Abstract
Alcoholism is associated with breast cancer incidence and progression, and moderate chronic consumption of ethanol is a risk factor. The mechanisms involved in alcohol's oncogenic effects are unknown, but it has been speculated that they may be mediated by acetaldehyde. We used the immortalized normal human epithelial breast cell line MCF-12A to determine whether short- or long-term exposure to ethanol or to acetaldehyde, using in vivo compatible ethanol concentrations, induces their oncogenic transformation and/or the acquisition of epithelial mesenchymal transition (EMT). Cultures of MCF-12A cells were incubated with 25 mM ethanol or 2.5 mM acetaldehyde for 1 week, or with lower concentrations (1.0–2.5 mM for ethanol, 1.0 mM for acetaldehyde) for 4 weeks. In the 4-week incubation, cells were also tested for anchorage-independence, including isolation of soft agar selected cells (SASC) from the 2.5 mM ethanol incubations. Cells were analyzed by immunocytofluorescence, flow cytometry, western blotting, DNA microarrays, RT/PCR, and assays for miRs. We found that short-term exposure to ethanol, but not, in general, to acetaldehyde, was associated with transcriptional upregulation of the metallothionein family genes, alcohol metabolism genes, and genes suggesting the initiation of EMT, but without related phenotypic changes. Long-term exposure to the lower concentrations of ethanol or acetaldehyde induced frank EMT changes in the monolayer cultures and in SASC as demonstrated by changes in cellular phenotype, mRNA expression, and microRNA expression. This suggests that low concentrations of ethanol, with little or no mediation by acetaldehyde, induce EMT and some traits of oncogenic transformation such as anchorage-independence in normal breast epithelial cells.
Keywords: breast cancer, MCF-12A, ethanol, alcohol, epithelial mesenchymal transition
Introduction
Alcoholism is a risk factor for breast cancer, with consumption of 3 or more drinks per day leading to a 40–50% increase in risk, and with approximately 50,000 alcohol-attributable cases per year worldwide (1–3). It is postulated that the effects of alcohol are not exerted in the early stage of the carcinogenic process, since alcohol effects are not associated with ductal carcinoma in situ of the breast in postmenopausal women. There is also a greater associated risk for lobular than for ductal breast cancer (4–6). Alcohol induced cancers may be restricted to estrogen receptor-positive tumors (2,7). Even low levels of alcohol consumption (3–6 drinks/week) are associated with a small increase in breast cancer risk, with the most consistent measure being cumulative alcohol intake throughout adult life combined with binge drinking (3–5).
Experimental studies in mice and rats have shown that ethanol consumption promotes mammary tumors and abnormal tissue development (8–13) at least partly via the estrogen pathway (8). In vitro, the estrogen association has mainly been shown using the breast cancer luminal epithelial cell line MCF7, which is estrogen and progesterone receptor positive and lacks ERBB2 gene amplification or Her2/neu protein overexpression (14–17). Studies, mostly with an ethanol exposure of less than 1 week and concentrations higher than 50 mM, produced modest stimulation of in vitro growth, invasiveness, and migration of MCF7 cells (18–23) and in other more malignant breast cancer cell lines (18–23).
In our recent preliminary study using MCF7 cells (see abstract) (24), we found that short-term exposure to ethanol was associated with modest transcriptional upregulation of the metallothionein family, but long-term exposure led to more substantial upregulation of Oct4, Nanog, and CEACAM6 protein expression, and results in a global oncogenic transcriptional signature as well as the stimulation of anchorage-independence. This process was not directly mediated by acetaldehyde, nor by observable estrogen responsiveness. These results suggested that ethanol may enhance the oncogenesis of breast cancer cells through the interplay of cancer-related genes and their regulatory miRs.
Surprisingly, few publications are available on the effects of exposure to ethanol on the MCF-12A cell line (25), which is derived from normal but immortalized breast epithelial cells, or on other epithelial breast cell cultures. Similar to MCF7, the MCF-12A cell line is both estrogen receptor-positive and progesterone receptor-positive (26). To our knowledge, there are no publications about MCF-12A cells regarding the transcriptional signatures that would help to characterize breast tissue related oncogenic transformation, neither any publications on the role of microRNAs (miRs) in ethanol treated MCF-12A, particularly with respect to the miRs that are known to regulate key mRNA levels in breast cancer oncogenic pathways (27–31). In turn, there are some related reports on other normal epithelial breast cell cultures (32,33).
The putative oncogenic effects of alcohol have been proposed to be mediated by stimulation of estrogen levels and/or estrogen responsiveness, and also potentially by other effects unrelated to estrogen (1,2,34–38). The latter may include the inhibition of DNA methylation, interaction with retinoid metabolism, or oxidative stress, and could operate either by ethanol effects or through the first ethanol metabolite, acetaldehyde, which is produced by alcohol dehydrogenase (ADH). Acetaldehyde effects might include the formation of stable DNA adducts or decreased glutathione. However, it is possible that ethanol, besides these direct or indirect oncogenic effects, could stimulate a later stage of tumor progression through increased cell invasiveness, detachment, and metastasis. A well studied process mediating these alterations in epithelial cancers is the epithelial mesenchymal transition (EMT), a process that has been extensively studied in breast cancer and MCF7 cells (39,40), but not under ethanol induction.
In a recent study, both non-tumorigenic (MCF 10A, MCF-12A) and tumorigenic (MCF7) breast epithelial cells exposed to cigarette smoke acquired mesenchymal properties. These properties included fibroblastoid morphology, increased anchorage-independent growth, motility, and invasiveness. For the MCF 10A cells, this may be related to the emergence of a CD44(high)/CD24(low) population, and in both normal cell types is associated with changes in gene expression related to EMT. The MCF 10A transplanted into mice, which were later treated with cigarette smoke extract, showed increased survival and colonization of the mammary ducts (41). In the only other report that we could find, the induction of the expression of transcription factor ESX in MCF-12A confers growth in soft agar and a transformed in vitro metastatic phenotype consistent with EMT (42). A few studies of ethanol or alcohol described EMT effects in other normal and malignant tissues and in breast cancer cells (43–45). No such effects on MCF-12A or other normal breast epithelial cells have been reported.
In the current study, the effects of short- and long-term exposures to various physiologically relevant concentrations of ethanol and relatively high dose acetaldehyde were studied using MCF-12A monolayers. In one experiment, ethanol-induced cells were selected for anchorage independence by survival in soft agar. Analysis of stem cell markers and global transcriptional gene expression signatures including miRs, with particular reference to EMT, were carried out to better understand the mechanism of action of alcohol on the putative induction of malignant features on normal breast epithelial cells, in order to better clarify early effects of alcohol in breast cancer.
Materials and methods
Cell lines
MCF-12A (catalog CRL-10782) was obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA) and grown according to the recommendations provided by the supplier, in medium containing DMEM/F12 in the presence of 2.5% horse serum and supplemented with epidermal growth factor, hydrocortisone, bovine insulin, cholera toxin, and antibiotic/antimycotic. Cells were carried in 6 well plastic plates at 37°C, 5% CO2 and passaged as required at 90% confluence or less. Ethanol, molecular grade (Fisher Scientific, Pittsburgh, PA, USA) and acetaldehyde ACS grade (Sigma Aldrich, St. Louis, MO, USA) were added to cell culture wells as described and replaced when culture medium was changed, typically at 3 day intervals. Mammospheres were generated by disaggregating monolayer cultures and applying 50,000 cells per well to Corning Inc. (Corning, NY, USA) ultra low attachment plastic plates in the presence of mammary epithelial cell growth medium (Fisher, MEBM) plus 2% v/v B27 supplement (Invitrogen, Carlsbad, CA, USA) and 0.01 mg/ml bovine insulin.
Western blots
Blots were accomplished using standard methods: Cells were dissolved in boiling buffer (1% SDS, 1 mM sodium orthovanadate, 10 mM Tris pH 7.4, and protease inhibitors) by scraping cells from wells and passing the lysate several times through a 26 gauge needle to reduce viscosity. Protein concentrations and recoveries were determined using the Pierce protein determination kit (Thermo Fisher, Waltham, MA, USA). Protein samples were applied to 4–15% linear gradient gels (Bio-Rad, Richmond, CA, USA) and electrophoresed, electrophoretically transferred to PVDF membranes (Bio-Rad), and analyzed for specific proteins by antibody binding using standard methods including enzyme bound second antibodies and Super Signal West Pico luminescent solution (Pierce).
RNA expression analysis
Cell cultures in 6-well plates were washed once with PBS, and RNA was extracted and purified using the Qiagen RNeasy Plus Micro kit (Qiagen Sciences Inc., Germantown, MD, USA). RNA concentration and recovery was determined using the Nanodrop apparatus (Thermo Fisher), and RNA integrity was determined by Bioanalyzer (Agilent, Santa Clara, CA, USA). All RNA samples were determined to have an RNA Integrity Number (RIN) of 8 or higher. Gene expression was determined from RNA samples by DNA microarray analysis using the Affymetrix (Santa Clara, CA, USA) Human Gene 1.0 ST carried out by the UCLA DNA Microarray Core facility, or by the Affymetrix Human Gene 1.1 ST by the UCLA Pathology Department DNA Laboratory. Relative expression values from different treatments were compared using Microsoft Excel. Some samples were subjected to polymerase chain reaction (PCR) as follows: RNA samples of 1 μg were reverse transcribed using the Invitrogen SuperScript III first-strand synthesis Supermix according to the manufacturer's recommendation. cDNA derived from the 1 week ethanol treated samples and untreated control cDNA samples from the same 6-well plate were subjected to polymerase chain reaction assays for MT1X and results were compared with GAPDH controls, according to standard methods using the Clontech Advantage 2 PCR kit (Mountain View, CA, USA) according to the manufacturer's recommendations. Primers: for MT1X forward TCATCTGTCCCGCTGCGTGT and reverse GGCACAGGAGCCAACAGGCG. For GAPDH, forward GTCGCCAGCCGAGCCACACT and reverse TGACCTTGGCCAGGGGTGCT. Gene identification labels are according to the NCBI (National Center for Biotechnology Information) Gene database: http://www.ncbi.nlm.nih.gov/gene and are referred to as NCBI:Gene in figure and table descriptions.
MicroRNA analysis
Cells were grown on 6-well plates in the presence or absence of either ethanol or acetaldehyde as described in Results. Total RNA including microRNAs (miRs) was purified using the Mirvana microRNA purification kit (Ambion; Life Technologies, Austin, TX, USA). Samples of RNA were analyzed for miR content by LC Sciences, Houston, TX, USA. Relative miR levels are expressed on an arbitrary scale following normalization. MicroRNA results and DNA micro-array results are deposited in the Gene Expression Omnibus (GEO) archive under accession number GSE76953. MicroRNA names and sequences are available at www.mirbase.org.
Flow cytometry
Cells were grown and treated with ethanol or acetaldehyde as described, washed twice with Hanks Balanced Salt Solution (HBSS), disaggregated by repeated pipeting in Cell Stripper (Mediatech, Manassas, VA, USA), pelleted, and resuspended in staining buffer consisting of PBS plus 3% FBS (SB). Cells were incubated in the presence of fluorescent conjugated antibodies for 30 min. on ice, washed twice with SB, and finally resuspended in SB for flow cytometry on an LSR II (BD Biosciences, San Diego, CA, USA). Controls using either no antibody or all possible combinations of antibodies were used to validate specificity of cell staining. Data analysis and plotting were done using FACSDiva Version 6.1.1 software. All fluorophore-conjugated antibodies and isotype controls were from eBioscience (San Diego, CA, USA).
Soft agar growth
Cells were trypsinized and subjected to the soft agar tumorigenic/anchorage-independent cell selection procedure: Cells were suspended in 1 ml/well of warm (37°C) 0.3–0.5% agar in culture medium (soft agar layer) and 10,000 cells/ml were deposited in duplicate or triplicate wells above a layer of 1 ml of 1% agar (in the same medium) that had been allowed to solidify on 6-well plates at 4°C (hard layer agar). Cultures were allowed to grow for 4 weeks and when foci were visible, they were stained with 0.005% crystal violet in Hanks' solution for 1 h, and colonies were counted. In certain cases, separate foci from the soft agar cultures not subjected to staining were transferred to culture medium in T-25 flasks, grown as monolayers in the absence of ethanol or acetaldehyde, and used for further experiments including gene expression analysis.
Results
Short-term continuous exposure of MCF-12A cells to high dose ethanol causes changes in the global transcriptional signature that are not, however, induced by acetaldehyde, and does not affect anchorage-independence
Monolayer cultures of MCF-12A cells were subjected to 1 week incubations with 25 mM ethanol, roughly equivalent to the peak alcohol concentration in serum after 4–5 glasses of wine, or to 2.5 mM acetaldehyde replaced daily, a concentration several-fold higher than would be expected from this level of alcohol ingestion in either humans or rats (46,47). These concentrations were similar to the ones used by us in a preliminary study of 1- and 4-week exposures of MCF7 breast epithelial cancer cells to these agents (24). No obvious effects on cell culture growth or morphology were observed in the MCF-12A cells, and the extracted RNAs were subjected to DNA microarray assays. Table I shows that mRNA levels for the metallothionein genes, known to be affected by ethanol, were increased by ethanol in these experiments, particularly the 1F, 1L and 1X, but the same genes were largely unaffected by acetaldehyde. This result was confirmed for MT1X by quantitative RT/PCR (Fig. 1).
Table I.
MCF-12A monolayers were exposed to ethanol or acetaldehyde for 1 week.a
| Ethanol (Eth) | Acetaldehyde (Act) | |||
|---|---|---|---|---|
|
|
|
|||
| Gene | Eth/C ratio | C value | Act/C ratio | C value |
| MT1A | 1.32 | 178 | 1.13 | 1039 |
| MT1B | 1.25 | 97 | 1.02 | 168 |
| MT1F | 2.24 | 2248 | 1.09 | 3193 |
| MT1G | 1.33 | 3777 | 0.94 | 4279 |
| MT1H | 1.31 | 176 | 1.11 | 221 |
| MT1L | 2.01 | 1806 | 0.99 | 3975 |
| MT1X | 2.14 | 1816 | 0.99 | 7219 |
| MT2A | 1.24 | 13940 | 0.96 | 20349 |
| MT4 | 1.53 | 128 | 1.25 | 147 |
RNA samples were subjected to analysis by DNA microarray. The ratios for treated (Eth or Act) vs. untreated control (C) are shown. Also shown are the C values from the DNA microarrays. Each C value represents the normalized gene expression value from the DNA microarray, representing the respective level of expression. Gene: gene symbols are according to the Gene database in the NCBI section of the National Library of Medicine, NIH, USA. Results for different metallothionein genes are shown.
Figure 1.
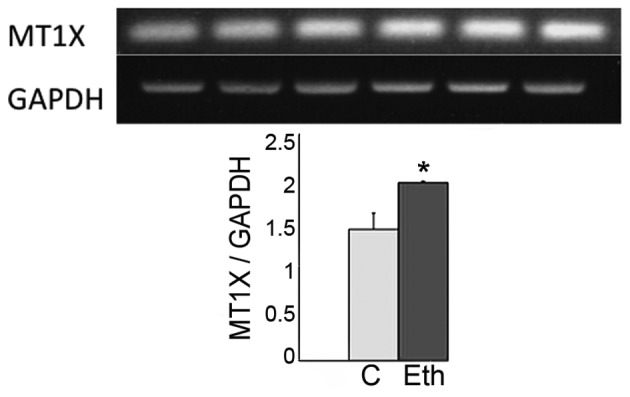
The short-term exposure of MCF-12A cells to a high dose of ethanol upregulates the transcriptional expression of the metallothionein (MT) gene family. Cultures were maintained for 7 days in the presence of 25 mM ethanol, 2.5 mM acetaldehyde (fresh daily), or no addition. Above: RNA was isolated from triplicate cultures of control cells (left side) and ethanol treated cells (right side) and submitted to RT/PCR. Below: Densitometry for triplicate samples *p<0.05.
In total, 129 MCF-12A genes were upregulated by >2.0 and 257 were downregulated to <0.5 by 25 mM ethanol, as compared to control MCF-12A cells grown in the absence of these agents. By contrast, 2.5 mM acetaldehyde only upregulated 19 genes by >2.0 and downregulated 58 genes by <0.5. Within these ranges, and considering genes that are related to alcohol metabolism, ethanol upregulated AOX1 and downregulated ALDH3B2, and acetaldehyde similarly affected to a lesser extent both genes and increased ALDH1A3 (Table II). However, the expression of the stem cell/Aldefluor related isoform, ALDH1A1, was not affected by either treatment.
Table II.
The short-term exposure of MCF-12A cells to high dose ethanol or acetaldehyde changes the transcriptional expression balance among members of the serpin, ankyrin, and alcohol metabolism gene families.a
| Ethanol (Eth) | Acetaldehyde (Act) | ||||
|---|---|---|---|---|---|
|
|
|
||||
| Gene name | Gene symbol | Eth/cont ratio | C value | Act/cont ratio | C value |
| Alcohol metabolism | |||||
| Aldehyde Dhd1A3 | ALDH1A3 | 0.80 | 1191 | 1.91 | 376 |
| Aldehyde Dhd3B2 | ALDH3B2 | 0.19 | 477 | 0.69 | 226 |
| Aldehyde Ox 1 | AOX1 | 1.91 | 138 | 1.86 | 213 |
| Serpins | |||||
| Serpin A1 | A1 | 4.41 | 314 | 0.98 | 504 |
| Serpin B2 | B2 (PAI 2) | 0.14 | 2625 | 2.04 | 495 |
| Serpin B3 | B3 | 0.10 | 2267 | 1.14 | 754 |
| Serpin B7 | B7 | 0.11 | 886 | 0.76 | 41 |
| Ankyrins | |||||
| Ankyrin 36B | ANKRD 36Bb | 2.07 | 104 | 1.60 | 152 |
| Ankyrin 22 | ANKRD 22 | 0.17 | 1027 | 0.72 | 67 |
| Epithelial/mesenchymal transition | |||||
| Cadherin 11 | CDH11 | 3.25 | 153 | 0.97 | 42 |
| Interleuk 31 rec A | IL3IRA | 2.72 | 125 | 1.49 | 62 |
| Connect tiss gf | CTGF | 2.67 | 284 | 0.91 | 748 |
| Integrin sub b6 | ITGB6 | 2.53 | 236 | 1.06 | 118 |
| Laminin β3 | LAMB3 | 2.46 | 1103 | 1.50 | 1822 |
| Matrix metl pep 2 | MMP2 | 2.34 | 210 | 1.09 | 495 |
| Thrombospondin 1 | THBS1 | 2.15 | 2410 | 1.36 | 1045 |
| Met assoc lung ad | MALAT1b | 1.86 | 388 | 1.67 | 2942 |
| Interleukin 6 | IL6 | 2.12 | 81 | 1.42 | 119 |
| Trans GFβ2 | TGFB2 | 2.09 | 413 | 1.36 | 856 |
| Cadherin 4 | CDH4 | 2.00 | 213 | 0.88 | 145 |
| Keratin 17 | KRT 17b | 0.51 | 10650 | 0.55 | 5277 |
| Keratin 78 | KRT 78 | 0.27 | 314 | 1.14 | 101 |
| Keratin 23 | KRT 23 | 0.24 | 228 | 0.76 | 127 |
| Keratin 80 | KRT 80 | 0.22 | 727 | 0.98 | 100 |
| Keratin 16 | KRT 16b | 0.28 | 2923 | 0.49 | 1850 |
| Keratin 4 | KRT 4 | 0.11 | 1197 | 0.71 | 176 |
Cultures were maintained for 7 days in the presence of 25 mM ethanol, 2.5 mM acetaldehyde (fresh daily), or no addition (cont). RNA was subjected to DNA microarray analysis by Affymetrix human Gene 1.0 ST.
A few genes have multiple probe sets in this system, so results were averaged. Details as in Fig. 1. Eth/cont ratio: ratio of DNA microarray values for ethanol treated vs. untreated cells. Act/cont ratio: as in ethanol vs. control treated, but the treatment was with acetaldehyde. C value: the normalized DNA microarray value for each gene's expression in the control specimens. Gene symbols are according to NCBI:Gene.
Alcohol substantially affected the transcription of 4 serpins, upregulating A1 and downregulating B3, B7, and B2 (PAI2) but acetaldehyde only affected SerpinB2. One ankyrin mRNA was increased by both agents (37), and one was decreased by both (22). Remarkably, a number of other genes related to EMT were upregulated by ethanol (e.g., CTGF, LAMB3, TGFB2 and others) or downregulated (KRT17, KRT16, KRT4, and other keratins) as expected for the EMT process. Again, acetaldehyde affected only some of these genes and to a much lesser extent.
These transcriptional alterations elicited by 1 week incubation did not translate into phenotypic changes related to malignancy, since neither ethanol nor acetaldehyde treatment caused anchorage-independence as judged by the lack of foci formation in soft agar, neither was there any effect regarding mammosphere formation or increased stem cell content as judged by immunocytochemistry of the embryonic stem cell genes Oct4 or nanog, or by quantitative western blots (data not shown).
Long-term continuous exposure of MCF-12A cells to lower doses of ethanol or acetaldehyde than the ones applied in the short-term incubations induce changes in morphology, cell growth, and anchorage-independence
In order to examine the impact of more prolonged exposure to ethanol or acetaldehyde, so as to better model the long term processes affecting tissues in the alcohol-drinking human, MCF-12A monolayer cultures were incubated in the presence of one or the other of these compounds for 4 weeks. However, in contrast to our previous experience with MCF7 cells, whose proliferation was stimulated by 25 mM ethanol (24), this high concentration led, strikingly, to MCF-12A cell death after 2 weeks, with only a very small fraction of cells surviving. Parallel incubations with lower concentrations (10 and 5 mM) allowed growth but caused a considerable slowing of cell proliferation, and only even lower concentrations (2.5 and 1.0 mM) allowed normal replication of MCF-12A cells.
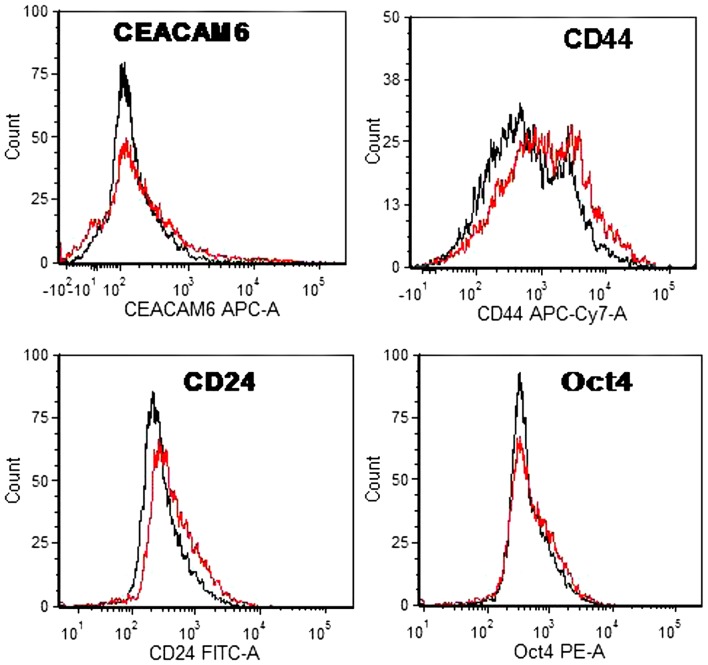
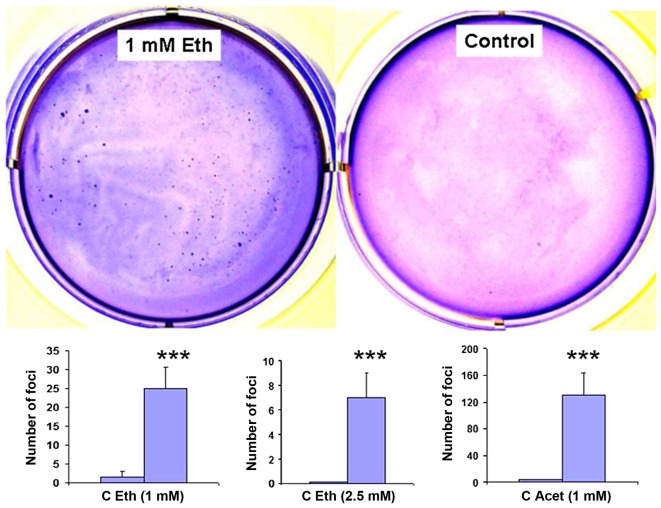
Flow cytometry of the cells exposed to 2.5 mM ethanol failed to show an enrichment of CEACAM6+ cells which had been observed in the case of the MCF7 cells incubated with ethanol at 25 mM (24). Flow cytometry also failed to show Oct4+ cells that would denote stem cells (Fig. 2), but showed the presence of CD44+/CD24(low) cells that are suspected as breast cancer stem cells (48). This finding prompted us to investigate whether ethanol exposure was associated with the induction of anchorage-independence as would be shown by foci formation in soft agar. Fig. 3 shows that this is the case, with 2.5 mM and as low as 1 mM ethanol, and that in contrast to the 1 week incubations with 2.5 mM acetaldehyde, which had not caused any oncogenic phenotype change, acetaldehyde exposure for 4 weeks at 1.0 mM increased the number of foci considerably.
Figure 2.
Flow cytometry shows an enrichment of CD44/CD24(low) cells in long-term 2.5 mM ethanol treated MCF-12A cells within an otherwise homogenous population for other markers. MCF-12A monolayers incubated 4 weeks with 2.5 mM ethanol in red and their untreated controls in black (no gate) for the markers above.
Figure 3.
The long-term exposure of MCF-12A cells to low dose ethanol or to acetaldehyde induces anchorage-independent growth as assessed by the soft agar assay. Monolayer cultures were maintained in triplicate for 4 weeks in the presence of or absence of 1 or 2.5 mM ethanol, or 1 mM acetaldehyde, as indicated, and then cells were transferred to soft agar for 2–3 weeks growth in triplicate on Petri dishes in the absence of added ethanol or acetaldehyde. Foci were stained with methylene blue (top, representing 1 mM ethanol cultures), photographed, and then counted (bottom). ***p<0.001.
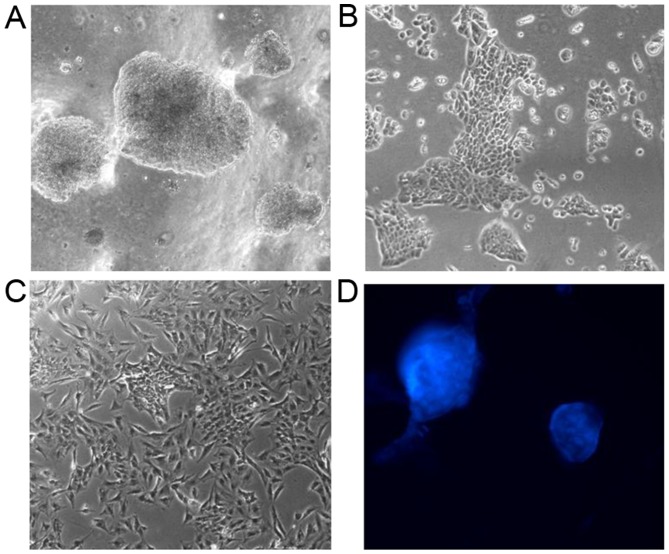
The foci from the 2.5 mM ethanol incubations (Fig. 4A) are able to grow in monolayer and although initially they show the cobblestone morphology of standard MCF-12A cells (Fig. 4B), after growth for approximately 4 weeks they develop typical mesenchymal morphology (Fig. 4C). This is suggestive of the EMT that the 1 week incubation with 25 mM alcohol suggested, as shown in Table II. Moreover, these cloned cells coalesced to form multiple mammospheres in the appropriate medium (Fig. 4D). The cells persist for several months in culture and can be stored stably at liquid nitrogen temperature.
Figure 4.
Long-term exposure of MCF-12A cells to low dose ethanol induces a gradual morphological EMT, and the induced soft agar foci are able to continue growing in monolayer and lead to spontaneous mammosphere formation. The 2.5 mM ethanol incubations of the MCF-12A monolayers that originated as the soft agar foci depicted in Fig. 2, one of them magnified in (A; ×200), continue to show the cobblestone shape (B; ×200), but upon 3 weeks of culture they undergo the spindle shape transformation indicative of EMT (C; ×200). These originated as multiple mammospheres in regular medium and culture flasks (D; ×200).
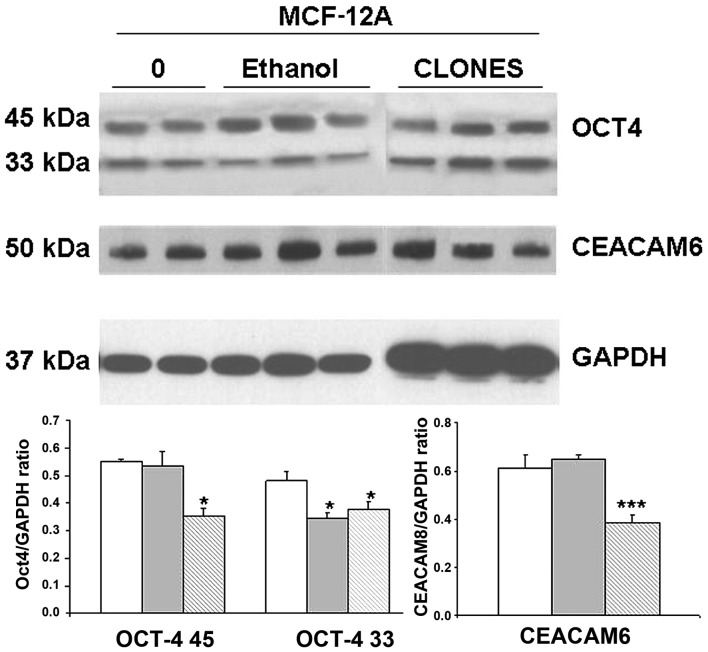
We expected, based on preliminary results with MCF7 cells, that CEACAM6, indicative of oncogenesis, or Oct4, indicative of stemness, would be overexpressed after the 4-week exposure to 2.5 mM ethanol and particularly in the soft agar selected cells (SASC). However, western blot analysis showed either no change or a significant decrease (Fig. 5).
Figure 5.
Long-term exposure of MCF-12A cells to low dose ethanol does not upregulate Oct4a or CEACAM6 protein expression, but the induced foci in soft agar have lower levels of expression of both genes. The 2.5 mM ethanol-incubated monolayer cultures and the resulting soft agar foci amplified in monolayer, as in Figs. 3 and 4, were used for quantitative western blot assay in triplicate. Blank columns: MCF-12A incubated in the absence of ethanol. Gray columns: MCF-12A cultured in the presence of 2.5 mM ethanol. Double hatched columns: soft agar clones from the previously ethanol exposed cells amplified in monolayer under 2.5 mM ethanol. *p<0.05; ***p<0.001.
Continuous exposure of MCF-12A cells to low dose ethanol is associated with significant alterations in the global transcriptional signature, including changes consistent with induction of EMT
DNA microarray assays were performed on RNAs isolated from the MCF-12A cells growing in monolayer in the absence of treatment (control) and from the 2.5 mM ethanol and 1.0 mM acetaldehyde incubations, as well as from the soft agar selected cells (SA clone) described above. Table III shows six families of genes that are targeted by or markers of EMT: claudin, integrin, keratin, cadherin, serpin, and laminin. Incubation with ethanol substantially downregulated six keratins and two claudins, with none upregulated, whereas in the case of integrins, cadherins, serpins and laminins, some were upregulated and some downregulated. Remarkably, out of the 26 substantial changes caused by ethanol, only 5 genes (keratin 10, cadherin related member 1, cadherin 2, serpin E2 and laminin 4) were similarly affected by acetaldehyde.
Table III.
Gene expression analysis of MCF-12A grown in monolayer in the presence or absence of ethanol, or as soft agar selected cell clones compared with control MCF-12A.a
| Monolayer | SA Clone | |||
|---|---|---|---|---|
|
|
|
|||
| Gene ID | Gene description | Eth/cont | Act/cont | SASC/cont |
| Claudin and Integrin families | ||||
| CLDN8 | Claudin 8 | 0.95 | 0.95 | 0.25 |
| CLDN4 | Claudin 4 | 0.9 | 1.0 | 0.21 |
| CLDN7 | Claudin 7 | 0.17 | 0.8 | 0.15 |
| CLDN 1 | Claudin 1 | 0.09 | 0.9 | 0.07 |
| ITGB8 | Integrin, β8 | 1.2 | 1.0 | 3.5 |
| ITGBL1 | Integrin β like 1 | 1.3 | 1.0 | 3.3 |
| ITGA3 | Integrin, α3 | 1.3 | 1.0 | 2.5 |
| ITGB4 | Integrin, β4 | 0.33 | 0.8 | 0.2 |
| ITGB6 | Integrin, β6 | 0.46 | 1.0 | 0.18 |
| ITGA6 | Integrin, α6 | 0.36 | 0.9 | 0.16 |
| Serpin family | ||||
| SERPINE2 | Serpin E2 | 22 | 7.7 | 9 |
| SERPINB1 | Serpin B1 | 1.7 | 1.6 | 3 |
| SERPINH1 | Serpin H1 | 1.5 | 1.2 | 2.9 |
| SERPINB5 | Serpin B5 | 0.6 | 1.0 | 0.3 |
| SERPINA3 | Serpin A3 | 0.5 | 1.0 | 0.2 |
| SERPINB13 | Serpin B13 | 0.1 | 0.7 | 0.06 |
| SERPINB3 | Serpin B3 | 0.05 | 1.0 | 0.02 |
| Keratin and Cadherin families | ||||
| KRT5 | Keratin 5 | 0.6 | 1.0 | 0.32 |
| KRT6C | Keratin 6c | 0.6 | 1.0 | 0.19 |
| KRT16P3 | Keratin 16 pseudogene 3 | 0.3 | 1.1 | 0.1 |
| KRt17 | Keratin 17 | 0.1 | 1.2 | 0.05 |
| KRT16 | Keratin 16 | 0.1 | 1.2 | 0.02 |
| KRT10 | Keratin 10 | 0.1 | 0.3 | 0.01 |
| CDHR1 | Cad rel-member 1 | 22 | 16 | 0.66 |
| CDH2 | Cad 2, t1, N-cadherin | 3 | 1.9 | 4.9 |
| CDH13 | Cad 13, H-cadherin | 1.7 | 1.2 | 0.43 |
| CDH1 | Cad1, type 1, E-cadher | 0.1 | 1.1 | 0.05 |
| CDH3 | Cad 3, t1, P-cadherin | 0.13 | 0.9 | 0.02 |
| Laminin family | ||||
| LAMA4 | α4 | 8.1 | 2.2 | 25 |
| LAMC1 | γ1 | 2.1 | 1.5 | 2.3 |
| LAMA3 | α3 | 0.38 | 0.9 | 0.33 |
| LAMB3 | β3 | 0.32 | 0.7 | 0.07 |
| LAMC2 | γ2 | 0.43 | 1.0 | 0.06 |
Gene expression analysis was carried out using Affymetrix human Gene 1.1 ST assays. Eth/cont: ratio of ethanol treated to control (untreated) DNA microarray values. Act/cont: ratio of acetaldehyde to control DNA microarray values. SASC/cont: ratio of soft agar selected clone gene expression to control gene expression. Gene ID: gene identifications according to NCBI:Gene.
The most striking changes were seen in the SASC clone, where virtually every change induced by ethanol in the monolayer was magnified to a remarkable extent, except for a few genes (serpinE2, LAMC1, CLDN7, CLDN1, and LAMA3). There were a few other genes not changed in the ethanol treated monolayer which appeared in the SASC changed population. This suggests that most of the alterations seen in the monolayer were due to changes to a subfraction of cells which are reflected in the cloned foci selected by soft agar growth.
The EMT signature was confirmed by changes in the expression of key gene families triggering EMT, such as inter-leukins, TGFβ, and IGF families, as well as the Twist1/Snail pathways (Table IV). Twist1 is of particular interest since it was also induced by acetaldehyde, and the SASC clone over-expression was much lower than that seen in monolayer. Some similar patterns described in Table III were seen: ethanol inducing in monolayer interleukin up- and down-regulation, upregulated genes in the TGFβ and IGF families, and most of these changes amplified in the SASC clone. However, a few genes did not follow this trend in the SASC clone as compared with the monolayer, such as IL1R2, CD36 and IGFBP4. Of note, acetaldehyde treated cells differed from the results shown in Table III.
Table IV.
Effects of long-term ethanol exposure of MCF-12A cells on the expression of key gene families triggering EMT.a
| SA Clone | Monolayer | |||
|---|---|---|---|---|
|
|
|
|||
| Gene ID | Gene description | SASC/cont | Eth/cont | Act/cont |
| Interleukin family | ||||
| IL7R | Interleukin 7 receptor | 92.1 | 13.5 | 9.1 |
| IL1R1 | Interleukin 1 receptor, type 1 | 10.5 | 8.1 | 5.5 |
| IL6ST | Interleukin 6 signal transducer | 5.1 | 2.2 | 1.4 |
| IL1R2 | Interleukin 1 receptor type II | 3.4 | 10.3 | 10.0 |
| CXCR1 | Chemokine (C-X-C motif) receptor 1 | 0.75 | 1.0 | 1.0 |
| IL22RA1 | Interleukin 22 receptor α1 | 0.44 | 1.0 | 1.0 |
| IL1A | Interleukin 1 | 0.18 | 0.18 | 0.64 |
| IL1RAP | Interleukin 1 receptor accessory protein | 1.29 | 0.59 | 0.83 |
| IL18 | Interleukin 18 (ifn γ inducing factor) | 0.13 | 0.22 | 1.0 |
| IGF family | ||||
| CTGF | Connective tissue growth factor | 11.9 | 10 | 4.6 |
| IGFBP4 | Insulin-like growth factor binding protein 4 | 6.9 | 6.0 | 4.3 |
| IGFBP3 | Insulin-like growth factor binding protein 3 | 5.0 | 4.5 | 2.7 |
| IGFBP2 | Insulin-like growth factor binding protein 2 | 4.0 | 1.7 | 1.0 |
| IGF2R | Insulin-like growth factor 2 receptor | 3.7 | 1.6 | 1.1 |
| IGF2BP3b | Insulin-like GF2 mRNA binding protein 3 | 2.0 | 1.0 | 1.0 |
| Others | ||||
| ANGPT1 | Angiopoieitin | 47 | 15 | 9 |
| FN1b | Fibronectin 1 | 8.7 | 5.8 | 4.5 |
| ZEB2 | Zinc finger E-box binding homeobox 2 | 5.5 | 5.2 | 2.6 |
| EGFR | Epidermal growth factor receptor | 0.49 | 0.44 | 0.76 |
| PPARgC1A | PPARγ coactivator 1α | 3.4 | 1.2 | 1.0 |
| ZEB1 | Zinc finger E-box binding homeobox 1 | 3.3 | 3 | 1.9 |
| TWIST1 | Twist basic helix-loop-helix transcription factor 1 | 2.4 | 6.7 | 4.5 |
| TGFβ and IGF families | ||||
| TGFb2 | Transforming growth factor β2 | 34 | 19 | 7.1 |
| FSL1 | Follistatin-like 1 | 33.6 | 11.5 | 9.3 |
| THBS1 | Thrombospondin 1 | 26.9 | 2.4 | 1.9 |
| CD36 | CD36 molecule (thrombospondin receptor) | 9.5 | 21.1 | 3.9 |
| TGFBR2 | Transforming growth factor, β receptor 2 | 8.6 | 2.4 | 1.3 |
| BMPR1Ab | Bone morphogenetic protein receptor, type 1A | 3.9 | 0.9 | 0.9 |
| BMPR2 | Bone morphogenetic protein receptor, type II | 3.7 | 1.1 | 1.1 |
| TGFBRAP1 | TGFβ associated receptor assoc protein 1 | 3.1 | 0.9 | 0.8 |
| SMAD7 | Smad family member 7 | 0.49 | 1.0 | 1.0 |
| GDF15 | Growth differentiation factor 15 | 0.4 | 0.5 | 0.7 |
| TGFB1 | Transforming growth factor β1 | 0.3 | 1.5 | 1.3 |
| THBD | Thrombomodulin | 0.07 | 0.4 | 0.9 |
| Aldehyde/Alcohol metabolism | ||||
| AOX1 | Aldehyde oxidase 1 | 18.5 | 7.9 | 4.2 |
| ALDH1B1 | Aldehyde dehydrogenase 1 family, member b1 | 4.9 | 1.2 | 1.0 |
| AKR1B1 | Aldo-keto reductase family 1, member B1 | 2.9 | 1.9 | 1.5 |
| ADH5 | Alcohol dehydrogenase 5 | 2.56 | 1.05 | 1.00 |
| ALDH1L2 | Aldhyde dehydrogenase 1 family, member L2 | 2.10 | 0.47 | 0.75 |
| ALDH1A3 | Aldehyde dehydrogenase 1 family, member A3 | 0.43 | 0.75 | 0.86 |
| ALDH3B2 | Aldehyde dehydrogenase 3 family, member B2 | 0.17 | 0.70 | 1.00 |
| AKR1B10 | Aldo-keto reductase family 1, member B10 | 0.01 | 0.14 | 1.46 |
Gene expression analysis was carried out using the Affymetrix human Gene 1.1 ST assay.
A few genes have multiple probe sets in this assay, so results were averaged. These are indicated after the Gene ID. Gene ID: gene identification according to NCBI:Gene. SASC/cont: ratio of gene expression for soft agar selected cells vs. control cells. Eth/cont and Act/cont refer to ethanol vs. control ratio and acetaldehyde vs. control ratio as in previous tables.
Table IV (bottom) shows changes that are to be expected in the transcription of genes related to aldehyde and alcohol metabolism, although acetaldehyde (with the exception of aldehyde oxidase 1) did not change their expression substantially. As in the other cases, the changes induced by ethanol were much higher in the SASC clone than in the monolayer culture. Other isolated gene expression alterations are compiled, particularly the increase in angiopoietin and fibronectin1 (Table IV: others).
MCF-12A transcriptional signature changes caused by long-term exposure to low doses of ethanol also include changes in cancer-related gene families and microRNAs
Ethanol also affects genes directly related to breast cancer as well as some genes with a possible relationship to oncogenesis (Table V). This was only observed in the current study in the SASC, and not in the parent monolayer exposed to ethanol or acetaldehyde. Upregulation is observed in 5 members of the BRCA family of tumor-suppressor genes, particularly related to breast cancer, such as BRCA1/BRCA2 and their complex BRCC3, at levels of 2.3- to 4.5-fold. Some members of the myc family (such as RLF, MYCBP2 and MINA) are upregulated from 2.9- to 3.3-fold. Within a group of genes still not well defined in relation to cancer, 9 members of the neuroblastoma breakpoint family (NBPFs) are all upregulated from 2.2- to 2.7-fold, and three members of the small nucleolar RNA family (SNORD) are upregulated by 2.0- to 2.5-fold, versus four others down-regulated (0.12 to 0.32).
Table V.
Effects of long-term ethanol exposure of MCF-12A cells on the expression of key cancer related gene families.a
| SA Clone | Monolayer | |||
|---|---|---|---|---|
|
|
|
|||
| Gene ID | Gene description | SASC/cont | Eth/cont | Act/cont |
| BRCA family | ||||
| BRCC3 | BRCA1/BRCA2-containing complex, subunit 3 | 4.5 | 0.85 | 1.0 |
| BRCA2 | Breast cancer 2, early onset | 2.6 | 0.80 | 0.88 |
| PALB2 | Partner and localizer of BRCA2 | 2.6 | 0.9 | 0.9 |
| BRCA1 | Breast cancer 1, early onset | 2.3 | 1.0 | 1.0 |
| BAP1 | BRCA1 associated protein-1 | 2.2 | 1.1 | 0.96 |
| Neuroblastoma breakpoint family | ||||
| NBPF10b | Neuroblastoma breakpoint family, member 10 | 2.6 | 1.0 | 1.0 |
| NBPF15 | Neuroblastoma breakpoint family, member 15 | 2.7 | 1.0 | 1.0 |
| NBPF16b | Neuroblastoma breakpoint family, member 16 | 2.6 | 1.0 | 1.0 |
| NBPF11b | Neuroblastoma breakpoint family, member 11 | 2.6 | 1.0 | 1.0 |
| NBPF1 | Neuroblastoma breakpoint family, member 1 | 2.6 | 1.0 | 1.0 |
| NBPF9 | Neuroblastoma breakpoint family, member 1 | 2.5 | 1.0 | 1.1 |
| NBPF14 | Neuroblastoma breakpoint family, member 14 | 2.4 | 1.0 | 1.1 |
| NBPF3b | Neuroblastoma breakpoint family, member 3 | 2.2 | 1.0 | 1.0 |
| Others | ||||
| MCAM | Melanoma cell adhesion molecule | 5.7 | 2.1 | 1.5 |
| ERBB2 | v-erb-b2 eryth leuk vir onc homol | 2.6 | 1.0 | 1.0 |
| TPD52L2 | Tumor protein D52-like | 1.8 | 1.0 | 1.0 |
| EHF | Ets homologous factor | 0.1 | 0.2 | 1.0 |
| MYC family | ||||
| RLF | Rearranged L-myc fusion | 3.3 | 1.1 | 1.0 |
| MYCBP2 | MYC binding protein 2 | 3.2 | 1.1 | 1.1 |
| MINAb | Myc induced nuclear antigen | 2.4 | 1.0 | 1.0 |
| MAX | Myc associated factor X | 2.0 | 1.0 | 1.0 |
| RBBP7 | Retinoblastoma binding protein 7 | 2.1 | 1.1 | 1.0 |
| Small nucleolar RNA family | ||||
| SNORD78 | Small nucleolar RNA, C/D box 78 | 2.5 | 0.9 | 1.0 |
| SNORD22 | Small nucleolar RNA, C/D box 22 | 2.4 | 1.0 | 1.0 |
| SNORD4B | Small nucleolar RNA, C/D box 4B | 2.0 | 0.9 | 0.9 |
| SNORD82 | Small nucleolar RNA, C/D box 82 | 0.32 | 1.0 | 1.0 |
| SNORD5 | Small nucleolar RNA, C/D box 5 | 0.31 | 1.2 | 1.5 |
| SNORD6 | Small nucleolar RNA, C/D box 6 | 0.31 | 0.9 | 0.9 |
| SNORD14E | Small nucleolar RNA, C/D box 14E | 0.12 | 1.0 | 1.0 |
Gene expression analysis was carried out using the Affymetrix human Gene 1.1 ST assay.
A few genes have multiple probe sets in this assay, so results were averaged. These are indicated after the Gene ID. Gene ID, SASC/cont, Eth/cont, and Act/cont as in previous tables.
Considering the observed changes in mRNA levels, it is logical to investigate what changes occur in their potential regulators, the microRNAs (miRs). Surprisingly, ethanol in monolayer cultures of MCF-12A did not cause substantial changes in the global miR expression. Acetaldehyde was somewhat more active but changes were still marginal. However, as in the DNA microarray data, the SASC clone derived from ethanol exposure showed 15 miRs upregulated by 2- to 3-fold, and 28 downregulated between 2- and 100-fold.
In particular, the miR-200 family, which modulates EMT and MET, is considerably altered in MCF-12A exposed to ethanol
Of particular interest are the members of the miR-200 family which were highly downregulated in the SASC clone. This family regulates EMT through its effects on EMT-provoking peptides including Zeb1, Zeb2, and Twist1, and is allegedly involved in cancer progression, including the EMT phase, which involves downregulated miR-200s, and later the mesenchymal to epithelial transition (MET) which involves return of miR-200 expression (49). In this study, the miR-200 family members miR-200b, miR-200c, and miR-141 were down-regulated by 25–100-fold in the SASC clone (Table VI). These results are consistent with the gene expression analysis for the EMT inducing peptides Zeb1, Zeb2 and Twist1, which were all upregulated in the SASC clone (see Table IV).
Table VI.
Effects of long-term ethanol exposure of MCF-12A cells on the miRNA global profile.a
| SA Clone | Monolayer | SA Clone | Monolayer | ||||
|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||
| miR | SASC/cont | Eth/cont | Act/cont | miR | SASC/cont | Eth/cont | Act/cont |
| 99a-5p | 2.89 | 1.37 | 2.15 | 24-3p | 0.34 | 0.98 | 1.44 |
| 125a-5p | 2.66 | 0.94 | 0.92 | 103a-3p | 0.34 | 1 | 1.75 |
| 320d | 2.64 | 1.18 | 0.62 | 27a-3p | 0.32 | 0.88 | 1.37 |
| 320e | 2.6 | 1.15 | 0.6 | 106b-5p | 0.28 | 0.97 | 1.56 |
| 320c | 2.55 | 1.13 | 0.66 | 19a-3p | 0.23 | 1.11 | 1.77 |
| 99b-5p | 2.53 | 0.82 | 1.04 | 29b-3p | 0.22 | 1.16 | 1.44 |
| 320a | 2.51 | 1.27 | 0.65 | 20b-5p | 0.22 | 0.85 | 1.47 |
| 125b-5p | 2.46 | 1.17 | 1.75 | 106a-5p | 0.22 | 0.84 | 1.29 |
| 320b | 2.45 | 1.15 | 0.68 | 20a-5p | 0.21 | 0.9 | 1.34 |
| let-7b-5p | 2.44 | 1.12 | 0.43 | 17-5p | 0.21 | 0.86 | 1.33 |
| 100-5p | 2.37 | 1.22 | 1.56 | 494 | 0.17 | 0.53 | 0.84 |
| 181b-5p | 2.35 | 0.99 | 1.04 | 101-3p | 0.17 | 1.01 | 2.14 |
| let-7e-5p | 2.28 | 1.13 | 0.37 | 183-5p | 0.16 | 0.96 | 0.51 |
| let-7c | 1.96 | 1.09 | 0.35 | 182-5p | 0.16 | 0.94 | 1.37 |
| let-7d-5p | 1.92 | 1.09 | 0.4 | 19b-3p | 0.15 | 1.24 | 1.42 |
| 22-3p | 0.69 | 0.96 | 1.84 | 200c-3p | 0.04 | 0.94 | 0.85 |
| 31-5p | 0.63 | 1.21 | 2.17 | 200b-3p | 0.03 | 0.9 | 0.85 |
| 29c-3p | 0.54 | 0.87 | 1.47 | 141-3p | 0.01 | 0.92 | 1.92 |
| 23a-3p | 0.53 | 0.93 | 1.24 | 96-5p | 0.01 | 0.72 | 1.8 |
| 15a-5p | 0.5 | 0.91 | 1.18 | 203a | 0.01 | 0.65 | 1.88 |
| 93-5p | 0.44 | 1.01 | 1.43 | 205-5p | 0.01 | 0.89 | 1.13 |
| 425-5p | 0.4 | 0.84 | 1.58 | ||||
| 16-5p | 0.37 | 0.84 | 1.08 | ||||
| 107 | 0.34 | 1.02 | 1.7 | ||||
Expression of microRNAs was assayed as described in Materials and methods. MicroRNAs that were substantially upregulated or downregulated are presented. miR: name of each microRNA according to mirbase. SASC/cont, Eth/cont, Act/cont as in previous tables.
Importantly, in addition to the miRs involved in EMT, there are several miRs that are known to function as tumor suppressors in some systems, and which were strongly downregulated in the SASC clone. These include miR-205 (which can function either as a tumor suppressor or in an oncogenic role) (50,51), miR-203a (52) which is reported to be upregulated in primary tumors but downregulated in metastatic growth, and involved in SNAI2 induction of EMT (53), miR-101 (54,55), and others (Table VI). By contrast, some miRs thought to be oncogenic such as miR-19a, miR-19b (56) and miR-103 (57) were down-regulated in the SASC clone. The microRNAs miR-22 and mir-31 have been reported to be involved in cancer suppression (58,59) and miR-31 may function both in suppression and oncogenesis (60). In the current study, mir-22 showed modest downregulation in the SASC sample, essentially no change after ethanol treatment in monolayers, and a modest upregulation after acetaldehyde treatment of monolayers. The mir-31 results were similar to the mir-21 results.
Clinical laboratory testing
There are several clinical laboratory tests available for gene expression in tissue samples taken from breast cancers (see Discussion). Some of the genes whose transcription is altered in MCF-12A by ethanol exposure are accepted markers for breast cancer in the clinical setting. Table VII shows the results for our SASC clone for the gene set from one of those tests. Ki67 and STK15 show considerable upregulation, consistent with the oncogenic phenotype displayed by these cells.
Table VII.
Genes used in clinical laboratory analysis of breast cancer and analyzed for gene expression in the soft agar selected cell (SASC) clones.a
| Gene | Cont | SASC/cont | Gene ID | Description |
|---|---|---|---|---|
| Ki67 | 58.8 | 6.73 | MKi67 | Nuc protein |
| STK15 | 194.2 | 2.23 | AURKA | Aurora kinase A |
| Survivin | 226 | 0.99 | BIRC5 | Inhibits apoptosis |
| CCNB1 | 174.1 | 2.05 | CCNB1 | Cyclin B1 |
| MYBL2 | 127.1 | 1.69 | MYBL2 | V-myb oncogene homology |
| MMP11 | 135.2 | 1.12 | MMP11 | Matrix metallopr 11 aka stromelysin 3 |
| CTSL2 | 74.4 | 0.49 | CTSL2 | Cathepsin L2 |
| GRB7 | 951.4 | 0.07 | GRB7 | Growth factor rec bound protein 7 |
| HER2 | 336.5 | 0.9 | ERBB2 | HER2 |
| GSTM1 | 86.3 | 1.33 | GSTM1 | Glutathione S-transferase mu 1 |
| CD68 | 1045 | 0.94 | CD68 | CD68 molecule |
| BAG1 | 204.1 | 0.77 | BAG1 | BCL2 associated athanogene |
| ERb | 45.1 | 0.83 | ESR1 | Estrogen rec 1 |
| PGR | 23.3 | 0.61 | PGR | Progesterone rec |
| BCL2 | 113.7 | 1.07 | BCL2 | B-cell CLL/lymphoma 2/blocks apoptosis |
| SCUBE2 | 49.1 | 1.09 | SCUBE2 | Signal peptide, cub domain, EGF-like 2 |
Gene expression was analyzed using the Affymetrix human Gene 1.1 ST assay.
The DNA microarray used more than one probe set for the estrogen receptor gene, and the result presented is the average. Gene: gene name commonly used in clinical laboratory assays. Cont: the normalized DNA microarray value for each gene in control cells, indicating relative levels of gene expression. Gene ID, SASC/cont as in previous tables.
Discussion
To our knowledge this is the first report on normal human epithelial breast cells in monolayer culture involving continuous exposure for 1 week to a moderate concentration of ethanol (25 mM) which: i) causes changes in the global transcriptional signature, showing induction of the metallothionein gene family and changes in alcohol metabolism genes, and in other genes suggesting the initiation of EMT, but without evident related phenotypic changes; ii) these transcriptional changes do not seem to be mediated by the main ethanol metabolite, acetaldehyde, since concentrations (2.5 mM) considerably higher than the ones that would result in vivo from this alcohol exposure, are virtually inactive; iii) the alteration in the expression of the metallothionein genes resembles that which was previously reported by us in preliminary experiments for similar incubations of the breast cancer epithelial cell line MCF7 (24) but no upregulation of stem cell genes like Oct4 was observed in the MCF-12A cells; iv) these changes suggest that early initiation by exposure to 25 mM ethanol provokes an EMT process in normal breast epithelial cells.
Even more significantly, this is also the first report that longer continuous exposure (4 weeks) of these normal breast epithelial cells to ethanol or acetaldehyde induces a late transformation consistent not only with EMT but also with oncogenesis, since: v) ethanol at 25 mM or acetaldehyde at 2.5 mM arrests cell growth and eventually causes cell death, in contrast to MCF7 (24); but vi) much lower concentrations of ethanol (2.5 or 1.0 mM) or acetaldehyde (1.0 mM) not only induce EMT but also result in oncogenic phenotypic changes that lead to the selection by 2.5 mM ethanol of anchorage independent cells able to grow in soft agar.
These growth and morphological changes induced by long-term exposure are paralleled by profound alterations in the global transcriptional signatures, particularly in the soft agar-selected cells, such as: vii) the silencing of metallothionein changes observed in the early exposure, and the amplification of the previous alcohol metabolism changes; viii) mRNA level alterations consistent not only with the induction of EMT but with the oncogenic phenotype, as well as changes in the levels of key related miRs; ix) these changes are particularly amplified in the ethanol-induced soft agar selected cells, suggesting that it is a subset of cells with the same changes as shown in soft agar selected cells that is responsible for the EMT and oncogenic transformation seen in the parent monolayer cultures; x) with the exception of the stimulation of cell growth in soft agar, the alterations induced by long-term incubation with 2.5 mM ethanol on MCF-12A cells are considerably more significant and rather different from the ones induced by 25 mM ethanol in MCF7 cells (24), specifically on the transcriptional signatures; xi) the long-term effects suggest that low concentrations of ethanol, with little or no mediation by acetaldehyde, induce the EMT and oncogenic transformation of normal breast epithelial cells, specifically MCF-12A cells, in contrast to the much higher concentrations of ethanol required to stimulate the growth and oncogenesis of MCF7 breast cancer epithelial cells (24), and act possibly by different mechanisms.
These in vitro results establish a proof of concept for potential EMT and carcinogenic effects of alcohol but, as such, are difficult to translate directly to actual human exposure to ethanol from drinking alcoholic beverages. The 2.5 mM ethanol (approximately 0.01%) is roughly equivalent to the peak level in blood after about 1/6 drink in a 50–60 kg woman, with one full drink being 0.06% (24). However, in the in vitro studies reported here, this concentration was maintained at a nearly constant level during a 28 day exposure, versus probably only around 30 min to 1 h in women who have consumed alcoholic beverages. It is impossible to extrapolate to the in vivo human situation, but speculatively we consider that the constant in vitro exposure of MCF-12A to the low 0.01% ethanol concentration may be even lower than that which occurs in an alcoholic woman drinking 3–4 drinks a day during the same 4 week period. However, this equivalence needs to be ascertained in laboratory animals to achieve a meaningful translation from cell culture to human blood and breast tissue. However, the current study suggests for the first time a new molecular paradigm for the potential oncogenic effects of excessive alcohol consumption. This is in agreement with what epidemiological studies of breast cancer incidence in alcoholic women suggest about ethanol and breast cancer risks (50). In addition, our study opens up multiple potential mechanisms by which alcohol targets certain gene families and their miR regulators.
It is interesting that, as in the case of MCF7 (24), ethanol induced an early upregulation of metallothioneins in the MCF-12A cells that was not present at 4 weeks (although for MCF-12A cells, long-term incubation was carried out with 2.5 mM ethanol rather than 25 mM ethanol, as in the previous study). It is likely that this increased expression of metallothioneins is due to the known fact that they are induced by ethanol (61). However, there may be a possibility that metallothioneins also play an early role in the oncogenic transformation of MCF-12A by ethanol, since MT-I and MT-II are antiapoptotic, proliferative, angiogenic, and oncogenic (62), and are increased in breast cancer and other tumors, correlating with higher tumor grade/stage, increased recurrence and poor survival in the highly malignant invasive ductal breast carcinomas, and predictive of poor prognosis in estrogen receptor-negative patients.
The early upregulation of aldehyde oxidase 1 (AOX1) and the decrease of aldehyde dehydrogenase 3B2 (ALDH3B2), maintained after 4 weeks, also observed in MCF7 (24), may be related to alcohol metabolism. AOX1 is a xenobiotic metabolizing protein that is found in the liver and produces reactive oxygen species (ROS), but paradoxically is reduced by heavy chronic alcohol consumption (63), and has not been related to breast cancer. Very little is known regarding the specific ALDH isoform ALDH3B2 other than being associated with alcohol dependence (64), but no relationship with cancer has been reported, in contrast to other isoforms.
The induction by ethanol of EMT associated changes in gene expression after long-term exposure to 2.5 mM ethanol, particularly in the soft agar-selected cells (SASC), is remarkable, since very little is available in the literature on the experimental effects of ethanol on EMT (or of alcoholic beverages in literature searches under the more general term alcohol). Our results showing increased levels of the transcription factors and key EMT inducers, Twist1, Snail1, Zeb1 and Zeb2, but not of Snail2 (Slug) are in agreement with the higher expression of Snail in immortalized human pancreatic ductal epithelial cells, with lower induction of Slug (44). Another study showed that alcohol upregulated the signature EMT phenotypic marker vimentin via Snail, as well as matrix metalloproteases MMP-2, MMP-7, and MMP-9 in colon and breast cancer cells (55). Snail siRNA knockdown prevented alcohol-stimulated vimentin expression, and in vivo Snail expression was significantly elevated in colonic mucosal biopsies from alcoholics (45).
The ethanol-induced SASC cultures showed strikingly different expression of some other EMT inducers, such as TGFβ2 (upregulated 34-fold) and TGFβ1 (downregulated 3-fold), when in general they act synergistically in inducing EMT (65,66), although in some cases, one or the other predominates (67). Similar situations occurred with IL7R vs IL1R, and THBS1 vs THBD, although the pattern of the joint involvement of the overall IL, IGF, and TGFβ families is in agreement with their cooperative role as EMT inducers (68). The considerable upregulation by ethanol of CTGF is also in agreement with its well known role in EMT (69), but the concerted upregulation of IGFBPs 2, 3 and 4 is more intriguing, considering the paucity of studies and some conflicting evidence for their pro- versus anti-EMT effects (70–72). The upregulation of IGFR2 is in agreement with the higher levels observed in breast cancer tissue (73).
The pattern of EMT occurring in the ethanol-induced SASC is typical, since it shows the expected downregulation of keratins, known markers of epithelial cells, with 6 members affected, whereas 3 integrins and 2 laminins, mesenchymal markers, were upregulated. Specifically, the upregulation of laminin α4 is present in EMT, involving a switch from laminin-5 to laminin-4 expression, which may be directly controlled by Snai (74).
As in the case of members of the TGFß family, some integrins such as integrin β4 go in the opposite direction of some of the other integrins, such as β8 or A3, but in addition, these effects may be associated with potential metastatic ability, since the combination of low ITGβ4, exclusively expressed in polarized epithelial cells, with high miR-21 and low PDCD4 expression is able to predict the presence of metastasis (75). This agrees with the downregulation of claudin 1, since the loss of apical cell adhesions (tight junctions) has been associated with malignant transformation, a process most often accompanied by a concomitant loss of claudin expression (76).
The EMT gene expression changes induced in MCF-12A by 4 week of incubation with 2.5 mM ethanol, and particularly manifested in the resulting SASC, are concordant with the acquisition of anchorage-independence, evidenced by their selection in soft agar, although they can also continue growing in monolayer after acquiring the mesenchymal phenotype, similarly to what was previously observed with keratinocytes undergoing EMT (77). This would suggest, as occurs with many cells undergoing EMT, that this would be associated with invasiveness, i.e., a potential tumor progression/metastatic phenotype, but no in vitro assays of invasion or in vivo tests of tumorigenesis and metastasis have been performed to test this point. In fact, the upregulation, rather than downregulation, of the BRCA1/2 family is difficult to reconcile with oncogenic progression because of their nature as tumor suppressors, unless they represent a cell defense response (78,79). Since no investigation of BRCA mutations was performed, this issue remains unresolved. Similarly, the relationship of some of the MYC, NBPF, or SNORD members with cancer, and specifically breast cancer, is not well defined.
Among the changes to RNA abundance resulting from exposure to ethanol, some of the most dramatic involve members of the miR-200 family, which, among other things, are involved in controlling the induction of EMT and its opposite, the mesenchymal epithelial transition (MET), both of which are critical elements in the metastatic process. As such, these miRs are involved in more than one phase of oncogenesis. The EMT phase represents a danger of progression following tumor formation, but prior to the colonization by tumor cells at potential sites of metastasis.
The most upregulated and downregulated miRs are presented in Table VI. They represent not only miRs which are involved in EMT, but also miRs which appear to be oncogenic in other ways, as well as miRs which would appear to be, at least potentially, capable of tumor suppression. Taken as a whole, the results suggest that the influence of long-term, low concentration ethanol on normal epithelial breast cell miRs is of a variable nature. The immediate mechanism(s) by which ethanol and acetaldehyde influence miR abundance is unclear at present, but is worthy of further study.
Another interesting aspect is the implication of our results for clinical assays of breast cancer gene expression in the pathology lab based on different gene sets. For example, the 21 gene recurrence score has been evaluated in terms of prognosis with respect to chemotherapy (80,81). In addition to reference gene expression values, the assay set includes proliferation genes Ki67, STK15, Survivin, CCNB1 (cyclin B1), MYBL2, invasive genes MMP11 (Stromolysin 3), CTSL2 (cathepsin L2), Her 2 genes GRB7 and HER2, as well as GSTM1, CD68, BAG1, and estrogen related genes ER, PGR, BCL2, and SCUBE2. In the SASC clone, Ki67 was upregulated in comparison with control MCF-12A cells by a factor of 6.73. STK15 (aka aurora kinase A) was upregulated by 2.23 fold, cyclin B1 by 2.05, and MYBL2 by 1.69. By comparison, reference genes showed SASC/control ratios closer to 1, with the maximal being beta actin, with a ratio of 1.55, GAPDH at 1.21, the ribosomal protein gene RPLPO at 0.99, and the transferrin receptor gene at 1.33.
These results suggest that a subset of the 16 oncogenes and tumor suppressors that are represented in this clinical assay are upregulated by ethanol treatment as selected by soft agar growth. In contrast, two genes, cathepsin L2 and the growth factor receptor bound protein 7 were substantially downregulated. The majority of the 16 genes were essentially unchanged between the control and SASC clone cultures.
In conclusion, our study suggests that a prolonged exposure of epithelial breast cells to alcohol in vivo may induce EMT and oncogenic changes, but the clinical translation of our findings still requires confirmation in animal models of breast cancer.
Acknowledgements
Financial support by a pilot grant to NGC from the NIH/NCI U54 CA14393-01 program grant to J.V., from U54 MD007598 to J.V. and N.G.-C., as well as from grant NIH/NIEHS 1U01ES020887-01 to N.G.-C. covering some DNA microarray and miR work, as well as support from CRECD R25 MD007610 to R.G. are gratefully acknowledged. We thank Dr David Heber for invaluable advice.
Abbreviations
- miR
microRNA
- SASC
soft agar selected clone
References
- 1.Seitz HK, Pelucchi C, Bagnardi V, La Vecchia C. Epidemiology and pathophysiology of alcohol and breast cancer: Update 2012. Alcohol Alcohol. 2012;47:204–212. doi: 10.1093/alcalc/ags011. [DOI] [PubMed] [Google Scholar]
- 2.Coronado GD, Beasley J, Livaudais J. Alcohol consumption and the risk of breast cancer. Salud Publica Mex. 2011;53:440–447. [PubMed] [Google Scholar]
- 3.Pelucchi C, Tramacere I, Boffetta P, Negri E, La Vecchia C. Alcohol consumption and cancer risk. Nutr Cancer. 2011;63:983–990. doi: 10.1080/01635581.2011.596642. [DOI] [PubMed] [Google Scholar]
- 4.Chen WY, Rosner B, Hankinson SE, Colditz GA, Willett WC. Moderate alcohol consumption during adult life, drinking patterns, and breast cancer risk. JAMA. 2011;306:1884–1890. doi: 10.1001/jama.2011.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narod SA. Alcohol and risk of breast cancer. JAMA. 2011;306:1920–1921. doi: 10.1001/jama.2011.1589. [DOI] [PubMed] [Google Scholar]
- 6.Saxena T, Lee E, Henderson KD, Clarke CA, West D, Marshall SF, Deapen D, Bernstein L, Ursin G. Menopausal hormone therapy and subsequent risk of specific invasive breast cancer subtypes in the California Teachers Study. Cancer Epidemiol Biomarkers Prev. 2010;19:2366–2378. doi: 10.1158/1055-9965.EPI-10-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kabat GC, Kim M, Shikany JM, Rodgers AK, Wactawski-Wende J, Lane D, Powell L, Stefanick ML, Freiberg MS, Kazlauskaite R, et al. Alcohol consumption and risk of ductal carcinoma in situ of the breast in a cohort of postmenopausal women. Cancer Epidemiol Biomarkers Prev. 2010;19:2066–2072. doi: 10.1158/1055-9965.EPI-10-0388. [DOI] [PubMed] [Google Scholar]
- 8.Wong AW, Dunlap SM, Holcomb VB, Nunez NP. Alcohol promotes mammary tumor development via the estrogen pathway in estrogen receptor alpha-negative HER2/neu mice. Alcohol Clin Exp Res. 2012;36:577–587. doi: 10.1111/j.1530-0277.2011.01654.x. [DOI] [PubMed] [Google Scholar]
- 9.Wang S, Xu M, Li F, Wang X, Bower KA, Frank JA, Lu Y, Chen G, Zhang Z, Ke Z, et al. Ethanol promotes mammary tumor growth and angiogenesis: The involvement of chemoattractant factor MCP-1. Breast Cancer Res Treat. 2012;133:1037–1048. doi: 10.1007/s10549-011-1902-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masso-Welch PA, Tobias ME, Vasantha Kumar SC, Bodziak M, Mashtare T, Jr, Tamburlin J, Koury ST. Folate exacerbates the effects of ethanol on peripubertal mouse mammary gland development. Alcohol. 2012;46:285–292. doi: 10.1016/j.alcohol.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Hong J, Holcomb VB, Tekle SA, Fan B, Núñez NP. Alcohol consumption promotes mammary tumor growth and insulin sensitivity. Cancer Lett. 2010;294:229–235. doi: 10.1016/j.canlet.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castro GD, de Castro CR, Maciel ME, Fanelli SL, de Ferreyra EC, Gómez MI, Castro JA. Ethanol-induced oxidative stress and acetaldehyde formation in rat mammary tissue: Potential factors involved in alcohol drinking promotion of breast cancer. Toxicology. 2006;219:208–219. doi: 10.1016/j.tox.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 13.Watabiki T, Okii Y, Tokiyasu T, Yoshimura S, Yoshida M, Akane A, Shikata N, Tsubura A. Long-term ethanol consumption in ICR mice causes mammary tumor in females and liver fibrosis in males. Alcohol Clin Exp Res. 2000;24(Suppl):S117–S122. [PubMed] [Google Scholar]
- 14.Singletary KW, Frey RS, Yan W. Effect of ethanol on proliferation and estrogen receptor-alpha expression in human breast cancer cells. Cancer Lett. 2001;165:131–137. doi: 10.1016/S0304-3835(01)00419-0. [DOI] [PubMed] [Google Scholar]
- 15.Etique N, Chardard D, Chesnel A, Merlin JL, Flament S, Grillier-Vuissoz I. Ethanol stimulates proliferation, ERalpha and aromatase expression in MCF-7 human breast cancer cells. Int J Mol Med. 2004;13:149–155. [PubMed] [Google Scholar]
- 16.Etique N, Flament S, Lecomte J, Grillier-Vuissoz I. Ethanol-induced ligand-independent activation of ERalpha mediated by cyclic AMP/PKA signaling pathway: An in vitro study on MCF-7 breast cancer cells. Int J Oncol. 2007;31:1509–1518. [PubMed] [Google Scholar]
- 17.Etique N, Grillier-Vuissoz I, Lecomte J, Flament S. Crosstalk between adenosine receptor (A2A isoform) and ERalpha mediates ethanol action in MCF-7 breast cancer cells. Oncol Rep. 2009;21:977–981. doi: 10.3892/or_00000311. [DOI] [PubMed] [Google Scholar]
- 18.Przylipiak A, Rabe T, Hafner J, Przylipiak M, Runnebaum R. Influence of ethanol on in vitro growth of human mammary carcinoma cell line MCF-7. Arch Gynecol Obstet. 1996;258:137–140. doi: 10.1007/s004040050114. [DOI] [PubMed] [Google Scholar]
- 19.Meng Q, Gao B, Goldberg ID, Rosen EM, Fan S. Stimulation of cell invasion and migration by alcohol in breast cancer cells. Biochem Biophys Res Commun. 2000;273:448–453. doi: 10.1006/bbrc.2000.2942. [DOI] [PubMed] [Google Scholar]
- 20.Luo J, Miller MW. Ethanol enhances erbB-mediated migration of human breast cancer cells in culture. Breast Cancer Res Treat. 2000;63:61–69. doi: 10.1023/A:1006436315284. [DOI] [PubMed] [Google Scholar]
- 21.Izevbigie EB, Ekunwe SI, Jordan J, Howard CB. Ethanol modulates the growth of human breast cancer cells in vitro. Exp Biol Med (Maywood) 2002;227:260–265. doi: 10.1177/153537020222700406. [DOI] [PubMed] [Google Scholar]
- 22.Etique N, Chardard D, Chesnel A, Flament S, Grillier-Vuissoz I. Analysis of the effects of different alcohols on MCF-7 human breast cancer cells. Ann N Y Acad Sci. 2004;1030:78–85. doi: 10.1196/annals.1329.010. [DOI] [PubMed] [Google Scholar]
- 23.Etique N, Grillier-Vuissoz I, Flament S. Ethanol stimulates the secretion of matrix metalloproteinases 2 and 9 in MCF-7 human breast cancer cells. Oncol Rep. 2006;15:603–608. [PubMed] [Google Scholar]
- 24.Vernet D, Gelfand R, Sarkissyan S, Heber D, Vadgama J, Gonzalez-Cadavid NF. Long-term exposure of breast cell lines to ethanol affects the transcriptional signature for some oncogenic gene families, but has little effect on this phenotype in mammospheres or on the expression of stem cell markers. Cancer Res. 2011;71(Suppl 8):5559. doi: 10.1158/1538-7445.AM2011-5559. [DOI] [Google Scholar]
- 25.Zhang Q, Jin J, Zhong Q, Yu X, Levy D, Zhong S. ERα mediates alcohol-induced deregulation of Pol III genes in breast cancer cells. Carcinogenesis. 2013;34:28–37. doi: 10.1093/carcin/bgs316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai J, Jian J, Bosland M, Frenkel K, Bernhardt G, Huang X. Roles of hormone replacement therapy and iron in proliferation of breast epithelial cells with different estrogen and progesterone receptor status. Breast. 2008;17:172–179. doi: 10.1016/j.breast.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feifei N, Mingzhi Z, Yanyun Z, Huanle Z, Fang R, Mingzhu H, Mingzhi C, Yafei S, Fengchun Z. MicroRNA expression analysis of mammospheres cultured from human breast cancers. J Cancer Res Clin Oncol. 2012;138:1937–1944. doi: 10.1007/s00432-012-1272-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cortez MA, Welsh JW, Calin GA. Circulating microRNAs as noninvasive biomarkers in breast cancer. Recent Results Cancer Res. 2012;195:151–161. doi: 10.1007/978-3-642-28160-0_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krell J, Frampton AE, Jacob J, Castellano L, Stebbing J. miRNA sin breast cancer: Ready for real time? Pharmacogenomics. 2012;13:709–719. doi: 10.2217/pgs.12.15. [DOI] [PubMed] [Google Scholar]
- 30.Shore AN, Herschkowitz JI, Rosen JM. Noncoding RNAs involved in mammary gland development and tumorigenesis: There's a long way to go. J Mammary Gland Biol Neoplasia. 2012;17:43–58. doi: 10.1007/s10911-012-9247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valastyan S. Roles of microRNAs and other non-coding RNAs in breast cancer metastasis. J Mammary Gland Biol Neoplasia. 2012;17:23–32. doi: 10.1007/s10911-012-9241-9. [DOI] [PubMed] [Google Scholar]
- 32.Fernandez SV. Estrogen, alcohol consumption, and breast cancer. Alcohol Clin Exp Res. 2011;35:389–391. doi: 10.1111/j.1530-0277.2010.01355.x. [DOI] [PubMed] [Google Scholar]
- 33.Fernandez-Cobo M, Holland JF, Pogo BG. Transcription profiles of non-immortalized breast cancer cell lines. BMC Cancer. 2006;6:99. doi: 10.1186/1471-2407-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seitz HK, Stickel F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nat Rev Cancer. 2007;7:599–612. doi: 10.1038/nrc2191. [DOI] [PubMed] [Google Scholar]
- 35.Hirano T. Alcohol consumption and oxidative DNA damage. Int J Environ Res Public Health. 2011;8:2895–2906. doi: 10.3390/ijerph8072895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balbo S, Meng L, Bliss RL, Jensen JA, Hatsukami DK, Hecht SS. Time course of DNA adduct formation in peripheral blood granulocytes and lymphocytes after drinking alcohol. Mutagenesis. 2012;27:485–490. doi: 10.1093/mutage/ges008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seitz HK, Stickel F. Acetaldehyde as an underestimated risk factor for cancer development: Role of genetics in ethanol metabolism. Genes Nutr. 2010;5:121–128. doi: 10.1007/s12263-009-0154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jelski W, Chrostek L, Szmitkowski M, Markiewicz W. The activity of class I, II, III and IV alcohol dehydrogenase isoenzymes and aldehyde dehydrogenase in breast cancer. Clin Exp Med. 2006;6:89–93. doi: 10.1007/s10238-006-0101-z. [DOI] [PubMed] [Google Scholar]
- 39.Guttilla IK, Adams BD, White BA. ERα, microRNAs, and the epithelial-mesenchymal transition in breast cancer. Trends Endocrinol Metab. 2012;23:73–82. doi: 10.1016/j.tem.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 40.Jain P, Alahari SK. Breast cancer stem cells: A new challenge for breast cancer treatment. Front Biosci (Landmark Ed) 2011;16:1824–1832. doi: 10.2741/3824. [DOI] [PubMed] [Google Scholar]
- 41.Di Cello F, Flowers VL, Li H, Vecchio-Pagán B, Gordon B, Harbom K, Shin J, Beaty R, Wang W, Brayton C, et al. Cigarette smoke induces epithelial to mesenchymal transition and increases the metastatic ability of breast cancer cells. Mol Cancer. 2013;12:90. doi: 10.1186/1476-4598-12-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schedin PJ, Eckel-Mahan KL, McDaniel SM, Prescott JD, Brodsky KS, Tentler JJ, Gutierrez-Hartmann A. ESX induces transformation and functional epithelial to mesenchymal transition in MCF-12A mammary epithelial cells. Oncogene. 2004;23:1766–1779. doi: 10.1038/sj.onc.1207391. [DOI] [PubMed] [Google Scholar]
- 43.Chan IS, Guy CD, Machado MV, Wank A, Kadiyala V, Michelotti G, Choi S, Swiderska-Syn M, Karaca G, Pereira TA, et al. Alcohol activates the hedgehog pathway and induces related procarcinogenic processes in the alcohol-preferring rat model of hepatocarcinogenesis. Alcohol Clin Exp Res. 2014;38:787–800. doi: 10.1111/acer.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward ST, Dangi-Garimella S, Shields MA, Collander BA, Siddiqui MA, Krantz SB, Munshi HG. Ethanol differentially regulates snail family of transcription factors and invasion of premalignant and malignant pancreatic ductal cells. J Cell Biochem. 2011;112:2966–2973. doi: 10.1002/jcb.23215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forsyth CB, Tang Y, Shaikh M, Zhang L, Keshavarzian A. Alcohol stimulates activation of Snail, epidermal growth factor receptor signaling, and biomarkers of epithelial-mesenchymal transition in colon and breast cancer cells. Alcohol Clin Exp Res. 2010;34:19–31. doi: 10.1111/j.1530-0277.2009.01061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reed TE, Kalant H, Gibbins RJ, Kapur BM, Rankin JG. Alcohol and acetaldehyde metabolism in Caucasians, Chinese and Amerinds. Can Med Assoc J. 1976;115:851–855. [PMC free article] [PubMed] [Google Scholar]
- 47.Shimada J, Miyahara T, Otsubo S, Yoshimatsu N, Oguma T, Matsubara T. Effects of alcohol-metabolizing enzyme inhibitors and beta-lactam antibiotics on ethanol elimination in rats. Jpn J Pharmacol. 1987;45:533–544. doi: 10.1254/jjp.45.533. [DOI] [PubMed] [Google Scholar]
- 48.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hilmarsdottir B, Briem E, Bergthorsson JT, Magnusson MK, Gudjonsson T. Functional role of the microRNA-200 family in breast morphogenesis and neoplasia. Genes (Basel) 2014;5:804–820. doi: 10.3390/genes5030804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu H, Mo YY. Targeting miR-205 in breast cancer. Expert Opin Ther Targets. 2009;13:1439–1448. doi: 10.1517/14728220903338777. [DOI] [PubMed] [Google Scholar]
- 51.Orang AV, Safaralizadeh R, Hosseinpour Feizi MA. Insights into the diverse roles of miR-205 in human cancers. Asian Pac J Cancer Prev. 2014;15:577–583. doi: 10.7314/APJCP.2014.15.2.577. [DOI] [PubMed] [Google Scholar]
- 52.Wang C, Zheng X, Shen C, Shi Y. MicroRNA-203 suppresses cell proliferation and migration by targeting BIRC5 and LASP1 in human triple-negative breast cancer cells. J Exp Clin Cancer Res. 2012;31:58. doi: 10.1186/1756-9966-31-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Z, Zhang B, Li W, Fu L, Fu L, Zhu Z, Dong JT. Epigenetic silencing of miR-203 upregulates SNAI2 and contributes to the invasiveness of malignant breast cancer cells. Genes Cancer. 2011;2:782–791. doi: 10.1177/1947601911429743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X, Schulz R, Edmunds S, Krüger E, Markert E, Gaedcke J, Cormet-Boyaka E, Ghadimi M, Beissbarth T, Levine AJ, et al. MicroRNA-101 suppresses tumor cell proliferation by acting as an endogenous proteasome inhibitor via targeting the proteasome assembly factor POMP. Mol Cell. 2015;59:243–257. doi: 10.1016/j.molcel.2015.05.036. [DOI] [PubMed] [Google Scholar]
- 55.Liu X, Lei Q, Yu Z, Xu G, Tang H, Wang W, Wang Z, Li G, Wu M. MiR-101 reverses the hypomethylation of the LMO3 promoter in glioma cells. Oncotarget. 2015;6:7930–7943. doi: 10.18632/oncotarget.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jia Z, Wang K, Zhang A, Wang G, Kang C, Han L, Pu P. miR-19a and miR-19b overexpression in gliomas. Pathol Oncol Res. 2013;19:847–853. doi: 10.1007/s12253-013-9653-x. [DOI] [PubMed] [Google Scholar]
- 57.Xia W, Ni J, Zhuang J, Qian L, Wang P, Wang J. MiR-103 regulates hepatocellular carcinoma growth by targeting AKAP12. Int J Biochem Cell Biol. 2016;71:1–11. doi: 10.1016/j.biocel.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 58.Sibbesen NA, Kopp KL, Litvinov IV, Jønson L, Willerslev-Olsen A, Fredholm S, Petersen DL, Nastasi C, Krejsgaard T, Lindahl LM, et al. Jak3, STAT3, and STAT5 inhibit expression of miR-22, a novel tumor suppressor microRNA, in cutaneous T-Cell lymphoma. Oncotarget. 2015;6:20555–20569. doi: 10.18632/oncotarget.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Valastyan S, Weinberg RA. miR-31: A crucial overseer of tumor metastasis and other emerging roles. Cell Cycle. 2010;9:2124–2129. doi: 10.4161/cc.9.11.11843. [DOI] [PubMed] [Google Scholar]
- 60.Kim HS, Lee KS, Bae HJ, Eun JW, Shen Q, Park SJ, Shin WC, Yang HD, Park M, Park WS, et al. MicroRNA-31 functions as a tumor suppressor by regulating cell cycle and epithelial-mesenchymal transition regulatory proteins in liver cancer. Oncotarget. 2015;6:8089–8102. doi: 10.18632/oncotarget.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ono S, Ishizaki Y, Tokuda E, Tabata K, Asami S, Suzuki T. Different patterns in the induction of metallothionein mRNA synthesis among isoforms after acute ethanol administration. Biol Trace Elem Res. 2007;115:147–156. doi: 10.1007/BF02686026. [DOI] [PubMed] [Google Scholar]
- 62.Pedersen MO, Larsen A, Stoltenberg M, Penkowa M. The role of metallothionein in oncogenesis and cancer prognosis. Prog Histochem Cytochem. 2009;44:29–64. doi: 10.1016/j.proghi.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 63.Fu C, Di L, Han X, Soderstrom C, Snyder M, Troutman MD, Obach RS, Zhang H. Aldehyde oxidase 1 (AOX1) in human liver cytosols: Quantitative characterization of AOX1 expression level and activity relationship. Drug Metab Dispos. 2013;41:1797–1804. doi: 10.1124/dmd.113.053082. [DOI] [PubMed] [Google Scholar]
- 64.Zhang R, Miao Q, Wang C, Zhao R, Li W, Haile CN, Hao W, Zhang XY. Genome-wide DNA methylation analysis in alcohol dependence. Addict Biol. 2013;18:392–403. doi: 10.1111/adb.12037. [DOI] [PubMed] [Google Scholar]
- 65.Do TV, Kubba LA, Du H, Sturgis CD, Woodruff TK. Transforming growth factor-beta1, transforming growth factor-beta2, and transforming growth factor-beta3 enhance ovarian cancer metastatic potential by inducing a Smad3-dependent epithelial-to-mesenchymal transition. Mol Cancer Res. 2008;6:695–705. doi: 10.1158/1541-7786.MCR-07-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kimura C, Hayashi M, Mizuno Y, Oike M. Endothelium-dependent epithelial-mesenchymal transition of tumor cells: Exclusive roles of transforming growth factor β1 and β2. Biochim Biophys Acta. 18302013:4470–4481. doi: 10.1016/j.bbagen.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 67.Yu Y, Xiao CH, Tan LD, Wang QS, Li XQ, Feng YM. Cancer-associated fibroblasts induce epithelial-mesenchymal transition of breast cancer cells through paracrine TGF-β signalling. Br J Cancer. 2014;110:724–732. doi: 10.1038/bjc.2013.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maleszewska M, Moonen JR, Huijkman N, van de Sluis B, Krenning G, Harmsen MC. IL-1β and TGFβ2 synergistically induce endothelial to mesenchymal transition in an NFκB-dependent manner. Immunobiology. 2013;218:443–454. doi: 10.1016/j.imbio.2012.05.026. [DOI] [PubMed] [Google Scholar]
- 69.Yang Z, Sun L, Nie H, Liu H, Liu G, Guan G. Connective tissue growth factor induces tubular epithelial to mesenchymal transition through the activation of canonical Wnt signaling in vitro. Ren Fail. 2015;37:129–135. doi: 10.3109/0886022X.2014.967699. [DOI] [PubMed] [Google Scholar]
- 70.Natsuizaka M, Ohashi S, Wong GS, Ahmadi A, Kalman RA, Budo D, Klein-Szanto AJ, Herlyn M, Diehl JA, Nakagawa H. Insulin-like growth factor-binding protein-3 promotes transforming growth factor-{beta}1-mediated epithelial-to-mesenchymal transition and motility in transformed human esophageal cells. Carcinogenesis. 2010;31:1344–1353. doi: 10.1093/carcin/bgq108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vijayan A, Guha D, Ameer F, Kaziri I, Mooney CC, Bennett L, Sureshbabu A, Tonner E, Beattie J, Allan GJ, et al. IGFBP-5 enhances epithelial cell adhesion and protects epithelial cells from TGFβ1-induced mesenchymal invasion. Int J Biochem Cell Biol. 2013;45:2774–2785. doi: 10.1016/j.biocel.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 72.Mehta HH, Gao Q, Galet C, Paharkova V, Wan J, Said J, Sohn JJ, Lawson G, Cohen P, Cobb LJ, et al. IGFBP-3 is a metastasis suppression gene in prostate cancer. Cancer Res. 2011;71:5154–5163. doi: 10.1158/0008-5472.CAN-10-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kalla Singh S, Tan QW, Brito C, De León M, De León D. Insulin-like growth factors I and II receptors in the breast cancer survival disparity among African-American women. Growth Horm IGF Res. 2010;20:245–254. doi: 10.1016/j.ghir.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Takkunen M, Ainola M, Vainionpää N, Grenman R, Patarroyo M, García de Herreros A, Konttinen YT, Virtanen I. Epithelial-mesenchymal transition downregulates laminin alpha5 chain and upregulates laminin alpha4 chain in oral squamous carcinoma cells. Histochem Cell Biol. 2008;130:509–525. doi: 10.1007/s00418-008-0443-6. [DOI] [PubMed] [Google Scholar]
- 75.Ferraro A, Kontos CK, Boni T, Bantounas I, Siakouli D, Kosmidou V, Vlassi M, Spyridakis Y, Tsipras I, Zografos G, et al. Epigenetic regulation of miR-21 in colorectal cancer: ITGB4 as a novel miR-21 target and a three-gene network (miR-21-ITGβ4-PDCD4) as predictor of metastatic tumor potential. Epigenetics. 2014;9:129–141. doi: 10.4161/epi.26842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stebbing J, Filipović A, Giamas G. Claudin-1 as a promoter of EMT in hepatocellular carcinoma. Oncogene. 2013;32:4871–4872. doi: 10.1038/onc.2012.591. [DOI] [PubMed] [Google Scholar]
- 77.Geiger T, Sabanay H, Kravchenko-Balasha N, Geiger B, Levitzki A. Anomalous features of EMT during keratinocyte transformation. PLoS One. 2008;3:e1574. doi: 10.1371/journal.pone.0001574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang Q, Greenberg RA. Deciphering the BRCA1 tumor suppressor network. J Biol Chem. 2015;290:17724–17732. doi: 10.1074/jbc.R115.667931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee H. Cycling with BRCA2 from DNA repair to mitosis. Exp Cell Res. 2014;329:78–84. doi: 10.1016/j.yexcr.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 80.Albain KS, Barlow WE, Shak S, Hortobagyi GN, Livingston RB, Yeh IT, Ravdin P, Bugarini R, Baehner FL, Davidson NE, et al. Breast Cancer Intergroup of North America. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: A retrospective analysis of a randomised trial. Lancet Oncol. 2010;11:55–65. doi: 10.1016/S1470-2045(09)70314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]