Abstract
In breast cancer, the most frequent site of metastasis is bone. Disseminated tumor cells (DTCs) can be detected in the bone marrow of patients by their expression of epithelial or oncogenic markers [1], and the presence and frequency of these DTCs are associated with poor prognosis. However, many of the details behind this process remain elusive, including the biological properties and fates of these apparently indolent cancer cells. To provide pre-clinical models of DTCs, we have developed a procedure that allows for controlled and enhanced delivery of tumor cells to the bone in animal experiments via injection into the iliac artery of the hind limb [2]. To our surprise, we found that most cancer cells became integrated into the solid bone matrix shortly after arriving in the bone, and only a minority can be flushed out with the bone marrow. Here we describe a method that helps to retrieve DTCs homing to the bone in which we achieve an improved recovery of those tumor cells closely associated with the bone microenvironment. In our view it is especially important to analyze these tumor cell subpopulations, as they may take full advantage of growth-, survival- and immune-protective signals provided by neighbor cells. We also show a pilot study on how this approach may be applied to the analysis of cancer dormancy. Our study suggests that the detection and retrieval of DTCs in clinical studies are incomplete because they are conducted exclusively with bone marrow aspirates.
Keywords: Breast cancer, Bone metastasis, Disseminated tumor cells, Circulating tumor cells, Collagenase type 2, Cancer dormancy
Introduction
Circulating tumor cells (CTCs) and disseminated tumor cells (DTCs) have been increasingly investigated in recent years because of their potential to reflect biological and pathological properties of otherwise intractable diseases [3, 4]. For instance, the quantity and certain characteristics of CTCs have been proposed as an index of how tumors respond to adjuvant therapies [5, 6], and the genetic/epigenetic profiles of CTCs are considered as a snapshot of residual disease [7].
The biological origin and destination of CTCs and DTCs remain controversial. They could be metastases in-transit, or alternatively, dead-end product of tumor evolution passively shed into the circulation [3, 8, 9]. Patients carrying CTCs and DTCs have a worse prognosis in most studies to date [10]. However, a significant proportion of CTC- and/or DTC-positive patients do not develop recurrences for years. Thus, it seems that the mere presence of these cells is only weakly associated with clinical outcomes, and is not predictive. Better qualitative characterization has become possible with recent development of single-cell technologies as well as improved enrichment approaches. Pioneering studies toward this direction have suggested that some properties of CTCs or DTCs may be critical for their roles in tumor progression. For instance, CTCs expressing mesenchymal markers are linked to tumor progression [5], and those expressing certain adhesion molecules and traveling in clusters tend to colonize more efficiently in distant organs [11]. These seemingly contradictory results require further mechanistic studies, which are hindered by lack of robust animal models and sensitive isolation protocols.
A key phenomenon related to DTC biology is cancer dormancy [12, 13]. Metastatic relapse often occurs after an extended, apparently disease-free period following primary tumor removal. In breast cancer, these dormancy periods can last many years. This phenomenon raises a number of questions about the role of disseminated tumor cells, and answering these questions could reveal a window of opportunity to therapeutically remove tumor cells while they are in an indolent stage. How do DTCs survive for such long times in the distant metastatic site? What keeps DTCs in the dormant state? What events eventually trigger metastatic progression? Recent studies emphasize the role of the tissue microenvironment in controlling DTC properties. However, the challenge lies in the detection and isolation of those rare, functionally significant DTCs which are viable, but non-dividing in the distant organ. As with CTCs, research on DTCs will benefit from suitable animal models, which allow isolation of an increased quantity of such DTCs that are in a viable cell condition, in order to facilitate the analysis of their molecular and functional properties.
Here, we present experimental protocols that we have invented to study DTCs and CTCs using mouse models (tested with similar efficacies in BALB/C and nude mouse strains). We also report some associated findings reflecting the biological properties of these cells, and suggest how this protocol could be applied to the research on cancer dormancy. We generated DTC models in the bone marrow using two approaches: spontaneous bone metastasis and intra-iliac artery (IIA) injection [2]. The latter directly delivers cancer cells into hind limb tissues through the artery circulation, thereby enriching DTCs by at least one order of magnitude. Using IIA injection we discovered that the microenvironment niche of early stage bone micrometastases is predominantly comprised of cells of the osteogenic lineage. Interestingly, cancer cells and osteogenic cells form very tight cellular junctions, which are functionally important in driving tumor progression. These junctions, on the other hand, also make it difficult to dissociate DTCs from the bone tissues and be isolated. For this reason, traditional bone-flushing procedures could only retrieve a proportion of DTCs that have not been engaged in interaction with osteogenic cells [2]. Therefore we developed a dissociation protocol that is more stringent, while at the same time keeps the extracted cells in a viable state.
Methods: Tumor Cell Isolation from Bone
Bone Metastasis Models
-
1.)
BALB/C and nude mouse strains were used in our study, although the procedure should also be applicable to other strains of mice. Mice were injected with tumor cells via intra-iliac artery injections thereby targeting tumor cells to the hind limb bones as described in [2]. Typically we injected 500,000 tumor cells/mouse in PBS. In our experiments we used tumor cells that were labeled with luciferase and green fluorescence protein (GFP) to have different options in tracking the tumor cells. Results shown here were obtained with the human breast cancer cell line MCF7 in nude mice, and with mouse breast cancer cell line 4 T1 in BALB/C mice, respectively.
-
2.)
At different time-points after tumor cell-injection that allow for bone colonization to occur – typically day 7 or day 14 post injection - animals are sacrificed and positioned on a sterile surface. Legs are sprayed with 70 % ethanol for sterilization. Small incisions are made in the skin on the backside of the hind limbs. Grabbing on both sides of the incision, the hind limbs are stripped of the skin. Exposed femur and tibia bones are collected by carefully removing muscles and tendons, including a final step of wiping bones with gauze or paper towel to clear residual adherent non-bone tissue. Bones are placed in PBS in a petri dish (petri dish 1).
Optional: To speed up the process when working with larger numbers of mice, residual tendon/muscle tissue can also be removed using the tissue homogenizer (Precellys 24, Bertin Technologies): Bones are placed in 2 ml-screwcap-tubes with ceramic beads (no liquid solution added) and treated with a 5 s pulse at lowest frequency (shaking speed: 5,000 rpm). Bones are immediately returned to petri dish 1 with PBS to prevent drying. Please see below (section: equipment needed) for a description of the tissue homogenizer. We use the same tissue homogenizer for grinding bones with steel beads in later steps 6–11.
Bone Marrow Aspirate Fraction (BM)
-
3.)
To obtain the bone marrow aspirate fraction, ends of bones are cut and put in second petri dish (petri dish 2) with PBS. A 27 ½G needle attached to 10 ml-syringe with 5 ml PBS is inserted in the remaining bone shafts and bone marrow is flushed out from bones into petri dish 1. After this step, bone shafts are combined with the bone ends in petri dish 2.
-
4.)
Pull plunger of syringe to force flushed-out bone marrow pieces present in petri dish 1 through the needle into the syringe, then push plunger to passage cells again through needle.
-
5.)
Pipette bone marrow cell suspension of petri dish 1 up and down with 10 ml-pipette ten times. Steps 3 and 4 will disperse the bone marrow into single cells.
These steps generate the first bone fraction, “bone marrow aspirate”.
Collagenase Type 2 Fraction (C2)
-
6.)
To grind bones in tissue homogenizer and liberate cells by collagenase type 2 treatment, place bone ends and bone-shafts from petri dish 2 into 2 ml-screw cap tube with five steel beads per tube. We typically process one femur and one tibia together in the same tube. However, when processing two femura and two tibiae together in one tube, results were similar in terms of cell viability and cell yield.
-
7.)
Add 600ul collagenase type 2 (final concentration: 1 mg/ml in DMEM/F12 with 10 %FBS and antibiotic/anti-mycotic, Cat.No. 17101–015, Life Technologies).
-
8.)
To grind bones, close screw caps tightly, place tubes in tissue homogenizer and apply 6-second pulse at a shaking speed of 5,500 rpm.
-
9.)
Remove supernatant medium (not the bone fragments) and place into 6 cm petri dish.
-
10.)
Add another 600ul collagenase type 2 to the bone fragments and place tubes back in tissue homogenizer.
-
11.)
Complete bone-grinding by 8-second pulse in tissue homogenizer at a shaking speed of 5,500 rpm.
-
7.)
12.) Briefly vortex and immediately pour medium including bone fragments into petri-dish combining with medium from step 9.
-
13.)
Add 1.2 ml collagenase type 2, so the total volume of collagenase type 2 per two to four bones is 3 ml.
-
14.)
Shake the bone fragments in collagenase type 2 solution at low speed on a horizontal orbital shaker, (typical shaker speed: 100 rpm) at 37 ° C for 1 h to 1h20min. Indicator of proper digestion is when bone-fragments form “mushy” aggregate.
-
15.)
Add DMEM/F12 medium and complete liberating cells by pipetting up and down ten times.
-
16.)
Filter all medium, not bones, through cell-strainer into 50 ml-conical tube.
-
17.)
Repeat steps 15 and 16 two more times.
-
18.)
After passage of cells through cell strainer, spin 50 ml-conical tubes of steps 15–17 at 1,200 rpm for 5 min. Remove supernatant from cell pellet. Add 20 ml collagenase-free DMEM/F12 medium to cell pellet, spin again. Repeat this wash step two more times to remove all collagenase type 2.
Optional EDTA/Trypsin Treatment
-
Opt. 19.)
As an option for further retrieval of cells tightly associated with bone tissue, the collagenase type 2-extracted bone fragments are treated with EDTA/Trypsin (e.g., 5 ml of 0.25 % trypsin, 5 mM EDTA for 30 min at 37 ° C) followed by pipetting up and down ten times, filtering through cell strainer and two washes with DMEM/F12 to remove EDTA/Trypsin.
Tests for Viable Tumor Cells in Cell Fractions
-
20.)
Cell fractions including the extracted bone fragments remaining after cell retrieval procedures are cultured in tumor-cell specific medium in 6 well plates. If luciferase-labeled tumor cells were used in the procedure, bioluminescence measurements using in-vivo-imaging-system (IVIS) are applied to determine the quantities of viable tumor cells in bone marrow aspirate, collagenase type 2 and trypsin/EDTA fractions and residual bone fragments, respectively. For this, let cells settle in 6 well plate for several hours, add luciferase substrate D-luciferin at recommended concentration (150ug/ml) and immediately image cells in IVIS machine.
-
21.)
If fluorescently labeled tumor cells were used, liberated tumor cells can be quantified via fluorescence-activated cell sorting (FACS), including 7-Amino-actinomycin D (7-AAD) dye to distinguish live and dead cells. FACS is also used to detect retrieved dormant tumor cells. In this method, dormant tumor cells will be distinguished as cells that retain high levels of fluorescence of the cell tracker dye carboxyfluorescein succinimidyl ester (final labeling concentration: 1uM, CFSE), or of GFP-histone H2B fusion protein (GFP-H2B), respectively. CFSE is a cell tracker dye that is gradually diluted in proliferating cells as it is distributed equally to the daughter cells in cell division, while it is maintained at high concentration in dormant, non-dividing cells. A similar principle underlies the use of GFP-H2B in these studies.
Notes
We have used this procedure with BALB/C and nude mouse strains. However, we believe it should be applicable to other mouse strains as well.
The above procedures will extract about 30–60 % of cancer cells injected into the bone through intra-iliac artery injection. 1–5 mM EDTA will significantly improve the yield. Some groups also reported that trypsin can further digest the bone and extract cells. Therefore, adding another trypsin/EDTA treatment step (e.g., 0.25 % trypsin with 1–5 mM EDTA at 37° for 0.5–1 h) after step 18 significantly improves the yield.
Steps 6–11 are intended to break bones into small pieces. We do it using the tissue homogenizer from Precellys (Precellys24). Many other groups simply cut bones using scalpels.
For FACS analysis in most cases up to 500,000 or 1,000, 000 events should be acquired to reliably detect scarce cancer cells. Optional: Red blood cells can be removed from samples by treatment with red blood cell lysis buffer (eBioscience) before analyzing the samples with FACS machine to further enrich for the cells of interest.
These procedures will also dissociate osteoblasts and osteocytes. If cancer cells have antibiotic resistance genes or fluorescence labels, they can be separated by drug selection or FACS.
A useful application of this protocol is in tracking the proliferation history of disseminated tumor cells. Therein, the tumor cells injected to the bones of mice carry a label that is specifically retained in dormant cells, and its intensity reduced in proliferating cells: carboxyfluorescein succinimidyl ester (CFSE) or green fluorescent protein (GFP)-modified histone H2B (H2B-GFP).
Equipment Needed
Basic mouse surgery and euthanization equipment
Gauze, sterilized
Petri dishes (6 and 10 cm diameter)
Conical polypropylene tubes (15, 50 ml)
10 ml-syringe with 27 ½G needle
-
Tissue homogenizer (Precellys 24, Bertin Technologies)
This tissue homogenizer operates with steel or ceramic beads. The tissue to be processed is placed in tubes – we use 2 ml-tubes for two to four hind limb bones – with several beads. Tubes are tightly closed using screw-caps and placed in homogenizer. Homogenizer induces shaking of tubes at high speed similar to a vortex. Thereby beads are accelerated and grind or homogenize tissue. Shaking-speed can be precisely controlled and type of beads can be chosen according to experimental needs.
-
Homogenizer accessories:
2 ml-screwcap tubes with ceramic beads (Precellys CK14, Cat.Nr. KT03961-1-003.2)
2 ml-screwcap tubes with steel beads (Precellys MK28, Cat.Nr. KT03961-1-001.2)
Low-speed horizontal orbital shaker at 37 ° C
Cell strainer (70um Nylon, Falcon Cat.Nr. 352350)
Bright-field inverted microscope
Contessa cell counter (Invitrogen)
6 well-cell culture plates
-
IVIS Lumina II (Advanced Molecular Vision)
For in vivo imaging of bioluminescence signal from luciferase-labeled cells
-
FACS machine, e.g., LSR Fortessa (BD Biosciences)
For detection of cells labeled with fluorescent markers
Reagents
70 % Ethanol
PBS
Collagenase type 2 (Cat.No. 17101–015, Life Technologies)
DMEM/F12
DMEM with high glucose
Fetal bovine serum (FBS)
Antibiotic/anti-mycotic
Trypsin/EDTA
Trypan blue solution for cell counting
D-luciferin, luciferase substrate (final concentration: 150ug/ml, LUCNA-1G, Goldbio)
CFSE cell tracker dye (final concentration: 1uM, Molecular Probes)
7-AAD to exclude dead cells in FACS analysis (5ul/test, BD Pharmingen)
Doxycycline (dose range: 50 ng/ml to 1ug/ml Doxycycline hyclate, Sigma)
Plasmids for Labeling Tumor Cells
Firefly luciferase fused to GFP was expressed in tumor cells using PWIPZ lentiviral vector system [2].
GFP-tagged histone H2B (H2B-GFP) was placed under control of doxycycline-inducible promoter. Same plasmid also contained RFP encoding sequence under control of constitutive promoter. Doxycycline induction was optimized for dose and duration to achieve high-level but transient expression of H2B-GFP.
Application: A Model for Cancer Dormancy in Bone
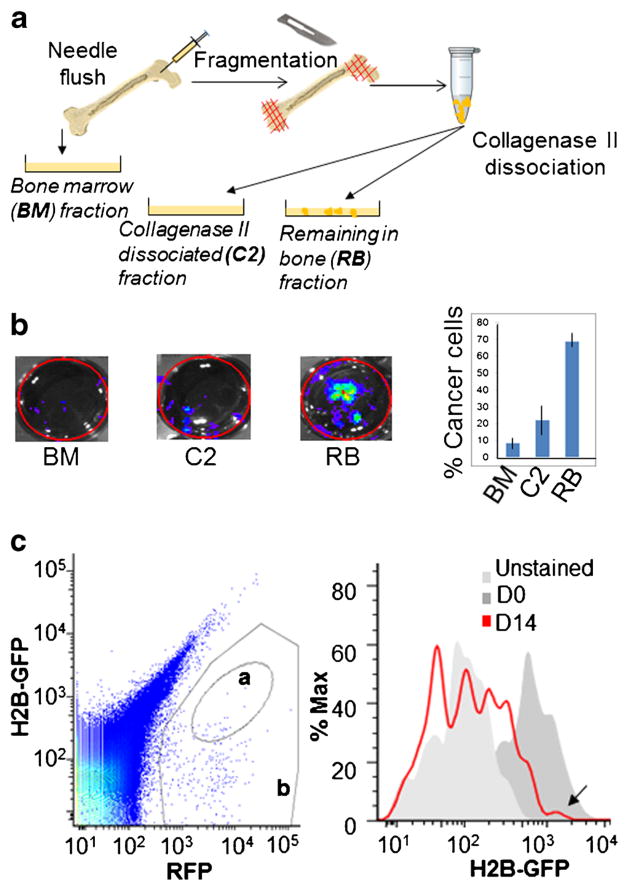
In experiments to optimize DTC collection from bones, MCF7 human breast cancer cells carrying a luciferase reporter plasmid were injected into nude mice using intra-iliac artery (IIA) technique to target delivery of tumor cells to hind limbs (500,000 tumor cells in 0.1 ml PBS injected per mouse). On day 0 and 7 and 14 days post injection, tumor cells residing in bones were isolated with different procedures, and recovery rates of viable tumor cells were assessed via bioluminescence measurement. Tissue homogenizer settings and enzymatic digestion conditions were adjusted to obtain highest tumor cell viability and release from bone tissue. Based on such pilot studies, we derived the experimental conditions described in Methods. Bioluminescence signals indicated that less than 10 % of tumor cells were present in the “flushed out bone marrow”, whereas an additional 20–50 % of cells could be liberated with collagenase type 2 treatment (Fig. 1). Further improved tumor cell recovery yields were obtained using EDTA/Trypsin (Note 2). Comparison of bioluminescence signals at different time-points suggested that after an initial drop of cancer cell numbers in the bone in the first few days post-injection, viable tumor cells in bones reached a steady-state level by d7.
Fig. 1.
Combination of mechanical disruption and collagenase type 2 treatment facilitates retrieval of DTCs from bone in cancer dormancy studies. a. Schematics of retrieval procedure. b. Luciferase-tagged MCF7 cells were delivered to bone via IIA injection, allowed to colonize for 7 days and retrieved from bone with procedure shown in A. Retrieved cells were cultured on 6 cm dish and the number of viable tumor cells in each extraction-fraction was assessed by bioluminescence measurement. c. MCF7 cells were tagged with a lentivirus that allows constitutive expression of RFP and doxycycline-inducible expression of GFP-Histone H2b fusion protein. Cells were treated in vitro with doxycycline to temporarily induce GFP-Histone H2b expression, and injected to bones of mice with IIA technique. On day 14 of bone colonization, tumor cells were isolated from bones with the technique shown in A and analyzed by FACS. In left panel, gate a denotes quiescent tumor cells (GFP-H2B+/RFP+), gate b denotes the total of tumor cells (RFP+). Right panel shows an overlay of fluorescence signal in GFP-H2B channel of MCF7 cells on day of injection to bones (D0) and upon isolation from bones on day 14 (D14). Arrow points to a cell population with same GFP-H2B fluorescence intensity on day 14 as on day 0
In order to track dormant tumor cell fraction, MCF7 cells were tagged with a construct that allows for constitutive expression of red fluorescent protein (RFP) and temporarily inducible (via doxycycline regulated promoter elements) expression of GFP-tagged histone H2B (H2B-GFP). Cells induced to express H2B-GFP were injected into hind limbs via IIA. Bone colonization step was allowed to proceed for 14 days. Then tumor cells were retrieved from bones using the collagenase type 2 extraction procedure. Extracted cells were analyzed by FACS and were shown to contain a subset with a similar H2B-GFP level as on day of injection (d0) indicating that these are a viable, dormant tumor cell subpopulation (Fig. 1c).
Conclusions
Our observations indicate that compared to CTCs, the sampling of DTCs faces an additional hurdle, as tumor cells become tightly associated with the bone tissue as colonization progresses. The difficulty to retrieve a representative spectrum of tumor cell heterogeneity in experimental studies on DTCs could result in missing cells that are biologically relevant in cancer progression, and we believe that the rigorous enzymatic and mechanical disruption steps presented in our method are necessary and will aid in isolating DTCs from bone. Taken together, we hope our protocols could contribute to a wider range of studies and provide unique insights into the interaction between DTC/CTCs and host tissues.
Acknowledgments
We would like to thank Aaron Muscarella for helpful suggestions. This project was supported by the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the NIH (P30 AI036211, P30 CA125123, and S10 RR024574) and the expert assistance of Joel M. Sederstrom. X. H.-F. Z. is supported by NCI CA183878, Breast Cancer Research Foundation, US Department of Defense DAMD W81XWH-13-1-0195, Susan G. Komen CCR14298445, and McNair Medical Institute. The authors acknowledge the joint participation by Diana Helis Henry Medical Research Foundation through its direct engagement in the continuous active conduct of medical research in conjunction with Baylor College of Medicine and its “Preclinical Modeling of Metastasis and Therapy Response of TNBC” Cancer Program.
Abbreviations
- CTC
Circulating tumor cells
- DTC
Disseminated tumor cells
- IIA
Intra-iliac artery
References
- 1.Braun S, Vogl FD, Naume B, Janni W, Osborne MP, Coombes RC, et al. A pooled analysis of bone marrow micrometastasis in breast cancer. N Engl J Med. 2005;353(8):793–802. doi: 10.1056/NEJMoa050434. [DOI] [PubMed] [Google Scholar]
- 2.Wang H, Yu C, Gao X, Welte T, Muscarella AM, Tian L, et al. The osteogenic niche promotes early-stage bone colonization of disseminated breast cancer cells. Cancer Cell. 2015;27(2):193–210. doi: 10.1016/j.ccell.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pantel K, Speicher MR. The biology of circulating tumor cells. Oncogene. 2015 doi: 10.1038/onc.2015.192. [DOI] [PubMed] [Google Scholar]
- 4.Pantel K, Alix-Panabieres C, Riethdorf S. Cancer micrometastases. Nat Rev Clin Oncol. 2009;6(6):339–51. doi: 10.1038/nrclinonc.2009.44. [DOI] [PubMed] [Google Scholar]
- 5.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339(6119):580–4. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smerage JB, Barlow WE, Hortobagyi GN, Winer EP, Leyland-Jones B, Srkalovic G, et al. Circulating tumor cells and response to chemotherapy in metastatic breast cancer: SWOG S0500. J Clin Oncol. 2014;32(31):3483–9. doi: 10.1200/JCO.2014.56.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polzer B, Medoro G, Pasch S, Fontana F, Zorzino L, Pestka A, et al. Molecular profiling of single circulating tumor cells with diagnostic intention. EMBO Mol Med. 2014;6(11):1371–86. doi: 10.15252/emmm.201404033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehes G, Witt A, Kubista E, Ambros PF. Circulating breast cancer cells are frequently apoptotic. Am J Pathol. 2001;159(1):17–20. doi: 10.1016/S0002-9440(10)61667-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W, Vivian CJ, Brinker AE, Hampton KR, Lianidou E, Welch DR. Microenvironmental influences on metastasis suppressor expression and function during a metastatic Cell’s journey. Cancer Microenviron. 2014;7(3):117–31. doi: 10.1007/s12307-014-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bidard FC, Peeters DJ, Fehm T, Nole F, Gisbert-Criado R, Mavroudis D, et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol. 2014;15(4):406–14. doi: 10.1016/S1470-2045(14)70069-5. [DOI] [PubMed] [Google Scholar]
- 11.Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158(5):1110–22. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quayle L, Ottewell PD, Holen I. Bone metastasis: molecular mechanisms implicated in tumour cell dormancy in breast and prostate cancer. Curr Cancer Drug Targets. 2015;15(6):469–80. doi: 10.2174/1568009615666150506092443. [DOI] [PubMed] [Google Scholar]
- 13.Pantel K, Alix-Panabieres C. Bone marrow as a reservoir for disseminated tumor cells: a special source for liquid biopsy in cancer patients. Bonekey Rep. 2014;3:584. doi: 10.1038/bonekey.2014.79. [DOI] [PMC free article] [PubMed] [Google Scholar]