Abstract
Monoubiquitinated histone H2B plays multiple roles in transcription activation. H2B is deubiquitinated by the Spt-Ada-Gcn5 acetyltransferase (SAGA) coactivator, which contains a four-protein subcomplex known as the deubiquitinating (DUB) module. The crystal structure of the Ubp8/Sgf11/Sus1/Sgf73 DUB module bound to a ubiquitinated nucleosome reveals that the DUB module primarily contacts H2A/H2B, with an arginine cluster on the Sgf11 zinc finger domain docking on the conserved H2A/H2B acidic patch. The Ubp8 catalytic domain mediates additional contacts with H2B, as well as with the conjugated ubiquitin. We find that the DUB module deubiquitinates H2B both in the context of the nucleosome and in H2A/H2B dimers complexed with the histone chaperone, FACT, suggesting that SAGA could target H2B at multiple stages of nucleosome disassembly and reassembly during transcription.
Histone modifications such as acetylation, methylation, phosphorylation, and ubiquitination regulate chromatin structure and interactions. Histone H2B monoubiquitination (H2B-Ub) at K123 in yeast, K120 in humans, is a transient modification that is a universal feature of genes that are actively transcribed by RNA polymerase II (1). H2B-Ub plays a nondegradative role in transcription activation, elongation, mRNA splicing, and mRNA export, as well as in DNA replication and damage repair (2). Histone H2B is monoubiquitinated by Rad6/Bre1 (3), which is required for H3K4 and H3K79 trimethylation (4, 5). H2B-Ub also enhances recruitment of FACT (facilitates chromatin transcription) (6), a histone chaperone that mediates H2A/H2B dimer eviction and nucleosome reassembly (7). H2B is deubiquitinated by the Spt-Ada-Gcn5 acetyltransferase (SAGA) coactivator complex (8). The minimal SAGA subcomplex with deubiquitinating (DUB) activity is called the SAGA DUB module, which in yeast contains the catalytic subunit Ubp8 as well as Sgf11, Sus1, and the Sgf73 N-terminal ~100 residues (8–10). The DUB module is an independently folding subcomplex that is connected to the remainder of the SAGA complex by the C-terminal portion of Sgf73 (11). The basic features of the yeast DUB module are conserved in human SAGA (12). The structure of the yeast DUB module in the presence and absence of bound ubiquitin (9, 10) revealed a highly intertwined arrangement for the four subunits, with Sgf11, Sgf73, and Sus1 serving in part as scaffolds that help to maintain the Ubp8 catalytic domain in an active conformation (13, 14).
We have determined the crystal structure of the DUB module bound to ubiquitinated nucleosomes at 3.9 Å resolution (table S1). The crystallized complex comprises two DUB module heterotetramers bound to a Xenopus laevis nucleosome core particle (NCP) containing two copies of H2B with ubiquitin attached to K120 of H2B (corresponding to K123 of yeast H2B) via a nonhydrolyzable dichloroacetone (DCA) linkage (details in materials and methods). Nucleosomes containing DCA-linked H2B-Ub inhibit DUB module cleavage of ubiquitin-AMC (7-amido-4-methylcoumarin) (fig. S1), indicating that the complex containing the DCA linkage mimics the catalytically competent complex. The DUB module in the crystals contains intact yeast Ubp8, Sgf11, and Sus1 and an N-terminal fragment of Sgf73 (residues 1 to 104) that is sufficient for full DUB module activity on nucleosomes (15). Substitution of the Ubp8 activesite cysteine with alanine (C146A) was required to promote binding to nucleosomes containing DCA-linked H2B-Ub (fig. S2). The structure was solved by molecular replacement using high-resolution structures of the DUB module bound to ubiquitin (9) [Protein Data Bank (PDB): 3MHS] and the X. laevis nucleosome core particle (16) (PDB: 3LZ0). We validated the solution by zinc anomalous difference Fourier maps and a composite omit map of Sgf11, and by observing DUB module electron density using molecular replacement phases from a partial model (figs. S3 to S5)
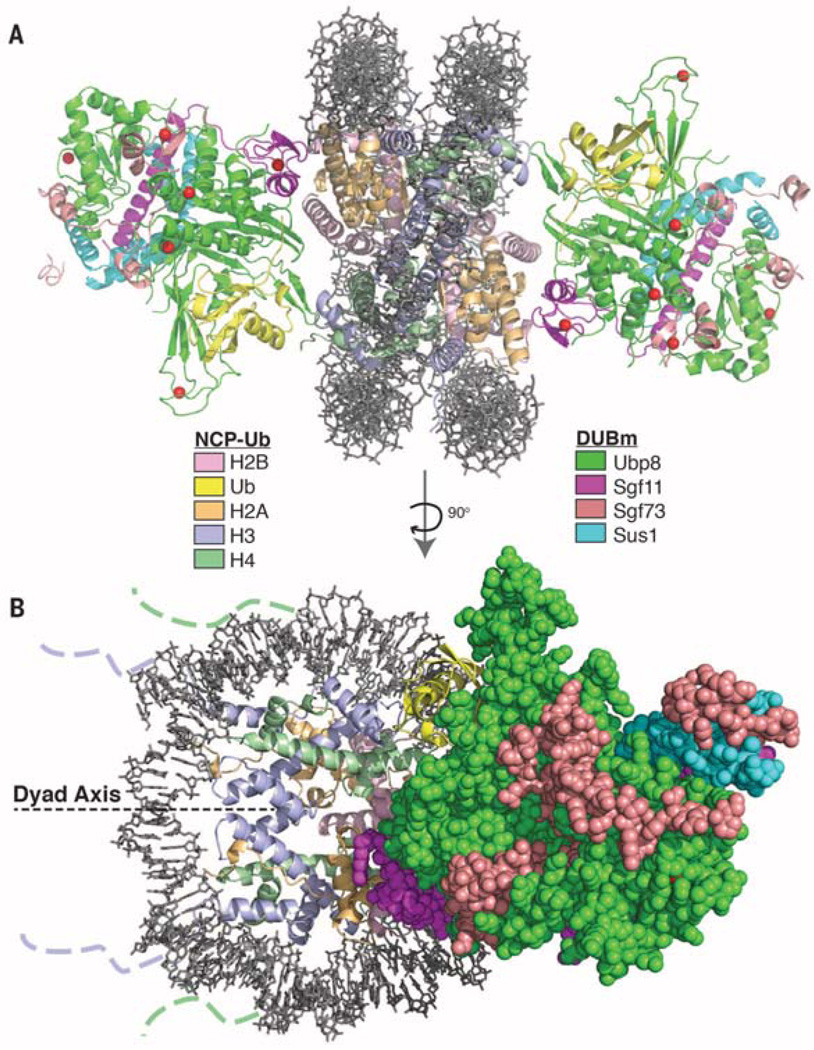
The structure contains one DUB module bound to each face of the nucleosome, distal to the entry and exit sites of the nucleosomal DNA (Fig. 1), with the overall structure of each DUB module essentially identical to that previously reported (9, 10). Each DUB module is docked on the nucleosome in a virtually identical manner, indicating that the observed mode of binding is not governed by crystal contacts. No electron density is seen corresponding to the DCA linkage or for ubiquitin residue G76C. The interaction interface between the DUB module and the nucleosome is about 1000 Å2, while the surface buried by the conjugated ubiquitin and the catalytic domain of Ubp8 is ~1650 Å2. The bound ubiquitin makes no apparent contacts with the nucleosome itself.
Fig. 1. Overview of the SAGA DUB module bound to the monoubiquitinated nucleosome core particle (NCP-Ub).
(A) Two DUB modules bind to each NCP-Ub. The catalytic Ubp8 USP domain and the Sgf11 zinc finger contact the substrate. (B) View of the complex perpendicular to (A). The nucleosome dyad axis is indicated. Dotted lines indicate where the N-terminal tails of histone H3 and H4 protrude from the DNA gyres.
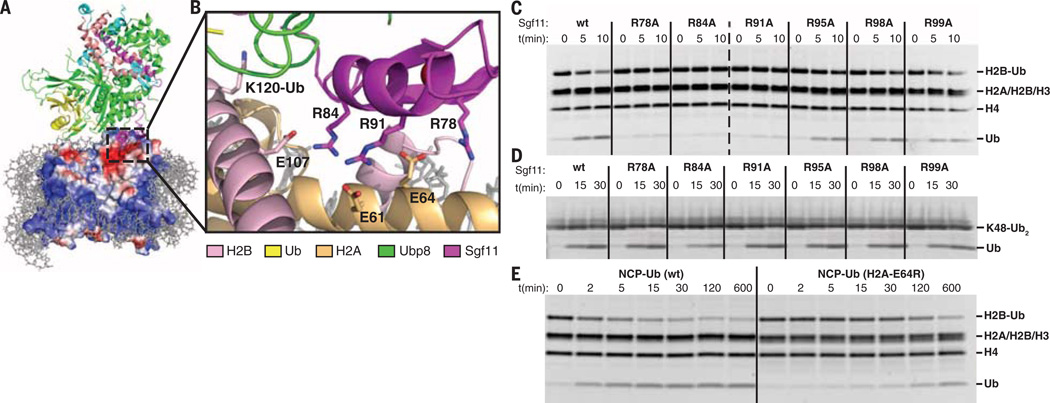
The DUB module engages the nucleosome almost exclusively through histones H2A and H2B (Fig. 2A). The catalytic lobe of the DUB module, which contains the ubiquitin-specific protease (USP) domain of Ubp8 and the Sgf11 zinc finger, contacts the C-terminal helix of H2B and the conserved acidic patch on H2A/H2B (17). The nucleosome acidic patch is contacted by the Sgf11-ZnF (Fig. 2A), which is located near the Ubp8 active site and contains a cluster of arginine residues implicated in DUB module interactions with chromatin (9, 10, 18). In contrast with proposed models in which the Sgf11 zinc finger contacts nucleosomal DNA (9, 18), the DUB module is oriented with the basic Sgf11-ZnF docked on the H2A/H2B acidic patch (Fig. 2B) with an interface area of 610 Å2. The Sgf11 arginine residues, R78, R84, and R91 (Fig. 2B), that face the nucleosome acidic patch are conserved in the Drosophila and human homologs, dSgf11 and ATXN7L3, respectively. Although the resolution of the structure determination is insufficient to show side-chain density for the arginine side chains, salt-bridge interactions can be modeled between Sgf11 residues R78, R84, and R91 and acidic patch residues H2A-E64, H2B-E107, and H2A-E61, respectively (Fig. 2B). The observed mode of nucleosome interactions, in which an arginine-rich motif interacts with the H2A/H2B acidic patch, has been observed in several complexes containing either a peptide or proteins bound to the nucleosome (17, 19–22). Despite this qualitative similarity between the DUB module and the nucleosome interactions mediated by Sir3-BAH (19), RCC1 (21), and the PRC1 E2-E3 complex (22), or by the LANA (17) and CENP-C (20) peptides, there appear to be no common structural features among these different complexes that specify binding to the acidic patch.
Fig. 2. The DUB module binds the acidic patch of the H2A/H2B heterodimer using the Sgf11 ZnF.
(A) View of interface between the DUB module and nucleosome showing the electrostatic surface potential of the histone octamer (red, negative; blue, positive). Dashed box indicates region shown in (B). (B) Modeled contacts between Sgf11 residues R78, R84, and R91 and acidic patch residues H2A-E64, H2B-E107, and H2A-E61. The ubiquitinated lysine, H2B-K120, is indicated. (C) Effect of Sgf11 ZnF mutations on deubiquitination of nucleosomal H2B (NCP-Ub). Monoubiquitinated nucleosomes (2 µM) were incubated with 200 nM wild-type or mutant DUB module for the indicated time. (D) Effect of DUB module mutations shown in (C) on cleavage of 2 µM K48-linked diubiquitin (K48-Ub2). (E) Effect of acidic patch mutation, H2A-E64R, on deubiquitination of nucleosomal H2B by the DUB module. Conditions as in (C).
To validate the role of the Sgf11 zinc finger in targeting the DUB module to nucleosomes, we assayed the effects of alanine substitutions at each of the six Sgf11-ZnF arginine residues on deubiquitination of nucleosomes. For these in vitro assays, we used semisynthetic methods to generate H2B-Ub containing a native isopeptide linkage (fig. S8). Substitutions of the three Sgf11 arginines located at the DUB module–nucleosome interface, R78A, R84A, and R91A, markedly reduce the rate at which the DUB module cleaves ubiquitin from H2B in nucleosomes (Fig. 2C). By contrast, Sgf11 substitutions R95A, R98A, and R99A, which are located in a disordered C-terminal tail and are not predicted to be involved in nucleosome binding (Fig. 2B), do not have a substantial effect on DUB module activity. The deleterious effect of the Sgf11 arginine substitutions specifically affects targeting to the nucleosome, as none of the Sgf11 mutations tested has a substantial effect on cleavage of K48-linked diubiquitin, a nonspecific DUB module substrate that is cleaved at roughly the rate ofH2B-Ub (Fig. 2D). Our results are consistent with previous studies showing that substitutions of Sgf11-ZnF residues R84 and R91 disrupt global H2B deubiquitination in vivo (10, 18). Moreover, yeast bearing mutations in Sgf11 residues R84, R91,or thedoubleR84/R91mutantdisplaygrowth defects when combined with gcn5 deletions (10), further consistent with the importance of these residues to DUB module activity.
To test the importance of the H2A/H2B acidic patch to the ability of the SAGA DUB module to deubiquitinate nucleosomal H2B, we assayed DUB module activity on ubiquitinated nucleosomes containing a mutation in the acidic patch. Deubiquitination of H2B in nucleosomes containing the mutant, H2A-E64R, is decreased by a factor of ~100 as compared to deubiquitination of H2B in nucleosomes containing wild-type H2A (Fig. 2E). The importance of the acidic patch is also consistent with a recent report that mutations in the H2A acidic patch give rise to defects in H2B ubiquitination downstream of ubiquitin conjugation (23).
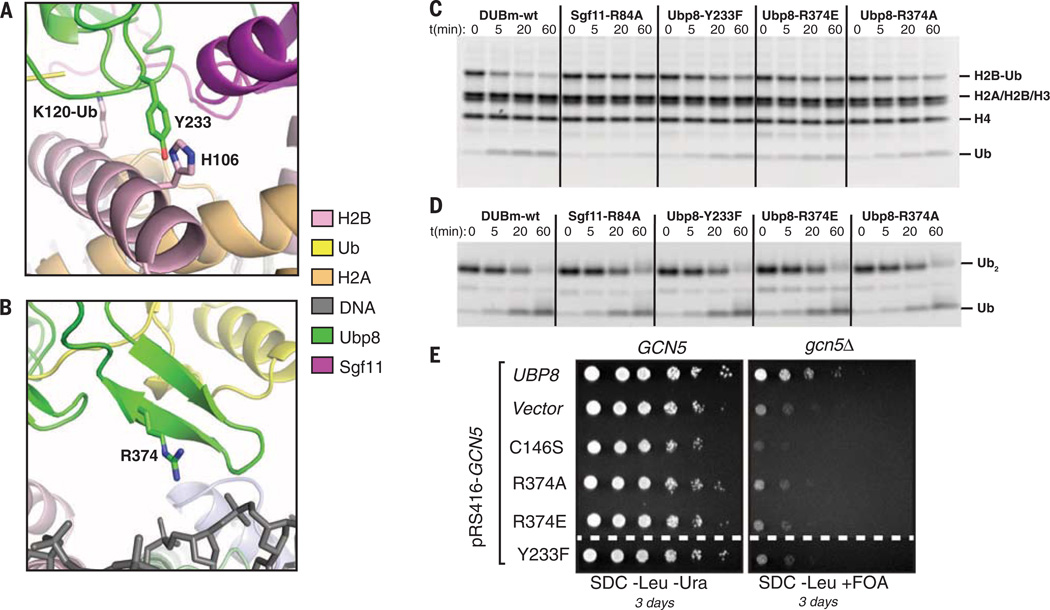
The C-terminal helix of H2B, which contains the ubiquitinated residue, K120 (corresponding to K123 in yeast H2B), lies in a depression centered at the Ubp8 active site (Fig. 3A). The close approach of Ubp8 to the C-terminal H2B helix and calculated interface area of 376 Å2 makes it likely that there are several intermolecular contacts along this interface. In particular, Ubp8 residue Y233 projects toward the C-terminal helix of H2B, with electron density corresponding to that of the tyrosine phenyl ring visible in all four copies of the interface in the crystallographic asymmetric unit (fig. S6D). Model building suggests that H2B residues H106 and S109 in the C-terminal helix of H2B could potentially form van der Waals or hydrogen-bonding interactions with Y233 (Fig. 3A). The remainder of the DUB module is positioned too far from the nucleosome to mediate any contacts, with the exception of the “fingers region” of the Ubp8 ubiquitin-binding pocket, which projects toward the nucleosomal DNA (Fig. 3B). Although no direct contacts can be observed due to resolution limits, model building suggests that Ubp8 residue R374 could form electrostatic contacts with the sugar-phosphate backbone of the nucleosomal DNA between bases −32 and −33 (Fig. 3B). BothY233 andR374 are conserved in fly and human homologs of Ubp8. Mutations Y233F and R374E or R374A in Ubp8 give rise to defects in module activity on nucleosomal H2B-Ub, but not K48-linked diubiquitin (Fig. 3, C and D), suggesting that these residues mediate nucleosome contacts. The effects of the Ubp8 mutations are, however, not as severe as the effects of Sgf11-Znf substitutions in the basic patch (R78, R84, and R91), indicating that Sgf11 contacts with the nucleosome have a greater contribution to substrate specificity. To test the effect of the Ubp8 mutations in yeast, we utilized an assay that was previously used to show that mutations in Sgf11 arginine patch residues give rise to synthetic growth defects (10). All three Ubp8 mutations show growth defects comparable to those of a Ubp8 catalytic mutation when combined with a gcn5 deletion, consistent with their proposed role in Ubp8 activity (Fig. 3E).
Fig. 3. Contribution of Ubp8 residues to DUB module activity on nucleosomes.
(A) Ubp8-Y233, which is in a position to form a potential interaction with H2B-H106. (B) Modeling showing a potential contact between Ubp8 residue R374 and nucleosomal DNA. (C) Activity assay as in Fig. 2C showing effect of Ubp8 mutations Y233F, R374E, and R374A on DUB module activity on H2B-Ub in nucleosomes. Mutant Sgf11-R84A is shown for comparison. (D) Activity of DUB module mutants shown in (C) on cleavage of 2 µM K48-linked diubiquitin (Ub2). (E) Synthetic growth assay for effect of Ubp8 mutations in combination with deletion of Gcn5.
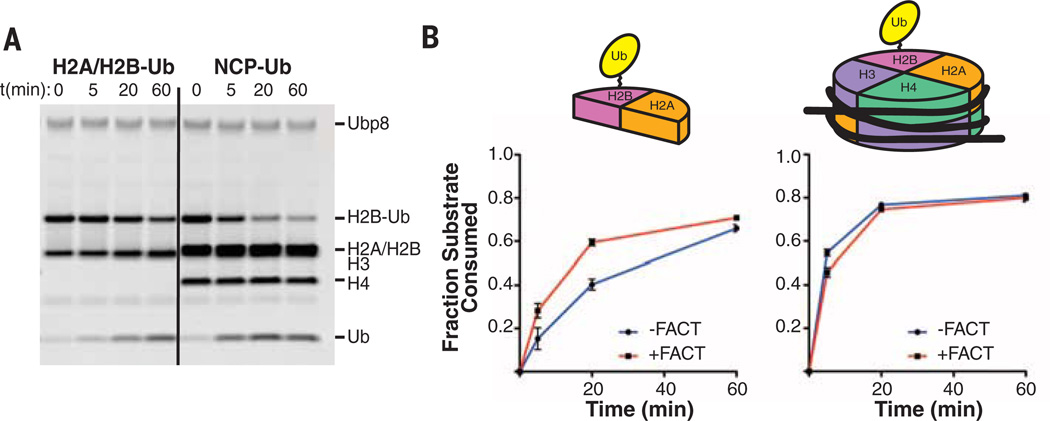
In genes regulated by yeast SAGA, FACT binds to H2A/H2Band facilitates nucleosome disassembly and reassembly (24), which is required to allow RNA polymerase to transcribe efficiently through chromatin. Recruitment of the FACT complex, Spt16/Pob3/Nhp6 (25), is enhanced by H2B monoubiquitination (6), although it is not known whether ubiquitin is cleaved from H2B that is still bound to FACT. Because the DUB module almost exclusively contactsH2A/H2B, we tested whether H2A/H2B-Ub alone is a substrate for the DUB module. As shown in Fig. 4A, the DUB module efficiently cleaves ubiquitin from H2A/H2B-Ub heterodimers, albeit about a factor of 5 to 10 more slowly than nucleosomes under the same conditions. We next asked whether FACT binding to the H2A/H2B-Ub heterodimer interferes with DUB module access to H2B-Ub. Structures of the FACT Spt16 subunit (26) and Spt16-C peptide bound to an H2A/H2B heterodimer (27) indicate that Spt16 does not overlap the DUB module docking surface (fig. S9), although these studies leave open the question of whether the full Spt16/Pob3/Nhp6 FACT complex could interfere with DUB module binding. We preincubated H2A/H2B-Ub heterodimers with FACT and found that the DUB module could cleave ubiquitin from H2B (Fig. 4B and fig. S10). The rate of H2A/H2B-Ub deubiquitination was somewhat higher in the presence of FACT and is comparable to the rate at which the DUB module deubiquitinates nucleosomes (Fig. 4B). FACT has no effect on the relative rate at which ubiquitin is cleaved from nucleosomes (Fig. 4B), thus ruling out a direct effect of FACT on intrinsic DUB module activity. Our results indicate that the SAGA DUB module can remove ubiquitin from either intact or disassembled nucleosomes and is therefore capable of acting at multiple points as RNA polymerase transcribes through a chromatin template.
Fig. 4. Activity of the DUB module on monoubiquitinated nucleosomes versus H2A/H2B-Ub in the presence and absence of FACT.
(A) Time course showing cleavage of H2B-Ub by the DUB module (200 nM) in H2A/H2B dimers (H2A/H2B-Ub, 2 µM) or nucleosomes containing H2B-Ub (NCP-Ub, 1 µM). (B) Time course comparing deubiquitination of H2A/H2B-Ub (left) and NCP-Ub (right) in the presence and absence of the FACT complex. Reactions were performed in triplicate; fraction of H2B-Ub consumed was determined by quantitating bands on gels shown in fig. S9).
Our study provides a framework for understanding global interactions between the SAGA complex and nucleosomes. In addition to the DUB module, SAGA also contains a ~180-kDa subcomplex called the histone acetyltransferase (HAT) module comprising Gcn5, Ada2, Ada3, and Sgf29 (28), which acetylates histone H3 N-terminal tails (29). The DUB module is located distal to the DNA entry and exit points, where the H3 tails targeted by the HAT module protrude, thus leaving much of the H3/H4 surface exposed and available for interactions with the HAT module or with other SAGA subunits (Fig. 1B). A recent cryo–electron microscopy study of SAGA shows the DUB and HAT modules adjacent to one another, where they could simultaneously engage the same nucleosome (30). The Sgf73 SCA7 domain (residues 209 to 280), a nonspecific DNA binding domain that is not present in the fragment used in our crystallographic study, is located between the DUB and HAT modules, where the SCA7 domain could potentially help to position a nucleosome. However, we note that the reported positioning of an unmodified nucleosome relative to the DUB module in intact SAGA (30) does not agree with our structure. Given the large contribution of the ubiquitin modification itself to the total interface between ubiquitinated nucleosomes and the DUB module, docking of the DUB module in a manner that mimics the catalytically competent complex is likely predicated on the presence of ubiquitinated H2B.
The structure of the yeast SAGA DUB module bound to ubiquitinated nucleosomes provides a basis for understanding how the human SAGA DUB module engages its H2B-Ub substrate in a chromatin context. The human DUB module comprises USP22, ATXN7L3, ATXN7, and ENY2, homologs of yeast Ubp8, Sgf11, Sgf73, and Sus1, respectively (31). The key residues implicated here in yeast DUB module function, including a three-arginine cluster on Sgf11 and residues Y233 and R374 in Ubp8, are conserved in ATXN7 and USP22, respectively. Our structure also accounts for the recent report that H2B deubiquitination by the SAGA DUB module is regulated in both humans and yeast by a newly discovered histone modification, phosphorylation of H2A-Y57/58 (human/yeast) (32). This H2A residue lies at the DUB module–nucleosome interface in our structure, where it could readily interfere with the observed docking of the DUB module on H2A/H2B (fig. S12). Further studies will be needed to interrogate the mechanistic details of how H2A–Y57/58 phosphorylation influences DUB module activity.
Supplementary Material
Acknowledgments
We thank T. Formosa for the generous gift of purified FACT and S. Tan for the plasmid, pST55-16x601. We thank X. Zhang for assistance with protein purification, P. Lombardi for assistance with data collection, and D. Leahy for helpful discussions. Supported by grant GM-095822 from the National Institute of General Medical Sciences (C.W.) and by a Ruth L. Kirschstein National Research Service Award (M.M.). A.B. is a Neubauer Professor and a Taub Foundation Fellow. GM/CA @ APS has been funded by the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract no. DE-AC02-06CH11357. Coordinates and data have been deposited in the Protein Data Bank with accession code 4ZUX. Plasmid pST55-16xNCP601 expressing the Widom 601 DNA fragment was obtained from S. Tan under a material transfer agreement with Pennsylvania State University.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/351/6274/725/suppl/DC1
Materials and Methods
Figs. S1 to S12
Table S1
References (33–48)
REFERENCES AND NOTES
- 1.Bonnet J, Devys D, Tora L. Drug Discov. Today. Technol. 2014;12:e19–e27. doi: 10.1016/j.ddtec.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Fuchs G, Oren M. Biochim. Biophys. Acta. 2014;1839:694–701. doi: 10.1016/j.bbagrm.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Wood A, et al. Mol. Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- 4.Sun ZW, Allis CD. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 5.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. Mol. Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 7.Pavri R, et al. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 8.Henry KW, et al. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samara NL, et al. Science. 2010;328:1025–1029. doi: 10.1126/science.1190049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Köhler A, Zimmerman E, Schneider M, Hurt E, Zheng N. Cell. 2010;141:606–617. doi: 10.1016/j.cell.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han Y, Luo J, Ranish J, Hahn S. EMBO J. 2014;33:2534–2546. doi: 10.15252/embj.201488638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spedale G, Timmers HT, Pijnappel WW. Genes Dev. 2012;26:527–541. doi: 10.1101/gad.184705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samara NL, Ringel AE, Wolberger C. Structure. 2012;20:1414–1424. doi: 10.1016/j.str.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan M, Wolberger C. J. Mol. Biol. 2015;427:1765–1778. doi: 10.1016/j.jmb.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Köhler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Nat. Cell Biol. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- 16.Vasudevan D, Chua EY, Davey CA. J. Mol. Biol. 2010;403:1–10. doi: 10.1016/j.jmb.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 17.Barbera AJ, et al. Science. 2006;311:856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- 18.Koehler C, et al. J. Biol. Chem. 2014;289:8989–8999. doi: 10.1074/jbc.M113.500868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. Science. 2011;334:977–982. doi: 10.1126/science.1210915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato H, et al. Science. 2013;340:1110–1113. doi: 10.1126/science.1235532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makde RD, England JR, Yennawar HP, Tan S. Nature. 2010;467:562–566. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGinty RK, Henrici RC, Tan S. Nature. 2014;514:591–596. doi: 10.1038/nature13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cucinotta CE, Young AN, Klucevsek KM, Arndt KM. PLOS Genet. 2015;11:e1005420. doi: 10.1371/journal.pgen.1005420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belotserkovskaya R, et al. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 25.Formosa T, et al. EMBO J. 2001;20:3506–3517. doi: 10.1093/emboj/20.13.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hondele M, et al. Nature. 2013;499:111–114. doi: 10.1038/nature12242. [DOI] [PubMed] [Google Scholar]
- 27.Kemble DJ, McCullough LL, Whitby FG, Formosa T, Hill CP. Mol. Cell. 2015;60:294–306. doi: 10.1016/j.molcel.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee KK, et al. Mol. Syst. Biol. 2011;7:503. doi: 10.1038/msb.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, Tan S. J. Biol. Chem. 2002;277:7989–7995. doi: 10.1074/jbc.M110849200. [DOI] [PubMed] [Google Scholar]
- 30.Durand A, Bonnet J, Fournier M, Chavant V, Schultz P. Structure. 2014;22:1553–1559. doi: 10.1016/j.str.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 31.Zhang XY, et al. Mol. Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basnet H, et al. Nature. 2014;516:267–271. doi: 10.1038/nature13736. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.