Abstract
The aim of this study was to determine whether a recombinant vesicular stomatitis virus (VSV) vector encoding a transgene could be used to infect and express a foreign gene in embryonic primary cell cultures derived from the freshwater microcrustacean Daphnia, the most widely used ecotoxicological model organism. To facilitate the evaluation of gene transfer, a reproducible method for establishing primary cultures from Daphnia embryonic tissues was developed. Within 24 hr after infection, transgene expression could be detected in cell culture. VSV was found to replicate in the cells with no apparent cytopathic effect. Here we report the first evidence of gene transfer and foreign gene expression in cultures of Daphnia embryonic cells using a recombinant viral vector.
The microcrustacean Daphnia has been widely used for ecological study for over a century (Birge, 1895; Edmonson, ’87) and continues to be the focus of diverse ecological research due to its central importance in freshwater ecosystems (McCauley et al., ’99; Rhode et al., 2001; Decaestecker et al., 2002; Ebert et al., 2002). This, plus laboratory convenience, parthenogenesis, and environmental sensitivity has led to its widespread adoption as an ecotoxicological model organism. Daphnia are intimately involved in the food web as primary consumers of phytoplankton and are an important food source of fish and other predators (Tessier and Woodruff, 2002). Their rapid life cycle and ability to replicate clonally make them ideal candidates for studies in evolutionary and ecological functional genomics (Feder and Mitchell-Olds, 2003). Furthermore, a concerted effort led by the Daphnia Genomics Consortium is underway to sequence the Daphnia genome (http://www.daphnia.cgb.indiana.edu). Despite the fact that Daphnia are one of the most widely used test organisms for ecological and toxicological studies, an understanding of many aspects of their molecular biology has been hindered by a lack of cell culture methods and the inability to introduce and express genes in this organism. With the expected rapid accumulation of Daphnia sequence data, the ability to introduce genes into Daphnia would greatly enhance its utility as an ecological model and sentinel of ecosystem health.
As a group, the vesiculoviruses have an extremely broad host range and can infect a wide variety of both insect and vertebrate species (Tesh et al., ’70). In invertebrate hosts, vesicular stomatitis virus (VSV) infection is typically non-cytopathic, and numerous insects have been implicated as reservoirs and vectors in enzootic areas (Tesh et al., ’72). Since natural VSV infections occur in various insect species including mosquitoes, midges, black flies, house flies, and sand flies (Shope and Tesh, ’87), we hypothesized that VSV might be useful in the transgenesis of the crustacean Daphnia. VSV is an enveloped negative-stranded RNA rhabdovirus of the genus Vesiculovirus. The virus enters target cells by receptor-mediated endocytosis followed by membrane fusion that releases a transcriptionally active viral core into the cytoplasm of host cells. Transcription of the five viral genes is polar and sequential and initiates directly at the beginning of the first viral gene to yield a gradient of monocistronic viral mRNAs (Abraham and Banerjee, ’76; Ball and White, ’76; Villarreal et al., ’76; Whelan and Wertz, 2002). Genomic replication initiates at the 3′ end of the viral genome and requires ongoing translation to provide a source of the viral nucleocapsid protein that coats the viral genome (Patton et al., ’84). Recently, infectious transgenic clones of VSV-containing additional open reading frames coding for a foreign reporter gene have been described (Whelan et al., 2000; Cherry et al., 2005). Here we present methods for the culture of embryonic Daphnia cells and the infection of these cultures with transgenic VSV, resulting in expression of a foreign transgene.
MATERIALS AND METHODS
Daphnia culture
Daphnia magna were maintained parthenogenetically in a high-hardness COMBO medium (Baer and Goulden, ’98) at 22–24 °C, a light/dark photo-period of 16:8 hr and fed green microalgae Scene-desmus auratus daily.
Primary embryonic Daphnia cell cultures
Approximately 120 gravid adults with 6–12 embryos in various stages of development were surface sterilized by sequential washing for 30 sec in 500 μl of the following solutions: 0.05% (w/v) NaHClO4 (× 1), sterile deionized H2O (dH2O) (× 2), 70% (v/v) ethanol (× 1), dH2O (× 2), and Daphnia cell culture medium (DCCM) (80% (v/v) Schneider's Drosophila Medium (Invitrogen Corporation, Carlsbad, CA) in dH2O, pH 7.0 with 10% (v/v) heat-inactivated fetal bovine serum (FBS), 10 μg/ml gentamicin, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2.5 μg/ml amphotericin B) (× 2). Under a dissecting microscope (Olympus SZ-60), embryos were removed from adults and surface sterilized in 200 μl of each solution followed by gentle homogenization using a sterile disposable tissue homogenizer micropestle (Fisher Scientific, Hampton, NH). Cells were collected by centrifugation at 201g for 5 min at 4 °C, resuspended in DCCM, and cultured at 22–24 °C. Cell viability was analyzed with nucleic acid stains SYBR 14 and propidium iodide (Molecular Probes, Eugene, OR) using fluorescence microscopy (Garner et al., ’94).
Propagation of VSV-GFP
An infectious cDNA clone of the Indiana serotype of VSV (Whelan et al., ’95) engineered to express green fluorescent protein (GFP), VSV-GFP (Whelan et al., 2000; Cherry et al., 2005), was propagated in baby hamster kidney (BHK-21) cells (American Type Culture Collection, Manassas, VA) in Dulbecco's Modified Eagle Medium (Invitrogen) supplemented with 10% (v/v) FBS, 2 mM L-alanyl-L-glutamine (Glutamax, Invitrogen), 100 units/ml penicillin, and 100 μg/ml streptomycin. Initial infection was made with a multiplicity of infection of 0.01 in serum-free medium for 1 hr at 37 °C. Eighteen hours post-infection, supernatants were collected, clarified by centrifugation at 3,220g for 5 min at 4 °C, and filtered through a 0.45 μm filter (Whatman, Florham Park, NJ). Virus was concentrated by centrifugation using an SW28 rotor at 72,000g for 90 min at 4 °C. Virus-containing pellets were resuspended in phosphate-buffered saline (PBS) pH 7.2 overnight on ice and stored at –80 °C. Ten-fold serial dilutions of virus were titered on BHK-21 cell monolayers. Fluorescent foci were counted and virus titers were expressed as focus-forming units per ml (FFU/ml).
Infection of embryonic Daphnia cultures with recombinant VSV
Primary embryonic Daphnia cells derived from approximately 250 embryos were infected with 1.5 × 107 FFU of VSV-GFP at a multiplicity of infection of approximately 0.1 (a spherical embryo (radius = 300 μm) was assumed containing 8 μm diameter cells) in 500 μl DCCM in a well of a 24-well tissue culture plate and cultured at 22–24 °C. GFP expression was analyzed by fluorescence microscopy and immunoblot assay.
Immunoblot of VSV-GFP infected embryonic Daphnia cultures
Seventy-two hours post-infection, VSV-GFP infected embryonic Daphnia cells were collected and lysed in 20 μl of cell lysis buffer (1% (v/v) Triton X-100, 20 mM Tris pH 7.5). Ten microliters of sample cell lysate was added to 5 μl of sample loading buffer (35 mM Tris-HCl pH 6.8, 2.8% (w/v) SDS, 20% (v/v) glycerol, 200 mM dithiothreitol, and 0.001% (w/v) bromophenol blue). Samples were heated to 100 °C, separated by SDS-PAGE in a 10% Tris-HCl gel (Bio-Rad, Hercules, CA), and transferred by electroblotting onto a PVDF membrane (Millipore, Billerica, MA) in 25 mM Tris base, 192 mM glycine, and 20% (v/v) methanol. Blots were probed overnight at 4 °C with a rabbit anti-GFP antibody (BD Living Colors A.v. Peptide Antibody; BD Biosciences Clontech, Mountain View, CA) diluted 1:100 in 5% nonfat dry milk in PBS–Tween (1% (v/v) Tween-20 in PBS). Horse-radish peroxidase-conjugated goat anti-rabbit antibody (Sigma, St. Louis, MO) was used as the secondary antibody at a 1:5,000 dilution in PBS-Tween, and blots were developed using ECL Western Blotting Detection Reagents (Amersham Pharmacia Biotech, Piscataway, NJ).
VSV-GFP replication in embryonic Daphnia cultures
Primary embryonic Daphnia cells derived from approximately 250 embryos were infected for 1 hr followed by five washes with 500 μl of DCCM to remove unbound input VSV-GFP. The fifth wash was collected to determine the amount of input virus that remained in the cultures. Cell culture supernatants were collected sequentially (500 μl each), filtered through a 0.45 μm filter at 1, 2, 4, 24, and 48 hr post-infection, and stored at –80 °C. BHK-21 cell monolayers in 96-well tissue culture plates were incubated with duplicate ten-fold serial dilutions of collected supernatants to determine viral titers as plaque-forming units per ml (pfu/ml). Cells were fixed with a 10% formalin solution (Formalde-Fresh, Fisher Scientific) and stained with crystal violet (1% (w/v) crystal violet in 70% ethanol) 5 days post-infection to quantitate cytopathic effects (CPE).
RESULTS AND DISCUSSION
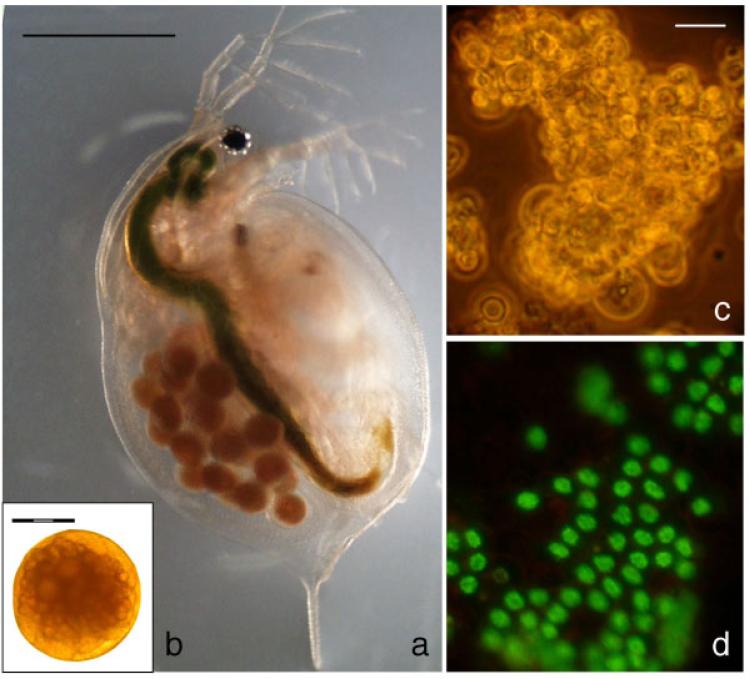
As there are no existing immortalized cell lines derived from Daphnia tissues to facilitate the testing of gene transfer vectors, we first developed a method for establishing primary cell cultures derived from embryos (Fig. 1a and b) that allowed the reproducible maintenance of viable cultures for at least 1 week. Daphnia embryos extracted from gravid adults were surface sterilized and gently homogenized. The resulting embryonic primary cell cultures were heterogeneous in morphology and included a large proportion of small spherical cells in grape-like clusters in suspension (Fig. 1c). Figure 1d demonstrates a population of viable Daphnia embryonic cells stained with the membrane-permeant nucleic acid stain, SYBR 14.
Fig. 1.
Daphnia embryonic primary cultures: (a) Gravid adult prior to dissection, (b) embryo removed from gravid adult, (c) bright field image of an embryonic primary culture, and (d) viability of primary embryonic cell cultures. Nuclear green staining with SYBR14 indicated that cells are viable whereas propidium iodide stained the nucleus of dead cells red. Scale bars: 1 mm (a), 300 μm (b), and 10 μm (c,d).
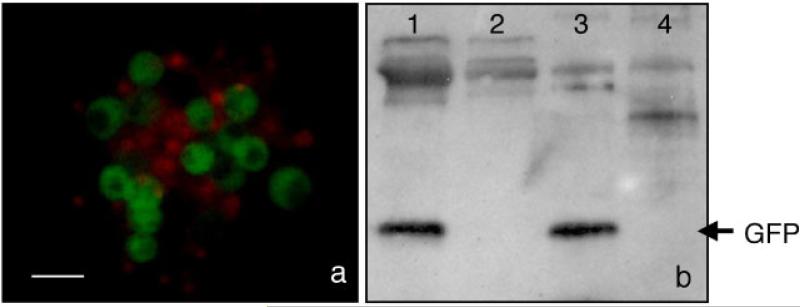
An infectious cDNA clone of VSV (Whelan et al., ’95) engineered to express the marker gene, GFP, VSV-GFP has been previously described (Whelan et al., 2000; Cherry et al., 2005). The coding sequence of GFP, flanked by the essential VSV gene-start and -end sequences, was inserted between the 3′ leader regulatory region and the VSV N gene. Owing to the sequential polar nature of viral mRNA synthesis, this design resulted in maximal expression of GFP. To determine whether VSV-GFP could infect Daphnia embryonic cultures and whether heterologous gene transfer could be achieved, primary cells derived from embryos were infected with VSV-GFP. Twenty-four hours post-infection, low levels of cytoplasmic GFP expression could be detected by fluorescence microscopy. At 48 hr, the level of GFP fluorescence markedly increased either due to protein expression or accumulation, and was maintained at this level for at least 7 days (Fig. 2a). The majority of cells expressed GFP by 48 hr. To confirm GFP expression, cells were collected 72 hr post-infection and a Western immunoblot assay was performed using an anti-GFP antibody with VSV-GFP infected and uninfected Daphnia cell lysates as well as control BHK-21 cells (Fig. 2b). A band was detected in the VSV-GFP infected Daphnia cell lysate that corresponded to the GFP band detected in the infected BHK control. These fluorescence microscopy and immunoblot results indicate that VSV-GFP can infect Daphnia embryonic primary cultures and express a heterologous protein, GFP.
Fig. 2.
GFP expression in VSV-GFP infected Daphnia embryonic primary cultures. (a) GFP expression in VSV-GFP infected Daphnia cells was detected by fluorescence microscopy 72 hr post-infection. The nuclei of nonviable cells were stained red with the membrane-impermeant dye, propidium iodide. (b) Immunoblot of VSV-GFP infected Daphnia cells. Lane 1, VSV-GFP infected Daphnia cells; lane 2, uninfected Daphnia cells; lane 3, VSV-GFP infected BHK-21 cells; and lane 4, uninfected BHK-21 cells. Scale bar, 10 μm (a).
To determine whether VSV induced CPE in Daphnia embryonic cells, VSV-GFP infected and uninfected cultures were stained with propidium iodide, a membrane-impermeant dye that stains the nuclei of dead cells red, each day for 3 days post-infection. As expected with primary cultures, the number of dead cells increased over time in both infected and uninfected cultures. No marked difference was observed between the viability of VSV-GFP infected and uninfected cultures. Therefore, CPE in Daphnia cell cultures that could be specifically attributed to virus infection was not observed. This is in agreement with previous observations that VSV does not produce CPE in most invertebrate cells (Gillies and Stollar, ’80).
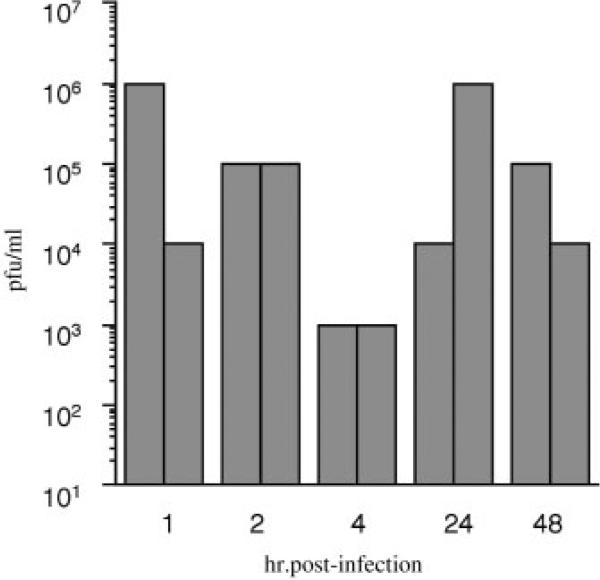
To determine whether VSV could replicate and produce progeny virus in Daphnia cells, supernatants from VSV-GFP infected Daphnia cells were sequentially collected at several time points post-infection and added to BHK-21 cells in tenfold serial dilutions. As VSV is stable at room temperature for several days, immediately following a 1-hr infection with VSV-GFP, Daphnia cells were washed five times with DCCM to reduce the amount of input virus that remained in the cultures. As expected, some input virus remained in the cultures, between 1 × 104 and 1 × 106 pfu/ml, as indicated by CPE in BHK-21 cells using the supernatant collected after the last wash 1 hr post-infection (Fig. 3). One hour later, cell culture supernatant from the same VSV-GFP infected Daphnia cultures was collected and replaced with fresh DCCM. Levels of input virus collected at 2 hr post-infection were at least 1 × 105 pfu/ml, indicating that input virus was still present. Four hours post-infection, viral titers from VSV-GFP infected Daphnia cultures decreased two orders of magnitude compared to titers at the 2 hr time point. This result is consistent with no virus replication occurring less than 4 hr post-infection, and additional input virus being washed off at the 2-hr time point. Of significance, viral titers increased 24 hr post-infection one to three orders of magnitude above titers at 4 hr post-infection. The amount of virus collected in the next 24 hr period at 48 hr post-infection remained one to two orders of magnitude above titers at 4 hr. These findings are consistent with de novo virus replication in Daphnia cells.
Fig. 3.
De novo replication of VSV-GFP in Daphnia embryonic primary cultures. Supernatant of VSV-GFP infected Daphnia cells collected at 1, 2, 4, 24, and 48 hr post-infection was titered on BHK-21 cells in ten-fold duplicate serial dilutions. Titers were scored in pfu/ml as the highest dilution which minimally yielded 1 pfu.
In this study, a recombinant VSV encoding GFP was able to infect primary cell cultures derived from Daphnia embryos and resulted in foreign gene expression. VSV replication in the cells caused no apparent CPE, a finding that is consistent with observations made in other invertebrate cells. In addition, infectious progeny virions were isolated from infected cells indicating that VSV can replicate in Daphnia cells. The ability to introduce genes into Daphnia cell lines permits the study of the molecular details of Daphnia cell biology and subsequent responses to environmental signals. Phenotypic studies of the effect of gene expression in embryonic Daphnia cell cultures may be limited by the finite lifespan of the cell cultures. In addition, gene transcription in the VSV vector is controlled by the viral promoter, and thus this vector cannot be used to study the effects of specific Daphnia promoters on gene expression. However, with the knowledge that Daphnia cells in culture are permissive to VSV infection, it is logical to propose that delivery of the viral vector into Daphnia adults or embryos may lead to adult transgenic Daphnia, facilitating additional molecular studies of in vivo effects. A similar system has been described in several mosquito and moth species using the positive stranded RNA alphavirus Sindbis (Foy et al., 2004). Sindbis has been delivered to adults either by microinjection into the thorax or feeding the virus to mosquitoes in blood meals. This system has been used both to express foreign transgenes as well as RNAi for gene silencing (Adelman et al., 2001). However, the usefulness of Sindbis virus vectors may be limited by the tropism of the parent virus (Nilkasson, ’88). VSV could similarly be exploited for both protein expression and RNAi studies where subgenomic mRNAs could silence genes in Daphnia allowing for the functional study of key regulatory genes. For some transgenic studies in Daphnia, a replication-defective VSV vector may be preferable. In these instances, a modified VSV vector could be employed where one or more structural genes has been deleted from the genome but is provided in trans in producer cells (Takada et al., ’97). The resulting virions would be capable of entry, replication, and gene expression, but without expression of additional structural proteins, progeny particles would be defective or not produced. Defective progeny virions would not be capable of infecting neighboring cells, eliminating additional spread of the vector after initial infection. Overall, we have shown the utility of a VSV-based system for expression of foreign trans-genes in Daphnia. To our knowledge, this is the first report of a noninsect arthropod cell line experimentally infected with VSV. The results of this work have broad implications for the genetic study of the microcrustacean Daphnia as well as other invertebrates of ecological, developmental, evolutionary, or medical relevance for which gene transfer technologies have not been successful to date.
ACKNOWLEDGMENTS
We are particularly grateful to Scott F. Michael for advice and members of the Daphnia Genomics Consortium for helpful discussions.
This work was supported by US National Science Foundation through an ADVANCE-Fellows award to S.I. (0521917) and an FIBR award to M.L. (0328516) and in part by the Coypu Foundation through a grant to the Tulane-Xavier Center for Bioenvironmental Research.
Footnotes
This publication is contribution #0011 for the Center for Biomolecular Science and Engineering, Florida Gulf Coast University.
LITERATURE CITED
- Abraham G, Banerjee AK. Sequential transcription of the genes of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1976;73:1504–1508. doi: 10.1073/pnas.73.5.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman ZN, Blair CD, Carlson JO, Beaty BJ, Olson KE. Sindbis virus-induced silencing of dengue viruses in mosquitoes. Insect Mol Biol. 2001;10:265–273. doi: 10.1046/j.1365-2583.2001.00267.x. [DOI] [PubMed] [Google Scholar]
- Baer KN, Goulden CE. Evaluation of a high-hardness COMBO medium and frozen algae for Daphnia magna. Ecotoxicol Environ Saf. 1998;39:201–206. doi: 10.1006/eesa.1997.1627. [DOI] [PubMed] [Google Scholar]
- Ball LA, White CN. Order of transcription of genes of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1976;73:442–446. doi: 10.1073/pnas.73.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birge EA. Plankton studies on Lake Medota. I. The vertical distribution of the pelagic Crustacea during July 1894. Transactions of the Wisconsin Academy of Sciences. Arts and Letters. 1895;11:274–451. [Google Scholar]
- Cherry S, Doukas T, Armknecht S, Whelan S, Wang H, Sarnow P, Perrimon N. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev. 2005;19:445–452. doi: 10.1101/gad.1267905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaestecker E, DeMeester L, Ebert D. In deep trouble: Habitat selection constrained by multiple enemies in zooplankton. Proc Natl Acad Sci USA. 2002;99:5481–5485. doi: 10.1073/pnas.082543099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert D, Haag C, Kirkpatrick M, Riek M, Hottinger JW, Pajunen VI. A selective advantage to immigrant genes in a Daphnia metapopulation. Science. 2002;295:485–488. doi: 10.1126/science.1067485. [DOI] [PubMed] [Google Scholar]
- Edmonson WT. Daphnia in experimental ecology: notes on historical perspectives. In: Peters RH, de Bernardi R, editors. Daphnia. Vol. 45. Memorie dell'Istituto di Idrobiologica; 1987. pp. 11–30. [Google Scholar]
- Feder ME, Mitchell-Olds T. Evolutionary and ecological functional genomics. Nat Rev Genet. 2003;4:649–655. doi: 10.1038/nrg1128. [DOI] [PubMed] [Google Scholar]
- Foy BD, Myles KM, Pierro DJ, Sanchez-Vargas I, Uhlirova M, Jindra M, Beaty BJ, Olson KE. Development of a new Sindbis virus transducing system and its characterization in three Culicine mosquitoes and two Lepidopteran species. Insect Mol Biol. 2004;13:89–100. doi: 10.1111/j.1365-2583.2004.00464.x. [DOI] [PubMed] [Google Scholar]
- Garner DL, Johnson LA, Yue ST, Roth BL, Haugland RP. Dual DNA staining assessment of bovine sperm viability using SYBR 14 and propidium iodide. J Androl. 1994;15:620–629. [PubMed] [Google Scholar]
- Gillies S, Stollar V. The production of high yields of infectious vesicular stomatitis virus in Aedes albopictus cells and comparison with replication in BHK-21 cells. Virology. 1980;107:509–513. doi: 10.1016/0042-6822(80)90317-7. [DOI] [PubMed] [Google Scholar]
- McCauley E, Nisbet RM, Murdock WW, de Roos AM, Gurney WSC. Large-amplitude cycles of Daphnia and it's algal prey in enriched environments. Nature. 1999;402:653–656. [Google Scholar]
- Nilkasson B. Sindbis and Sindbis-like viruses. In: Monath TP, editor. The Arboviruses: epidemics and ecology. IV. CRC Press; Boca Raton, FL: 1988. pp. 167–176. [Google Scholar]
- Patton JT, Davis NL, Wertz GW. N protein alone satisfies the requirement for protein synthesis during RNA replication of vesicular stomatitis virus. J Virol. 1984;49:303–309. doi: 10.1128/jvi.49.2.303-309.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode SC, Pawlowski M, Tollrian R. The impact of ultraviolet radiation on the vertical distribution of zoo-plankton of the genus Daphnia. Nature. 2001;412:69–72. doi: 10.1038/35083567. [DOI] [PubMed] [Google Scholar]
- Shope RE, Tesh RB. Wagner RR, editor. The ecology of rhabdoviruses that infect vertebrates. The Rhabdo-viruses. New York: Plenum Press. 1987:509–534. [Google Scholar]
- Takada A, Robison C, Goto H, Sanchez A, Murti KG, Whitt MA, Kawaoka Y. A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci USA. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesh RB, Peralta PH, Johnson KM. Ecologic studies of vesicular stomatitis virus II. Results of experimental infection in Panamanian wild animals. Am J Epidemiol. 1970;91:216–224. doi: 10.1093/oxfordjournals.aje.a121130. [DOI] [PubMed] [Google Scholar]
- Tesh RB, Chaniotis BN, Johnson KM. Vesicular stomatitis virus (Indiana serotype): transovarial transmission by phlebotomine sandflies. Science. 1972;175:1477–1479. doi: 10.1126/science.175.4029.1477. [DOI] [PubMed] [Google Scholar]
- Tessier AJ, Woodruff P. Cryptic trophic cascade along a gradient of lake size. Ecology. 2002;83:1263–1270. [Google Scholar]
- Villarreal LP, Breindl M, Holland JJ. Determination of molar ratios of vesicular stomatitis virus induced RNA species in BHK21 cells. Biochemistry. 1976;15:1663–1667. doi: 10.1021/bi00653a012. [DOI] [PubMed] [Google Scholar]
- Whelan SP, Wertz GW. Transcription and replication initiate at separate sites on the vesicular stomatitis virus genome. Proc Natl Acad Sci USA. 2002;99:9178–9183. doi: 10.1073/pnas.152155599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan SP, Ball LA, Barr JN, Wertz GW. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci USA. 1995;92:8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan SP, Barr JN, Wertz GW. Identification of a minimal size requirement for termination of vesicular stomatitis virus mRNA: implications for the mechanism of transcription. J Virol. 2000;74:8268–8276. doi: 10.1128/jvi.74.18.8268-8276.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]