Abstract
Cerebral cavernous malformations (CCMs) are common inherited and sporadic vascular malformations that cause stroke and seizures in younger individuals1. CCMs arise from endothelial cell loss of KRIT1, CCM2, or PDCD10, non-homologous proteins that form an adaptor complex2. How disruption of the CCM complex results in disease remains controversial, with numerous signaling pathways (including Rho3,4, SMAD5 and Wnt/β-catenin6) and processes such as endothelial-mesenchymal transition (EndMT)5 proposed to play causal roles. CCM2 binds MEKK37–11, and we have recently demonstrated that CCM complex regulation of MEKK3 is essential during vertebrate heart development12. Here, we investigate this mechanism in CCM disease pathogenesis. Using a neonatal mouse model of CCM disease, we find that expression of the MEKK3 target genes KLF2 and KLF4, as well as Rho and ADAMTS protease activity, are increased in the endothelial cells of early CCM lesions. In contrast, we find no evidence of EndMT or increased SMAD or Wnt signaling during early CCM formation. Endothelial-specific loss of Mekk3, Klf2, or Klf4 dramatically prevents lesion formation, reverses the increase in Rho activity, and rescues lethality. Consistent with these findings in mice, we demonstrate that endothelial expression of KLF2 and KLF4 is elevated in human familial and sporadic CCM lesions, and that a disease-causing human CCM2 mutation abrogates MEKK3 interaction without affecting CCM complex formation. These studies identify gain of MEKK3 signaling and KLF2/4 function as causal mechanisms for CCM pathogenesis that may be targeted to develop new CCM therapeutics.
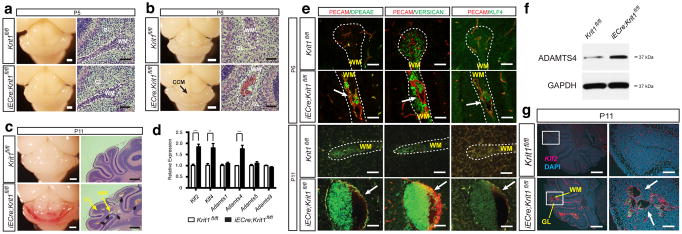
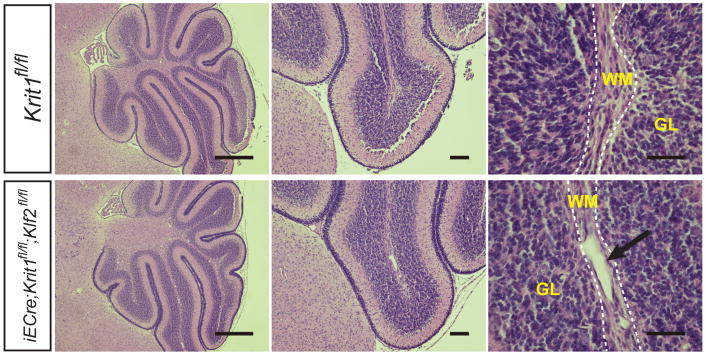
To understand the cellular and molecular mechanisms that underlie CCM formation, we first examined the temporal course of lesion formation in mice with induced, endothelial specific deletion of Krit1 immediately after birth (iECre;Krit1fl/fl termed “Krit1ECKO mice”). Vascular malformations were first detected at P6 as dilated venules in the cerebellar white matter of Krit1ECKO mice, with numerous mature lesions present by P11 (Fig. 1a–c and Extended Data Fig. 1)13. We recently demonstrated that loss of KRIT1, CCM2 or PDCD10 in endothelial cells of the developing heart upregulates expression of KLF2, KLF4, ADAMTS4, and ADAMTS5 due to increased activity of the MEKK3 signaling pathway12. Analysis of isolated cerebellar endothelial cells from neonatal Krit1ECKO and littermate control mice at P6 revealed increased ADAMTS4 mRNA and protein, but not Adamts5, in addition to elevated levels of both Klf2 and Klf4 (Fig. 1d, f). ADAMTS4 cleaves the proteoglycan versican to expose a neo-epitope (DPEAAE) that was detected immediately adjacent to the endothelial cells of both early and late CCM lesions (Fig. 1e). Elevated levels of nuclear KLF4 protein and Klf2 mRNA were also detected in the endothelial cells of CCM lesions and other vessels in the cerebellum (Fig. 1e, g). These findings reveal increased levels of KLF2, KLF4 and ADAMTS4 during the earliest phase of CCM lesion formation in vivo.
Figure 1. KLF2, KLF4 and ADAMTS4 are increased in the earliest CCM lesions.
a–b, Early CCM lesions (arrow) appear as hemorrhagic, dilated venules in the white matter of the cerebellum. White bars, 1mm; black bars, 50 μm. c, Mature CCM lesions are detected as blood-filled caverns in the hindbrain of P11 Krit1ECKO animals. White bars, 1mm; black bars, 500 μm. d, qPCR analysis reveals increased Klf2, Klf4 and Adamts4 expression in hindbrain endothelial cells. *** P<0.001; ** P<0.01. N=4. Error bars indicate SEM. e, Abluminal ADAMTS-cleaved versican (DPEAAE) and nuclear KLF4 are detected in nascent (P6) and mature (P11) CCM lesions (arrows). Scale bars, 50 μm. f, Elevated levels of ADAMTS4 detected by immunoblotting of P11 Krit1ECKO hindbrain lysate. Results are representative of 3 separate experiments. g, Increased Klf2 expression in CCM lesions (arrows) and meningeal vessels of Krit1ECKO animals detected using in situ hybridization. Scale bars, 500 μm (left) and 100 μm (right). WM, white matter; GL, granular layer. Dotted lines outline cerebellar WM.
Recent studies have implicated numerous signaling mechanisms as causal for CCM formation, including the Rho3,4,14, TGF-β/BMP5, Wnt/β-catenin6 and Notch15 pathways. To determine if changes in Rho activity are an early event in CCM pathogenesis we examined phospho-myosin light chain (pMLC), a ROCK substrate. pMLC levels were markedly elevated in the brain capillary and venous endothelial cells of P6 neonatal Krit1ECKO mice compared with littermate controls, including those lining the earliest detectable CCM lesions (Extended Data Fig. 2a). TGF-β/BMP signaling through SMAD1 and SMAD3 and a mechanism of EndMT has been proposed to explain gene expression changes in KRIT1-deficient brain endothelial cells, including increased KLF45,16. A recent study by the same group suggested that increased β-catenin signaling is a primary event that culminates in later EndMT6. To assess endothelial β-catenin signaling during CCM lesion formation, we generated neonatal Krit1ECKO mice on a TCF/Lef:H2B-GFP Wnt/β-catenin reporter background17. Immunostaining for GFP revealed β-catenin signaling in white matter vascular endothelium that was not increased in P6 CCM lesions (Extended Data Fig. 2b), while the levels of both GFP protein and the β-catenin target genes Axin2 and Lef1 were unchanged in cerebellar endothelial cells isolated from P6 or P11 neonatal Krit1ECKO mice (Extended Data Fig. 2c, d). Immunostaining of brain sections and immunoblotting of cerebellar endothelial cell protein also revealed no changes pSMAD3 (Extended Data Fig 2e, f). Finally, analysis of cerebellar endothelial gene expression analysis revealed no change in the expression of Notch target genes at P6, although an increase in Hes1 was noted at P11 (Extended Data Fig. 3). These studies reveal that primary CCM lesion formation is associated with increases in Klf2, Klf4 and Adamts4 expression and Rho/ROCK activity, but not in TGF-β/BMP, Wnt/β-catenin, or Notch signaling.
The above studies suggested that changes in KLF2/4 and ADAMTS4 expression may be causal for CCM formation. The CCM complex directly binds MEKK37–11, a MAP3 kinase known to regulate KLF2 and KLF4 expression in cultured endothelial cells12, and we previously found that Map3k3 haploinsufficiency rescues the loss of cardiac jelly associated with endocardial loss of CCM signaling12. Map3k3 haploinsufficiency was also found to rescue the early embryonic lethality conferred by pan-endothelial loss of KRIT1 (Extended Data Fig. 4a and18), suggesting that excess endothelial MEKK3 signaling may play a broad role in the cardiovascular phenotypes associated with loss of CCM signaling.
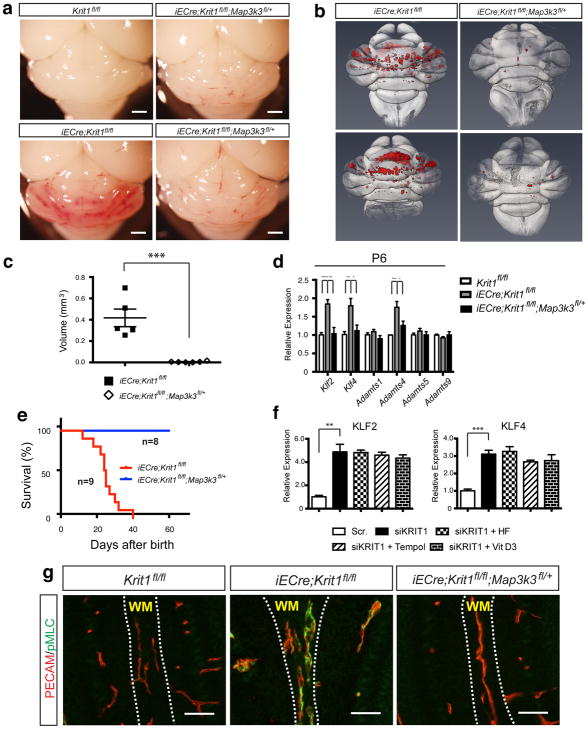
To determine whether this paradigm underlies CCM formation we generated iECre;Krit1fl/fl;Map3k3fl/+ mice (MEKK3HetRSQ). Visual inspection of the hindbrains of P11 MEKK3HetRSQ mice compared with neonatal Krit1ECKO littermate controls revealed a dramatic reduction in the number and size of vascular lesions (Fig. 2a). To precisely quantitate CCM formation we imaged P11 hindbrains using contrast-enhanced, high resolution X-ray micro tomography (microCT) and measured actual lesion volumes using semi-automated software. MEKK3HetRSQ mouse hindbrains exhibited nearly complete prevention of the lesion phenotype compared with neonatal Krit1ECKO littermates as assessed by hindbrain microCT imaging (Fig. 2b), blinded measurement of total CCM lesion volume (Fig. 2c), and measurement of Klf2, Klf4, and Adamts4 in P6 cerebellar endothelial cells (Fig. 2d). While almost all neonatal Krit1ECKO mice were dead by P30, all MEKK3HetRSQ animals remained alive (Fig. 2e), and exhibited normal growth and development. Finally, partial loss of MEKK3 also fully rescued CCM lesion formation in Ccm2ECKO mice (Extended Data Fig. 4b). These genetic findings support the conclusion that CCM lesions arise from gain of MEKK3 signaling and altered downstream gene expression in the endothelium.
Figure 2. Genetic rescue of CCM formation and elevated Rho activity with loss of MEKK3.
a, Visual detection of CCM lesions in the hindbrains of P11 Krit1fl/fl animals (top left), Krit1ECKO animals (bottom left), and MEKK3HetRSQ animals (right). Scale bars indicate 1mm. b, Composite microCT images of Krit1ECKO and MEKK3HetRSQ hindbrains. CCM lesions are shown in red. c, Quantitation of CCM lesion volumes. N=5 for each group. d, qPCR analysis of gene expression in cerebellar endothelial cells. N=4. e, Postnatal survival of Krit1ECKO animals with and without endothelial loss of one Map3k3 allele. P=0.0009. f, qPCR analysis of gene expression in human umbilical vein endothelial cells treated with scrambled (“Scr.”) or KRIT1 targeting siRNAs, alone and in the presence of the indicated Rho antagonists. HF, hydroxyfasudil. N=3. g, Normalization of Rho activity with loss of MEKK3. PECAM and pMLC staining of white matter vessels in the indicated P6 littermate brains is shown. Images are representative of 5 independent studies for each genotype. Scale bars, 50 μm. **** indicates P<0.0001; *** indicates P<0.001; ** indicates P<0.01; * indicates P<0.05. Error bars indicate SEM.
Elevated endothelial pMLC is coincident with increased Klf2, Klf4 and Adamts4 expression in the earliest CCM lesions (Figs. 1e, g and Extended Data Fig. 2a), suggesting either that changes in Rho/ROCK activity are downstream of changes in MEKK3 activity or vice versa. The Rho inhibiting agents hydroxyfasudil, Tempol and vitamin D319 failed to reverse the increase in KLF2 and KLF4 expression conferred by loss of KRIT1 in cultured endothelial cells (Fig. 2f), suggesting that Rho is not upstream of the KLF2/4 expression changes associated with loss of CCM function. In contrast, P6 MEKK3HetRSQ mice exhibited a complete normalization of endothelial pMLC staining (Fig. 2g), indicating that elevated Rho activity arises secondary to increased MEKK3 signaling during CCM formation.
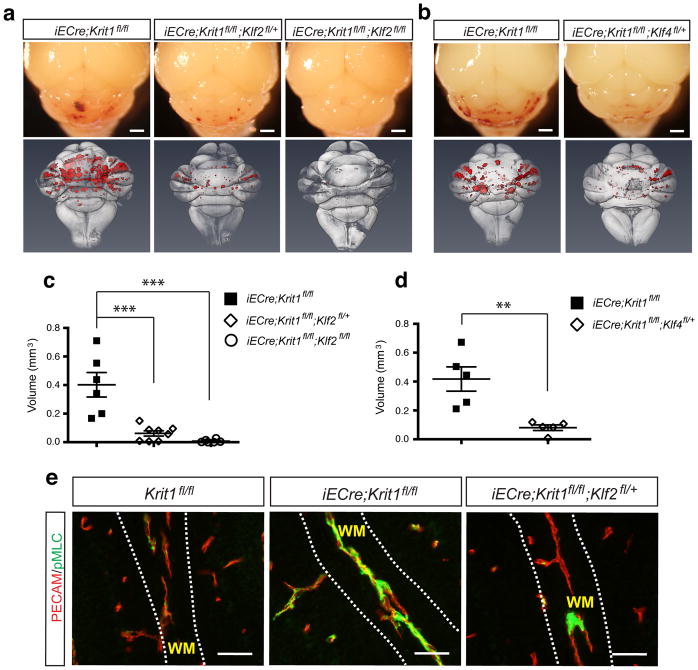
To test the roles of KLF2 and KLF4 in CCM pathogenesis, we measured lesion formation in Klf2HetRSQ mice (iECre;Krit1fl/fl;Klf2fl/+), Klf2HomoRSQ mice (iECre;Krit1fl/fl;Klf2fl/fl), and Klf4HetRSQ mice (iECre;Krit1fl/fl;Klf4fl/+) compared with littermate neonatal Krit1ECKO controls at P11. Klf2HetRSQ and Klf4HetRSQ mice exhibited a marked but incomplete prevention of lesion formation (75% and 80% rescue for Klf2HetRSQ and Klf4HetRSQ mice, respectively) based on visual inspection of cerebellar lesions and quantitation of CCM lesion volume following microCT imaging (Fig. 3a–d). Remarkably, Klf2HomoRSQ mice exhibited 99% rescue (Fig. 3a, c), with only a small amount of venule dilatation visible histologically (Extended Data Fig. 5). pMLC staining was normalized in P6 Klf2HetRSQ mice (Fig. 3e), indicating that elevated Rho/ROCK activity arises secondary to increased KLF2 expression. These findings identify gain of KLF2 and KLF4 as causal for CCM formation, and suggest that these transcription factors are the primary downstream targets of MEKK3 in this disease model. They also highlight the remarkable molecular conservation of this endothelial pathway: from zebrafish to mammals, and embryonic vascular endothelium and endocardium to postnatal brain endothelium.
Figure 3. Genetic rescue of CCM formation and increased Rho activity with loss of KLF2 and KLF4.
a–b, Visual appearance of CCM lesions in Krit1ECKO, Klf2HetRSQ, Klf2HomoRSQ and Klf4HetRSQ animals is shown above and composite microCT images of the same hindbrains shown below. Scale bars, 1mm. c–d, Volumetric quantitation of CCM lesions in Krit1ECKO littermates with and without endothelial loss of one or two Klf2 alleles (c) or one Klf4 allele (d). *** indicates P<0.001; ** indicates P<0.01. N=5–7. e, Rescue of increased Rho activity by loss of KLF2. PECAM and pMLC staining of white matter (“WM”) vessels in the indicated P6 littermate brains is shown. Images are representative of 4 independent studies for each genotype. Scale bars, 50 μm.
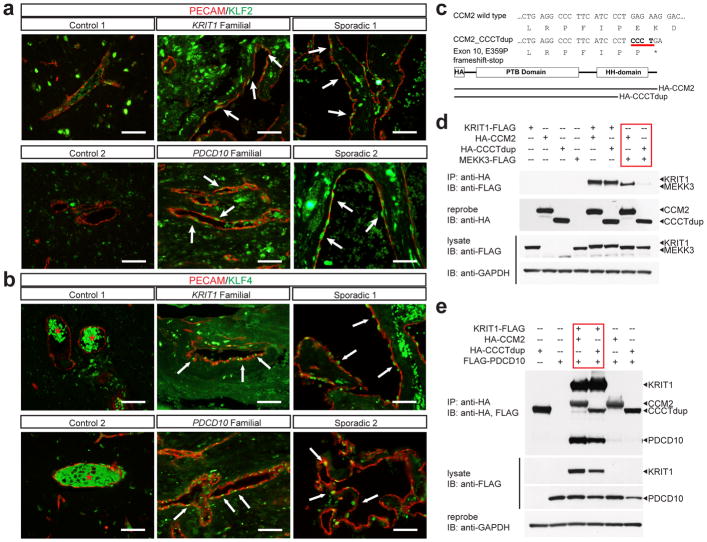
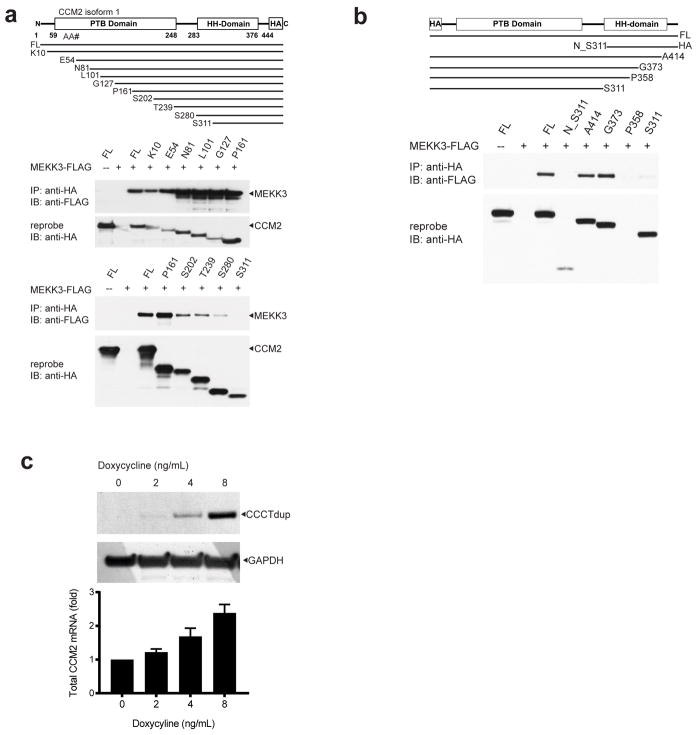
To determine if MEKK3-KLF2/4 signaling is increased in human CCMs we examined resected lesions from two familial CCM patients bearing KRIT1 and PDCD10 germline mutations, and two sporadic CCM patients lacking any prior genetic or molecular data. Markedly increased nuclear KLF2 and KLF4 was observed in the endothelial cells of both familial and sporadic human CCM lesions (Fig. 4a, b), findings consistent with increased MEKK3 signaling and studies performed using the mouse model. MEKK3 binds CCM2 through the C-terminal helical harmonin domain (HHD) of CCM2, and CCM2 truncation mutants lacking this domain do not bind MEKK3 (Extended Data Fig. 6a-b and10,11,20,21). A literature search identified a familial CCM patient with a four nucleotide duplication in the last exon of CCM2 (CCCTdup) predicted to delete most of the HHD (Fig. 4c)22. CCM2 CCCTdup expressed normally in HEK293T cells and bound KRIT1 and PDCD10 in a manner indistinguishable from wild-type CCM2, but failed to interact with MEKK3 (Fig. 4c–e and Extended Data Fig. 6c). These results suggest that specific disruption of the CCM2-MEKK3 interaction is sufficient to cause familial CCM disease, and that human CCMs also arise due to loss of MEKK3 regulation and increased expression of KLF2 and KLF4.
Figure 4. Human CCMs exhibit high levels of endothelial KLF2 and KLF4 and arise due to selective loss of CCM2-MEKK3 interaction.
a–b, Immunostaining for PECAM and KLF2 (a) or KLF4 (b) is shown for cerebral vessels in individuals without CCM disease (“control”, left), for CCM lesions arising due to germline mutations in KRIT1 or PDCD10 (“familial”, middle), and for sporadic CCM lesions from two individuals (“sporadic”, right). Arrows indicate nuclear KLF2-high and nuclear KLF4-high endothelial cells in the CCM lesions. The red asterisk indicates fluorescence due to trapped intravascular erythrocytes. Scale bars, 50 μm. c, Schematic representation of the CCM2 CCCT duplication (“CCM2 CCCTdup”). d, CCM2 CCCTdup does not bind MEKK3. The indicated proteins were expressed in HEK293T cells, immunoprecipitated using anti-HA antibodies, and immunoblotted using the indicated antibodies. e, CCM2 CCCT dup binds KRIT1 and PDCD10 like wild-type CCM2. The indicated proteins were expressed and co-immunoprecipitated as described in (c). The results shown are representative of at least 3 separate experiments.
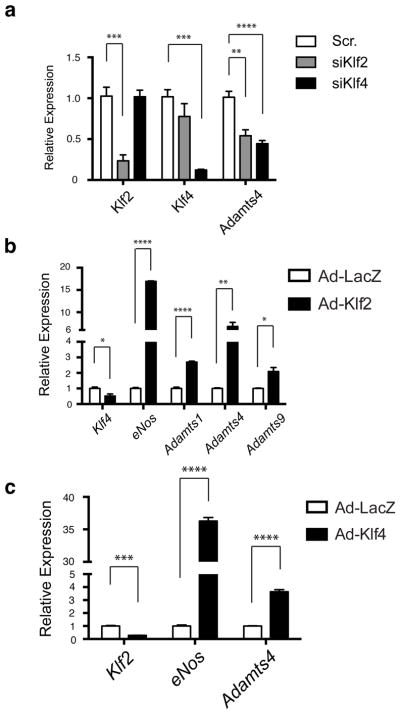
How does gain of MEKK3-KLF2/4 signaling confer CCM formation? It has been proposed that EndMT underlies CCM pathogenesis5,6,23, but we detect no evidence of a change in phenotype from endothelial to mesenchymal with loss of CCM signaling and demonstrate that loss of the non-mesenchymal transcription factor KLF2 is sufficient to fully rescue CCM formation. Our studies identify two effector pathways downstream of MEKK3-KLF2/4 signaling that may drive early CCM pathogenesis, Rho signaling and ADAMTS proteolytic activity. Elevated Rho activity has been linked to increased stress fiber formation, loosened junctions and decreased tube formation in cultured endothelial cells3,14,24 and loss of vascular integrity in mice4,25, but whether and how these changes might cause CCM formation is not yet clear. Increased ADAMTS activity is conferred by gain of KLF2 and KLF4 (Extended Data Fig. 7), and may confer CCM formation through breakdown of a proteoglycan matrix that is required specifically for the CNS vasculature (e.g versican (Extended Data Fig. 1b)). A proteolytic mechanism would also explain the autosomal dominant inheritance of this disease, as CCM-deficient endothelial cells generated by a rare second-hit mechanism26,27 could degrade the matrix supporting an entire vessel, thereby creating a cavernous malformation.
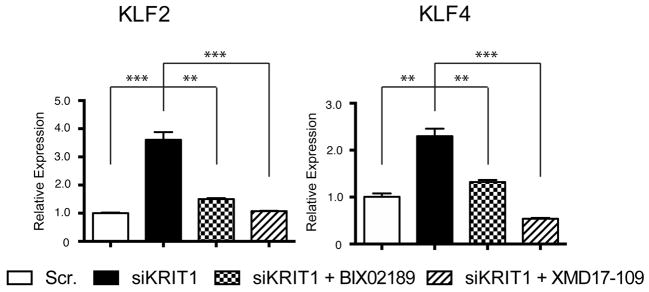
Finally, these studies may help identify new therapies for CCM disease. Since MEKK3 up-regulates KLF2 and KLF4 through the MEK5 and ERK5 downstream MAPKs12,28, we evaluated the effects of available inhibitors on CCM formation. BIX02189 (anti-MEK5) and XMD17-109 (anti-ERK5) reversed the increase in KLF2 and KLF4 expression associated with loss of CCM signaling in cultured endothelial cells (Extended Data Fig. 8), but failed to affect CCM formation at the low doses tolerated by neonatal Krit1ECKO mice (not shown). Therapies targeting Rho (e.g. Fasudil29 or other recently identified agents19) or the ADAMTS proteases may prove more effective, but future studies that rigorously test the causal role of these putative downstream effectors remain essential for the rational development of CCM disease therapies.
Methods
Mice
VECadCreERT2 (“iECre”) transgenic mice were a gift from Ralf Adams30. Tie2Cre, Krit1fl/fl, Ccm2fl/fl, Map3k3fl/fl, Klf2fl/fl and Klf4fl/fl animals have been previously described12,25,31–34. The TCF/LEF:H2B-GFP reporter line was obtained from Jackson Laboratories (#013752)17. All intercrossed animals were maintained on mixed strain backgrounds. Breeding pairs between two and eight months of age were used to generate the neonatal CCM mouse model pups. Male and female animals were used in equal numbers for all experiments. The University of Pennsylvania Institutional Animal Care and Use Committee approved all animal protocols, and all procedures were performed in accordance with these protocols.
Induction of the neonatal CCM mouse model
Pups were intragastrically injected with 25 μg of 4-hydroxytamoxifen (Sigma Aldrich, H7904) at intervals of 1 day, 3 days, and 5 days after birth. The 4-hydroxytamoxifen was dissolved in warm 10% Ethanol/Corn oil vehicle and 50 μL total volume was used per injection and delivered with a 30 gauge needle. Importantly, pups were injected in blinded fashion without knowledge of genotypes, and there were no phenotypic indications within the first five days post-birth. Pups were harvested at specified timepoints 6 days or 11 days after birth.
Mouse embryos for branchial arch artery rescue
Embryos were harvested at E10.5, gross images taken, and placed in 4% paraformaldehyde overnight. Samples were processed for histology as detailed below.
Histology
Embryos or mouse brains were fixed in 4% formaldehyde overnight, dehydrated in 100% ethanol, and embedded in paraffin. 8 μm thick sections were used for H&E and immunohistochemistry staining. Klf2 in situ hybridization was performed as previously reported32. The following antibodies were used for immunostaining: rat anti-PECAM (1:500, R&D MAB3628), rat anti-PECAM (1:20, Histo Bio Tech DIA-310), rabbit anti-versican (1:200, Millipore AB1033), rabbit anti-DPEAAE (1:200, Pierce-Antibodies PA1-1748A), mouse anti-KLF4 (1:100, R&D AF3158), rabbit anti-pSmad3 (1:25, Abcam ab52903), rabbit anti-pMLC2 (1:200, Cell Signaling 3674S).
Isolation of cerebellar endothelial cells
Cerebellar endothelial cells were isolated through enzymatic digestion followed by separation using magnetic-activated cell sorting (MACS MS system, Miltenyi Biotec). Mice were first anesthetized with Avertin (Sigma Aldrich, T48402) and perfused with sterile phosphate buffered saline (PBS). Cerebella of the mice were then digested by 1 mg/ml collagenase/dispase (Sigma) and 0.02 mg/ml DNase I (Sigma) in complete DMEM for 10 min at 37 °C with gentle shaking. The digestion was then passed through a 70 μm cell strainer (BD Biosciences). Cells were centrifuged, resuspended, and incubated with anti-mouse CD31 antibody conjugated microbeads for 15 min at 4 °C. Microbead-bound cells were then washed and separated using MACS MS columns according to vendor protocol. Cells bound to the magnetic column were eluted and centrifuged for downstream applications, including western blotting and qPCR analysis.
Western blotting of protein from isolated endothelial cells
Protein of freshly isolated cerebellar endothelial cells was purified using RIPA buffer with complete protease inhibitor cocktail (Roche) and PhosSTOP phosphatase inhibitor cocktail (Roche). The following antibodies were used for immunoblotting: rabbit anti-GAPDH (1:5000, Cell Signaling 2118), rabbit anti-ADAMTS4 (1:1000, Abcam ab28285), rabbit anti-SMAD3 (1:1000, Cell Signaling 9513), rabbit anti-pSMAD3 (1:1000, Cell Signaling C25A9), rabbit anti-GFP (1:1000, Abcam ab290).
Gene expression analysis
Total RNA of isolated endothelial cells was isolated with that RNeasy micro kit (Qiagen). For qPCR analysis, cDNA was synthesized from 150 ng total RNA using the Superscript III Reverse Transcriptase (Invitrogen). Real-time PCR was performed with Power SYBR Green PCR Master Mix (Applied Biosciences) using the primers listed:
mGapdh Forward - 5′-AAATGGTGAAGGTCGGTGTGAACG -3′
mGapdh Reverse - 5′-ATCTCCACTTTGCCACTGC -3′
mKlf2 Forward - 5′-CGCCTCGGGTTCATTTC -3′
mKlf2 Reverse - 5′-AGCCTATCTTGCCGTCCTTT -3′
mKlf4 Forward - 5′-GTGCCCCGACTAACCGTTG -3′
mKlf4 Reverse - 5′-GTCGTTGAACTCCTCGGTCT -3′
mAdamts1 Forward - 5′-CTCTCACCCTTCGGAATTTCTG -3′
mAdamts1 Reverse - 5′-GGAGCCACATAAATCCTGTCTG -3′
mAdamts4 Forward - 5′-CAGTGCCCGATTCATCACT -3′
mAdamts4 Reverse - 5′-GAGTCAGGACCGAAGGTCAG -3′
mAdamts5 Forward - 5′-CGACCCTCAAGAACTTTTGC -3′
mAdamts5 Reverse - 5′-CGTCATGAGAAAGGCCAAGT -3′
mAdamts9 Forward - 5′-TTGGGACCTGCTCAAGAACG -3′
mAdamts9 Reverse - 5′-ACCATTGATGTTGAAGTGTTTGC -3′
mAxin2 Forward - 5′-CAGCCCTTGTGGTTCAAGCT -3′
mAxin2 Reverse - 5′-GGTAGATTCCTGATGGCCGATGT -3′
mLef1 Forward - 5′-TAACGAGTCCGAAATCATCCCAGC -3′
mLef1 Reverse - 5′-TTCATCAGGGTGTTCTCTGGCCTT -3′
mHes1 Forward - 5′-AAAGCCTATCATGGAGAAGAGGCG -3′
mHes1 Reverse - 5′-GGAATGCCGGGAGCTATCTTTCTT -3′
mHey1 Forward - 5′-GCGCGGACGAGAATGGAAA -3′
mHey1 Reverse - 5′-TCAGGTGATCCACAGTCATGTG -3′
mHey2 Forward - 5′-AAGCGCCCTTGTGAGGAAAC -3′
mHey2 Reverse - 5′-GGTAGTTGTCGGTGAATTGGAC -3′
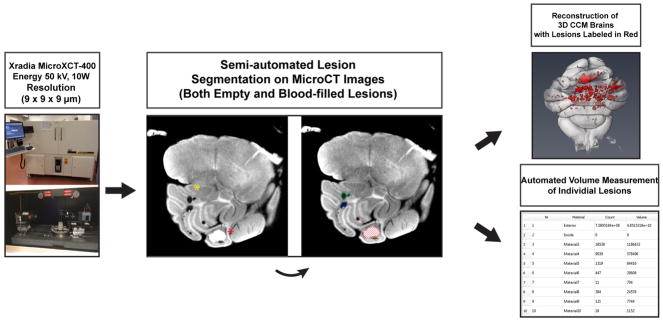
X-ray micro tomography-based quantification of neonatal, cerebellar lesions
11 day-old pups were anesthetized with Avertin and underwent intra-cardiac perfusion with PBS and 2% paraformaldehyde. Brains were quickly dissected and fixed overnight in a 2.5% glutaraldehyde, 4% formaldehyde 0.1 M sodium phosphate buffer. Then, brains were rinsed several times with 0.1 M sodium phosphate buffer and hindbrains were detached from mid/forebrain structures by severing the pons. Next, hindbrains were soaked overnight in 2% osmium tetroxide solution (tissue contrast agent) and washed several times with water. Hindbrains were randomized and scanned by a blinded operator using Xradia MicroCT system (Xradia MicroXCT-400, Xradia, CA, USA). Images were acquired at 50 kV, 10W, 721 projections, 3s integration per 180° rotation.
Raw image stacks from each scanned hindbrain were analyzed using Aviso 3D image processing software (Lite edition, FEI Visualization Sciences Group). Each image stack was imported and lesions were labeled in semi-automated, blinded fashion using a region-growing segmentation algorithm included with the software. Labeled lesions could then be analyzed for pixel volume and converted to cubic μm. Importantly, to prevent measurement bias, a single individual, without direct experimental involvement or knowledge of genotypes, was used to label lesions of randomly ordered brains. This individual was trained prior to beginning analysis of the hindbrains. New-user accuracy was confirmed by comparison of results from a test brain analysis with the results from an experienced user. A flow-chart of this process is shown in Extended Data Figure 8.
Post-lesion labeling, the hindbrain image stack was volume rendered and over-laced with the labeled lesions in the Avizo 3D environment. Orientation, projection depth, and shadow/lighting effects of each volume render were adjusted for publication quality images. Importantly, these changes were made post-lesion labeling, and did not affect lesion detection or their visual presentation.
We blinded samples at three distinct points in the analysis. First, neonatal CCM model pups were injected with 4-hydroxytamoxifen without knowledge of genotypes. Second, hindbrains from genotyped animals were given randomized labels to provide for blinded microCT scanning by an independent operator. Third, randomized microCT image stacks were analyzed in a blinded manner by a single operator not involved in any prior experimental steps.
cDNA constructs and transient expression in HEK293T cells
The human KRIT1-FLAG expression plasmid was a gift from Douglas Marchuk8. Human CCM2-FLAG, PDCD10-FLAG, and MEKK3-FLAG expression plasmids were purchased from Origene (RC201418, RC200235, and RC210317). Further epitope tag modifications and CCM2 truncations were generated through PCR and conventional cloning methods. The CCM2 CCCTdup mutant was generated by site directed mutagenesis to generate the exact four-nucleotide duplication (Agilent 200521). 1 μg of each construct was transfected in HEK293T cells using Fugene 6 (Promega E2691) and cells were harvested 48 hours later with gentle lysis buffer (Life Tech 87787) supplemented with protease and phosphatase inhibitors (Roche 11873580001 and 04906845001).
Co-immunoprecipitation (co-IP) and western blotting
Anti-HA antibody (5 μg per co-IP, Sigma H3663) conjugated to Protein G Dynabeads (50 μL per co-IP, Life Technologies 10003D) was used for all experiments. 250 μg of protein from HEK293T cells was used for each co-IP with an incubation period of 30 minutes at room temperature. HEK293T cells were obtained directly from the ATCC, authenticated and tested negative for mycoplasma. Anti-FLAG (1:5000 Santa Cruz sc-807) and anti-HA (1:5000 Santa Cruz sc-805) antibodies were used for western blotting and re-probed with anti-GAPDH antibody (1:10,000 Cell Signaling 2118). For co-IP blots of PDCD10-FLAG, HRP-conjugated anti-FLAG antibody (1:2000, Sigma A8592) was used to avoid antibody light chain detection.
Expression of tetracycline-inducible CCM2 CCCTdup lentivirus in cultured endothelial cells
N-terminal, HA-epitope tagged, human CCM2 CCCTdup was cloned into the pLVX-TRE3G lentiviral vector (Takara Clontech) and lentivirus was generated from co-transfection with packaging plasmids in HEK293T cells as previously described12. HUVECs were co-infected with CCM2 CCCTdup along with Tet-On 3G lentiviruses and varying doxycycline amounts were added as previously described12. Cells were harvested 48 hours post-doxycycline addition. Collected cells were pelleted, resuspended, then divided in half. One half was used for total RNA isolated by TRIzol (Life Technologies), and the other half was used for protein extracted by gentle lysis buffer as described above. cDNA was generated from 500 ng of total RNA using the Superscript III Reverse Transcriptase (Invitrogen). mRNA levels of the CCM2 CCCTdup lentivirus were assessed by qPCR using both N-terminal and C-terminal human CCM2 qPCR primers as follows:
hCCM2 n-term Forward: 5′-CATACCAGGATACCTGAATCCCT-3′
hCCM2 n-term Reverse: 5′-AGCTTGACGTTGTACGCAGAC-3′
hCCM2 c-term Forward: 5′-GCCTCTATCCACGAGTTCTGCA-3′
hCCM2 c-term Reverse: 5′-AAGTGCTGGCTGTCCTTCTCAG-3′
Western blotting was performed as previously described with 75 μg protein from HUVEC lysate for each condition.
siRNA knockdown and adenoviral overexpression in cultured endothelial cells
siRNA experiments in human umbilical vein endothelial cells (HUVECs) were performed as previously described12. HUVECs from Lonza were authenticated and tested negative for mycoplasma. siRNAs directed against KRIT1 (s2510, Invitrogen), KLF2 (s20270, Invitrogen), KLF4 (s17793, Invitrogen), STK24 (s15993, Invitrogen) or STK25 (s20570, Invitrogen) were used for the knockdown experiments. For overexpression studies, human microvascular endothelial cells (HMVECs) were infected with adenovirus encoding either mouse KLF2 (Penn Vector Core), human KLF4 (Vector BioLabs), or a LacZ control (Penn Vector Core). HMVECs from Lonza were authenticated and tested negative for mycoplasma. Cells were harvested 48 hours post-infection and total RNA was isolated using TRIzol Reagent (Life Technologies). cDNA was generated from 1 μg total RNA using SuperScript First-strand Synthesis System (Invitrogen) and real time qPCR was performed using Power SYBR Green PCR Master Mix (Applied Biosciences).
Primers for qPCR:
hKLF2 Forward: 5′-CTACACCAAGAGTTCGCATCTG-3′
hKLF2 Reverse: 5′-CCGTGTGCTTTCGGTAGTG-3′
hKLF4 Forward - 5′-AGAGTTCCCATCTCAAGGCA -3′
hKLF4 Reverse - 5′-GTCAGTTCATCTGAGCGGG -3′
hADAMTS4 Forward - 5′-CTGACTTCCTGGACAATGGC -3′
hADAMTS4 Reverse - 5′-GCGGTCAGCATCATAGTCCT -3′
Drug treatments in cultured endothelial cells with loss of KRIT1
After siRNA knockdown of KRIT1, HUVECs were treated with BIX02189 (10 uM), XMD17-109 (1 uM), Hydroxyfasudil (10 uM), Tempol (10 uM) or Vit D3 (10 uM) for 24 hours prior to harvest.
KLF2 and KLF4 immunostaining in human CCM lesions and control brain sections
Sections of human samples were obtained from CCM lesions resected at University of Chicago using IRB approved protocols. Control samples were obtained from two autopsy subjects. Rabbit anti-KLF2 (1:200 Abcam ab203591), rabbit anti-KLF4 (1:50 Cell Signaling 4038S) and mouse anti-PECAM-1 (1:1000 Cell Signaling 3528S) were used for immunostaining.
Patient information pertaining to the human CCM samples studied is summarized below.
KRIT1 Familial: female, age 11, with the CCM1 Common Hispanic Mutation, removed due to CCM hemorrhage.
PDCD10 Familial: female, age 19, PDCD10 474+5G>A mutation, removed due to CCM hemorrhage.
Sporadic 1: female, age 24, solitary sporadic lesion, removed due to headaches and lesion growth.
Sporadic 2: male, age 31, solitary sporadic lesion, removed due to seizures.
Statistics
Sample sizes were estimated based on our preliminary findings for rescue of the CCM phenotype. These were estimated from visual analysis of the hindbrains of P11 animals from which lesion numbers and sizes can be directly assessed, an observation corroborated by microCT-based volume rendering of lesions. These preliminary assessments suggested that there were very large differences between genetic rescue and non-rescue samples (conservatively eighty percent rescue) that would allow statistical interpretation with relatively small Ns. All experimentals and controls were littermates and none were excluded from analysis. P values were calculated using an unpaired 2-tailed Student’s t-test, ANOVA, or Chi Square analysis as indicated. The mean and standard error of mean (SEM) are shown in the bar graphs.
Extended Data
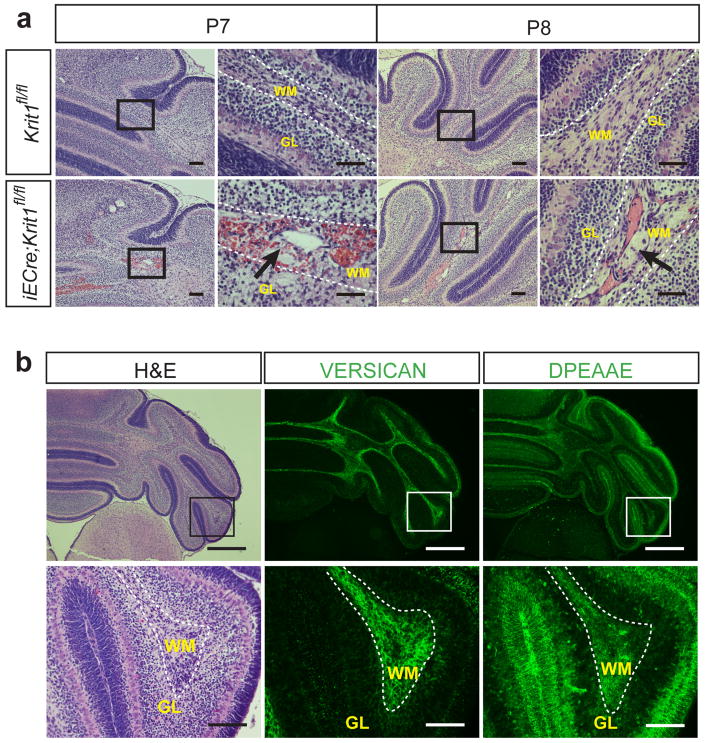
Extended Data Figure 1. Histologic characteristics of early CCM lesions and the cerebellar white matter in which they form.
a, P7 and P8 CCM lesions in the Krit1 model. Hindbrains from Krit1ECKO mice were harvested at P7 and P8 and sections stained with hematoxylin-eosin to detect vascular malformations. Images are representative of 4 independent experiments. Dotted lines indicate the cerebellar white matter. Arrows indicate CCM lesions. WM, white matter; GL, granular layer. b, Versican is abundant in the white matter of the P7 hindbrain, the site of primary CCM formation in the neonatal mouse model. H–E staining of the hindbrain at low (top left) and high (bottom left) magnification. Antibody staining reveals abundant versican protein in the white matter of the hindbrain (center), the site at which CCM lesions specifically appear at this timepoint in the Krit1 mouse model, and less abundant versican in the cortex. DPEAAE staining reveals a pattern of ADAMTS-mediated versican degradation that is the inverse of intact versican, i.e. higher in the cortex and lower in the white matter (right). Results representative of three independent experiments. Scale bars: top, 500 μm; bottom, 100 μm. Boxes indicate regions shown at higher magnification below. Dotted lines indicate the cerebellar white matter. WM, white matter; GL, granular layer.
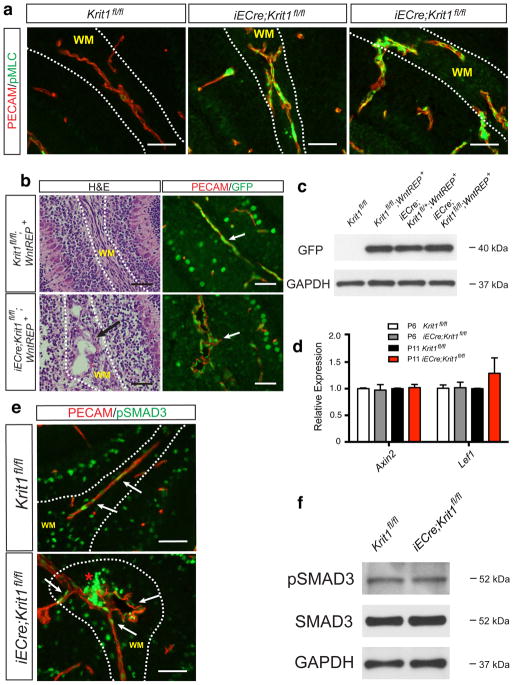
Extended Data Figure 2. Endothelial Rho activity, but not β-catenin or SMAD3 signaling, is increased during CCM formation.
a, Immunostaining for the endothelial marker PECAM and pMLC in the white matter (“WM”) of the cerebellum of P6 control and Krit1ECKO littermates is shown. Scale bars, 50 μm. b, Anti-GFP immunostaining to detect TCF/Lef:H2B-GFP Wnt/β-catenin reporter (“WntREP”) activity. Scale bars, 50 μm. c, Immunoblotting of P6 brain endothelial cell lysate for GFP. Results are representative of 3 separate experiments. d, qPCR analysis of β-catenin target genes in hindbrain endothelial cells. N=4–5; P>0.05 for comparison of all values. Error bars indicate SEM. e, Immunostaining for phospho-SMAD3 (“pSMAD3”) and PECAM. Scale bars, 50 μm. f, Total SMAD3 and pSMAD3 were detected by immunoblotting cerebellar endothelial cell lysate. Results are representative of 3 separate experiments.
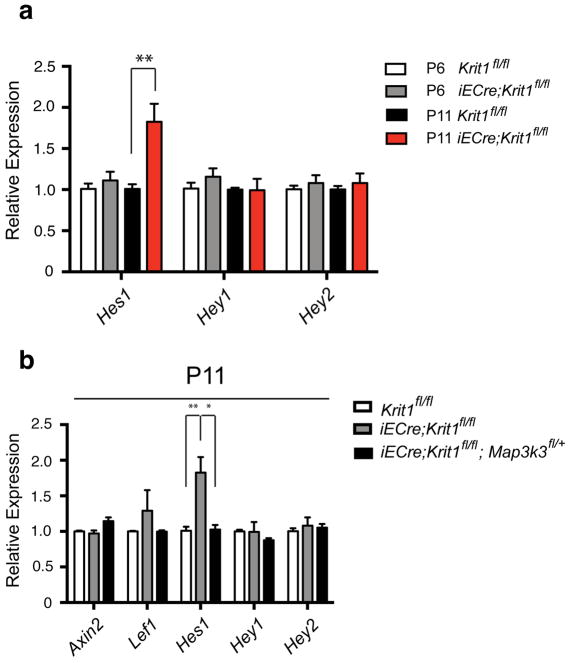
Extended Data Figure 3. Hes1 expression rises after primary CCM lesion formation in Krit1ECKO hindbrain endothelial cells and is reversed by Map3k3 haploinsufficiency.
a, qPCR analysis of Notch target genes in endothelial cells freshly harvested from the hindbrain of P6 or P11 Krit1ECKO animals compared with control, tamoxifen-treated littermates. ** indicates P<0.01. N=4. Error bars indicate SEM. b, qPCR analysis of gene expression in endothelial cells freshly harvested from the cerebellum of P11 Krit1fl/fl and Krit1ECKO animals with and without endothelial loss of one Map3k3 allele. ** indicates P<0.01. N=4. Error bars indicate SEM.
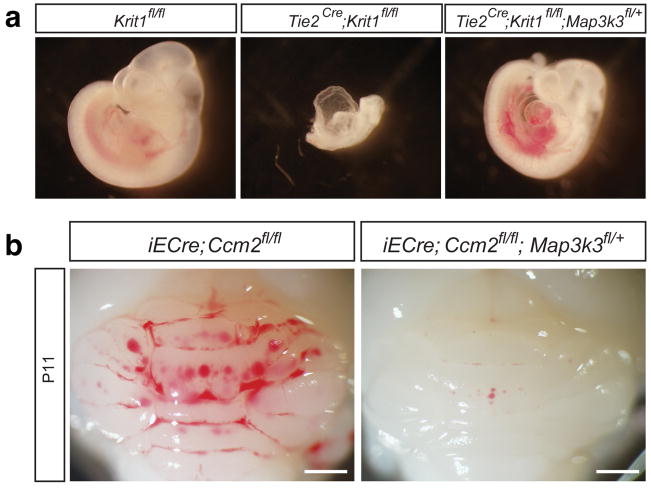
Extended Data Figure 4. Map3k3 haploinsufficiency rescues both early embryonic lethality due to endothelial loss of Krit1 and postnatal CCM lesion formation due to endothelial loss of Ccm2.
a, Partial loss of MEKK3 rescues early lethality associated with endothelial deletion of Krit1. E10.5 littermate embryos are shown. Tie2-Cre;Krit1fl/fl animals lacking endothelial KRIT1 die by E9.5 due to lack of patent branchial arch arteries (middle). In contrast, Tie2-Cre;Krit1fl/fl;Map3k3fl/+ animals develop patent arteries and survive past mid-gestation (right). Note the presence of pericardial edema due to a persistent cardiac defect in the Tie2-Cre;Krit1fl/fl;Map3k3fl/+ embryo. Images are representative of 3 independent experiments. b, Rescue of CCM formation in the Ccm2 model with loss of MEKK3. VE-cadherin CreERT2;Ccm2fl/fl; Map3k3+/+ (“Ccm2ECKO”) and VE-cadherin CreERT2;Ccm2fl/fl; Map3k3fl/+ neonates were induced with tamoxifen at P1 and lesion formation scored by visual detection in the hindbrain at P11 as done for Krit1ECKO animals. Images are representative of 4 independent experiments.
Extended Data Figure 5. Rescue of CCM formation in Krit1ECKO animals with homozygous loss of Klf2.
H-E sections through the hindbrain of P11 VE-cadherin CreERT2;Krit1fl/fl; Klf2fl/fl and control VE-cadherin CreERT2;Krit1fl/fl animals are shown. No true CCM lesions were observed but venous dilatation was evident (arrow). Images are representative of 3 independent experiments. Dotted lines indicate the cerebellar white matter. WM, white matter; GL, granular layer.
Extended Data Figure 6. CCM2 binds MEKK3 via its C-terminal helical harmonin domain.
a, MEKK3 binds the C-terminal domain of CCM2. A schematic of the CCM2 protein domains is shown above and the series of N-terminal truncated CCM2-HA proteins used to map the CCM2-MEKK3 binding region shown below. MEKK3-FLAG and the indicated N-terminally truncated CCM2-HA proteins were co-expressed in HEK293T cells prior to immunoprecipitation with anti-HA antibodies and immunoblotting with anti-FLAG antibodies to detect associated MEKK3 proteins (above). The immunoprecipitated HA-CCM2 was detected with anti-HA antibodies (below). b, MEKK3-FLAG and the indicated C-terminally truncated CCM2-HA proteins were co-immunoprecipitated to map the CCM2-MEKK3 binding region. c, CCM2 CCCTdup protein is stably expressed in cultured endothelial cells. CCM2 CCCTdup-HA protein was expressed in cultured HUVECs using a tetracycline-inducible lentiviral vector at varying doses of doxycycline. The total (i.e. endogenous CCM2 + viral CCM2 CCCTdup) CCM2 mRNA levels were measured using qPCR (shown below) and CCM2 CCCTdup protein detected using anti-HA immunoblotting (shown above). GAPDH immunoblots are shown for loading control.
Extended Data Figure 7. The versican-degrading ADAMTS proteases are regulated by KLF2 and KLF4.
a, siRNA knockdown of KLF2 or KLF4 in cultured HUVECs reduces expression of ADAMTS4 measured using qPCR. b, Adenoviral expression of KLF2 drives expression of the versican-degrading proteases ADAMTS1, ADAMTS4 and ADAMTS9 in addition to the known KLF2 target gene eNOS. KLF4 levels are reduced in KLF2-overexpressing HUVECs. Results are shown relative to expression following exposure to control adeno-LacZ virus. c, Adenoviral expression of KLF4 drives expression of ADAMTS4 in addition to the known KLF2/4 target gene eNOS. KLF2 levels are reduced in KLF4-overexpressing HUVECs. Results are shown relative to expression following exposure to control adeno-LacZ virus. N=4–5; ***P<0.001; **P<0.01;*P<0.05. Error bars indicate SEM.
Extended Data Figure 8. The MEK5 inhibitor BIX02189 and the ERK5 inhibitor XMD17–109 reverse the increase in KLF2 and KLF4 expression conferred by loss of KRIT1 in cultured endothelial cells.
HUVECs were treated with scrambled (Scr.) or KRIT1-targeted siRNAs with and without BIX02189 (10 μM) and XMD17-109 (1 μM) for 24 hours and the levels of KLF2 and KLF4 mRNA measured using qPCR. *** indicates P<0.001; ** indicates P<0.01. N=3. Error bars indicate SEM.
Extended Data Figure 9. Imaging and volume measurement of mouse hindbrain CCM lesions using microCT.
The imaging of P11 mouse hindbrains and analysis of the raw data to create composite CCM lesion images and volumes is shown.
Supplementary Material
Acknowledgments
We thank the members of the Kahn lab for their thoughtful comments during the course of this work. These studies were supported by National Institute of Health grants R01HL094326 (MLK), P01NS092521 (MK), VAMC 1IO1BX002976 (DL), R01NS075168 (KW), T32HL07439 (AT), P01NS092521 (IA), and Australian NHMRC project grant 161558 (XZ).
Footnotes
Author Contributions
ZZ, AT and WW designed and performed most of the experiments and helped write the manuscript. SB performed all of the microCT CCM lesion measurements in a blinded manner. SA, KW, RS, IA and DL provided critical reagents. SB, LG, SZ, JY, AW, MF, XZ and MK helped design and perform the experiments and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Cavalcanti DD, et al. Cerebral cavernous malformations: from genes to proteins to disease. J Neurosurg. 2012;116:122–132. doi: 10.3171/2011.8.JNS101241. [DOI] [PubMed] [Google Scholar]
- 2.Plummer NW, Zawistowski JS, Marchuk DA. Genetics of cerebral cavernous malformations. Curr Neurol Neurosci Rep. 2005;5:391–396. doi: 10.1007/s11910-005-0063-7. [DOI] [PubMed] [Google Scholar]
- 3.Whitehead KJ, et al. The cerebral cavernous malformation signaling pathway promotes vascular integrity via Rho GTPases. Nat Med. 2009;15:177–184. doi: 10.1038/nm.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stockton RA, Shenkar R, Awad IA, Ginsberg MH. Cerebral cavernous malformations proteins inhibit Rho kinase to stabilize vascular integrity. J Exp Med. 2010 doi: 10.1084/jem.20091258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maddaluno L, et al. EndMT contributes to the onset and progression of cerebral cavernous malformations. Nature. 2013;498:492–496. doi: 10.1038/nature12207. [DOI] [PubMed] [Google Scholar]
- 6.Bravi L, et al. Sulindac metabolites decrease cerebrovascular malformations in CCM3-knockout mice. Proc Natl Acad Sci U S A. 2015;112:8421–8426. doi: 10.1073/pnas.1501352112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uhlik MT, et al. Rac-MEKK3-MKK3 scaffolding for p38 MAPK activation during hyperosmotic shock. Nat Cell Biol. 2003;5:1104–1110. doi: 10.1038/ncb1071. [DOI] [PubMed] [Google Scholar]
- 8.Zawistowski JS, et al. CCM1 and CCM2 protein interactions in cell signaling: implications for cerebral cavernous malformations pathogenesis. Hum Mol Genet. 2005;14:2521–2531. doi: 10.1093/hmg/ddi256. [DOI] [PubMed] [Google Scholar]
- 9.Cullere X, Plovie E, Bennett PM, MacRae CA, Mayadas TN. The cerebral cavernous malformation proteins CCM2L and CCM2 prevent the activation of the MAP kinase MEKK3. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1510495112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher OS, et al. Structure and vascular function of MEKK3-cerebral cavernous malformations 2 complex. Nat Commun. 2015;6:7937. doi: 10.1038/ncomms8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, et al. Structural Insights into the Molecular Recognition between Cerebral Cavernous Malformation 2 and Mitogen-Activated Protein Kinase Kinase Kinase 3. Structure. 2015 doi: 10.1016/j.str.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Z, et al. The cerebral cavernous malformation pathway controls cardiac development via regulation of endocardial MEKK3 signaling and KLF expression. Dev Cell. 2015;32:168–180. doi: 10.1016/j.devcel.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boulday G, et al. Developmental timing of CCM2 loss influences cerebral cavernous malformations in mice. J Exp Med. 2011;208:1835–1847. doi: 10.1084/jem.20110571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glading A, Han J, Stockton RA, Ginsberg MH. KRIT-1/CCM1 is a Rap1 effector that regulates endothelial cell cell junctions. J Cell Biol. 2007;179:247–254. doi: 10.1083/jcb.200705175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wustehube J, et al. Cerebral cavernous malformation protein CCM1 inhibits sprouting angiogenesis by activating DELTA-NOTCH signaling. Proc Natl Acad Sci U S A. 2010;107:12640–12645. doi: 10.1073/pnas.1000132107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuttano R, et al. KLF4 is a key determinant in the development and progression of cerebral cavernous malformations. EMBO Mol Med. 2015 doi: 10.15252/emmm.201505433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrer-Vaquer A, et al. A sensitive and bright single-cell resolution live imaging reporter of Wnt/ss-catenin signaling in the mouse. BMC Dev Biol. 2010;10:121. doi: 10.1186/1471-213X-10-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitehead KJ, Plummer NW, Adams JA, Marchuk DA, Li DY. Ccm1 is required for arterial morphogenesis: implications for the etiology of human cavernous malformations. Development. 2004;131:1437–1448. doi: 10.1242/dev.01036. [DOI] [PubMed] [Google Scholar]
- 19.Gibson CC, et al. Strategy for identifying repurposed drugs for the treatment of cerebral cavernous malformation. Circulation. 2015;131:289–299. doi: 10.1161/CIRCULATIONAHA.114.010403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher OS, et al. Structural basis for the disruption of the cerebral cavernous malformations 2 (CCM2) interaction with Krev interaction trapped 1 (KRIT1) by disease-associated mutations. J Biol Chem. 2015;290:2842–2853. doi: 10.1074/jbc.M114.616433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Draheim KM, et al. CCM2-CCM3 interaction stabilizes their protein expression and permits endothelial network formation. J Cell Biol. 2015;208:987–1001. doi: 10.1083/jcb.201407129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spiegler S, et al. High mutation detection rates in cerebral cavernous malformation upon stringent inclusion criteria: one-third of probands are minors. Mol Genet Genomic Med. 2014;2:176–185. doi: 10.1002/mgg3.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchi S, et al. Defective autophagy is a key feature of cerebral cavernous malformations. EMBO Mol Med. 2015;7:1403–1417. doi: 10.15252/emmm.201505316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borikova AL, et al. Rho kinase inhibition rescues the endothelial cell cerebral cavernous malformation phenotype. J Biol Chem. 2010;285:11760–11764. doi: 10.1074/jbc.C109.097220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng X, et al. Dynamic regulation of the cerebral cavernous malformation pathway controls vascular stability and growth. Dev Cell. 2012;23:342–355. doi: 10.1016/j.devcel.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pagenstecher A, Stahl S, Sure U, Felbor U. A two-hit mechanism causes cerebral cavernous malformations: complete inactivation of CCM1, CCM2 or CCM3 in affected endothelial cells. Hum Mol Genet. 2009;18:911–918. doi: 10.1093/hmg/ddn420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akers AL, Johnson E, Steinberg GK, Zabramski JM, Marchuk DA. Biallelic somatic and germline mutations in cerebral cavernous malformations (CCMs): evidence for a two-hit mechanism of CCM pathogenesis. Hum Mol Genet. 2009;18:919–930. doi: 10.1093/hmg/ddn430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chao TH, Hayashi M, Tapping RI, Kato Y, Lee JD. MEKK3 directly regulates MEK5 activity as part of the big mitogen-activated protein kinase 1 (BMK1) signaling pathway. J Biol Chem. 1999;274:36035–36038. doi: 10.1074/jbc.274.51.36035. [DOI] [PubMed] [Google Scholar]
- 29.McDonald DA, et al. Fasudil decreases lesion burden in a murine model of cerebral cavernous malformation disease. Stroke. 2012;43:571–574. doi: 10.1161/STROKEAHA.111.625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, et al. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature. 2010;465:483–486. doi: 10.1038/nature09002. [DOI] [PubMed] [Google Scholar]
- 31.Mleynek TM, et al. Lack of CCM1 Induces Hypersprouting and Impairs Response to Flow. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JS, et al. Klf2 is an essential regulator of vascular hemodynamic forces in vivo. Dev Cell. 2006;11:845–857. doi: 10.1016/j.devcel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 33.Katz JP, et al. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kisanuki YY, et al. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.