore medically and industrially important compounds.3–5 Polyketides are industrially well-exploited class of compounds in microbes mainly due to their medicinal importance.6 Several Aspergillus polyketide products have been very well characterized and understood, eg, aflatoxins from Aspergillus flavus,7,8 melanin pigments from Aspergillus fumigatus,9 and lovastatin from Aspergillus terreus.10,11 In order to exploit the potential of Aspergillus species for the secretion of useful polyketides, it is necessary to examine and understand the PKS enzyme machinery.12
PKS enzymes of Aspergilli are iterative type I PKSs and are close structural and functional analogs to mammalian fatty acid synthases (FASs).13 This type of enzymes reuses their domains in cyclic fashion. In Aspergilli, PKSs catalyze the condensation of precursor, acetyl-CoA and malonyl-CoA(n), to produce polyketides.8,12 Three essential domains of PKSs are ketosynthase (KS), acyltransferase (AT), and acyl carrier protein (ACP). The presence of other domains such as β-ketoacyl reductase (KR), enoyl reductase (ER), dehydratase (DH), methyltransferase (MT), and Claisen cyclase/thioesterase (CLC/TE) is variable and depends on the type of PKSs in Aspergillus species.14 The functionality of reducing and nonreducing (NR) domains such as MT, DH, ER, and CLC/TE of PKS enzyme directs the types of end compounds produced. PKSs based on the presence of their domains may have differences in end product catalysis. Gene clusters containing PKS and nonribosomal peptide synthases (NRPSs) are putatively identified, and experimental studies are being conducted to link these putative clusters with the secreted compound in various in vitro conditions.15–17 Selecting the gene clusters with PKS gene based on in silico evidence and short-listing the candidates for further knockdown studies to explore the polyketide compound will narrow down the research efforts for possible SM exploration.
A comparative sequence analysis of the Aspergillus full-length PKS proteins has been carried out with functionally characterized fungal PKSs by a phylogenetic approach, to examine and assign the putative function to unexplored PKSs of Aspergillus species. In the current study, attempts have been made to characterize selected PKS genes from six Aspergillus species and to better understand the gene architecture and protein structures of PKS enzyme.
Methods and Materials
Fungal strains and cultural conditions
Aspergillus isolates were collected from the Indian Type Culture Collection, Indian Agricultural Research Institute, New Delhi, India, in BSL2 facility. These isolates were isolated from various agricultural and animal sources and were morphologically characterized as Aspergillus species and later verified by amplification using Internal transcribed spacer (ITS) region primers. All fungal isolates were handled in biosafety hood cabinet A2 as per the biosafety protocol of the institute. Cultures were grown on YGT media (0.5% yeast extract, 2% dextrose, and 1 mL of trace elements) at 27 °C for 24 hours at 125 rpm. Genomic DNA was extracted using the LETS buffer that contains 0.1 M lithium, 20 mM EDTA, and 0.5% SDS.18 DNA from fungal species (Aspergillus, Trichoderma spp., and Fusarium spp.) was isolated, as described earlier.18,19 The quality of DNA from the isolations was checked by gel electrophoresis, and DNA concentration was determined by NanoDrop.
Database search
Six Aspergillus species, A. fumigatus Af293, A. flavus NRRL3357, Aspergillus niger CBS 513.88, A. terreus NIH2624, Aspergillus oryzae RIB40, and Aspergillus clavatus NRRL1, were selected for this study based on their agricultural, medical, and industrial relevance. Aspergillus genome was searched for putative PKS sequences by subjecting the protein sequence of KS domain of PKSP (XP_756095.1) of A. fumigatus Af293 as a query into BLASTp. Domains in PKS were searched by subjecting each putative amino acid sequence to online tools SEARCHPKS,20 MapsiDB,21 and CDD (NCBI). A total of 190 Aspergilli PKS sequences were retrieved and analyzed in this study. Iterative type I PKS sequences from other fungi, where polyketide products are characterized, were also retrieved and included for comparison.22 These PKS proteins are from Penicillium chrysogenum Wisconsin 54-1255,23 Penicillium marneffei ATCC 18224,24 Pyrenophora tritici-repentis Pt-1C-BFP, Gibberella zeae PH-1,25 Penicillium citrinum,26 Gibberella moniliformis,27,28 Emericella nidulans,29 Monascus purpureus,30 Gibberella fujikuroi,27 and Cochliobolus heterostrophus.31 Type III PKSs from bacteria (Streptomyces coelicolor A3(2),32 Myxococcus xanthus DK 1622,33 and Mycobacterium tuberculosis H37Rv)34 and FASs (from Homo sapiens [AAC50259], Gallus gallus [P12276], and Caenorhabditis elegans [NP_492417]) were used in this study for comparison and outgrouping. Gene accession numbers for these PKSs are given in Supplementary Table 2.
Phylogenetic analysis
Full-length PKS protein sequences were used in multiple sequence alignments using Clustal X (2.0.12).35 The resulting data were saved as Clustal and PHYLIP format files, and alignments were written as postscript files for further analysis. The phylogenetic analyses were performed in PHYLIP (ver. 3.69) programs, SEQBOOT, PROTDIST, NEIGHBOR, and CONSENSE, in the order as described previously.36,37 The Jones–Taylor–Thornton amino acid substitution matrix was performed where input order of sequences for phylogenetic analysis was randomized. For generating the KS domain tree, maximum parsimony-based method with a bootstrap value of 1000 was used and the final consensus tree was selected from the 100 MP trees obtained. Alignment gaps were treated as fifth character state, parsimony uninformative characters were excluded, and all the characters were unordered and equal weight.38 Tree length and consistency index were also calculated. Phylogenetic tree CONSENSE files obtained from PHYLIP were viewed with TreeView and MEGA (5.0).39
Conserved motifs for KS, sequencing, and phylogeny verification
Sequence alignments were studied for consensus sequences in KS domain across the Aspergillus species. Amino acid change in KS sequences was analyzed for predicting the functionality of model proteins and type of end polyketide product it catalyzes. Multiple alignment files of KS sequences were used to identify homologous regions of amino acid from Aspergillus spp. Amino acid sequences were converted into gene sequences using ExPASy (www.expasy.org/translate/), and forward and reverse primers of length 20–28 bp spanning KS region were designed by using Primer3 software (www.bioinfo.ut.ee/primer3-0.4.0) to check Tm, delta G, and self-hybridization. Degenerate primers were checked for specificity by BLASTn at NCBI before synthesis. Primer set KS_F and KS_R was standardized to amplify the PCR product of same size from three Aspergillus species in one reaction. Degenerate primer sequences are:
KS_F:AGTCTTGCKGCYATYCAYWTGGSCTGCAAYKCSATCTGGAGRA, KS_R: TGAGTWCCTGKTCCRTGCATTTCMACGTAGC. As positive control, universal fungal primers – ITS1 (based on 18S rRNA) and ITS4 (based on 28S rRNA): TCCGTAGGTGAACCTGCGG and ITS4: TCCTCCGCTTATTGATATG – were used for amplification from Aspergillus DNA (data not shown here).24 PCR was standardized in a total reaction volume of 50 µL. The mixture contained 10 × reaction buffer (100 mM Tris, 500 mM KCl; pH 8.3), 10 mM dNTP mix, forward and reverse primer (20 µM), five units of Taq DNA Polymerase (Platinum® Taq DNA Polymerase; Invitrogen), 500 ng of template DNA, and autoclaved water in 50 µL of total volume. PCR conditions were initial denaturation at 95 °C for 5 minutes, followed by 35 cycles of denaturation at 95 °C for 30 seconds, annealing of 56 °C for 30 seconds, and extension at 72 °C for 1 minute, followed by a final extension at 72 °C for 10 minutes in a Mastercycler gradient (Eppendorf). Amplified DNA products were separated by electrophoresis in a 1% agarose gel with a 100-bp DNA ladder as a molecular size marker (Promega Corporation). PCR products were purified by Qiagen PCR purification column (Qiagen, Germany) and submitted for sequencing. End PCR product was sequenced by either KS_F or KS_R and searched for sequence homology by BLASTX at NCBI. The deduced amino acid sequence was determined using ExPASy server at EBI (http://web.expasy.org/translate/). Multiple alignments of annotated KS sequences with KS protein from Aspergillus species were carried out using Clustal X. Consensus sequences were observed with respect to earlier bioinformatics analysis of KS for the presence of motifs. Neighbor joining tree was generated using PHYLIP package. Sequences were submitted to NCBI (KT221846-KT221852 and KT213730-KT213740).
Structural modeling of KS domains
KS sequence from UniProt or program database (PDB) was retrieved, and PDB files were generated by subjecting sequences to SBSPKS software (http://www.nii.ac.in/∼pksdb/sbspks/master.html).40 KS domain sequences from Aspergillus PKSs were named in this study as PKSP (XP_756095.1/Q4 WZA8), 6-methylsalicylic acid (6MSA; BAE65442.1/Q2PIT2), lovastatin nonaketide synthase (LNKS; Q9Y8A5), Afl25 (XP_002384329.1), Afu1 (XP_753141.2), and Anr (XP_001402408.2), which were modeled using the template structure of beta-ketoacyl-ACP synthase II from Escherichia coli (PDB ID: 1KAS).41 Modeller9v7 was used to build protein structures (target-template alignment and model generation), which uses an automated approach for comparative protein structure modeling.42 For each of the target protein sequence, 20 independent structures were generated and the best model was selected on the basis of minimum Discrete Optimized Protein Energy score. Then, the top model was further minimized through 400 steps of the steepest descent, followed by 3000 steps of conjugate gradient using GROMACS 4.0.5.43 Six conserved motifs identified through sequence alignment of each clade have been mapped on these modeled structures. The area and volume of the active site conformations were measured through CASTp server.44
Results
Sequence analysis and annotation of putative PKS proteins of Aspergilli
Based on the amino acid sequence of KS domain, putative PKSs were retrieved from six Aspergillus species. We studied more than 224 PKS sequences: 190 sequences from Aspergillus species and the rest from other filamentous fungi. Supplementary Tables 1 and 2 summarize the characterized and annotated PKSs of Aspergilli.
Phylogeny of Aspergillus PKS proteins
With the aim to observe the sequence diversity of full-length PKS proteins in Aspergillus, a phylogenetic tree was constructed using a full-length PKS enzyme with characterized fungal PKS. Phylogeny was classified into eight different clades, mainly classified based on the presence and absence of NR and reducing domains (Fig. 1). Among the full-length PKS phylogenies, three distinct groups were observed with a domain architect: (i) SAT-KS-AT-PT-ACP-(ACP)-CYC/TE, present in NR PKS; (ii) KS-AT-(DH)-(MT)-TE/PP, present in partially reducing (PR) PKS; and (iii) KS-AT-DH-(MT)-ER-KR-TE/PP, present in highly reducing (HR) PKS. Based on the groups, Aspergillus PKS enzymes are classified into NR, PR, and HR enzymes. Phylogeny was performed with functionally characterized PKSs for a suggestive polyketide it may produce based on PKS protein sequence homology (Supplementary Figs. 1–6). KS domain in PKS protein is the most conserved domain across Aspergillus species.45 Phylogeny was also constructed with only KS sequences from PKSs to check if phylogeny is primarily driven by KS domain (Fig. 2). The resulting KS genealogy was evaluated to classify the arrangement of major clades and subclades of domains and compared with full-length PKS phylogeny. We found similar classification in KS and full-length PKS phylogeny except that few PKS proteins change their clade based on additional domains. To support the sequence homology of KS classification, structural modeling of selected KS was carried out. This study was performed to score PKS candidates for possible polyketide exploration by in vitro experiments. We have summarized the PKS list for Aspergillus, presence of domains using KS, and PKS phylogeny probability of pk compound production. This information is given in Supplementary Table 1, with domain architecture of each PKS. Each clade exhibited a unique domain arrangement, and each clade contained at least one characterized PKS, except clade V.
Figure 1.
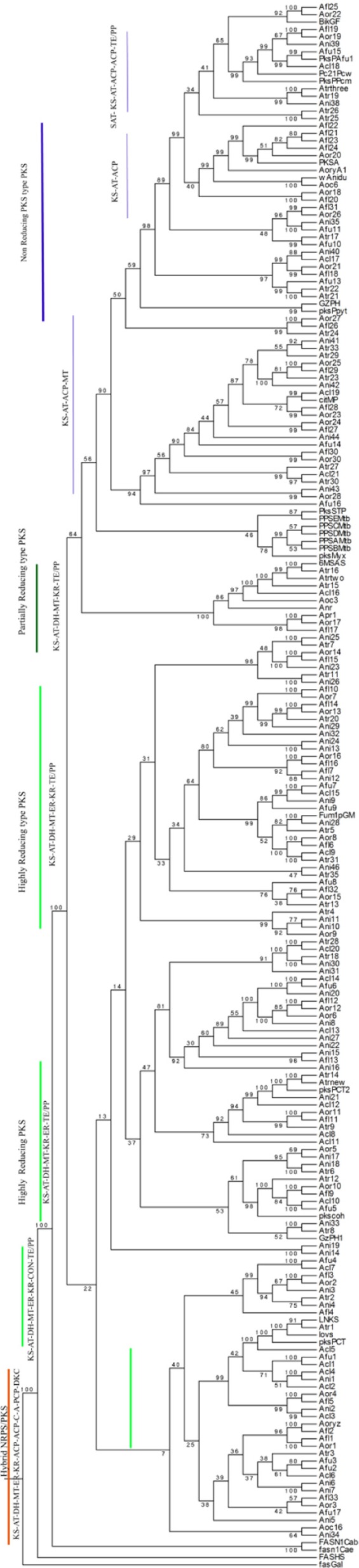
PKS genealogy of Aspergillus PKSs with characterized PKS (1000 boot strap value).
Notes: Genealogy of KS domain from type I PKSs of Aspergillus species is inferred by maximum parsimony analysis of the Aspergillus PKSs. Major clades and subclades are indicated by vertical bars that share a common organization of domains. Designations beginning with Afu, Afl, Ani, Atr, Aor, Apr, and Acl correspond to the A. fumigatus, A. flavus, A. niger, A. terreus, A. oryzae, A. parasiticus, and A. clavatus, respectively. Details of these PKS can be found in Supplementary Table 1. Protein FASs from G. gallus, H. sapiens, Caenorhabditis briggsae, and C. elegans served as an outgroup for this study. Bar colored in blue represents NR PKS with the domain architect SAT-KS-AT-ACP-ACP-TE/PP. Bar colored in green shows PR PKS with the domain architects KS-AT-ACP-MT-TE/PP. Bar with dark green color shows HR PKS with the domain architect KS-AT-DH-MT-ER-KR-TE/PP and also other HR domain containing PKS, such as KS-AT-DH-MT-KR-TE/PP and KS-AT-DH-MT-KR-ER-TE/PP. Hybrid PKS–NRPSs are noted with bar colored in orange with the domain architect of both PKS and NRPS: KS-AT-DH-MT-ER-KR-ACP-ACP-C-A-PCP-DKC.
Figure 2.
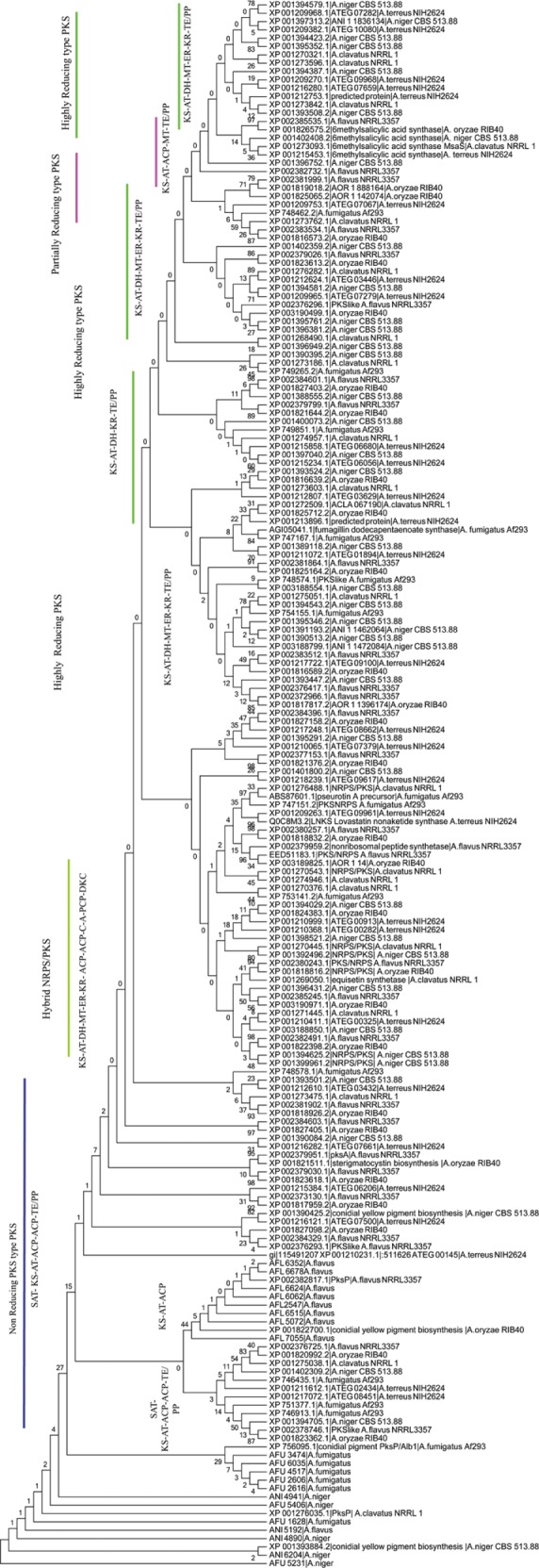
KS genealogy of Aspergillus PKSs (1000 boot strap value).
Notes: Phylogeny of KS domain from type I PKSs of Aspergillus species is inferred by maximum parsimony analysis. Major clades and subclades are indicated by vertical bars, which share a common organization of domains. Numbers below branches indicate percentage bootstrap values for each clade. KS domain from A. flavus, A. fumigatus, and A. niger sequenced in this study is also included, and details of which can be found in Supplementary Table 1.
NR PKSs – pigment
Sequences in clade I contain the domain architect SAT-KS-AT-ACP-ACP-TE/PP, known to be present in NR type of PKS, alb1. Alb1/pksP from A. fumigatus, enzyme known to be involved in the production of 1,3,6,8-tetrahydroxynaphthalene, a precursor for melanin,9 and red pigment Bikaverin-producing PKS4 from Fusarium spp., and melanin-producing PKS (Pc21Pcw, PksPPcm) from Penicillium spp. are classified in this group.24,46 These PKS enzymes lack ER, DH, and KR domains, required for reduction and dehydration steps, but they contain additional ACP domains.
NR PKSs – aflatoxin
Clade I also represents the domain architect, KS-AT-ACP-TE, which is present in NR PKS. Proteins in this subclade contain single ACP domain. Such domain architect is known to present in Polyketide Synthase A (PKSA) from A. flavus and Aspergillus parasiticus producing aflatoxin.7 With the help of hexA and hexB, PKSA catalyzes a hexanoyl unit and six iteratively derived malonyl units into a first stable polyketide compound, norsolonic acid, in the aflatoxin biosynthetic pathway.47,48
NR PKSs – others
Clade II represents the PKSs with the domain architect KS-AT-ACP-MT. Few proteins classified in this clade have extra MT domain; however, none of the characterized proteins of NR PKS has MT domain. This extra domain might be nonfunctional. PKS protein classified in this clade from A. fumigatus Af293 is Afu14, which is a part of cluster AFUA_3G02570 and contains putative phenol hydroxylase and C6 transcription factor, suggesting the synthesis of the product with an NR property of final compound.
Partially reducing PKSs – 6MSAS
Clade III shows the presence of PKS enzymes with the domain architect KS-AT-ACP-DH-KR. This group of PKSs has reducing domains DH and KR. One characterized PKS, atX from A. terreus, has been classified under this clade, which produces a partially reduced pk compound, 6MSA.49
Highly reducing PKSs
Clade IV is observed to have the domain architect KS-AT-ACP-DH-(MT)-ER-KR-(TE). The characterized protein classified in this clade is FUM1 PKS from G. moniliformis that is known to synthesize fumonisin, a mycotoxin that contaminates maize-based food and feed products.27,46 This clade is represented as HR PKS, with the domain architect KS-AT-ACP-DH-ER-KR. The known enzymes classified in this clade are PKS1 from C. heterostrophus and ZEA2/GzPH from G. zeae (anamorph: Fusarium graminearum), which are known to produce mycotoxin T-toxin and zearalenone, respectively.50,51 In this study, A. fumigatus, A. flavus, and A. clavatus show the presence of similar architect proteins designated as Afu5, Afl9, and Acl10, respectively (Fig. 1).
Clades V and VI classify PKSs containing HR domains. Such PKSs are known to produce a polyketide compound of HR nature, such as T-toxin. PKS classified in this clade is PKS PCT2 from M. purpureus that produces citrinin, a polyketide with multiple aromatic rings. Citrinin is found to have nephrotoxic activity in mammals and is bactericidal.30
Clade VII displays the domain architect KS-AT-ACP-DH-MT-ER-KR-CON, which is known to present in lovastatin-producing PKS. Two proteins LNKS and lovastatin diketide synthase (LDKS) have been characterized from A. terreus.11 LNKS is an HR iterative type I PKS with KS-AT-DH-MT-KR-ACP-CON domain but has an inactive ER domain. LDKS also has the same domain architect (KS-AT-ACP-DH-MT-ER-KR-CON) as LNKS, but its ER domain is active.52 Our analysis suggests that A. fumigatus has one such protein Afu1 similar to LNKS. Screening the upstream and downstream genes of Afu1 from A. fumigatus suggests the presence of enzymes such as enoylreductase, esterases, and cytochrome P450 oxygenases, known to be present in the lovastatin biosynthetic pathway in A. terreus (data not shown). The upstream genes of Afu1 also have a transcription factor that may be responsible for the activation of this cluster. A. niger has two proteins, Ani1 and Ani2. A. clavatus has three proteins, Acl1, Acl2, and Acl3, which are classified in this group. Acl2 and Acl4 are hybrid protein and PKS–NRPS enzyme, respectively, while Acl3 is characterized as equistein synthetase. Proteins Aor4 from A. oryzae and Afl5 from A. flavus are grouped in this clade.53
Clade VIII enzymes contain a hybrid PKS–NRPS domain structure KS-AT-ACP-DH-CON-A-T-E-TE (A, adenylation domain; T, thiolation domain; CON, condensation domain; and E, epimerase domain). A. fumigatus PKS Afu2 (ABS87601.1) producing pseurotin A, a reduced compound, is classified in this clade.54 This type of PKS is a hybrid PKS with nonribosomal PKS domains.
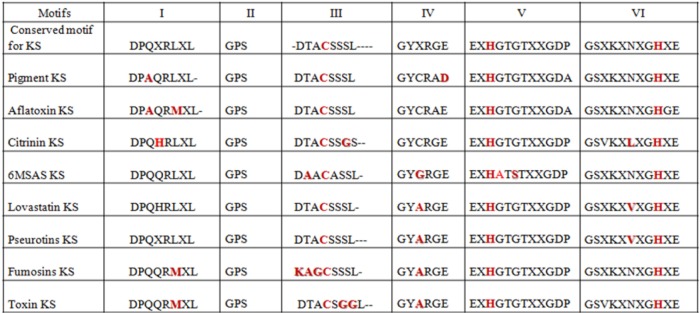
Modeling of KS domain and mapping of conserved motifs
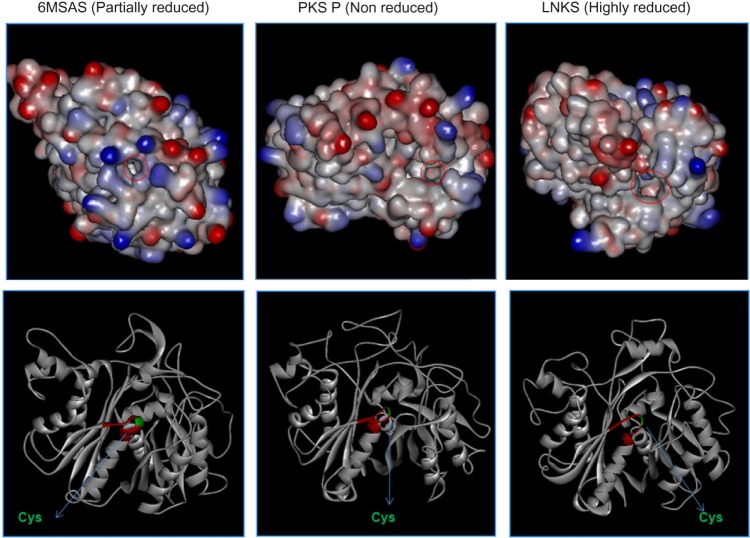
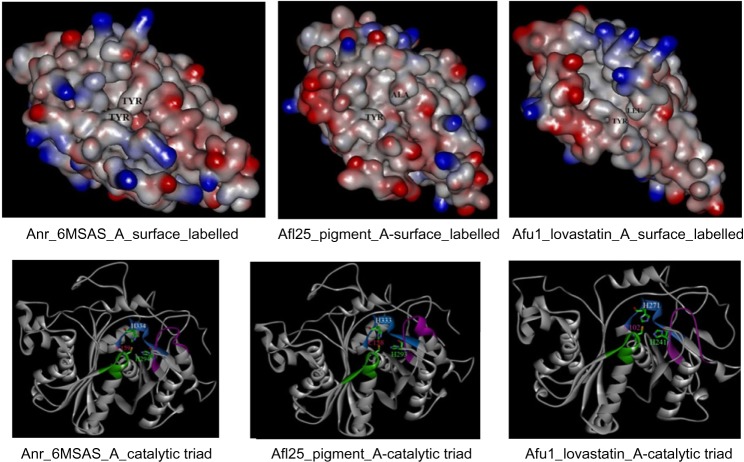
KS sequences from eight major clades were aligned, and six conserved consensus motifs were identified.55 Particularly, motif – DTACSSSL – in KS carries Cys residue in the active site, which is a signature conserved amino acid across the KS domain in the PKSs from other species.56 The two His residues, known to be in KS catalytic triad, were also found to be conserved in motifs EXHGTGTXXGDP and GSXKXNXGHXE in the KS sequences. A careful analysis of conserved motifs shows the amino acid changes with respect to the specific type of pk. Figure 3 shows the conserved motifs in KS sequences and variations among them with respect to the type of PKSs. To further verify our grouping of NR, PR, and HR based on phylogeny, we used modeling study for KS proteins. Our hypothesis was that NR, PR, and HR types of KS may have their differences in how they dock the substrate at the active site and thus impacts the catalytic activity of the enzyme. In order to find the structural changes in the three types of PKS enzymes (NR, PR, and HR), KS domain structures have been predicted by homology modeling. Model proteins used for this study were PKSP from A. fumigatus for NR PKS, 6MSA from A. terreus for PR PKS, and LNKS from A. terreus for HR PKS. Three Aspergillus PKSs classified and observed in the current study as NR, PR, and HR were also taken to test our hypothesis. These hypothetical proteins were Afl25 (XP_002384329.1) from A. flavus as NR, Anr (XP_001402408.2) from A. niger as PR, and Afu1 (XP_753141.2) from A. fumigatus as HR type. The percentage identities of the template sequence of 1KAS (used as a standard reference for genera ting the model) with KS sequences of Afl25, Anr, and Afu1 are 28%, 26%, and 27%, respectively. As reported in the crystal structure of KS domain of FAS of E. coli, the catalytic triad Cys-His-His is observed to be conserved in all these structures. The area and volume of these active site conformations were measured through CASTp server. The active site pocket volume of these different types of proteins varies according to the type of PKS and the nature of chain elongation reactions (NR, PR, and HR; Fig. 4). Chain elongations eventually determine the diversity of chemical compounds in iterative PKSs. The alignments of KS domain sequences from NR type KS, HR type KS, and PR type KS are shown in Figure 5. The conserved motifs are highlighted in yellow color, and two Tyr residues that are found conserved in all three types of PKSs are marked in the box. PR PKS, 6MSA from A. terreus, contains the smallest cavity volume (365.8 Å3), as the iteration reactions in this enzyme are reported to be only 3. Similar results were found with the modeling of protein Anr from A. niger. NR PKS such as PKSP from A. fumigatus shows medium cavity volume (749.8 Å3) and is known to have five iteration cycles. Modeled structure of Afl25 from A. flavus classified in clade I (pigment) is also found to have moderate cavity volume (∼750 Å3). HR type of PKS such as LNKS from A. terreus is found to have the largest cavity volume (1408.3 Å3) with the variable (<20) iteration reactions. Similar results were found in modeled protein Afu1 from A. fumigatus. In these proteins, the number of iteration cycles was observed to be dependent on the size of the cavity in the active site of the PKS enzyme in a particular type with products such as pigment, 6MSA, and lovastatin, as described in Figure 5.
Figure 3.
Conserved motifs on type I KS sequences.
Notes: Conserved motifs observed in the type I KS for Fungal PKSs are presented in the table.
Figure 4.
Three-dimensional modeled structures of NR, PR, and HR type of KS.
Notes: Structures have been modeled for KS domain from protein designated as 6MSAS producing 6-mehtlysalisylic acid (PR), PKS P producing pigment (NR), and LNKS producing lovastatin (HR). The cavities of the modeled structures have been shown in surface rendering and circled in red. Each model has been superimposed with the structural template of 1KAS.
Figure 5.
Three-dimensional modeled structure from Aspergillus KSs.
Notes: NR type KS Afl25 (XP_002384329.1) predicated to produce pigment, HR type KS Afu1 (XP_753141.2) predicated for lovastatin, and PR type KS Anr (XP_001402408.2) predicated for 6-metylsalicylic acid were modeled using the template structure of beta-ketoacyl-ACP synthase II from Escherichia coli (PDB ID: 1KAS). Tyr and Ala residues have been marked on the surface topology. Catalytic triad Cys-His-His are also marked in the modeled structure for validation in stick model in different colors, namely, purple, green, and blue.
The conserved motif amino acid sequences identified as pigment KS were converted to DNA sequences, and degenerate primers were designed for amplification for specific detection of Aspergillus species. Primers based on KS region from PKS genes were used for PCR amplification using the DNA of Aspergillus species, such as A. fumigatus, A. flavus, A. parasiticus, A. niger, and A. oryzae (Supplementary Fig. 6). A PCR product of 450 bp was observed with the DNA template of A. fumigatus (5 isolates), A. flavus (18 isolates), and A. niger (6 isolates) specifically. DNA from Trichoderma, Fusarium, and Bacillus species and human DNA were used for PCR controls, which did not give any amplification in the expected range. Sequences were confirmed with multiple alignments of pksP (gene) from A. fumigatus, A. flavus, and A. niger and submitted to NCBI (KT221846-KT221852 and KT213730-KT213740; Supplementary Fig. 6).
Discussion
PKSs play a major role in contributing to the diversity of polyketide products produced in Aspergilli. Polyketide biosynthesis mainly uses acetyl-CoA and malonyl-CoA as precursors and synthesizes hexanoyl-CoA or pentaketide as the end product. This is used by PKS to generate the stable polyketide precursor with various lengths, which will eventually be processed by the remaining biosynthetic enzymes in the pathway to generate polyketide compound with different applications, such as toxins, antibiotics, and pigment compounds. To gain insight into the synthesis of diverse product by observing the diversity in the PKS enzyme, full-length Aspergillus PKSs and only KS domain from six important Aspergillus species with functionally characterized fungal PKS enzymes were used for comparative sequence analysis. This also correlates the unknown PKSs of Aspergillus with known polyketides they produce. Phylogenetic analysis of Aspergillus PKSs based on the domain architecture facilitated the classification of PKS proteins into three different types of enzymes, ie, NR, PR, and HR PKSs grouped into eight clades.
Earlier, phylogenetic studies were performed for type I PKSs of Ascomycota group of fungi, which divided PKSs into 18 clades, indicating that the grouping was based on the presence/absence of reducing and NR domains in PKS.22,57 Distribution of PKS enzymes in Aspergilli is studied by phylogeny,57 and we have attempted to assign Aspergilli PKS protein to the probable chemical nature of the compound based on sequence homology. In the current study, Aspergillus PKSs fall into eight clades corresponding to the probable chemistry of the end compound they may synthesize based on the rationale of homology. The NR fungal clade contains PKSs that synthesize unreduced, and usually aromatic, PKs that are precursors to toxins, eg, aflatoxins and pigments.7,58 All PKSs within this clade lacked ER, DH, and KR domains, which are interpreted as a loss of reducing domains, compared to the domain structure of type I PKSs. The NR fungal PKSs are predicted to synthesize PKs in which the keto groups are either not reduced or reduced by enzymes other than PKS. Unreduced PKs are typically synthesized from acetyl- and malonyl-CoA. PKS proteins with an additional TE/PP domain were scattered throughout the NR PKS clade, as was the case for reducing PKS subclade III. The functional significance of these duplicated PP domains is not known. Aspergillus PKSs are iterative type I class of enzymes, and exceptions to these are recently found type III PKSs in A. flavus and A. oryzae. To outgroup any other type of enzymes, such as closely homologous proteins in FASs, and also type III and modular type I PKSs, representative proteins were also included in our study. M. tuberculosis PKSs are known to have type I modular and type III PKS and are characterized to be a part of gene cluster producing virulent lipids, such as phthiocerol and phenol-phthiocerol.59,60 Modular type I PKS from S. coelicolor is known to produce antibiotics.61 These enzymes were classified into different subclades, and none of Aspergillus PKSs are categorized in this clade. This confirms that Aspergillus species do not have modular PKSs that may produce lipids such as polyketides.
Conserved motifs in the KS domain have been identified that are specific to pk they produce. Three-dimensional structures also reported the changes at the active site confirmations with respect to the type of compound they produce and the cavity volume in their active sites. Change in the cavity groove volume can be linked to the malonyl starter units fitted at the enzymatic site and thus may help predict substrate utilization by the enzyme. In the modeled 6MSAS KS structure, Yadav et al has observed that two tyrosine residues are protruding into the cavity blocking the downward flow of the cavity.62 In the current study, Aspergillus-modeled proteins (annotated as NR, PR, and HR) were also found to have two Tyr residues highly conserved in all KSs and were also aligned on the structure. This finding supports the hypothesis that putative Aspergillus PKS may be functionally active in appropriate in vitro conditions. These three proteins can be selected for further in vitro studies and explored for polyketide exploration studies using advance methods.63 The presence of certain amino acids at their active site pocket and their alignment in a particular fashion to accommodate the substrate clearly suggest that the diversity in the end product is related to the substrate size and the number of molecules of substrate it can fit for condensation reaction. Recent experiments involving the generation of altered fatty acid-polyketide hybrid products by the rational manipulation of benastatin biosynthetic pathway also suggest that the number of chain elongations is dependent on the size of the PKS enzyme cavity.64 The in silico analysis of the sequence and structural features of iterative KS domains reported in this study may provide logical selection of residues to be mutated and help in exploring the effect of substrate specificity and the end product. Strategic site-directed mutagenesis studies can be planned; knockout and overexpression studies using molecular cassette in reference strains can be performed to identify which products are accumulating in a reasonable amount at certain cultural conditions compared to wild type. Such bioinformatics studies will be helpful in providing pilot results to choose best PKSs candidates for the exploration of novel polyketide compounds for knockout studies. No experimental studies on the modeled Aspergillus proteins analyzed in the current study have yet been reported; therefore, these proteins can be top PKS candidates to explore pk compound. The present in silico analysis gives key leads for such experiments.
We report the unexplored repertoire of PKSs in Aspergillus species. We predicted three proteins of Aspergillus spp. that can be explored for their reducing or NR pk products, namely, XP_002384329.1, XP_753141.2, and XP_001402408.2. Among Aspergillus spp., A. flavus, predominantly an agricultural pathogen and mycotoxin producer, is often reported in immunosuppressed patients.65 A. flavus genome contains 25 PKSs.49 It has been reported by various comparative genomics studies that A. flavus has 55 gene clusters associated with secondary metabolism. However, only handful metabolites with their pathways have been assigned to these clusters.66 Our analysis suggests that A. flavus has the potential to produce numerous polyketides of different natures. This remains to be explored by various expression studies. In the current study, A. flavus protein Afl19 has been linked with pigment production, which has been explored for DHN melanin pathway, while this manuscript was under preparation.67 Aspergillus ochraceus is known to produce ochratoxin A and penicillic acid, which cause significant problems in animal and human health.47 It also produces other metabolites derived from these mycotoxins, such as diaporthin, orthosporin, and asperlactone. The biological activity of these SMs has not been characterized so far, and many PKSs have not been functionally characterized in this species. These molecules may be beneficial (antibiotics) or harmful (mycotoxins) to human health.68 The analysis of this report may find helpful to start with selective pickup for PKs to study.
Utility of KS region diversity can be exploited in the detection of three important Aspergilli, together with one degenerate primer pair designed in this study. Such detection method can be standardized further to identify Aspergillus species, A. flavus, A. fumigatus, and A. niger, in one reaction for screening agricultural and clinical samples.
In conclusion, the sequence-based analysis reported in this study highlights the diversity of the compounds that may probably be produced by Aspergillus species. This study shall serve as a platform to screen the particular type of metabolite synthesized by an Aspergillus species. These in silico analyses of PKSs have facilitated the understanding of the biosynthetic pathways for unknown SMs from Aspergillus species, as well as its utility for unique identification.
Supplementary Material
Supplementary Figure 1. Closer view of clade I, where Non reducing Aspergillus PKSs are classified and predicated to produce Aflatoxin, pigment or other non reduced compound.
Supplementary Figure 2. Closer view of clade II, where Non reducing Aspergillus PKSs are classified and predicated to produce citrinin like non reduced compound and also clade III, of which partially reduced Aspergillus PKS are predicted to produce 6 methylsalisylic acid type compound.
Supplementary Figure 3. Closer view of clade IV, Highly reduced Aspergillus PKSs classified in clade IV, predicted to produce fumonisin type highly reduced compound.
Supplementary Figure 4. Closer view of clade V and VI where Aspergillus PKSs classified in Highly reduced PKSs and predicted to produce highly reduced compound/T-toxin type compound.
Supplementary Figure 5. Closer view of clade VII, where PKSs are classified as Highly reduced Aspergillus and predicted to produce Lovastatin type highly reduced compound. Clade VIII shows the closer view of Aspergillus PKSs predicted to produce hybrid type of PKS-NRPS compound.
Supplementary Figure 6. KS amplification from Aspergillus and other fungal species and bacteia.
Supplementary Figure 7. Alignment of the sequenced putative KS sequences from Aspergillus in this study with characterized conidial pigment PKS from Aspergillus flavus, A. fumigatus and A. niger.
Supplementary Table 1. Annotation and classification of Aspergillus PKSs analyzed in this study.
Supplementary Table 2. Gene Accession no. of PKSs and other proteins.
Acknowledgments
Authors thank Dr. T. Prameela Devi, Principal Scientist and In-charge Indian Type Culture Collection, Division of Plant Pathology, Indian Agricultural Research Institute New Delhi, India at for providing Aspergillus isolates.
Footnotes
ACADEMIC EDITOR: Jike Cui, Associate Editor
PEER REVIEW: Five peer reviewers contributed to the peer review report. Reviewers’ reports totaled 2349 words, excluding any confidential comments to the academic editor.
FUNDING: We are grateful to the Council of Scientific and Industrial Research and Department of Science and Technology, Government of India, for the financial support. PJB and AB and MP are the recipients of Senior Research Fellowship and Junior Research Fellowship of the Council of Scientific and Industrial Research, Government of India, respectively. RSM thanks the ICMR for the funding support [IRIS ID: 2013–1551G]. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: PJB, YS, AV, PUS and TM disclose a patent pending, US20130266941 A1, for Diagnostic Assays for the Detection and Identification of Aspergilli. Other authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: PB, MP, PUS. Work was carried out under the project conceived by PUS. Analyzed the data: PB, AB, RM. Wrote the first draft of the manuscript: PB, PUS. Contributed to the writing of the manuscript: PB, AB, RM. Made critical revisions and approved final version: PB, PUS, TM, YS, AV. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Chiang YM, Oakley BR, Keller NP, Wang CC. Unraveling polyketide synthesis in members of the genus Aspergillus. Appl Microbiol Biotechnol. 2010;86(6):1719–36. doi: 10.1007/s00253-010-2525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khaldi N, Seifuddin FT, Turner G, et al. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47(9):736–41. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szewczyk E, Chiang YM, Oakley CE, Davidson AD, Wang CC, Oakley BR. Identification and characterization of the asperthecin gene cluster of Aspergillus nidulans. Appl Environ Microbiol. 2008;74(24):7607–12. doi: 10.1128/AEM.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahuja M, Chiang YM, Chang SL, et al. Illuminating the diversity of aromatic polyketide synthases in Aspergillus nidulans. J Am Chem Soc. 2012;134(19):8212–21. doi: 10.1021/ja3016395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai HF, Wheeler MH, Chang YC, Kwon-Chung KJ. A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J Bacteriol. 1999;181(20):6469–77. doi: 10.1128/jb.181.20.6469-6477.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, Sivonen K, Fewer DP. Genomic insights into the distribution, genetic diversity and evolution of polyketide synthases and nonribosomal peptide synthetases. Curr Opin Genet Dev. 2015;35:79–85. doi: 10.1016/j.gde.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Feng GH, Leonard TJ. Characterization of the polyketide synthase gene (pksL1) required for aflatoxin biosynthesis in Aspergillus parasiticus. J Bacteriol. 1995;177(21):6246–54. doi: 10.1128/jb.177.21.6246-6254.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu J, Bhatnagar D, Ehrlich KC. Aflatoxin biosynthesis. Rev Iberoam Micol. 2002;19(4):191–200. [PubMed] [Google Scholar]
- 9.Langfelder K, Jahn B, Gehringer H, Schmidt A, Wanner G, Brakhage AA. Identification of a polyketide synthase gene (pksP) of Aspergillus fumigatus involved in conidial pigment biosynthesis and virulence. Med Microbiol Immunol. 1998;187(2):79–89. doi: 10.1007/s004300050077. [DOI] [PubMed] [Google Scholar]
- 10.Ma SM, Tang Y. Biochemical characterization of the minimal polyketide synthase domains in the lovastatin nonaketide synthase LovB. FEBS J. 2007;274(11):2854–64. doi: 10.1111/j.1742-4658.2007.05818.x. [DOI] [PubMed] [Google Scholar]
- 11.Hendrickson L, Davis CR, Roach C, et al. Lovastatin biosynthesis in Aspergillus terreus: characterization of blocked mutants, enzyme activities and a multifunctional polyketide synthase gene. Chem Biol. 1999;6(7):429–39. doi: 10.1016/s1074-5521(99)80061-1. [DOI] [PubMed] [Google Scholar]
- 12.Bhetariya PJ, Madan T, Basir SF, Varma A, Usha SP. Allergens/Antigens, toxins and polyketides of important Aspergillus species. Indian J Clin Biochem. 2011;26(2):104–19. doi: 10.1007/s12291-011-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujii I. Functional analysis of fungal polyketide biosynthesis genes. J Antibiot (Tokyo) 2010;63(5):207–18. doi: 10.1038/ja.2010.17. [DOI] [PubMed] [Google Scholar]
- 14.Weissman KJ. Biochemistry. Anatomy of a fungal polyketide synthase. Science. 2008;320(5873):186–7. doi: 10.1126/science.1157677. [DOI] [PubMed] [Google Scholar]
- 15.Perrin RM, Fedorova ND, Bok JW, et al. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 2007;3(4):e50. doi: 10.1371/journal.ppat.0030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergmann S, Schümann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol. 2007;3(4):213–7. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- 17.Cary JW, Harris-Coward PY, Ehrlich KC, et al. Functional characterization of a veA-dependent polyketide synthase gene in Aspergillus flavus necessary for the synthesis of asparasone, a sclerotium-specific pigment. Fungal Genet Biol. 2014;64:25–35. doi: 10.1016/j.fgb.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 18.Bir N, Paliwal A, Muralidhar K, Reddy P, Sarma PU. A rapid method for the isolation of genomic DNA from Aspergillus fumigatus. Prep Biochem. 1995;25(4):171–81. doi: 10.1080/10826069508010119. [DOI] [PubMed] [Google Scholar]
- 19.Duran RM, Gregersen S, Smith TD, et al. The role of Aspergillus flavus veA in the production of extracellular proteins during growth on starch substrates. Appl Microbiol Biotechnol. 2014;98(11):5081–94. doi: 10.1007/s00253-014-5598-6. [DOI] [PubMed] [Google Scholar]
- 20.Yadav G, Gokhale RS, Mohanty D. SEARCHPKS: a program for detection and analysis of polyketide synthase domains. Nucleic Acids Res. 2003;31(13):3654–8. doi: 10.1093/nar/gkg607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tae H, Sohng JK, Park K. MapsiDB: an integrated web database for type I polyketide synthases. Bioprocess Biosyst Eng. 2009;32(6):723–7. doi: 10.1007/s00449-008-0296-3. [DOI] [PubMed] [Google Scholar]
- 22.Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc Natl Acad Sci U S A. 2003;100(26):15670–5. doi: 10.1073/pnas.2532165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van den Berg MA, Albang R, Albermann K, et al. Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat Biotechnol. 2008;26(10):1161–8. doi: 10.1038/nbt.1498. [DOI] [PubMed] [Google Scholar]
- 24.Woo PC, Tam EW, Chong KT, et al. High diversity of polyketide synthase genes and the melanin biosynthesis gene cluster in Penicillium marneffei. FEBS J. 2010;277(18):3750–8. doi: 10.1111/j.1742-4658.2010.07776.x. [DOI] [PubMed] [Google Scholar]
- 25.Gaffoor I, Brown DW, Plattner R, Proctor RH, Qi W, Trail F. Functional analysis of the polyketide synthase genes in the filamentous fungus Gibberella zeae. Eukaryot Cell. 2005;4(11):1926–33. doi: 10.1128/EC.4.11.1926-1933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abe Y, Suzuki T, Ono C, Iwamoto K, Hosobuchi M, Yoshikawa H. Molecular cloning and characterization of an ML-236B (compactin) biosynthetic gene cluster in Penicillium citrinum. Mol Genet Genomics. 2002;267(5):636–46. doi: 10.1007/s00438-002-0697-y. [DOI] [PubMed] [Google Scholar]
- 27.Proctor RH, Desjardins AE, Plattner RD, Hohn TM. A polyketide synthase gene required for biosynthesis of fumonisin mycotoxins in Gibberella fujikuroi mating population A. Fungal Genet Biol. 1999;27(1):100–12. doi: 10.1006/fgbi.1999.1141. [DOI] [PubMed] [Google Scholar]
- 28.Proctor RH, Brown DW, Plattner RD, Desjardins AE. Co-expression of 15 contiguous genes delineates a fumonisin biosynthetic gene cluster in Gibberella moniliformis. Fungal Genet Biol. 2003;38(2):237–49. doi: 10.1016/s1087-1845(02)00525-x. [DOI] [PubMed] [Google Scholar]
- 29.Yu JH, Leonard TJ. Sterigmatocystin biosynthesis in Aspergillus nidulans requires a novel type I polyketide synthase. J Bacteriol. 1995;177(16):4792–800. doi: 10.1128/jb.177.16.4792-4800.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimizu T, Kinoshita H, Ishihara S, Sakai K, Nagai S, Nihira T. Polyketide synthase gene responsible for citrinin biosynthesis in Monascus purpureus. Appl Environ Microbiol. 2005;71(7):3453–7. doi: 10.1128/AEM.71.7.3453-3457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawrence DP, Kroken S, Pryor BM, Arnold AE. Interkingdom gene transfer of a hybrid NPS/PKS from bacteria to filamentous Ascomycota. PLoS One. 2011;6(11):e28231. doi: 10.1371/journal.pone.0028231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotowska M, Pawlik K, Smulczyk-Krawczyszyn A, Bartosz-Bechowski H, Kuczek K. Type II thioesterase ScoT, associated with Streptomyces coelicolor A3(2) modular polyketide synthase Cpk, hydrolyzes acyl residues and has a preference for propionate. Appl Environ Microbiol. 2009;75(4):887–96. doi: 10.1128/AEM.01371-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paitan Y, Alon G, Orr E, Ron EZ, Rosenberg E. The first gene in the biosynthesis of the polyketide antibiotic TA of Myxococcus xanthus codes for a unique PKS module coupled to a peptide synthetase. J Mol Biol. 1999;286(2):465–74. doi: 10.1006/jmbi.1998.2478. [DOI] [PubMed] [Google Scholar]
- 34.Chopra T, Gokhale RS. Polyketide versatility in the biosynthesis of complex mycobacterial cell wall lipids. Methods Enzymol. 2009;459:259–94. doi: 10.1016/S0076-6879(09)04612-6. [DOI] [PubMed] [Google Scholar]
- 35.Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 36.Bhaduri A, Misra R, Maji A, et al. Mycobacterium tuberculosis cyclophilin A uses novel signal sequence for secretion and mimics eukaryotic cyclophilins for interaction with host protein repertoire. PLoS One. 2014;9(2):e88090. doi: 10.1371/journal.pone.0088090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narayan A, Sachdeva P, Sharma K, Saini AK, Tyagi AK, Singh Y. Serine threonine protein kinases of mycobacterial genus: phylogeny to function. Physiol Genomics. 2007;29(1):66–75. doi: 10.1152/physiolgenomics.00221.2006. [DOI] [PubMed] [Google Scholar]
- 38.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 39.Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12(4):357–8. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 40.Anand S, Prasad MV, Yadav G, et al. SBSPKS: structure based sequence analysis of polyketide synthases. Nucleic Acids Res. 2010;38(Web Server issue):W487–96. doi: 10.1093/nar/gkq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang W, Jia J, Edwards P, Dehesh K, Schneider G, Lindqvist Y. Crystal structure of beta-ketoacyl-acyl carrier protein synthase II from E.coli reveals the molecular architecture of condensing enzymes. EMBO J. 1998;17(5):1183–91. doi: 10.1093/emboj/17.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2008;426:145–59. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 43.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26(16):1701–18. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 44.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34(Web Server issue):W116–8. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hopwood DA. Genetic contributions to understanding polyketide synthases. Chem Rev. 1997;97(7):2465–98. doi: 10.1021/cr960034i. [DOI] [PubMed] [Google Scholar]
- 46.Linnemannstöns P, Schulte J, del Mar Prado M, Proctor RH, Avalos J, Tudzynski B. The polyketide synthase gene pks4 from Gibberella fujikuroi encodes a key enzyme in the biosynthesis of the red pigment bikaverin. Fungal Genet Biol. 2002;37(2):134–48. doi: 10.1016/s1087-1845(02)00501-7. [DOI] [PubMed] [Google Scholar]
- 47.Cary JW, Ehrlich KC, Beltz SB, Harris-Coward P, Klich MA. Characterization of the Aspergillus ochraceoroseus aflatoxin/sterigmatocystin biosynthetic gene cluster. Mycologia. 2009;101(3):352–62. doi: 10.3852/08-173. [DOI] [PubMed] [Google Scholar]
- 48.Chang PK, Cary JW, Yu J, Bhatnagar D, Cleveland TE. The Aspergillus parasiticus polyketide synthase gene pksA, a homolog of Aspergillus nidulans wA, is required for aflatoxin B1 biosynthesis. Mol Gen Genet. 1995;248(3):270–7. doi: 10.1007/BF02191593. [DOI] [PubMed] [Google Scholar]
- 49.Pazoutová S, Linka M, Storková S, Schwab H. Polyketide synthase gene pksM from Aspergillus terreus expressed during growth phase. Folia Microbiol (Praha) 1997;42(5):419–30. doi: 10.1007/BF02826548. [DOI] [PubMed] [Google Scholar]
- 50.Baker SE, Kroken S, Inderbitzin P, et al. Two polyketide synthase-encoding genes are required for biosynthesis of the polyketide virulence factor, T-toxin, by Cochliobolus heterostrophus. Mol Plant Microbe Interact. 2006;19(2):139–49. doi: 10.1094/MPMI-19-0139. [DOI] [PubMed] [Google Scholar]
- 51.Gaffoor I, Trail F. Characterization of two polyketide synthase genes involved in zearalenone biosynthesis in Gibberella zeae. Appl Environ Microbiol. 2006;72(3):1793–9. doi: 10.1128/AEM.72.3.1793-1799.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kennedy J, Auclair K, Kendrew SG, Park C, Vederas JC, Hutchinson CR. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science. 1999;284(5418):1368–72. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- 53.Kiyota T, Hamada R, Sakamoto K, Iwashita K, Yamada O, Mikami S. Aflatoxin non-productivity of Aspergillus oryzae caused by loss of function in the aflJ gene product. J Biosci Bioeng. 2011;111(5):512–7. doi: 10.1016/j.jbiosc.2010.12.022. [DOI] [PubMed] [Google Scholar]
- 54.Maiya S, Grundmann A, Li X, Li SM, Turner G. Identification of a hybrid PKS/NRPS required for pseurotin A biosynthesis in the human pathogen Aspergillus fumigatus. Chembiochem. 2007;8(14):1736–43. doi: 10.1002/cbic.200700202. [DOI] [PubMed] [Google Scholar]
- 55.Ponting CP, Schultz J, Milpetz F, Bork P. SMART: identification and annotation of domains from signalling and extracellular protein sequences. Nucleic Acids Res. 1999;27(1):229–32. doi: 10.1093/nar/27.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reid R, Piagentini M, Rodriguez E, et al. A model of structure and catalysis for ketoreductase domains in modular polyketide synthases. Biochemistry. 2003;42(1):72–9. doi: 10.1021/bi0268706. [DOI] [PubMed] [Google Scholar]
- 57.Lin SH, Yoshimoto M, Lyu PC, Tang CY, Arita M. Phylogenomic and domain analysis of iterative polyketide synthases in Aspergillus species. Evol Bioinform Online. 2012;8:373–87. doi: 10.4137/EBO.S9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takano Y, Kubo Y, Shimizu K, Mise K, Okuno T, Furusawa I. Structural analysis of PKS1, a polyketide synthase gene involved in melanin biosynthesis in Colletotrichum lagenarium. Mol Gen Genet. 1995;249(2):162–7. doi: 10.1007/BF00290362. [DOI] [PubMed] [Google Scholar]
- 59.Sankaranarayanan R, Saxena P, Marathe UB, Gokhale RS, Shanmugam VM, Rukmini R. A novel tunnel in mycobacterial type III polyketide synthase reveals the structural basis for generating diverse metabolites. Nat Struct Mol Biol. 2004;11(9):894–900. doi: 10.1038/nsmb809. [DOI] [PubMed] [Google Scholar]
- 60.Gokhale RS, Saxena P, Chopra T, Mohanty D. Versatile polyketide enzymatic machinery for the biosynthesis of complex mycobacterial lipids. Nat Prod Rep. 2007;24(2):267–77. doi: 10.1039/b616817p. [DOI] [PubMed] [Google Scholar]
- 61.Ichinose K, Ozawa M, Itou K, Kunieda K, Ebizuka Y. Cloning, sequencing and heterologous expression of the medermycin biosynthetic gene cluster of Streptomyces sp. AM-7161: towards comparative analysis of the benzoisochromanequinone gene clusters. Microbiology. 2003;149(pt 7):1633–45. doi: 10.1099/mic.0.26310-0. [DOI] [PubMed] [Google Scholar]
- 62.Yadav G, Gokhale RS, Mohanty D. Towards prediction of metabolic products of polyketide synthases: an in silico analysis. PLoS Comput Biol. 2009;5(4):e1000351. doi: 10.1371/journal.pcbi.1000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang PK, Scharfenstein LL, Wei Q, Bhatnagar D. Development and refinement of a high-efficiency gene-targeting system for Aspergillus flavus. J Microbiol Methods. 2010;81(3):240–6. doi: 10.1016/j.mimet.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 64.Parvatkar RR, D’Souza C, Tripathi A, Naik CG. Aspernolides A and B, butenolides from a marine-derived fungus Aspergillus terreus. Phytochemistry. 2009;70(1):128–32. doi: 10.1016/j.phytochem.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 65.Hedayati MT, Pasqualotto AC, Warn PA, Bowyer P, Denning DW. Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiology. 2007;153(pt 6):1677–92. doi: 10.1099/mic.0.2007/007641-0. [DOI] [PubMed] [Google Scholar]
- 66.Georgianna DR, Fedorova ND, Burroughs JL, et al. Beyond aflatoxin: four distinct expression patterns and functional roles associated with Aspergillus flavus secondary metabolism gene clusters. Mol Plant Pathol. 2010;11(2):213–26. doi: 10.1111/j.1364-3703.2009.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jørgensen TR, Park J, Arentshorst M, et al. The molecular and genetic basis of conidial pigmentation in Aspergillus niger. Fungal Genet Biol. 2011;48(5):544–53. doi: 10.1016/j.fgb.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 68.Awad G, Mathieu F, Coppel Y, Lebrihi A. Characterization and regulation of new secondary metabolites from Aspergillus ochraceus M18 obtained by UV mutagenesis. Can J Microbiol. 2005;51(1):59–67. doi: 10.1139/w04-117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Closer view of clade I, where Non reducing Aspergillus PKSs are classified and predicated to produce Aflatoxin, pigment or other non reduced compound.
Supplementary Figure 2. Closer view of clade II, where Non reducing Aspergillus PKSs are classified and predicated to produce citrinin like non reduced compound and also clade III, of which partially reduced Aspergillus PKS are predicted to produce 6 methylsalisylic acid type compound.
Supplementary Figure 3. Closer view of clade IV, Highly reduced Aspergillus PKSs classified in clade IV, predicted to produce fumonisin type highly reduced compound.
Supplementary Figure 4. Closer view of clade V and VI where Aspergillus PKSs classified in Highly reduced PKSs and predicted to produce highly reduced compound/T-toxin type compound.
Supplementary Figure 5. Closer view of clade VII, where PKSs are classified as Highly reduced Aspergillus and predicted to produce Lovastatin type highly reduced compound. Clade VIII shows the closer view of Aspergillus PKSs predicted to produce hybrid type of PKS-NRPS compound.
Supplementary Figure 6. KS amplification from Aspergillus and other fungal species and bacteia.
Supplementary Figure 7. Alignment of the sequenced putative KS sequences from Aspergillus in this study with characterized conidial pigment PKS from Aspergillus flavus, A. fumigatus and A. niger.
Supplementary Table 1. Annotation and classification of Aspergillus PKSs analyzed in this study.
Supplementary Table 2. Gene Accession no. of PKSs and other proteins.