Abstract
A dedicated network of cellular factors ensures that proteins translocated into the endoplasmic reticulum (ER) are folded correctly before they exit this compartment en route to other cellular destinations or for secretion. When proteins misfold, selective ER-resident enzymes and chaperones are recruited to rectify the protein-misfolding problem in order to maintain cellular proteostasis. However, when a protein becomes terminally misfolded, it is ejected into the cytosol and degraded by the proteasome via a pathway called ER-associated degradation (ERAD). Strikingly, toxins and viruses can hijack elements of the ERAD pathway to access the host cytosol and cause infection. This review focuses on emerging data illuminating the molecular mechanisms by which these toxic agents co-opt the ER-to-cytosol translocation process to cause disease.
Keywords: bacterial toxin, chaperone, ERAD, infection, membrane transport, polyomavirus, cholera toxin, ubiquitin-proteasome system
Introduction
Central to maintaining cellular proteostasis is the assurance that proteins fold accurately, enabling them to execute their dedicated functions. This is most relevant in the endoplasmic reticulum (ER) where approximately 30% of all cellular proteins fold and mature. When a nascent protein is translocated from the cytosol into the ER, a network of ER-resident factors ensures that the client attains its native three-dimensional conformation and/or assembly state. When this is achieved, the client is transported to other cellular organelles or for secretion. However, despite the presence of enzymes and chaperones that promote folding and assembly, nascent proteins often misfold or misassemble due to either genetic lesions, environmental conditions, or simply the high volume of clients undergoing folding (Buchberger et al., 2010). To rectify this situation, a cohort of ER-resident enzymes and chaperones attempt to re-direct the misfolded protein along the productive folding path (Hartl et al., 2011). When these efforts fail, the terminally misfolded ER protein is triaged in the cytosol using a pathway called ER-associated degradation (ERAD).
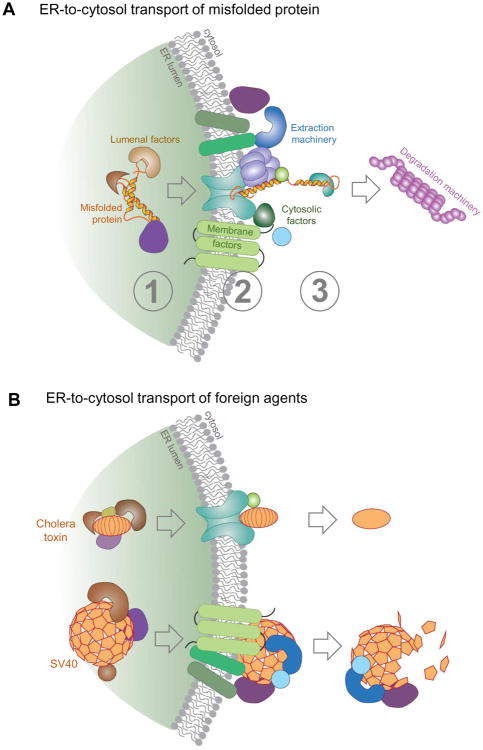
ERAD is an ER protein quality control pathway that identifies and retro-translocates a misfolded (or misassembled) ER protein to the cytosol for ubiquitin-dependent proteasomal degradation (Olzmann et al., 2013, Ruggiano et al., 2014, Hampton and Garza, 2009). This pathway is vital for maintaining cellular integrity as impairment of the ERAD system has been linked to more than 60 human diseases, including neurological illnesses such as Alzheimer's and Parkinson's diseases, cystic fibrosis, diabetes, and cancer (Guerriero and Brodsky, 2012). Intense efforts over the past two decades have unraveled a plethora of ERAD components that coordinately propel a misfolded substrate from the ER into the cytosol (Olzmann et al., 2013) (Table 1). This process can be generally divided into three distinct steps. First, ER lumenal factors recognize and target the misfolded substrate to the ERAD membrane complex (Figure 1A, step 1). Second, the substrate is physically transported across the ER membrane by traversing a retro-translocon to reach the cytosol where it is ubiquitinated (Figure 1A, step 2). In the final third step, cytosolic factors that bind to the retro-translocon engage the ubiquitinated substrate, ejecting it into the cytosol so that it can be delivered to the proteasome for degradation (Figure 1A, step 3).
Table 1. ERAD factors that recognize and extract toxins/viruses from the ER into the cytosol.
| Protein family | Factors (synonyms) | Cellular function | Bacterial toxin | Polyomavirus |
|---|---|---|---|---|
| ER lumenal | ||||
|
| ||||
| Hsp70 | BiP (Grp78, HspA5) | Lumenal Hsp70 | Masks hydrophobic region, stabilizes toxin | Masks hydrophobic region |
| Colocalizes in virus-induced foci | ||||
|
| ||||
| NEF | Sil1 (BAP) | HspBP1-like NEF, releases BiP from substrates | Releases BiP from toxin | - |
| Grp 170 (Hyou1) | Hsp 110 family NEF, releases BiP from substrates | Releases BiP from toxin | Releases BiP from SV40, Colocalizes in virus-induced foci | |
|
| ||||
| Redox enzymes | PDI | Oxidizes disulfide bonds, acts as a chaperone | Unfolds CTA1 | Remodels VP1 |
| ERdj5 (DnaC10) | Reduces disulfide bonds, also acts as a J-protein | Stimulates BiP to capture CT | Reduces disulfide bonds | |
| ERp57 (Grp58) | Acts as an isomerase | - | Isomerizes disulfide bonds | |
| ERp29 | Acts as a chaperone | - | Remodels VP1 C-terminal arm | |
| Ero1 | A PDI oxidase | Releases unfolded CTA1 from PDI | - | |
|
| ||||
| J-proteins | ERdj5 (DnaC10) | Reduces disulfide bonds and acts as a J-protein | Stimulates BiP to capture CT | Reduces disulfide bonds |
| ERdj2 (DnaC23) | Acts as a J-protein | - | - | |
| ERdj3 (DnaJC1) | Acts as a J-protein | Facilitates retro-translocation of CTA1 and Stx | Recruits BiP to SV40 | |
|
| ||||
| AAA+ ATPase | Torsin A (TORIA) | - | Facilitates CTA1 retro-translocation | - |
|
| ||||
| ER membrane | ||||
|
| ||||
| Hsp40 (type II) | DnaJB14 | Role in ERAD | - | Recruit and anchor cytosolic Hsc70 and co-chaperones to ER membrane |
| DnaJB12 | Role in ERAD | - | ||
| DnaJC 18 | No clear cellular function | - | ||
|
| ||||
| Bap family | Bap29 (BCAP29) | Probable role in transport of membrane proteins | - | Colocalizes in virus-induced foci |
| Bap31 (BCAP31) | Role in ERAD and export of secretory proteins | - | Interacts with VP2 and colocalizes in foci | |
|
| ||||
| Membrane translocon | Derlin-1 | Component of the ERAD membrane complex | Transfers CTA1 from PDI to Hrd1 | Likely confer stability to PyV within the ER membrane |
| Derlin-2 | Component of the ERAD membrane complex | Mediates retrotranslocation of CTD | ||
|
| ||||
| Hrd1 (SYVN1) | Component of the ERAD with E3 ligase activity | Component of the ERAD with E3 ligase activity | - | |
| Sel 1L | Adaptor protein of Hrd1 | Facilitates capture of unfolded CTA1 | - | |
|
| ||||
| Yod1 | Deubiquintinase involved in ERAD | Negatively regulates CTA1 retrotranslocation | - | |
|
| ||||
| Sec61 | Entry of cytosolic nascent polypeptides into ER | Postulated channel for CTA1 and Stx | - | |
|
| ||||
| Cytosol | ||||
|
| ||||
| Hsp70 | Hsp70 (HSPA1) | ATPase, chaperone in protein folding | - | - |
| Hsc70 (HSPA8) | Constitutively expressed Hsp70 | - | ER-cytosol translocation and disassembly | |
|
| ||||
| Hsp90 | - | Assists protein folding and stabilization | Putative role in toxin intoxication | - |
|
| ||||
| NEF (Hsp110) | Hsp105 (HspH1) | NEF, protein folding, disaggregase | - | NEF, component of cytosolic extraction machinery |
| Apg2 (HspH2) | NEF, protein folding, disaggregase | - | ||
| Apg1 (HspH3) | NEF, protein folding, disaggregase | - | ||
|
| ||||
| SGTA | - | Involved in tail-anchor protein biosynthesis | - | Part of cytosolic extraction machinery |
|
| ||||
| AAA+ ATPase | p97 (VCP1) | Extracts misfolded ER proteins into the cytosol | Involved in cytosol arrival of CDT, not CTA1 | - |
Figure 1. ER-associated degradation.
A. ER-to-cytosol transport of misfolded protein. ERAD can be broadly divided into three distinct steps. Step 1: ER lumenal factors recognize and target the misfolded substrate to the ERAD membrane complex. Step 2: The substrate is physically transported across the ER membrane by traversing a retro-translocon to reach the cytosol where it is ubiquitinated. Step 3: Specific cytosolic factors anchored to the retro-translocon engage the ubiquitinated substrate, ejecting it into the cytosol which is then delivered to the proteasome for degradation. B. ER-to-cytosol transport of foreign agents. Foreign agents such as a bacterial toxin (cholera toxin) and a virus (SV40) can hijack the existing ER-to-cytosol gateway to access the cytosol during infection.
Provided that pathogens succeed in reaching the ER from the cell surface, the existence of a gateway from the ER into the cytosol for misfolded proteins creates the intriguing opportunity that such a pathway may be usurped by pathogens to access the host cytosol. Whether the cell surface-to-ER transport pathway exists constitutively in cells (Geiger et al., 2013) or is induced by the invading pathogen (Romer et al., 2007, Ewers et al., 2010) remains unclear, and is a topic beyond the scope of this review. However, what is clear is that selective toxins and viruses are able to penetrate the ER membrane to enter the host cytosol as part of their infection routes (Figure 1B). In this review, we detail the ER-to-cytosol translocation mechanism experienced by a bacterial toxin and a non-enveloped virus during entry.
Bacterial toxin
To intoxicate host cells, certain toxins including the bacterial toxins cholera toxin (CT) (Wernick et al., 2010) and shiga toxin (Stx) (Spooner and Lord, 2012), as well as the plant toxin ricin (Spooner and Lord, 2015), co-opt the ERAD machinery to retro-translocate into the cytosol. We will primarily focus on CT's retro-translocation mechanism because it is by far the best-studied bacterial toxin. We will also compare the retro-translocation mechanism of CT to that of other toxins and endogenous misfolded clients, with the objective of revealing common principles.
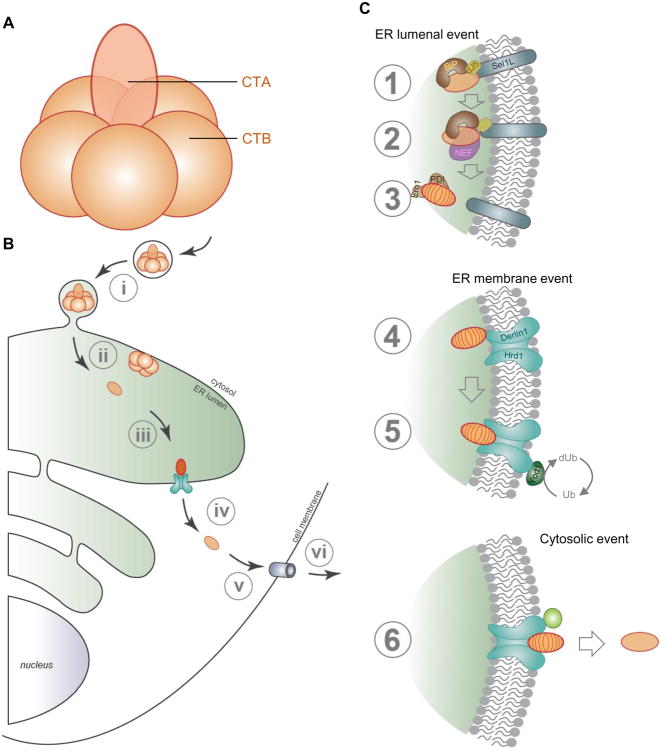
CT produced by the bacterium Vibrio cholerae is the virulence factor responsible for cholera, a disease characterized by massive secretory diarrhea that remains a global health threat (Spangler, 1992). The CT holotoxin consists of a toxic A (CTA) subunit associated with the pentameric receptor-binding B (CTB) subunit (Figure 2A). Proteolytic cleavage of CTA, which can occur during host entry (Lencer et al., 1997), generates the catalytic A1 (CTA1) and A2 (CTA2) subunits that remain attached by a disulfide bond. To enter cells, CTB binds to the GM1 ganglioside receptor on the intestinal epithelial cell surface. The toxin-receptor complex then undergoes retrograde transport to the Golgi apparatus, from where it reaches the ER (Figure 2B, step i-iii) (Fujinaga et al., 2003, Wernick et al., 2010, Cho et al., 2012). Once in the ER, CTA's disulfide bond is reduced to liberate the toxic CTA1 chain (Orlandi, 1997). CTA1 in turn masquerades as a misfolded protein, co-opting the ERAD machinery to reach the cytosol (Figure 2B, step iv) (Hazes and Read, 1997). In the cytosol, this toxic peptide escapes proteasomal degradation and instead triggers a G protein-dependent signaling cascade that leads to loss of intracellular chloride ion followed by massive water secretion (Figure 2B, steps v and vi), causing the cholera disease (Muanprasat and Chatsudthipong, 2013). Arguably the least understood step in CT's host entry pathway is how the toxin retro-translocates across the ER membrane to access the cytosol.
Figure 2. Cholera toxin co-opts the ERAD machinery to retro-translocate into the cytosol.
A. Structure of cholera toxin. The holotoxin consists of a toxic A (CTA) subunit associated with the receptor-binding B homo-pentameric (CTB) subunit. B. Cholera toxin intoxication pathway. CT intoxicates cells by binding to host surface GM1 receptor, which delivers the holotoxin to the ER by retrograde transport (step i). In the ER, CTA's disulfide bond is reduced (step ii) to liberate the toxic CTA1 chain (step iii). By disguising as a misfolded protein, CTA1 hijacks ERAD machinery and reaches the cytosol (step iv), where it triggers a host-signaling cascade (step v) to activate the chloride channel to cause disease (step vi). C. Detailed overview of CT ER-to-cytosol transport. Step 1: Once in the ER, CT is captured by BiP when the J-protein ERdj5 stimulates BiP's ATPase activity. This reaction occurs proximal to the ERAD machinery because ERdj5 is anchored to the ER membrane by Sel1L. Step 2: To release toxin from BiP, ER-resident NEFs (Grp170 and/or Sil1) induces nucleotide exchange of BiP, converting BiP from a high to low substrate-binding affinity state. Step 3: Upon release, CTA is subsequently handed off to the redox chaperones (PDI-Ero1), which unfold the toxin to prime it for retro-translocation. Step 4: Unfolded CTA1 is delivered to Derlin-1 that is part of the proposed Hrd1 membrane translocon complex. Step 5: The toxin navigates through the retro-translocon in a process regulated by the ubiquitin ligase activity of Hrd1 and the cytosolic deubiquitinase activity of YOD1. Step 6: Upon arrival at the cytosol, CTA1 refolds to an active conformation, triggering a downstream signaling pathway that causes disease.
ER lumenal events
Similar to retro-translocation of misfolded cellular clients, CT must first be identified as a “misfolded” substrate and targeted to the ERAD membrane machinery to cross the ER membrane. This targeting phase for CT requires the BiP ATPase along with its co-chaperones. BiP is the ER-resident Hsp70 that normally binds to hydrophobic moieties displayed in a misfolded ER substrate, preventing it from aggregation (Gething, 1999, Behnke et al., 2015). This reaction preserves the solubility of various cellular misfolded substrates prior to retro-translocation (Nishikawa et al., 2001, Ushioda et al., 2013). The ability of BiP to interact with a substrate depends strictly on its ATP/ADP bound states: ATP-BiP displays a low affinity for substrates, while ADP-BiP exhibits a high substrate-binding affinity. These two opposing states are in turn regulated by co-chaperones of BiP including J-proteins (Kampinga and Craig, 2010) and nucleotide exchange factors (NEFs) (Weitzmann et al., 2007, Andreasson et al., 2010, Yan et al., 2011, Hale et al., 2010). A J-protein harbors a J-domain that stimulates BiP's ATPase activity, converting ATP-BiP to ADP-BiP, which binds tightly to a substrate. In contrast, a NEF exchanges ADP for ATP in ADP-BiP to form ATP-BiP, which releases the substrate.
When CT reaches the ER from the cell surface, BiP captures the toxin, maintaining it in a soluble, retro-translocation-competent state (Winkeler et al., 2003). Although this finding suggests that CT possesses structural features mimicking a misfolded protein, the precise recognition signal remains unclear (Teter et al., 2006). As Stx (Falguieres and Johannes, 2006) and the viral A/B K28 toxin (Heiligenstein et al., 2006) also rely on BiP during their ER-to-cytosol membrane transport, a common structural motif or conformation may be present in these toxins that triggers BiP recruitment. Because BiP is located throughout the entire ER, it is enigmatic how BiP-bound CT is selectively targeted to the retro-translocation site on the ER membrane. Our group provided a potential solution to this question by demonstrating that ERdj5, a specific ER-resident J-protein, localizes to the ERAD membrane machinery by interacting with an adapter of this machinery called Sel1L (Williams et al., 2013) (Figure 2C, step 1). This juxtaposition enables ERdj5 to stimulate BiP to capture CT via activating BiP's ATPase activity, thereby recruiting CT to the retro-translocation site. While ERdj5 also plays a role during ERAD of cellular clients (Ushioda et al., 2013, Ushioda et al., 2008, Hagiwara et al., 2011), it instead acts as a reductase in these instances, reducing non-native disulfide bonds in the clients by using the catalytic cysteines within its thioredoxin domains. Interestingly, ERdj5 does not reduce CTA's disulfide bond (Williams et al., 2013), implicating other reductases (see below) in breaking CTA's lone disulfide bond. Another J-protein called ERdj3 has similarly been implicated in facilitating retro-translocation of CTA1 (Massey et al., 2011) as well as Stx (Yu and Haslam, 2005). However, whether ERdj3 is positioned proximally to the ERAD membrane machinery remains unknown. As ERdj5 and ERdj3 are reported to facilitate CTA1 retro-translocation, further investigations are required to determine if these two J-proteins exert non-overlapping or redundant functions during this translocation process. While another J-protein called ERdj4 binds to the ERAD membrane component Derlin-1 (Lai et al., 2012) and can facilitate retro-translocation of cellular ERAD substrates (Buck et al., 2010, Dong et al., 2008), there is no evidence that this J-protein play any function during CT retro-translocation.
Once BiP captures CT in an ERdj5-dependent manner, the toxin must be released from BiP so that it can be processed and primed for translocation to the cytosol. Indeed, both ER-resident NEFs Grp170 and Sil1 were recently reported to promote toxin release from BiP by stimulating nucleotide exchange on BiP (Williams et al., 2015) (Figure 2C, step 2). In this reaction, Grp170 and Sil1 appear to function interchangeably as depletion of one NEF is compensated by the other (Williams et al., 2015). It is tempting to speculate that these NEFs may also be localized to the ERAD machinery to strategically induce substrate release at the retro-translocation site.
Upon release from BiP, CT is subsequently handed off to the protein disulfide isomerase (PDI)-ER oxidoreductin 1 (Ero1) redox chaperone system that is crucial for further processing of CTA (Figure 2C, step 3) (Tsai et al., 2001, Forster et al., 2006, Moore et al., 2010, Wernick et al., 2010). First, its disulfide bond is reduced to generate the catalytic CTA1 peptide. To date, the identity of the ER-resident reductase responsible for breaking CTA's disulfide bond remains controversial (Orlandi, 1997, Majoul et al., 1997). Once generated, CTA1 is bound and unfolded by the reduced form of PDI unfoldase (Tsai et al., 2001). Subsequent oxidation of PDI by Ero1 alters PDI's conformation, releasing the unfolded CTA1 peptide (Tsai and Rapoport, 2002, Moore et al., 2010, Wernick et al., 2010). While another model posits that PDI displaces CTA1 from the holotoxin enabling CTA1 to “spontaneously” unfold (Taylor et al., 2014), this model is difficult to reconcile with the observation that PDI can unfold the isolated CTA1 chain in the absence of the holotoxin (Tsai et al., 2001). Although toxin unfolding precedes its translocation into the cytosol, substrate unfolding may not be a general prerequisite during retro-translocation (Tirosh et al., 2003). Regardless, once generated, the unfolded CTA1 peptide is poised for translocation across the retro-translocon.
ER membrane events
In yeast, two recent reports provide compelling evidence that the retro-translocon is composed of the multi-transmembrane proteins Der1p (Derlin-1/2 in mammals) (Mehnert et al., 2014) and the Hrd1p E3 ubiquitin ligase (also Hrd1 in mammals (Stein et al., 2014). In this model, a misfolded substrate in the ER lumen first engages Der1/Derlin-1, which is then delivered to Hrd1. Hrd1 likely serves as the channel, threading the misfolded client across the ER membrane, although additional studies are required to conclusively demonstrate that Hrd1 is sufficient to act as the retro-translocation channel. Regardless, when the substrate emerges into the cytosol, it becomes ubiquitinated by the Hrd1 ligase domain located in the cytosolic side. Importantly, CTA1 uses the mammalian Derlin-1/Hrd1 complex (Bernardi et al., 2008, Bernardi et al., 2010). A separate finding however suggests that Derlin-1 and the related Derlin-2 are not required in this process (Saslowsky et al., 2010), suggesting that Derlins may simply enable substrate delivery to Hrd1 more efficiently. Thus, we propose that once the unfolded CTA1 peptide is released from PDI, it is transferred to Derlin-1 which then passes the toxin to Hrd1 (Figure 2C, step 4); direct recruitment of CTA1 to Hrd1 may be possible, but is likely less efficient. In contrast to CTA1, the cytolethal distending toxin (CDT) relies on the Derlin-2/Hrd1 complex to gain access into the cytosol during entry, with two distinct Derlin-2 domains serving as critical determinants in guiding the toxin across the ER membrane (Eshraghi et al., 2014). These findings suggest that distinct toxins selectively interact with different cellular factors during ER-to-cytosol transport. In addition, another E3 ligase gp78 is also found to be associated with CT and PDI in a Derlin-1-dependent manner (Bernardi et al., 2010), but further investigations are needed to fully understand its functional relationship to Hrd1.
Strikingly, while CTA1 is not ubiquitinated (Rodighiero et al., 2002, Bernardi et al., 2013), its retro-translocation requires an intact Hrd1 ubiquitin ligase activity (Bernardi et al., 2010). This finding challenges a central assumption in ERAD, which postulates that Hrd1 directly ubiquitinates the misfolded client to promote its extraction into the cytosol. As the toxin is not ubiquitinated, we hypothesize that Hrd1 must ubiquitinate a trans factor in order to regulate translocation of the toxin into the cytosol (Figure 2C, step 5). This idea has been suggested previously in the case of T cell receptor α, a lysine-less ERAD membrane substrate, where expression of a dominant-negative ubiquitin mutant was still able to impair retro-translocation of this mutant receptor (Yu and Kopito, 1999) (Lysine residues in clients are the major ubiquitination sites). Thus Hrd1 not only serves as the physical channel promoting retro-translocation, but its enzymatic activity is also crucial in controlling toxin transport into the cytosol. Consistent with this idea, a deubiquitinase called YOD1 binds to Hrd1 and negatively regulates CTA1 retro-translocation (Bernardi et al., 2013). Ultimately, how ubiquitination and deubiquitination of a trans factor control cytosolic arrival of CTA1 remains mysterious. In one scenario, Hrd1 might auto-ubiquitinate itself to regulate its channel activity, as has been suggested (Bays et al., 2001, Stein et al., 2014, Carvalho et al., 2010). Alternatively, Hrd1 auto-ubiquitination may recruit additional cellular components that influence Hrd1's channel function. In both of these scenarios, YOD1 would deubiquitinate Hrd1. Hrd1's catalytic activity is also required during cytosolic entry of the Escherichia coli Shiga-like toxin 1 (Li et al., 2012). Thus there may be a conserved mechanism by which Hrd1-dependent ubiquitination generally regulates toxin retro-translocation. It should be noted that Sec61, the ER membrane channel that supports translocation of nascent polypeptide chains from the cytosol into the ER (Park and Rapoport, 2012), was initially implicated as the retro-translocon for CT (Schmitz et al., 2000) and Stx (Yu and Haslam, 2005), but this channel's role in guiding toxin retro-translocation has not since been confirmed or clarified.
Cytosolic events
Once CTA1 emerges from the cytosolic surface of the ER membrane, it must be released into the cytosol. In principle, this release step can result from “pushing” of the toxin from the ER lumenal side, “pulling” of the toxin from the cytosolic side, or both. In this context, an ER lumenal AAA+ ATPase called torsin A was found to facilitate CTA1 retro-translocation (Nery et al., 2011). Although the precise mechanism by which torsin A promotes toxin membrane translocation is unclear, it might provide the motor to push the toxin into the cytosol.
On the cytosolic side, the AAA+ ATPase called p97 along with its cofactors Ufd1 and Npl4 is considered the primary engine pulling canonical ERAD substrates into the cytosol (Ye et al., 2001). However, p97 does not play a significant role, if any, in mediating CTA1 extraction into the cytosol (Kothe et al., 2005, Abujarour et al., 2005, Moore et al., 2013). And because p97 normally couples substrate extraction with delivery to the proteasome for degradation, the minimal use of this chaperone by CTA1 may explain how it escapes degradation upon reaching the cytosol. Conversely, p97 was implicated in cytosolic arrival of CDT (Eshraghi et al., 2014), raising the question of how this toxin evades proteasomal degradation.
What factor then might extract CTA1 into the cytosol? Although it has been claimed that Hsp90 is a candidate for the extraction reaction (Taylor et al., 2010, Burress et al., 2014), an unbiased fractionation approach suggested that Hsp90 does not mediate this step (Moore et al., 2013). Instead, a GTP-dependent activity might provide the energy to dislocate the toxin from the ER membrane (Moore et al., 2013). Because the Ran GTPase is implicated in the ERAD process (Zhong et al., 2011), it is important to evaluate whether Ran participates in CTA1 retro-translocation. In an entirely different model, CTA1's inherent ability to spontaneously refold was postulated to prevent toxin backsliding, thereby favoring its forward movement into the cytosol (Rodighiero et al., 2002). As the proteasome typically degrades unfolded substrates (Finley, 2009), CTA1's propensity for spontaneous refolding may enable it to escape degradation. Thus, based on the available data, we propose that CTA1 uses its refolding capacity coupled with the energy provided by a cytosolic p97-independent factor to eject into the cytosol from the ER membrane (Figure 2C, step 6). Identifying this cytosolic factor requires additional experimentation.
Polyomavirus
Another well-studied toxic agent that translocates across the ER membrane to reach the cytosol as part of its infection pathway is the non-enveloped polyomavirus (PyV) (Figure 1B) (Kartenbeck et al., 1989, Tsai et al., 2003, Qian et al., 2009). This virus family includes the human BK PyV that causes Py-associated nephropathy and hemorrhagic cystitis (Bennett et al., 2012), and JC PyV responsible for progressive multifocal leukoencephalopathy (Maginnis et al., 2014). Strikingly within the past nine years, ten new human Pys have been discovered (DeCaprio and Garcea, 2013, White et al., 2013). Of these, the Merkel cell PyV has already been implicated as the causative agent of an aggressive skin cancer called Merkel cell carcinoma (Feng et al., 2008). The archetype PyV is the simian virus 40 (SV40), which has historically been used to study PyV entry due to its genetic and structural similarity to human PyVs.
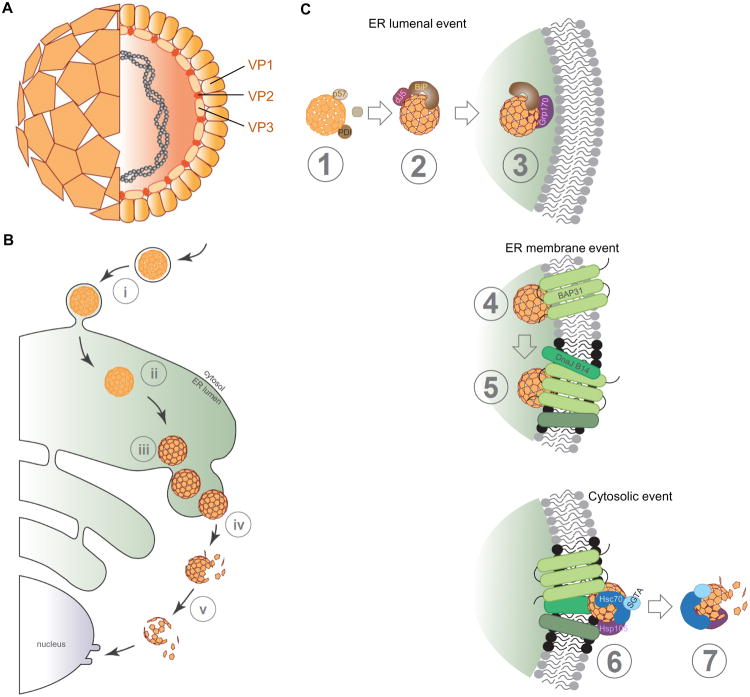
Structurally, SV40 consists of 72 homo-pentamers formed by the coat protein VP1, with each pentamer encasing either a VP2 or VP3 internal protein (Figure 3A) (Liddington et al., 1991, Stehle et al., 1994). VP2 and VP3 are hydrophobic viral components hidden in the native virion (Chen et al., 1998) that execute important roles during many stages of the host entry process. The VP1 pentameric shell in turn encloses its closed-circular double-stranded DNA genome. Of the 72 pentamers, 12 are five-coordinated while 60 are six-coordinated (Liddington et al., 1991). Individual VP1 within a pentamer inserts its C-terminal arm to a neighboring pentamer to stabilize the overall viral architecture (Liddington et al., 1991, Stehle et al., 1994), and a complex network of disulfide bonds along with virus-bound calcium ions further reinforce the viral structure (Stehle and Harrison, 1996, Stehle and Harrison, 1997). When properly assembled, SV40 displays a near-spherical geometry with an approximate diameter of 45 nm. To infect cells, Py binds to ganglioside receptors on the plasma membrane (Tsai et al., 2003, Low et al., 2006, Ravindran et al., 2013). After internalization, the virus-receptor complex is first transferred to the endolysosomes (Eash et al., 2004, Querbes et al., 2006, Engel et al., 2011) and then to the ER via a lipid-dependent sorting mechanism (Qian et al., 2009, Gilbert and Benjamin, 2004). Once the virus reaches this organelle, it co-opts elements of the ERAD machinery to penetrate the ER membrane and access the cytosol (Figure 3B, step i-v). In contrast to CT, SV40's voyage does not finish in the cytosol but instead continues into the nucleus where ensuing transcription and replication of the viral genome cause lytic infection or cellular transformation. ER-to-cytosol membrane transport remains SV40's most enigmatic entry step, although recent reports have begun to unravel some of this step's mysteries.
Figure 3. The non-enveloped virus SV40 co-opts elements of the ERAD machinery to translocate into the cytosol.
A. Structure of SV40. SV40 consists of 72 homo-pentamers formed by the coat proteins VP1, with each pentamer encasing either a VP2 or VP3 internal protein (see cross-section). Within this proteinaceous shell, the viral circular double-stranded DNA genome is enclosed. B. SV40 infection pathway. To infect cells, the virus recognizes the host surface glycolipid (GM1) receptor. The virus-receptor complex undergoes endocytosis (step i) to reach the ER (step ii). In the ER lumen, several ER-resident factors isomerizes/reduces the virus disulfide bonds to generate conformationally-altered hydrophobic particles (step iii). These viral particles penetrate the ER membrane to reach cytosol with the aid of several ER-membrane and cytosolic factors (step iv). During cytosolic extraction, the virus is simultaneously disassembled (step v) to prepare a subviral intermediate (containing the genome) for transport into the nucleus for infection. C. Detailed overview of SV40 ER-to-cytosol transport. Step 1: Once SV40 reaches the ER lumen, ER-resident PDI family members including ERp57, ERdj5, and PDI act on the viral particle to trigger critical conformation changes. These structural rearrangements expose the internal proteins VP2 and VP3, generating a hydrophobic viral particle. Step 2: The hydrophobic viral particle recruits BiP via the action of the J-protein ERdj3. Step 3: The NEF Grp170 induces BiP release from SV40 proximal to the ER membrane. Step 4: The hydrophobic viral particle initiates membrane penetration by interacting with and integrating into the ER membrane via engaging the membrane protein BAP31. Step 5: BAP31 in turn recruits other membrane proteins (including the ER membrane J-proteins) to the vicinity to assemble a dense platform within the ER membrane (called foci, black dots) essential for membrane penetration. Step 6: At the cytosolic interface, several cytosolic factors (including Hsc70, SGTA, and Hsp105) anchored to the ER membrane engage the viral particle, extracting it into the cytosol in an energy dependent manner. Step 7: During cytosolic extraction, the virus is simultaneously disassembled to prepare for its journey into the nucleus.
ER lumenal events
Once SV40 reaches the ER lumen, it exploits ER-resident PDI family members to undergo critical conformational changes essential for membrane penetration. For instance, the ER-resident ERp57 isomerizes a subset of SV40's disulfide bonds, breaking and forming non-native disulfide bonds to initiate partial disassembly of the viral particle (Figure 3C, step 1) (Schelhaas et al., 2007); in this reaction, ERp57 acts on disulfide bonds involved in stabilizing the 12 five-coordinated pentamers. In addition to isomerization, disulfide bonds present in SV40 are also reduced by ERdj5 (Inoue et al., 2015); whether this reaction is imposed on disulfide bonds surrounding the 60 six-coordinated pentamers is unclear. It is interesting to note that ERdj5 functions as a J-protein and not as a reductase during ERAD of CT. Canonical PDI neither isomerizes nor reduces SV40's disulfide bonds, but probably uses its chaperone function to unfold the virus (Schelhaas et al., 2007, Inoue et al., 2015), reminiscent of PDI's role as an unfoldase during CTA1 retro-translocation. Thus, despite being in the same protein family, distinct PDI members execute unique roles in priming SV40 for ER membrane transport. In the case of the murine PyV, a non-catalytic PDI family member called ERp29 acts in concert with ERp57 and PDI to remodel the VP1 C-terminal arm (Walczak and Tsai, 2011), destabilizing the viral structure to generate a hydrophobic virus (Magnuson et al., 2005); the human JC PyV was also reported to hijack the activities of the ERp57-PDI-ERp29 triad during infection (Nelson et al., 2012). Another virus reported to reach the ER during infection is the human papillomavirus (HPV) (Disbrow et al., 2005, Zhang et al., 2014), raising the possibility that HPV might also penetrate the ER membrane to access the cytosol during entry. Interestingly, PDI and another PDI family member called ERp72 were found to facilitate HPV infection (Campos et al., 2012), suggesting that the redox and chaperone activities inherent in these factors may trigger HPV conformational changes essential for its ER membrane translocation.
Architectural rearrangements imparted by the PDI family proteins lead to formation of a hydrophobic PyV particle due, in part, to exposure of its internal hydrophobic proteins VP2 and VP3 (Figure 3C, step 1). These structural changes have been documented biochemically (Daniels et al., 2006, Rainey-Barger et al., 2007, Geiger et al., 2011), as well as by imaging approaches when the invading virus reaches the ER from the cell surface (Norkin et al., 2002, Goodwin et al., 2011). While the structural mechanism by which VP2/3 exposure is achieved is unknown, a recent report using the human JC PyV suggests that the pore formed by the VP1 pentamer may enable these internal proteins to slip out (Nelson et al., 2015). Regardless of the mechanism, the hydrophobic virus not only binds to artificial liposomes (Magnuson et al., 2005, Geiger et al., 2011), but can also interact with and integrate into the more physiologically-relevant ER membrane-derived microsomes (Rainey-Barger et al., 2007). These data suggest that PyVs penetrate the ER membrane by traversing its lipid bilayer, in agreement with the finding that SV40 does not use the Hrd1 retro-translocon during infection (Geiger et al., 2011). This view is further supported by the observation that SV40 remains largely intact in the ER (approximately 45 nm in diameter) despite structural alterations imposed on the virus (Inoue and Tsai, 2011, Inoue et al., 2015) – the relatively large SV40 particle prevents it from crossing a typical channel unless the virus is able to induce formation of a massive proteinaceous conduit. Although it has been suggested that SV40 undergoes partial uncoating to generate a 34 nm virion in the ER (Geiger et al., 2011), this particle is still likely too bulky to cross a channel.
Prior to engaging the ER lipid bilayer, BiP is recruited to hydrophobic SV40 to mask its exposed VP2/3, thereby maintaining the virus in a soluble, transport-competent state (Figure 3C, step 2). SV40's recruitment of BiP is catalyzed by the ER-resident J-protein ERdj3 (Goodwin et al., 2011). BiP serves a similar purpose during ER-to-cytosol transport of CTA1 (Winkeler et al., 2003). When the SV40-BiP complex is proximal to the ER membrane, BiP is presumably released from the viral particle, allowing the virus to initiate penetration across this membrane barrier. Acting as an energy-dependent NEF, Grp170 (but not Sil1) stimulates BiP release from SV40 (Figure 3C, step 3) (Inoue and Tsai, 2015). Why there is selectivity in this step is unclear, but it could be due to presence of Grp170's C-terminal holdase domain essential for a NEF to bind to and position the virus for efficient nucleotide release.
ER membrane events
Once BiP disengages from the virus, SV40 is poised to insert into the ER membrane. During insertion, the N-terminal region of the exposed VP2 interacts with an ERAD membrane factor called BAP31 (Figure 3C, step 4) (Geiger et al., 2011). This interaction likely stabilizes the virus, and recruits other transmembrane chaperones to maintain the membrane-integrated virus in an extraction-competent state (Figure 3C, step 5). Other ERAD membrane components including Derlin-1 and Derlin-2 have been implicated in facilitating PyV infection (Lilley et al., 2006, Schelhaas et al., 2007, Jiang et al., 2009). In addition to serving as retro-translocons, Derlin-1/Derlin-2 may confer stability to the viral intermediate within the ER membrane. An shRNA screen led to the identification of three different membrane J-proteins (DNAJ B12, DNAJ B14, and DNAJ C18) as playing critical roles in facilitating ER membrane translocation of SV40 (Goodwin et al., 2011). These J-proteins may directly engage the virus within the membrane to stabilize the integrated virus, as well as to promote their extraction into the cytosol (see below). There is in fact precedence for J-proteins to bind to their substrates before delivering it to the Hsp70 chaperone (Kampinga and Craig, 2010, Shen and Hendershot, 2005). Alternatively, the J-proteins might enlist other membrane components that in turn stabilize the membrane-penetrating virus.
In a rather intriguing set of observations, many of the ER membrane proteins that support SV40 ER membrane penetration including BAP31, BAP29, B12, B14, and C18 were found to reorganize within the ER membrane upon SV40 infection, mobilizing to discrete ER subdomains called foci where the virus also accumulates (Figure 3C, step 5) (Geiger et al., 2011, Walczak et al., 2014, Bagchi et al., 2015). In fact, VP2/3-exposed SV40 that represent membrane penetration-competent virus also co-localizes with the foci (Bagchi et al., 2015), suggesting that the foci may serve as the cytosol entry site for the viral particle. This idea is further supported by the findings that the kinetics of foci formation temporally coincides with SV40 cytosol entry (Walczak et al., 2014), and that SV40 mutants which fail to penetrate the ER membrane to reach the cytosol also cannot trigger foci formation (Geiger et al., 2011, Bagchi et al., 2015). Why does the virus induce the rearrangement of host components to the foci structure? A potential answer is that increasing the local concentration of host factors surrounding a viral particle within the ER membrane enhances the stability of the viral particle in this environment, thereby increasing the efficiency of the penetration process. Alternatively, reorganization of membrane components might alter physical properties of the ER membrane itself (e.g. tubules versus sheets) that allow the virus to be released into the cytosol more readily. Indeed, our understanding of the physiological significance of the foci is at its infancy. What is clearly needed is a high-resolution image of the foci that might provide deeper insights into the virus-induced structure.
Cytosolic events
The identification of B12, B14, and C18 with their functional J-domain oriented towards the cytosol as executing crucial roles in ER-to-cytosol transport of SV40 (Goodwin et al., 2011) immediately raises the possibility that the cytosolic Hsc70 chaperone system is recruited to these J-proteins to promote viral release into the cytosol. This hypothesis is in agreement with a report demonstrating that SV40 employs a p97-independent mechanism to reach the cytosol (Geiger et al., 2011). Indeed, using a conventional biochemical approach, an Hsc70 co-chaperone called SGTA was found to form a complex with the ER membrane J-proteins, and facilitate SV40 extraction into the cytosol (Figure 3C, step 6) (Walczak et al., 2014, Bagchi et al., 2015). Interestingly, SGTA was also reported to play a role during conventional ERAD (Xu et al., 2012) - it cooperates with the Bag6-Ubl4A-Trc35 holdase complex and delivers misfolded substrates from p97 to the proteasome (Wang et al., 2011). However, as cytosol extraction of SV40 does not apparently require p97 and Bag6 (Walczak et al., 2014), Hsc70-bound SGTA must operate in a different manner during this process.
We hypothesize that iterative cycles of binding to and release of SV40 by Hsc70 is likely required to “pull” the virus into the cytosol. In this scenario, we envision that a cytosol-localized NEF is necessary to complete the cycle. A recent report from our laboratory in fact identified the cytosolic Hsp110 NEFs as critical in extracting SV40 into the cytosol (Ravindran et al., 2015) (Figure 3C, step 6). Previous reports documented that Hsp110s, acting in concert with Hsc70 and a J-protein, function as a cellular disaggregase (Dragovic et al., 2006, Mattoo et al., 2013, Oh et al., 1997, Rampelt et al., 2012, Shorter, 2011). Strikingly, our analyses found that the Hsp110-Hsc70-B14 triad disassembles the ER membrane-penetrating SV40, suggesting that viral disassembly and extraction may be coupled events (Figure 3C, step 7). Hence, the Hsp110-triggered disassembly activity may be harnessed to drive a viral membrane translocation process.
Conclusion
The ER is one of the most active intracellular organelles, harboring channels, enzymes, and chaperones essential for promoting protein translocation and folding. Moreover, when proteins misfold, this compartment also contains a dynamic quality control machinery which ensures that misfolded proteins are properly triaged via the ERAD pathway. As this degradative pathway contains a portal for a protein to reach the cytosol from the ER, it can be co-opted by different toxins and viruses to gain access into the host cytosol during infection. Our review focused on how the well-characterized bacterial toxin CT and the DNA tumor virus PyV hijack elements of the ERAD pathway to accomplish this feat (see Table 1). Despite being vastly different, these two toxic agents co-opt common ER lumenal components to prime themselves for membrane penetration. However, their membrane transport and cytosol extraction differ significantly. CTA1 crosses the ER membrane through the existing retro-translocation channel, whereas PyV penetrates the lipid bilayer. Furthermore, while PyV utilizes a membrane-tethered cytosolic Hsc70 chaperone/co-chaperone system to be propelled into the cytosol, ejection of CTA1 to the cytosol remains relatively unclear. In this context, although CTA1 is not ubiquitinated, intact ubiquitination machinery is nonetheless important for controlling its dislocation. Clarifying how ubiquitination impacts retro-translocation of this non-ubiquitinated substrate deserves further attention. In addition to promoting pathogen entry, the ER also supports replication and assembly of different viruses – a topic reviewed elsewhere (Inoue and Tsai, 2013; Byun et al., 2014). Thus while the ER is the site of protein biosynthesis vital for maintaining basic cellular integrity, this organelle is ironically targeted by pathogens to support their entire life cycle. Beyond the field of infectious disease, understanding how two pathogenic agents hijack a protein quality control pathways normally dedicated to the disposal of cellular misfolded proteins to cause infection is likely to reveal important insights into the pathogenesis of other human diseases where protein misfolding is at the root of their pathology.
Acknowledgments
Authors would like to thank members of Tsai lab for discussions and suggestions. We also thank Christopher Walczak (Stanford University) and Paul Moore (UCSF) for critical review of this manuscript.
This work is funded by the National Institutes of Health (RO1 AI064296-08 and RO1 GM113722).
Footnotes
Declaration of interest: None of the authors have a conflict of interest.
References
- Abujarour RJ, Dalal S, Hanson PI, Draper RK. p97 Is in a complex with cholera toxin and influences the transport of cholera toxin and related toxins to the cytoplasm. J Biol Chem. 2005;280:15865–71. doi: 10.1074/jbc.M406316200. [DOI] [PubMed] [Google Scholar]
- Andreasson C, Rampelt H, Fiaux J, Druffel-Augustin S, Bukau B. The endoplasmic reticulum Grp170 acts as a nucleotide exchange factor of Hsp70 via a mechanism similar to that of the cytosolic Hsp110. J Biol Chem. 2010;285:12445–53. doi: 10.1074/jbc.M109.096735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi P, Walczak CP, Tsai B. The endoplasmic reticulum membrane J protein C18 executes a distinct role in promoting simian virus 40 membrane penetration. J Virol. 2015;89:4058–68. doi: 10.1128/JVI.03574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001;3:24–9. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- Behnke J, Feige MJ, Hendershot LM. BiP and its nucleotide exchange factors Grp170 and Sil1: mechanisms of action and biological functions. J Mol Biol. 2015;427:1589–608. doi: 10.1016/j.jmb.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett SM, Broekema NM, Imperiale MJ. BK polyomavirus: emerging pathogen. Microbes Infect. 2012;14:672–83. doi: 10.1016/j.micinf.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi KM, Forster ML, Lencer WI, Tsai B. Derlin-1 facilitates the retro-translocation of cholera toxin. Molecular Biology of the Cell. 2008;19:877–884. doi: 10.1091/mbc.E07-08-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi KM, Williams JM, Inoue T, Schultz A, Tsai B. A deubiquitinase negatively regulates retro-translocation of nonubiquitinated substrates. Mol Biol Cell. 2013;24:3545–56. doi: 10.1091/mbc.E13-06-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi KM, Williams JM, Kikkert M, Van Voorden S, Wiertz EJ, Ye YH, Tsai B. The E3 Ubiquitin Ligases Hrd1 and gp78 Bind to and Promote Cholera Toxin Retro-Translocation. Molecular Biology of the Cell. 2010;21:140–151. doi: 10.1091/mbc.E09-07-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol Cell. 2010;40:238–52. doi: 10.1016/j.molcel.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Buck TM, Kolb AR, Boyd CR, Kleyman TR, Brodsky JL. The endoplasmic reticulum-associated degradation of the epithelial sodium channel requires a unique complement of molecular chaperones. Mol Biol Cell. 2010;21:1047–58. doi: 10.1091/mbc.E09-11-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burress H, Taylor M, Banerjee T, Tatulian SA, Teter K. Co- and post-translocation roles for HSP90 in cholera Intoxication. J Biol Chem. 2014;289:33644–54. doi: 10.1074/jbc.M114.609800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos SK, Chapman JA, Deymier MJ, Bronnimann MP, Ozbun MA. Opposing effects of bacitracin on human papillomavirus type 16 infection: enhancement of binding and entry and inhibition of endosomal penetration. J Virol. 2012;86:4169–81. doi: 10.1128/JVI.05493-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Stanley AM, Rapoport TA. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell. 2010;143:579–91. doi: 10.1016/j.cell.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XS, Stehle T, Harrison SC. Interaction of polyomavirus internal protein VP2 with the major capsid protein VP1 and implications for participation of VP2 in viral entry. EMBO J. 1998;17:3233–40. doi: 10.1093/emboj/17.12.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JA, Chinnapen DJ, Aamar E, Te Welscher YM, Lencer WI, Massol R. Insights on the trafficking and retro-translocation of glycosphingolipid-binding bacterial toxins. Front Cell Infect Microbiol. 2012;2:51. doi: 10.3389/fcimb.2012.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels R, Rusan NM, Wadsworth P, Hebert DN. SV40 VP2 and VP3 insertion into ER membranes is controlled by the capsid protein VP1: implications for DNA translocation out of the ER. Mol Cell. 2006;24:955–66. doi: 10.1016/j.molcel.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Decaprio JA, Garcea RL. A cornucopia of human polyomaviruses. Nat Rev Microbiol. 2013;11:264–76. doi: 10.1038/nrmicro2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disbrow GL, Hanover JA, Schlegel R. Endoplasmic reticulum-localized human papillomavirus type 16 E5 protein alters endosomal pH but not trans-Golgi pH. J Virol. 2005;79:5839–46. doi: 10.1128/JVI.79.9.5839-5846.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M, Bridges JP, Apsley K, Xu Y, Weaver TE. ERdj4 and ERdj5 are required for endoplasmic reticulum-associated protein degradation of misfolded surfactant protein C. Mol Biol Cell. 2008;19:2620–30. doi: 10.1091/mbc.E07-07-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25:2519–28. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eash S, Querbes W, Atwood WJ. Infection of vero cells by BK virus is dependent on caveolae. J Virol. 2004;78:11583–90. doi: 10.1128/JVI.78.21.11583-11590.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel S, Heger T, Mancini R, Herzog F, Kartenbeck J, Hayer A, Helenius A. Role of endosomes in simian virus 40 entry and infection. J Virol. 2011;85:4198–211. doi: 10.1128/JVI.02179-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshraghi A, Dixon SD, Tamilselvam B, Kim EJ, Gargi A, Kulik JC, Damoiseaux R, Blanke SR, Bradley KA. Cytolethal distending toxins require components of the ER-associated degradation pathway for host cell entry. PLoS Pathog. 2014;10:e1004295. doi: 10.1371/journal.ppat.1004295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers H, Romer W, Smith AE, Bacia K, Dmitrieff S, Chai W, Mancini R, Kartenbeck J, Chambon V, Berland L, Oppenheim A, Schwarzmann G, Feizi T, Schwille P, Sens P, Helenius A, Johannes L. GM1 structure determines SV40-induced membrane invagination and infection. Nat Cell Biol. 2010;12:11–8. doi: 10.1038/ncb1999. sup pp 1-12. [DOI] [PubMed] [Google Scholar]
- Falguieres T, Johannes L. Shiga toxin B-subunit binds to the chaperone BiP and the nucleolar protein B23. Biol Cell. 2006;98:125–34. doi: 10.1042/BC20050001. [DOI] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster ML, Sivick K, Park YN, Arvan P, Lencer WI, Tsai B. Protein disulfide isomerase-like proteins play opposing roles during retrotranslocation. Journal of Cell Biology. 2006;173:853–859. doi: 10.1083/jcb.200602046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga Y, Wolf AA, Rodighiero C, Wheeler H, Tsai B, Allen L, Jobling MG, Rapoport T, Holmes RK, Lencer WI. Gangliosides that associate with lipid rafts mediate transport of cholera and related toxins from the plasma membrane to endoplasmic reticulm. Mol Biol Cell. 2003;14:4783–93. doi: 10.1091/mbc.E03-06-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger R, Andritschke D, Friebe S, Herzog F, Luisoni S, Heger T, Helenius A. BAP31 and BiP are essential for dislocation of SV40 from the endoplasmic reticulum to the cytosol. Nat Cell Biol. 2011;13:1305–14. doi: 10.1038/ncb2339. [DOI] [PubMed] [Google Scholar]
- Geiger R, Luisoni S, Johnsson K, Greber UF, Helenius A. Investigating endocytic pathways to the endoplasmic reticulum and to the cytosol using SNAP-trap. Traffic. 2013;14:36–46. doi: 10.1111/tra.12018. [DOI] [PubMed] [Google Scholar]
- Gething MJ. Role and regulation of the ER chaperone BiP. Semin Cell Dev Biol. 1999;10:465–72. doi: 10.1006/scdb.1999.0318. [DOI] [PubMed] [Google Scholar]
- Gilbert J, Benjamin T. Uptake pathway of polyomavirus via ganglioside GD1a. J Virol. 2004;78:12259–67. doi: 10.1128/JVI.78.22.12259-12267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin EC, Lipovsky A, Inoue T, Magaldi TG, Edwards AP, Van Goor KE, Paton AW, Paton JC, Atwood WJ, Tsai B, Dimaio D. BiP and multiple DNAJ molecular chaperones in the endoplasmic reticulum are required for efficient simian virus 40 infection. MBio. 2011;2:e00101–11. doi: 10.1128/mBio.00101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero CJ, Brodsky JL. The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol Rev. 2012;92:537–76. doi: 10.1152/physrev.00027.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara M, Maegawa K, Suzuki M, Ushioda R, Araki K, Matsumoto Y, Hoseki J, Nagata K, Inaba K. Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5. Mol Cell. 2011;41:432–44. doi: 10.1016/j.molcel.2011.01.021. [DOI] [PubMed] [Google Scholar]
- Hale SJ, Lovell SC, De Keyzer J, Stirling CJ. Interactions between Kar2p and its nucleotide exchange factors Sil1p and Lhs1p are mechanistically distinct. J Biol Chem. 2010;285:21600–6. doi: 10.1074/jbc.M110.111211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY, Garza RM. Protein quality control as a strategy for cellular regulation: lessons from ubiquitin-mediated regulation of the sterol pathway. Chem Rev. 2009;109:1561–74. doi: 10.1021/cr800544v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–32. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hazes B, Read RJ. Accumulating evidence suggests that several AB-toxins subvert the endoplasmic reticulum-associated protein degradation pathway to enter target cells. Biochemistry. 1997;36:11051–4. doi: 10.1021/bi971383p. [DOI] [PubMed] [Google Scholar]
- Heiligenstein S, Eisfeld K, Sendzik T, Jimenez-Becker N, Breinig F, Schmitt MJ. Retrotranslocation of a viral A/B toxin from the yeast endoplasmic reticulum is independent of ubiquitination and ERAD. EMBO J. 2006;25:4717–27. doi: 10.1038/sj.emboj.7601350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Dosey A, Herbstman JF, Ravindran MS, Skiniotis G, Tsai B. The ERdj5 reductase cooperates with PDI to promote SV40 ER membrane translocation. J Virol. 2015 doi: 10.1128/JVI.00941-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Tsai B. A large and intact viral particle penetrates the endoplasmic reticulum membrane to reach the cytosol. PLoS Pathog. 2011;7:e1002037. doi: 10.1371/journal.ppat.1002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Tsai B. A nucleotide exchange factor promotes endoplasmic reticulum-to-cytosol membrane penetration of the nonenveloped virus simian virus 40. J Virol. 2015;89:4069–79. doi: 10.1128/JVI.03552-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Abend JR, Tsai B, Imperiale MJ. Early events during BK virus entry and disassembly. J Virol. 2009;83:1350–8. doi: 10.1128/JVI.02169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–92. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartenbeck J, Stukenbrok H, Helenius A. Endocytosis of simian virus 40 into the endoplasmic reticulum. J Cell Biol. 1989;109:2721–9. doi: 10.1083/jcb.109.6.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe M, Ye Y, Wagner JS, De Luca HE, Kern E, Rapoport TA, Lencer WI. Role of p97 AAA-ATPase in the retrotranslocation of the cholera toxin A1 chain, a non-ubiquitinated substrate. J Biol Chem. 2005;280:28127–32. doi: 10.1074/jbc.M503138200. [DOI] [PubMed] [Google Scholar]
- Lai CW, Otero JH, Hendershot LM, Snapp E. ERdj4 protein is a soluble endoplasmic reticulum (ER) DnaJ family protein that interacts with ER-associated degradation machinery. J Biol Chem. 2012;287:7969–78. doi: 10.1074/jbc.M111.311290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencer WI, Constable C, Moe S, Rufo PA, Wolf A, Jobling MG, Ruston SP, Madara JL, Holmes RK, Hirst TR. Proteolytic activation of cholera toxin and Escherichia coli labile toxin by entry into host epithelial cells. Signal transduction by a protease-resistant toxin variant. J Biol Chem. 1997;272:15562–8. doi: 10.1074/jbc.272.24.15562. [DOI] [PubMed] [Google Scholar]
- Li SY, Spooner RA, Hampton RY, Lord JM, Roberts LM. Cytosolic Entry of Shiga-Like Toxin A Chain from the Yeast Endoplasmic Reticulum Requires Catalytically Active Hrd1p. Plos One. 2012;7 doi: 10.1371/journal.pone.0041119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddington RC, Yan Y, Moulai J, Sahli R, Benjamin TL, Harrison SC. Structure of simian virus 40 at 3.8-A resolution. Nature. 1991;354:278–84. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- Lilley BN, Gilbert JM, Ploegh HL, Benjamin TL. Murine polyomavirus requires the endoplasmic reticulum protein Derlin-2 to initiate infection. J Virol. 2006;80:8739–44. doi: 10.1128/JVI.00791-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low JA, Magnuson B, Tsai B, Imperiale MJ. Identification of gangliosides GD1b and GT1b as receptors for BK virus. J Virol. 2006;80:1361–6. doi: 10.1128/JVI.80.3.1361-1366.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maginnis MS, Nelson CD, Atwood WJ. JC polyomavirus attachment, entry, and trafficking: unlocking the keys to a fatal infection. J Neurovirol. 2014 doi: 10.1007/s13365-014-0272-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson B, Rainey EK, Benjamin T, Baryshev M, Mkrtchian S, Tsai B. ERp29 triggers a conformational change in polyomavirus to stimulate membrane binding. Mol Cell. 2005;20:289–300. doi: 10.1016/j.molcel.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Majoul I, Ferrari D, Soling HD. Reduction of protein disulfide bonds in an oxidizing environment. The disulfide bridge of cholera toxin A-subunit is reduced in the endoplasmic reticulum. FEBS Lett. 1997;401:104–8. doi: 10.1016/s0014-5793(96)01447-0. [DOI] [PubMed] [Google Scholar]
- Massey S, Burress H, Taylor M, Nemec KN, Ray S, Haslam DB, Teter K. Structural and functional interactions between the cholera toxin A1 subunit and ERdj3/HEDJ, a chaperone of the endoplasmic reticulum. Infect Immun. 2011;79:4739–47. doi: 10.1128/IAI.05503-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo RU, Sharma SK, Priya S, Finka A, Goloubinoff P. Hsp110 is a bona fide chaperone using ATP to unfold stable misfolded polypeptides and reciprocally collaborate with Hsp70 to solubilize protein aggregates. J Biol Chem. 2013;288:21399–411. doi: 10.1074/jbc.M113.479253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehnert M, Sommer T, Jarosch E. Der1 promotes movement of misfolded proteins through the endoplasmic reticulum membrane. Nat Cell Biol. 2014;16:77–86. doi: 10.1038/ncb2882. [DOI] [PubMed] [Google Scholar]
- Moore P, Bernardi KM, Tsai B. The Ero1alpha-PDI redox cycle regulates retro-translocation of cholera toxin. Mol Biol Cell. 2010;21:1305–13. doi: 10.1091/mbc.E09-09-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P, He K, Tsai B. Establishment of an in vitro transport assay that reveals mechanistic differences in cytosolic events controlling cholera toxin and T-cell receptor alpha retro-translocation. Plos One. 2013;8:e75801. doi: 10.1371/journal.pone.0075801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muanprasat C, Chatsudthipong V. Cholera: pathophysiology and emerging therapeutic targets. Future Med Chem. 2013;5:781–98. doi: 10.4155/fmc.13.42. [DOI] [PubMed] [Google Scholar]
- Nelson CD, Derdowski A, Maginnis MS, O'hara BA, Atwood WJ. The VP1 subunit of JC polyomavirus recapitulates early events in viral trafficking and is a novel tool to study polyomavirus entry. Virology. 2012;428:30–40. doi: 10.1016/j.virol.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CD, Stroh LJ, Gee GV, O'Hara BA, Stehle T, Atwood WJ. Modulation of a pore in the capsid of JC polyomavirus reduces infectivity and prevents exposure of the minor capsid proteins. J Virol. 2015;89:3910–21. doi: 10.1128/JVI.00089-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery FC, Armata IA, Farley JE, Cho JA, Yaqub U, Chen P, Da Hora CC, Wang Q, Tagaya M, Klein C, Tannous B, Caldwell KA, Caldwell GA, Lencer WI, Ye Y, Breakefield XO. TorsinA participates in endoplasmic reticulum-associated degradation. Nat Commun. 2011;2:393. doi: 10.1038/ncomms1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa SI, Fewell SW, Kato Y, Brodsky JL, Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J Cell Biol. 2001;153:1061–70. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norkin LC, Anderson HA, Wolfrom SA, Oppenheim A. Caveolar endocytosis of simian virus 40 is followed by brefeldin A-sensitive transport to the endoplasmic reticulum, where the virus disassembles. J Virol. 2002;76:5156–66. doi: 10.1128/JVI.76.10.5156-5166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh HJ, Chen X, Subjeck JR. Hsp110 protects heat-denatured proteins and confers cellular thermoresistance. J Biol Chem. 1997;272:31636–40. doi: 10.1074/jbc.272.50.31636. [DOI] [PubMed] [Google Scholar]
- Olzmann JA, Kopito RR, Christianson JC. The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a013185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandi PA. Protein-disulfide isomerase-mediated reduction of the A subunit of cholera toxin in a human intestinal cell line. J Biol Chem. 1997;272:4591–9. [PubMed] [Google Scholar]
- Park E, Rapoport TA. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu Rev Biophys. 2012;41:21–40. doi: 10.1146/annurev-biophys-050511-102312. [DOI] [PubMed] [Google Scholar]
- Qian M, Cai D, Verhey KJ, Tsai B. A lipid receptor sorts polyomavirus from the endolysosome to the endoplasmic reticulum to cause infection. PLoS Pathog. 2009;5:e1000465. doi: 10.1371/journal.ppat.1000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querbes W, O'Hara BA, Williams G, Atwood WJ. Invasion of host cells by JC virus identifies a novel role for caveolae in endosomal sorting of noncaveolar ligands. J Virol. 2006;80:9402–13. doi: 10.1128/JVI.01086-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey-Barger EK, Magnuson B, Tsai B. A chaperone-activated nonenveloped virus perforates the physiologically relevant endoplasmic reticulum membrane. J Virol. 2007;81:12996–3004. doi: 10.1128/JVI.01037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampelt H, Kirstein-Miles J, Nillegoda NB, Chi K, Scholz SR, Morimoto RI, Bukau B. Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 2012;31:4221–35. doi: 10.1038/emboj.2012.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran MS, Bagchi P, Inoue T, Tsai B. A Non-enveloped Virus Hijacks Host Disaggregation Machinery to Translocate across the Endoplasmic Reticulum Membrane. PLoS Pathog. 2015;11:e1005086. doi: 10.1371/journal.ppat.1005086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran MS, Tanner LB, Wenk MR. Sialic acid linkage in glycosphingolipids is a molecular correlate for trafficking and delivery of extracellular cargo. Traffic. 2013;14:1182–91. doi: 10.1111/tra.12100. [DOI] [PubMed] [Google Scholar]
- Rodighiero C, Tsai B, Rapoport TA, Lencer WI. Role of ubiquitination in retro-translocation of cholera toxin and escape of cytosolic degradation. EMBO Rep. 2002;3:1222–7. doi: 10.1093/embo-reports/kvf239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer W, Berland L, Chambon V, Gaus K, Windschiegl B, Tenza D, Aly MR, Fraisier V, Florent JC, Perrais D, Lamaze C, Raposo G, Steinem C, Sens P, Bassereau P, Johannes L. Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature. 2007;450:670–5. doi: 10.1038/nature05996. [DOI] [PubMed] [Google Scholar]
- Ruggiano A, Foresti O, Carvalho P. Quality control: ER-associated degradation: protein quality control and beyond. J Cell Biol. 2014;204:869–79. doi: 10.1083/jcb.201312042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saslowsky DE, Cho JA, Chinnapen H, Massol RH, Chinnapen DJ, Wagner JS, De Luca HE, Kam W, Paw BH, Lencer WI. Intoxication of zebrafish and mammalian cells by cholera toxin depends on the flotillin/reggie proteins but not Derlin-1 or -2. J Clin Invest. 2010;120:4399–4409. doi: 10.1172/JCI42958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelhaas M, Malmstrom J, Pelkmans L, Haugstetter J, Ellgaard L, Grunewald K, Helenius A. Simian Virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell. 2007;131:516–29. doi: 10.1016/j.cell.2007.09.038. [DOI] [PubMed] [Google Scholar]
- Schmitz A, Herrgen H, Winkeler A, Herzog V. Cholera toxin is exported from microsomes by the Sec61p complex. J Cell Biol. 2000;148:1203–12. doi: 10.1083/jcb.148.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Hendershot LM. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP's interactions with unfolded substrates. Mol Biol Cell. 2005;16:40–50. doi: 10.1091/mbc.E04-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS One. 2011;6:e26319. doi: 10.1371/journal.pone.0026319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler BD. Structure and function of cholera toxin and the related Escherichia coli heat-labile enterotoxin. Microbiol Rev. 1992;56:622–47. doi: 10.1128/mr.56.4.622-647.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner RA, Lord JM. How ricin and Shiga toxin reach the cytosol of target cells: retrotranslocation from the endoplasmic reticulum. Curr Top Microbiol Immunol. 2012;357:19–40. doi: 10.1007/82_2011_154. [DOI] [PubMed] [Google Scholar]
- Spooner RA, Lord JM. Ricin trafficking in cells. Toxins (Basel) 2015;7:49–65. doi: 10.3390/toxins7010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehle T, Harrison SC. Crystal structures of murine polyomavirus in complex with straight-chain and branched-chain sialyloligosaccharide receptor fragments. Structure. 1996;4:183–94. doi: 10.1016/s0969-2126(96)00021-4. [DOI] [PubMed] [Google Scholar]
- Stehle T, Harrison SC. High-resolution structure of a polyomavirus VP1-oligosaccharide complex: implications for assembly and receptor binding. EMBO J. 1997;16:5139–48. doi: 10.1093/emboj/16.16.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehle T, Yan Y, Benjamin TL, Harrison SC. Structure of murine polyomavirus complexed with an oligosaccharide receptor fragment. Nature. 1994;369:160–3. doi: 10.1038/369160a0. [DOI] [PubMed] [Google Scholar]
- Stein A, Ruggiano A, Carvalho P, Rapoport TA. Key Steps in ERAD of Luminal ER Proteins Reconstituted with Purified Components. Cell. 2014;158:1375–88. doi: 10.1016/j.cell.2014.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M, Burress H, Banerjee T, Ray S, Curtis D, Tatulian SA, Teter K. Substrate-induced unfolding of protein disulfide isomerase displaces the cholera toxin A1 subunit from its holotoxin. PLoS Pathog. 2014;10:e1003925. doi: 10.1371/journal.ppat.1003925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M, Navarro-Garcia F, Huerta J, Burress H, Massey S, Ireton K, Teter K. Hsp90 is required for transfer of the cholera toxin A1 subunit from the endoplasmic reticulum to the cytosol. J Biol Chem. 2010;285:31261–7. doi: 10.1074/jbc.M110.148981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teter K, Jobling MG, Sentz D, Holmes RK. The cholera toxin A1(3) subdomain is essential for interaction with ADP-ribosylation factor 6 and full toxic activity but is not required for translocation from the endoplasmic reticulum to the cytosol. Infect Immun. 2006;74:2259–67. doi: 10.1128/IAI.74.4.2259-2267.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh B, Furman MH, Tortorella D, Ploegh HL. Protein unfolding is not a prerequisite for endoplasmic reticulum-to-cytosol dislocation. J Biol Chem. 2003;278:6664–72. doi: 10.1074/jbc.M210158200. [DOI] [PubMed] [Google Scholar]
- Tsai B, Gilbert JM, Stehle T, Lencer W, Benjamin TL, Rapoport TA. Gangliosides are receptors for murine polyoma virus and SV40. EMBO J. 2003;22:4346–55. doi: 10.1093/emboj/cdg439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B, Rapoport TA. Unfolded cholera toxin is transferred to the ER membrane and released from protein disulfide isomerase upon oxidation by Ero1. J Cell Biol. 2002;159:207–16. doi: 10.1083/jcb.200207120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B, Rodighiero C, Lencer WI, Rapoport TA. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell. 2001;104:937–948. doi: 10.1016/s0092-8674(01)00289-6. [DOI] [PubMed] [Google Scholar]
- Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–72. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- Ushioda R, Hoseki J, Nagata K. Glycosylation-independent ERAD pathway serves as a backup system under ER stress. Mol Biol Cell. 2013;24:3155–63. doi: 10.1091/mbc.E13-03-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CP, Ravindran MS, Inoue T, Tsai B. A cytosolic chaperone complexes with dynamic membrane J-proteins and mobilizes a nonenveloped virus out of the endoplasmic reticulum. PLoS Pathog. 2014;10:e1004007. doi: 10.1371/journal.ppat.1004007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CP, Tsai B. A PDI family network acts distinctly and coordinately with ERp29 to facilitate polyomavirus infection. J Virol. 2011;85:2386–96. doi: 10.1128/JVI.01855-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Liu Y, Soetandyo N, Baek K, Hegde R, Ye Y. A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Mol Cell. 2011;42:758–70. doi: 10.1016/j.molcel.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzmann A, Baldes C, Dudek J, Zimmermann R. The heat shock protein 70 molecular chaperone network in the pancreatic endoplasmic reticulum - a quantitative approach. FEBS J. 2007;274:5175–87. doi: 10.1111/j.1742-4658.2007.06039.x. [DOI] [PubMed] [Google Scholar]
- Wernick NL, Chinnapen DJ, Cho JA, Lencer WI. Cholera toxin: an intracellular journey into the cytosol by way of the endoplasmic reticulum. Toxins (Basel) 2010;2:310–25. doi: 10.3390/toxins2030310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MK, Gordon J, Khalili K. The rapidly expanding family of human polyomaviruses: recent developments in understanding their life cycle and role in human pathology. PLoS Pathog. 2013;9:e1003206. doi: 10.1371/journal.ppat.1003206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JM, Inoue T, Banks L, Tsai B. The ERdj5-Sel1L complex facilitates cholera toxin retrotranslocation. Molecular Biology of the Cell. 2013;24:785–795. doi: 10.1091/mbc.E12-07-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JM, Inoue T, Chen G, Tsai B. The nucleotide exchange factors Grp170 and Sil1 induce cholera toxin release from BiP to enable retro-translocation. Mol Biol Cell. 2015 doi: 10.1091/mbc.E15-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkeler A, Godderz D, Herzog V, Schmitz A. BiP-dependent export of cholera toxin from endoplasmic reticulum-derived microsomes. FEBS Lett. 2003;554:439–42. doi: 10.1016/s0014-5793(03)01217-1. [DOI] [PubMed] [Google Scholar]
- Xu Y, Cai M, Yang Y, Huang L, Ye Y. SGTA recognizes a noncanonical ubiquitin-like domain in the Bag6-Ubl4A-Trc35 complex to promote endoplasmic reticulum-associated degradation. Cell Rep. 2012;2:1633–44. doi: 10.1016/j.celrep.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Li J, Sha B. Structural analysis of the Sil1-Bip complex reveals the mechanism for Sil1 to function as a nucleotide-exchange factor. Biochem J. 2011;438:447–55. doi: 10.1042/BJ20110500. [DOI] [PubMed] [Google Scholar]
- Ye YH, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Yu H, Kopito RR. The role of multiubiquitination in dislocation and degradation of the alpha subunit of the T cell antigen receptor. J Biol Chem. 1999;274:36852–8. doi: 10.1074/jbc.274.52.36852. [DOI] [PubMed] [Google Scholar]
- Yu M, Haslam DB. Shiga toxin is transported from the endoplasmic reticulum following interaction with the luminal chaperone HEDJ/ERdj3. Infect Immun. 2005;73:2524–32. doi: 10.1128/IAI.73.4.2524-2532.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Kazakov T, Popa A, Dimaio D. Vesicular trafficking of incoming human papillomavirus 16 to the Golgi apparatus and endoplasmic reticulum requires gamma-secretase activity. MBio. 2014;5:e01777–14. doi: 10.1128/mBio.01777-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang Y, Yang H, Ballar P, Lee JG, Ye Y, Monteiro MJ, Fang S. Importin beta interacts with the endoplasmic reticulum-associated degradation machinery and promotes ubiquitination and degradation of mutant alpha1-antitrypsin. J Biol Chem. 2011;286:33921–30. doi: 10.1074/jbc.M111.272906. [DOI] [PMC free article] [PubMed] [Google Scholar]