Abstract
The probabilistic record linkage (PRL) is based on a likelihood score that measures the degree of similarity of several matching variables. Screening test results for different diseases are available for the blood donor population. In this paper, we describe the accuracy of a PRL process used to track blood donors from the Fundação Pró-Sangue (FPS) in the Mortality Information System (SIM), in order that future studies might determine the blood donor’s cause of death. The databases used for linkage were SIM and the database made up of individuals that were living (200 blood donors in 2007) and dead (196 from the Hospital das Clinicas de São Paulo that died in 2001–2005). The method consists of cleaning and linking the databases using three blocking steps comparing the variables “Name/Mother’s Name/ Date of Birth” to determine a cut-off score. For a cut-off score of 7.06, the sensitivity and specificity of the method is 94.4% (95%CI: 90.0–97.0) and 100% (95%CI: 98.0–100.0), respectively. This method can be used in studies that aim to track blood donors from the FPS database in SIM.
Keywords: Medical Record Linkage, Blood Donors, Mortality Rate
Introduction
Record linkage is a process that aims to accurately identify whether two or more records relate to the same individual. In Brazil, it has been used increasingly for studies involving the health information system databases managed by the Brazilian Ministry of Health, including the Mortality Information System (SIM). The process is used not only to detect and remove duplicate records relating to the same individual from within a database1 but also to integrate records across health information system databases and in between them and other databases2,3,4,5,6,7. By doing so, these studies help to improve data quality and integrity, allow for the reuse of existing data that can be used for answering a broader range of study questions, reduce costs and efforts related to data acquisition and also help to improve record linkage methodology and its assessment8.
Our team has been involved with studies of blood banking and transfusion medicine for many years. Some of our studies have made exclusive use of the Fundação Pró-Sangue Hemocentro de São Paulo (FPS) blood bank donors’ and recipients’ routine secondary data, while others have made use of such data in conjunction with primary data collected within the context of the international collaboration Retrovirus Epidemiology Donor Study (REDS II)9,10 and The Recipient Epidemiology and Donor Evaluation Study-III (REDS-III). We are now particularly interested in performing longitudinal studies in which we need to ascertain some long term outcomes (e.g. vital status) of donors and transfused recipients included in our primary and secondary databases. This will allow us to investigate their disease progression and case fatality rates, therefore contributing to a better characterization of the natural history of many transfusion-related diseases, as well as their history under certain controlled circumstances.
Such long-term data is not contained in our databases, so we depend on tracking our patients from other secondary sources that are known to include the relevant information, such as the SIM. There are two major complications in doing so. First, the number of records in these databases is huge. The SIM database, for example, has over 1 million records per year. Secondly, these databases do not share a unique personal identifier that would enable us to simply join them (like the US social security number or the long awaited “cartão do Sistema Único de Saúde (SUS)” number) and they do have many inexorable errors, abbreviations and missing or incorrectly formatted values in the personal identification variables (like name, mother’s name and date of birth). In such a scenario, a more sophisticated record linkage process is needed. As our intention is to repeat the linkage process several times using the same and also other similar data sources over time, the process needs to be robust and reproducible, and should preferably not be overly dependent on human intervention. In transfusion medicine, just a few longitudinal studies have been done which were made possible by the consolidation of different data sources through record linkage methods11, but many such studies have greatly contributed in other topics12,13,14,15.
There are two main types of record linkage: the deterministic record linkage in which the linkage of records is based on exact agreement of one or several matching variables, and the probabilistic linkage, which is based on a likelihood score that is calculated taking into consideration how similar each of several matching variables are. Pairs with high scores have higher probabilities of being true matches, and pairs with low scores have lower probabilities. The problem lies in pairs in the intermediate “grey” zone, which usually require manual review in order to be satisfactorily classified16. Having to depend on manual review often involves a high cost for probabilistic linkage exercises in terms of the engagement of skilled human resources in order to perform such a task, and it also raises privacy concerns since these individuals have access to nominal information. Alternatively, the need for manual review can be removed by explicitly selecting a single cut-off value above which all pairs are declared true matches.
In studies that aim to assess the performance of a probabilistic record linkage process, such as this one, the results of the manual review or of a database with known true matches and non-matches are often considered the “gold-standard” against which different values of the likelihood score are compared. Among the different measures that have been used for assessing the accuracy of record linkage processes are sensitivity, positive predictive value and specificity. Sensitivity is the capacity of the linkage process to recognize the true matches, while specificity is the capacity of the linkage process to recognize non-matches. Positive predictive is the proportion of matches found by the linkage process that are true. Sensitivity and positive predictive value are often respectively called recall and precision in the linkage field17. As in studies of diagnostic tests, there is often a trade-off between recall and precision, in which high precision can normally only be attained at the cost of low recall, and vice-versa.
In this paper, we aimed to study the accuracy of a probabilistic linkage process used to track our blood bank patients in the SIM databases. Our goal was to define cut-off scores that would maximize sensitivity and minimize as much as possible the need to perform manual review of future studies that aimed to describe mortality rates of blood transfusion transmitted diseases in Brazilian individuals.
Materials and methods
Data sources
The reference file was created by appending 200 randomly selected records of the FPS database with 196 randomly selected hospital discharge records of the Hospital das Clínicas, Faculdade de Medicina, Universidade de São Paulo (HC-FMUSP) database. In the former database, 200 records were randomly chosen among the records of individuals aged 18 and above that had donated blood in 2007 and were, therefore, known to be alive during the period from 2001 to 2005. In the latter case, 196 records were randomly chosen among records of individuals aged 18 and above who were known to have died during their hospitalization in the period from 2001 to 2005, as reported in the database’s outcome variable. Given the ratio of 200 to 196 individuals, we were artificially creating a scenario in which the death ratio was close to 50%.
The comparison file was the SIM database (version 19/Apr/2007) for the period of 2001 to 2006, containing records of all Brazilian individuals who had died at the age of 18 and above. Due to notification delays, as the database extraction had been done on 19 April 2007, the 2006 data were still incomplete.
The number of 196 deceased individuals to be included in our sample size was estimated taking into account an expected sensitivity of 85% (P), a 95% confidence interval (Zα = 1.96) with a total width of 10% (W) and using the equation N = 4 Z2αP(1−P) ÷ W2 18.
Pre-processing
Both the reference and the comparison files contained the variables identity number, name, mother’s name, gender, date of birth, among others. They underwent an extensive pre-processing step aiming to clean and standardize the data to be used in the linkage process. The pre-processing was done using SQL-Analysis Server (Microsoft SQL Server 2008), and involved the following data transformations: the value 9999 was defined for all non-numeric values for age and for records in which age was less than 18, and were excluded (496,056 records); all records with missing mother’s name were kept but “**” was filled in (222,044); the value 9 was defined for all missing or invalid information for gender; date of birth null values were transformed into “18000101” (47,533). Importantly, in the SIM database, records with missing names or with invalid names (477,174) or with missing death certificate IDs (0) were excluded from subsequent linkage steps. The excluded records represented 16.9% of the total number of records. It is important to note that for this version of SIM, 27.9% of 2001 records were excluded because of missing names or invalid names, whereas for the years of 2002, 2003, 2004, 2005 and 2006 only 4% of the records were excluded for the same reason. After the cleaning steps the SIM database used for the linkage had 4,775,164 records as described in Figure 1.
Figure 1.
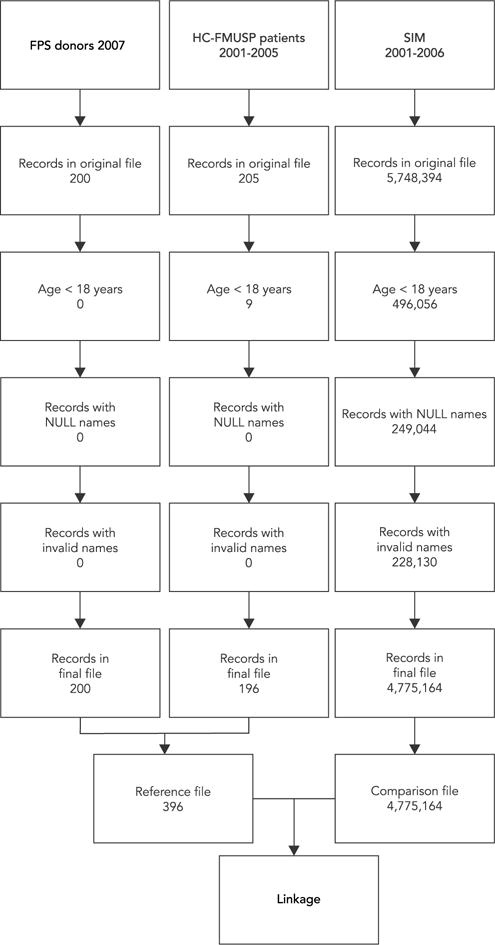
Cleaning description of the reference and comparison files used on the linkage process.
FPS: Fundação Pró-Sangue Hemocentro de São Paulo; HC-FMUSP: Hospital das Clínicas, Faculdade de Medicina, Universidade de São Paulo; SIM: Mortality Information System.
Linkage strategy
The record linkage was performed using a software called Record Linkage III – version 3.0.4 4005 (RECLINK-III; http://www.iesc.ufrj.br/reclink/RecLink_arquivos/RecLinkdl.html), an open-source probabilistic linkage software specially developed to associate records taking into consideration the Portuguese language phonetics19. Standardization of common fields was performed where correction of upper/lower case variations was done, removal of accents and prepositions inside names (e.g. ”Maria Conceição dos Santos → ”Maria Conceicao Santos), standardization of date formats was performed and finally, removal of commas and other punctuation marks. All missing values were correctly classified as such. Using the variable name, four other secondary variables were generated: first name, last name and first and last names phonetic codes. Three blocking steps strategy were used from a combination of the phonetic codes of the variables first name, last name and gender20. By allowing comparisons of record pairs only within blocks, the computational task is much reduced. The first blocking step used first name phonetic code, last name phonetic code and gender. The second blocking step used only first name phonetic code and gender and the third blocking step used only last name phonetic code and gender.
The matching variables used for pairing were name, mother’s name and date of birth. RECLINK-III estimates scores for each pair of records, with a higher score representing a higher likelihood of the pair being a true match. Scores are proportional to log-likelihood ratios, which are derived in a standard probabilistic linkage approach. In this approach, m is the probability that the characters sequence of a particular matching variable agree for records that are true matches (m is analogous to sensitivity), and u is the probability that the characters sequence of a particular matching variable agree for records that are false matches (u is analogous to 1-specificity), i.e. by chance alone. When the contents of the variable do agree in between the records being compared, the score is proportional to logarithm (m/u), and when they disagree to logarithm ((1−m)/(1−u)). Name and mother’s name had their characters’ sequence compared using the Levenshtein distance, which returned values between 1 (total concordance) and 0 (total discordance) with 92% sensitivity (m) and 1% 1-specificity (u), with a minimum proportion of concordance of 85%. Date of birth had its sequences of digits, ignoring separators, compared using the character algorithm which returned values between 1 (total concordance) and 0 (total discordance), with 90% sensitivity (m) and 5% 1- specificity (u), with a minimum concordance of 65%. The scores for each matching variable are added to determine a composite score 16. Total concordance of all three matching variables resulted in a maximum score of log (0.92/0.01) + log (0.92/0.01) + log (0.9/0.05) = 17.22, while total discordance of all three matching variables resulted in a minimum score of log (0.08/0.99) + log (0.08/0.99) + log (0.1/0.95) = −10.51, using base 2 logarithms.
Post-processing
The manual review of the pairs formed during each blocking step was performed by one of the authors (L.C.) in the module “Combine” of the RECLINK-III software. In this module, the user is able to see the score assigned to each pair and to visually compare variables from both records, including matching variables as well as additional ones that did not participate in the linking process. All pairs were manually reviewed except those in the last score level (−10.51), which comprised the majority of pairs (97.5% out of 709,550 for the first blocking step, 99,9% out of 1,448,313 for the second blocking step) and were directly considered non-matches. After classifying pairs into matches and non-matches, RECLINK-III automatically created three files: a file with matched pairs of records from both the reference and the comparison files, a file with all non-matched records from the reference file and another with all non-matched records from the comparison file. These two latter files were then used in the next blocking steps. The three files with the matched pairs of records found at each of the blocking steps were then appended for analysis. All steps in the linkage process were done blind to the vital status of individuals in our reference file.
Data analysis
In order to test the performance of our record linkage strategy, sensitivity and specificity and their 95% binomial confidence intervals were calculated assuming different cut-off points for score. Sensitivity was defined as the proportion of true matches among the 196 individuals known to be dead (which should have corresponding records in SIM), and specificity was defined as the proportion of non-matches among the 200 individuals known to be alive (which should not have corresponding records in SIM). For the chosen cut-off point, the positive predictive value was also calculated as the proportion of true matches among the sum of true and false matches, along with its 95% binomial confidence interval. Sensitivity and positive predictive value are often respectively called recall and precision in the linkage field17.
As expected, for every record in the reference file many matches were generated, but only the pair with the highest score was considered in the sensitivity and specificity calculations. In other words, we only considered the highest score in which a record from our reference file appeared as a pair. When two or more pairs were formed with the same score, which included the same record from the reference file, only the most comparable one, as classified by the “gold standard” manual review, was considered to be a true match (for the 196 individuals known to be dead) or a false match (for the 200 individuals known to be alive).
The choice of the cut-off point took into consideration the need to maximize the specificity of the matching strategy, i.e. to minimize the possibility of making false matches while if necessary accepting some level of failures to find true matches.
Ethical approval for this study was obtained from the Ethics Research Committee of HC-FMUSP (CAAE: 0543.0.015.000-08).
Results
The reference file, which was composed of an extraction of 200 records from the FPS database and 196 records from the HC-FMUSP database, had 100% completion for the matching variables name, mother’s name and data of birth, while the fields name and mother’s name had no abbreviations. The comparison file, which was the SIM database for the period of 2001 to 2006 containing 4,775,164 records of individuals who had died at aged 18 and above, had 100% completion for name (as this was a prerequisite for SIM records to be included in the comparison file), 95.35% completion for mother’s name and 99% completion for date of birth.
In the first blocking step, 179 true matches were formed and in the second blocking step, this number increased to 185 out of the total 196 possible true matches. As no new true match was found in the third blocking step, we will only report results from the first two.
Table 1 shows the totality of pairs formed in between the reference and the comparison databases, given the limitations imposed by the blocking variables. Pairs with high scores were mostly found in step 1. When considering the number of pairs formed in step 1 from the highest to the lowest scores, we can see that at score 9.8, even though five pairs were found, only one of them referred to the first time a specific record from the reference file was found in a pair. The other four referred to records from the reference file that had already formed pairs with the same or higher scores. The same happened at many lower scores, increasing as the score decreased. Most true matches were found at score 17.22. Only one to four true matches were found at scores ranging from 17.01 to 9.8. Then no true matches were found up to score 7.06, where 16 were found. The first six false matches appeared at score 6.02, and from there numbers increased very slowly. It was only at the very lowest scores that the majority of the false matches were found. Sensitivity at score 9.08 was 86.2% (95%CI: 80.5–90.7) and at score 7.06 was 94.4% (95%CI: 90.2–97.2), the maximum value obtained. Specificity was 100% (95%CI: 98.2–100.0) up to score 6.02. Such results would point to a natural choice of a score cut-off value able to discriminate pairs into true and false matches, as the last true matches were found at score 7.06 and the first false matches appeared only at score 6.02. Using a cut-off inside this range, the positive predictive value was 100% (95%CI: 98.2–100.0).
Table 1.
Sensitivity, specificity and positive predictive value of several scores of the probabilistic linkage process.
| Score | Step 1 | Step 2 | Steps 1 + 2 | Steps 1 + 2 | Steps 1 + 2 | Steps 1 + 2 | Steps 1 + 2 | ||
|---|---|---|---|---|---|---|---|---|---|
| All pairs | First time pairs* | All pairs | First time pairs* | True matches | False matches | Sensitivity [% (95%CI)] | Specificity [% (95%CI)] | Positive predictive value [% (95%CI)] | |
| 17.22 | 147 | 147 | 0 | 0 | 147 | 0 | 75.0 (68.0–80.0) | 100.0 (98.0–100.0) | 100.0 (97.0–100.0) |
| 17.01 | 2 | 2 | 0 | 0 | 2 | 0 | 76.0 (69.0–82.0) | 100.0 (98.0–100.0) | 100.0 (98.0–100.0) |
| 16.99 | 1 | 1 | 0 | 0 | 1 | 0 | 76.5 (70.0–82.0.0) | 100.0 (98.0–100.0) | 100.0 (98.0–100.0) |
| 16.97 | 2 | 2 | 0 | 0 | 2 | 0 | 77.6 (71.0–83.0) | 100.0 (98.0–100.0) | 100.0 (98.0–100.0) |
| 16.95 | 1 | 1 | 0 | 0 | 1 | 0 | 78.1 (72.0–84.0) | 100.0 (98.0–100.0) | 100.0 (98.0–100.0) |
| 16.93 | 2 | 2 | 0 | 0 | 2 | 0 | 79.1 (73.0–85.0) | 100.0 (98.0–100.0) | 100.0 (98.0–100.0) |
| 16.91 | 1 | 1 | 0 | 0 | 1 | 0 | 79.6 (73.0–85.0) | 100.0 (98.0–100.0) | 100.0 (98.0–100.0) |
| 16.89 | 1 | 1 | 0 | 0 | 1 | 0 | 80.1 (74.0–85.0) | 100.0 (98.0–100.0) | 100.0 (98.0–100.0) |
| 16.87 | 3 | 3 | 1 | 1 | 4 | 0 | 82.1 (76.0–87.0) | 100.0 (98.0–100.0) | 100.0 (98.0–100.0) |
| 16.78 | 1 | 1 | 0 | 0 | 1 | 0 | 82.7 (77.0–88.0) | 100.0 (98.0–100.0) | 100.0 (98.0–100.0) |
| 16.70 | 4 | 4 | 0 | 0 | 4 | 0 | 84.7 (79.0–89.0) | 100.0 (98.0–100.0) | 100.0 (98.0–100.0) |
| 16.60 | 2 | 2 | 0 | 0 | 2 | 0 | 85.7 (80.0–90.0) | 100.0 (98.0–100.0) | 100.0 (98.0–100.0) |
| 9.80 | 5 | 1 | 0 | 0 | 1 | 0 | 86.2 (81.0–91.0) | 100.0 (98.0–100.0) | 100.0 (98.0–100.0) |
| 9.50 | 1 | 0 | 0 | 0 | 0 | 0 | 86.2 (81.0–91.0) | 100.0 (98.0–100.0) | 100.0 (98.0–100.0) |
| 9.03 | 1 | 0 | 0 | 0 | 0 | 0 | 86.2 (81.0–91.0) | 100.0 (98.0–100.0) | 100.0 (98.0–100.0) |
| 8.98 | 1 | 0 | 0 | 0 | 0 | 0 | 86.2 (81.0–91.0) | 100.0 (98.0–100.0) | 100.0 (98.0–100.0) |
| 8.83 | 1 | 0 | 0 | 0 | 0 | 0 | 86.2 (81.0–91.0) | 100.0 (98.0–100.0) | 100.0 (98.0–100.0) |
| 8.55 | 1 | 0 | 0 | 0 | 0 | 0 | 86.2 (81.0–91.0) | 100.0 (98.0–100.0) | 100.0 (98.0–100.0) |
| 8.26 | 1 | 0 | 0 | 0 | 0 | 0 | 86.2 (81.0–91.0) | 100.0 (98.0–100.0) | 100.0 (98.0–100.0) |
| 7.98 | 1 | 0 | 0 | 0 | 0 | 0 | 86.2 (81.0–91.0) | 100.0 (98.0–100.0) | 100.0 (98.0–100.0) |
|
| |||||||||
| 7.06 | 12 | 11 | 5 | 5 | 16 | 0 | 94.4 (90.0–97.0) | 100.0 (98.0–100.0) | 100.0 (98.0–100.0) |
|
| |||||||||
| 6.54 | 2 | 0 | 0 | 0 | 0 | 0 | 94.4 (90.0–97.0) | 100.0 (98.0–100.0) | 100.0 (98.0–100.0) |
| 6.23 | 2 | 0 | 0 | 0 | 0 | 0 | 94.4 (90.0–97.0) | 100.0 (98.0–100.0) | 100.0 (98.0–100.0) |
|
| |||||||||
| 6.02 | 34 | 6 | 0 | 0 | 0 | 6 | 94.4 (90.0–97.0) | 97.0 (94.0–99.0) | 97.0 (93.0–99.0) |
|
| |||||||||
| 5.86 | 1 | 0 | 0 | 0 | 0 | 0 | 94.4 (90.0–97.0) | 97.0 (94.0–99.0) | 97.0 (93.0–99.0) |
| 5.78 | 1 | 0 | 0 | 0 | 0 | 0 | 94.4 (90.0–97.0) | 97.0 (94.0–99.0) | 97.0 (93.0–99.0) |
| 5.74 | 1 | 0 | 0 | 0 | 0 | 0 | 94.4 (90.0–97.0) | 97.0 (94.0–99.0) | 97.0 (93.0–99.0) |
| 5.71 | 14 | 1 | 0 | 0 | 0 | 1 | 94.4 (90.0–97.0) | 96.5.0 (93.0–99.0) | 96.0 (93.0–98.0) |
| 5.70 | 1 | 1 | 0 | 0 | 0 | 1 | 94.4 (90.0–97.0) | 96.0 (92.0–98.0) | 96.0 (92.0–98.0) |
| 5.65 | 1 | 1 | 0 | 0 | 0 | 1 | 94.4 (90.0–97.0) | 95.5.0 (92.0–98.0) | 95.0 (91.0–98.0) |
| 5.64 to −10.51 | 709284 | 106 | 1448307 | 85 | 0 | 191 | 94.4 (90.0–97.0) | 95.5 to 0.0 | 48 to 0.0 |
| Total | 709532 | 294 | 1448313 | 91 | 185 | 200 | |||
95%CI: 95% confidence interval.
“First time pairs” refers to the first time a record from the reference file was found in a pair, searching from the highest to the lowest score.
The manual review revealed why 16 true matches were found only at score 7.06. The problem was in the variable name. In one of the records of these pairs, one element of the name was missing, e.g. in one record one woman possibly had her maiden name (Maria Antonia Lima) and in the other her married name (Maria Antonia Lima Castro – this is a fictitious name).
Discussion
The probabilistic linkage process used in the present study aims to help track blood bank patients from the FPS database in the SIM. In order to assess the accuracy of this linkage process, we assembled a file in which we knew beforehand the death status of its recorded individuals. By doing so, we were able to know which of the obtained pairs were true or false matches. This strategy of using a “known vital status” in order to ascertain linkage accuracy has been used in a number of studies, including a few Brazilian ones2,3.
The accuracy of our linkage process was very high, with a sensitivity of 94% and a specificity of 100% using a score in between 7.06 and 6.02 as the cut-off point. Our results are slightly better than those of a Brazilian study that used RECLINK-III and the same matching variables in order to link the AIDS surveillance database with SIM, where they found a sensitivity of 87.6% and a specificity of 99.6%.2 However, apart from the differences in the size of our reference file and their surveillance database, we have to bear in mind that while our reference file did not have any missing or incomplete values in the matching variables, theirs was a much bigger database that was likely to have many records with these types of errors. The use of large databases obviously also makes the manual review process a real challenge. Our results are comparable to those of another Brazilian study that used an in-house deterministic linkage process in order to link data from two study cohorts of HIV-infected patients with SIM, where they found a sensitivity of 96.5% and a specificity of 99.6%2.
Although sensitivity was high, the linkage process was still unable to find 11 individuals known to be dead from the reference file in the SIM database. There are at least four possible explanations for this finding: (1) the death of these individuals had not been registered in the SIM database, (2) the death of these individuals had been registered in the SIM database, but their records were among those that were discarded before the linkage process either because they had missing data for the variable name, or had been wrongly recorded as belonging to an individual younger than 18 years of age or had missing data for death certificate IDs, (3) the death of these individuals had been registered in the SIM database, but information available at the three matching variables differed to such an extent between the reference and the comparison files that the pair obtained was classified with the last possible score, i.e. −10.52, in which case it was not submitted to manual review, and therefore was not found, and (4) pairs were not formed because records were blocked by the variable gender, present in all three of our blocking steps.
The former explanation suggests that there is under notification in SIM. SIM has undergone significant improvements in coverage and overall data quality over the last decade21, but mostly these improvements were needed for remote rural areas of the Northeastern and Northern regions of the country, and for individuals who had died outside hospitals and were at the extremes of age22. This was not the case for the 196 individuals from our reference file who were known to have died during their hospitalization at the biggest public university hospital in the city of São Paulo, where they had certainly been granted a death certificate. Another possibility is that these deaths had not yet been registered at the SIM database due to notification delays. But we also think this is unlikely because they happened from 2002 to 2005, and the SIM database was extracted in April, 2007.
The second and third explanations refer to quality issues in the SIM database, which may have prevented us from reaching a higher sensitivity. The sensitivity of record linkage studies is very much dependent on the quality of its data sources. We believe it is likely that the 11 missing individuals were included in the 16.7% of SIM records that had been discarded from our linkage process due to missing data on the variables name, or that had been wrongly recorded as belonging to an individual younger than 18. On the other hand, considering the nature of the probabilistic linkage and the fact that the last score represents pairs that had total discordance of all three matching variables, it is most unlikely that any true matches for these 11 individuals would be found at the score −10.51, which was not manually reviewed.
Gender was chosen as a good blocking variable to be used in combination with others, as we believed not many records were likely to be wrongly classified or to have missing values in this field. However rare, such mistakes may have happened, even though it is unlikely that this possibility alone would explain all of the 11 missing individuals. In any case, in our future linkages, we will not have the same variable participating in all of our blocking steps.
With only 396 individuals coming from two data sources (FPS and HC-FMUSP databases), our reference file was relatively small and therefore not very heterogeneous. If we were to repeat our linkage process using the whole or a more representative sample of the FPS database, the bigger number of records would obviously increase the variability of the values presented in our matching variables, which would mostly increase the number of false matches, and possibly increase the scores of these false matches3,17. As this could decrease the specificity observed for a given score value, perhaps a natural choice of a score cut-off point, as we had in the present study, would not be presented. The immediate consequence would be the need to check whether the same cut-off point used in the present study could apply to the new exercise. This would be accomplished by manually reviewing at least some of the pairs in the so-called grey zone. Importantly, while linking records from primary or secondary data sources to a mortality database, we are dealing with a situation in which we obviously expect that only one true match will be found per individual, unless duplicates exist in one or both files. Today, Reclink removes from both databases the records that are part of a pair considered a true pair in a blocking step and on the next blocking step these records will not form pairs. As a suggestion to improve and accelerate the manual review in each blocking step, Reclink could automatically remove from the list of pairs that will be submitted to the manual review all pairs with lower scores that contain one or the other of two records of pairs that have reached a higher score. This strategy would significantly decrease the burden of the manual review.
Another consequence of repeating our linkage process using the whole or a more representative sample of the FPS database, which would very likely have a different death ratio, would naturally be the change of the positive predictive value for the chosen cut-off point18. Most importantly, such accuracy changes would also impart on how the longitudinal studies that will depend on the linkage results would be interpreted. As mentioned in other similar studies3, it is best to use a more stringent cut-off point so as to maximize the specificity of the matching process, avoiding false matches even while accepting some level of failures to find true matches. The reason for this is that if we are to compare risk factors for death among blood donors or recipients, false matches obtained as a non-differential error (i.e. the same error rate across the risk factor levels) would bias both the risk difference and the risk ratio towards the null hypothesis, whereas failing to find some true matches would only bias the risk difference but not the risk ratio19.
In conclusion, we believe our record linkage process can be used in studies that aim to track blood bank patients from the FPS database and, if necessary, from other blood bank databases that collect similar data, in the SIM, if special care is taken in the selection of the cut-off point, so as to guarantee that a high specificity be maintained.
Acknowledgments
This work is supported and funded by the National Institute of Allergy and Infectious Disease – Grant Number 1P50AI098461-01 REVISED.
Footnotes
Contributors
L. Capuani contributed to the conception and design of the study; acquisition, analysis, and interpretation of data for the study; drafting the study, and revising it for important intellectual content; final approval of the version to be published; agreement to be accountable for all aspects of the work. A. L. Bierrenbach contributed to the conception and design of the study; analysis and interpretation of data for the study; drafting the study and critically revising it for important intellectual content; final approval of the version to be published; agreement to be accountable for all aspects of the study. F. Abreu contributed to the drafting the study and critically revising it for important intellectual content; final approval of the version to be published; agreement to be accountable for all aspects of the study. P. L. Takecian contributed to the analysis and interpretation of data for the study; critically revising it for important intellectual content; final approval of the version to be published; agreement to be accountable for all aspects of the study. J. E. Ferreira and E. C. Sabino contributed to the conception and design of the study; analysis and interpretation of data for the study; critically revising it for important intellectual content; final approval of the version to be published; agreement to be accountable for all aspects of the study.
References
- 1.Bierrenbach AL, de Oliveira GP, Codenotti S, Gomes AB, Stevens AP. Duplicates and misclassification of tuberculosis notification records in Brazil, 2001–2007. Int J Tuberc Lung Dis. 2010;14:593–9. [PubMed] [Google Scholar]
- 2.Pacheco AG, Saraceni V, Tuboi SH, Moulton LH, Chaisson RE, Cavalcante SC, et al. Validation of a hierarchical deterministic record-linkage algorithm using data from 2 different cohorts of human immunodeficiency virus-infected persons and mortality databases in Brazil. Am J Epidemiol. 2008;168:1326–32. doi: 10.1093/aje/kwn249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fonseca MGP, Coeli CM, Lucena FFA, Veloso VG, Carvalho MS. Accuracy of a probabilistic record linkage strategy applied to identify deaths among cases reported to the Brazilian AIDS surveillance database. Cad Saúde Pública. 2010;26:1431–8. doi: 10.1590/s0102-311x2010000700022. [DOI] [PubMed] [Google Scholar]
- 4.Migowski A, Chaves RB, Coeli CM, Ribeiro AL, Tura BR, Kuschnir MC, et al. Accuracy of probabilistic record linkage in the assessment of high-complexity cardiology procedures. Rev Saúde Pública. 2011;45:269–75. doi: 10.1590/s0034-89102011005000012. [DOI] [PubMed] [Google Scholar]
- 5.de Oliveira GP, Pinheiro RS, Coeli CM, Barreira D, Codenotti SB. Mortality information system for identifying underreported cases of tuberculosis in Brazil. Rev Bras Epidemiol. 2012;15:468–77. doi: 10.1590/s1415-790x2012000300003. [DOI] [PubMed] [Google Scholar]
- 6.Balabram D, Turra CM, Gobbi H. Survival of patients with operable breast cancer (Stages I–III) at a Brazilian public hospital: a closer look into cause-specific mortality. BMC Cancer. 2013;13:434. doi: 10.1186/1471-2407-13-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliveira SB, Merchán-Hamann E, Amorim LDAF. HIV/AIDS coinfection with the hepatitis B and C viruses in Brazil. Cad Saúde Pública. 2014;30:433–8. doi: 10.1590/0102-311X00010413. [DOI] [PubMed] [Google Scholar]
- 8.Lynch J, Stuckler D. In God we trust, all others (must) bring data. Int J Epidemiol. 2012;41:1503–6. doi: 10.1093/ije/dys204. [DOI] [PubMed] [Google Scholar]
- 9.Goncalez TT, Sabino EC, Capuani L, Liu J, Wright DJ, Walsh JH, et al. Blood transfusion utilization and recipient survival at Hospital das Clinicas in Sao Paulo, Brazil. Transfusion. 2012;52:729–38. doi: 10.1111/j.1537-2995.2011.03387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabino EC, Ribeiro AL, Salemi VM, Di Lorenzo Oliveira C, Antunes AP, Menezes MM, et al. Ten-year incidence of Chagas cardiomyopathy among asymptomatic Trypanosoma cruzi-seropositive former blood donors. Circulation. 2013;127:1105–15. doi: 10.1161/CIRCULATIONAHA.112.123612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Busch M, Custer B. Health outcomes research using large donor-recipient databases: a new frontier for assessing transfusion safety and contributing to public health. Vox Sang. 2006;91:282–4. doi: 10.1111/j.1423-0410.2006.00847.x. [DOI] [PubMed] [Google Scholar]
- 12.van Hest NA, Smit F, Baars HW, De Vries G, De Haas PE, Westenend PJ, et al. Completeness of notification of tuberculosis in The Netherlands: how reliable is record-linkage and capture-recapture analysis? Epidemiol Infect. 2007;135:1021–9. doi: 10.1017/S0950268806007540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fournel I, Schwarzinger M, Binquet C, Benzenine E, Hill C, Quantin C. Contribution of record linkage to vital status determination in cancer patients. Stud Health Technol Inform. 2009;150:91–5. [PubMed] [Google Scholar]
- 14.Roberts SE, Williams JG, Meddings D, Goldacre MJ. Incidence and case fatality for acute pancreatitis in England: geographical variation, social deprivation, alcohol consumption and aetiology – a record linkage study. Aliment Pharmacol Ther. 2008;28:931–41. doi: 10.1111/j.1365-2036.2008.03809.x. [DOI] [PubMed] [Google Scholar]
- 15.Donati S, Senatore S, Ronconi A. Maternal mortality in Italy: a record-linkage study. BJOG. 2011;118:872–9. doi: 10.1111/j.1471-0528.2011.02916.x. [DOI] [PubMed] [Google Scholar]
- 16.World Health Organization. Assessing tuberculosis under-reporting through inventory studies. Geneva: World Health Organization; 2013. [Google Scholar]
- 17.Christen P, Goiser C. Quality and complexity measures for data linkage and deduplication. In: Guillet F, Hamilton HJ, editors. Quality measures in data mining. Berlin: Springer Berlin Heidelberg; 2007. pp. 127–51. [Google Scholar]
- 18.Hulley SB, Cummings SR, Browner WS, Grady DG, Newman TB. Delineando a pesquisa clínica: uma abordagem epidemiológica. 3a. Porto Alegre: Editora Artmed; 2008. [Google Scholar]
- 19.Camargo KR, Jr, Coeli CM. Reclink: aplicativo para o relacionamento de bases de dados, implementando o método probabilistic record linkage. Cad Saúde Pública. 2000;16:439–47. doi: 10.1590/s0102-311x2000000200014. [DOI] [PubMed] [Google Scholar]
- 20.Jaro MA. Probabilistic linkage of large public health data files. Stat Med. 1995;14:491–8. doi: 10.1002/sim.4780140510. [DOI] [PubMed] [Google Scholar]
- 21.Jorge MH, Laurenti R, Di Nubila HB. Death and its epidemiological investigation: considerations about some relevant aspects. Rev Bras Epidemiol. 2010;13:561–76. doi: 10.1590/s1415-790x2010000400002. [DOI] [PubMed] [Google Scholar]
- 22.Franca E, de Abreu DX, Rao C, Lopez AD. Evaluation of cause-of-death statistics for Brazil, 2002–2004. Int J Epidemiol. 2008;37:891–901. doi: 10.1093/ije/dyn121. [DOI] [PubMed] [Google Scholar]


