Abstract
Objective:
Histone modifications set transcriptional competency and can perpetuate pathologic expression patterns. We defined systemic lupus erythematosus (SLE)-specific changes in H3K4me3 and K3K27me3, histone marks of gene activation and repression, respectively.
Methods:
We used ChIP-seq to define histone modifications in monocytes from SLE patients and controls.
Results:
Both promoters and enhancers exhibited significant changes in histone methylation in SLE. Regions with differential H3K4me3 in SLE were significantly enriched in potential interferon-related transcription factor binding sites and pioneer transcription factor sites.
Conclusion:
Enhancer activation defines the character of the cell and our data support extensive disease effects in monocytes, a particularly plastic lineage. Type I interferons not only drive altered gene expression but may also alter the character of the cell through chromatin modifications.
Keywords: : enhancer, epigenome, histone methylation, interferon, IRF1, lupus
Systemic lupus erythematosus (SLE) is a systemic autoimmune disease characterized by antibodies directed to nuclear antigens and chronic inflammation typically including skin, kidneys and joints. In spite of significant advances in treatment, the majority of patients have life-long disease with loss of economic potential and quality of life [1–3]. A consistent feature of studies designed to investigate the immunologic features of SLE has been the identification of an interferon signature [4–6]. This is reflected in the activation of genes downstream of type I interferons. The mechanism by which type I interferons drive the disease process is not yet fully defined, but they contribute to antigen presentation and loss of B cell tolerance.
The type I interferon signature is seen in over half of SLE patients in spite of infrequent detection of the interferons themselves. Whether this relates to the sensitivity of the assays, episodic production of type I interferons or other mechanisms is not understood. One potential explanation is that the interferons reset the epigenetic landscape which facilitates the expression of downstream genes. To date, most investigations of the epigenome in SLE have focused on DNA methylation [7–10]. Those studies have demonstrated that SLE is associated with a dramatic change in the overall DNA methylation pattern. DNA demethylation drives pathologic gene expression and may integrate diet, infection and hereditary influences [8,11–12]. Furthermore, demethylated DNA is more immunogenic, contributing to autoantibody formation, thus participating in disease at multiple levels. Less attention has been paid to histone modifications. Our group and others [13] have begun to assess specific histone modification patterns in SLE and have found a landscape that is highly dysregulated, as was seen for DNA methylation. Therefore, the epigenome is widely altered in SLE and additional studies are required to establish mechanisms and consequences. Our previous data demonstrated that H4 acetylation was significantly different in SLE monocytes with increased H4 acetylation seen at potential IRF1 binding sites and we went on to show that IRF1 binding is increased at loci with increased expression in SLE. The IRF1 binding sites seemed to be embedded in peaks of H3K4me3. This study was performed to examine whether the H3K4me3 pattern overall was impacted by SLE and whether examination of sequences within the H3K4me3 peaks altered in SLE could define pathways that for therapeutic targeting.
The myeloid lineage has been extensively examined because monocytes and macrophages are highly plastic cells that modulate their character depending on developmental signals and environmental cues [14–16]. Myeloid cell subsets are initially defined by the binding of pioneer transcription factors to enhancer elements. These set the enhancers to a poised state, defining a set of genes competent for expression in that lineage [16]. Enhancers become reversibly activated upon additional environmental cues with distinct sets of enhancers defining the character of the cell. This model has recently been shown to be true in all terminally differentiated cells [17]. The regulatory motifs define the character of the cells and recent studies have found enrichment of disease-associated genetic variants in regulatory regions, leading to speculation that some variants contribute to disease susceptibility by altering binding of critical transcription factors [18]. The current study was designed to investigate histone H3 methylation using ChIP-seq to gain further insights into the altered epigenome of SLE patients. We found that H3 methylation at both promoters and enhancers was highly altered in SLE. Increased expression was associated with increased transcription start site (TSS) H3K4me3, activation of monocyte-specific enhancers and a higher number of enhancers. Enhancers largely determine cell-state specific gene expression and an altered enhancer landscape would be expected to contribute to aberrant cell behavior in SLE.
Methods
Patients & cells
Monocytes were purified from healthy female controls (with no medical conditions other than inactive allergies) and female SLE patients by adherence as previously described [19–23]. CD14 staining confirmed >90% purity. The main identifiable contaminant was CD19 B cells; however, the majority of non-CD14+ cells were not identifiable using standard flow staining. The controls and the SLE samples were comparable in the average of non-CD14+ cells in the preparations. Clinical characteristics of the patients used for ChIP-seq have been previously reported and are summarized in Supplementary Table 1 [24]. Patients were selected for low disease activity and minimal medications to limit confounders related to medications. The organ system involvement and autoantibodies given in the table are cumulative to the date of the sample and the SLEDAI is from the day of the sample. Samples for qRT-PCR also came from Johns Hopkins and clinical characteristics are summarized in Supplementary Table 2. All patients and controls were female and were age-matched. This study was approved by the Johns Hopkins Institutional Review Board.
THP1 cells were used as a monocytic cell line. They were maintained in 10% cosmic calf serum. Stimulations were performed with IFN-α2 (PBL Assay Science, NJ, USA)
Chromatin immunoprecipitation & qRT-PCR
Immunoprecipiation of histones was carried out as previously described [19–20,25–26] and utilized H3K4me3 (Active Motif, Carlsbad, CA, USA) and H3K27me3 (Millipore, Temecula, CA, USA). Immunoprecipitation with anti-GST (Invitrogen, Camarillo, CA, USA) and input were used to define background. The same samples had parallel RNA-seq performed, which has been previously reported [24]. The library preparation utilized the SOLiD ChIP-seq kit and was performed according to the manufacturer's instructions. ChIP assays were performed to investigate target genes using the same antibodies. The signal was normalized to 10% input using the following formula 2-ΔCT. Primer sequences for IFITM1 and IL18R ChIP assays were:
IFITM1-TSS-Forward: TGCACAAGGAGGAACATGAG;
IFITM1-TSS-Probe: CAGCACCATCCTTCCAAGGTCCA;
IFITM1-TSS-Reverse: GTCTCGCTGTGGATGTTGAT;
IFITM1-3801FWD: CTTCATCCAGGACCGAAGTAAG;
IFITM1-3801Probe: TGCAGGAAATGAGTGTTTCACAGCT;
IFITM1-3801Rev: TGACCAACCTGCTCATGAAA;
IL18R1-6604F1: CCCAGACCGCTACACTC;
IL18R1-6604Probe: TTAGCAGCCAGGAGCTGCCA;
IL18R1-6604 R1: GGAACAGAAAGAAGAATGTCAGG.
qRT-PCR was used to define quantitative differences in RNA abundance. The Clontech Advantage RT for PCR kit (Clontech, CA, USA) was used to generate cDNA. Gene expression was detected by real-time PCR using the TaqMan 7900. The primers for each target were purchased from Life technologies (NY, USA). Transcript levels were normalized to the 18S signal.
Bioinformatics
Both histone modifications were measured by ChIP-seq in six SLE patients and six control samples. The binding of transcription factor IRF1 and the transcriptome of the same samples have been previously measured by ChIP-seq and RNA-seq, respectively [24,27]. The ChIP-seq reads of both histone modifications were mapped to human reference genome (hg19), after which the sequencing depth around 27,588 uniquely located TSSs annotated by RefSeq was summarized to obtain a measurement for each TSS and sample. The measurements were first normalized between samples and then adjusted by subtracting background signal measured by input controls. We used the CHOP-seq pipeline we developed with further refinements [28]. We defined promoters and TSS as -250-+250 bp from the TSS and distal promoters as -250–1 kb from the TSS. Enhancers were required to be >1 kb from the nearest TSS. Additional information is available in the Supplementary methods.
Results
Characteristics of H3K4me3 & H3K27me3 patterns
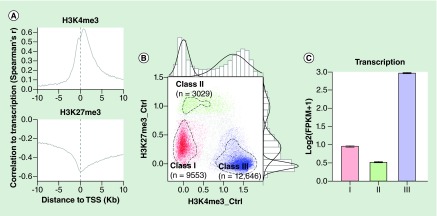
We first evaluated our data for conformity with expected patterns as a validation strategy. H3K4me3 is a mark of gene activation that has been widely used to define regulatory regions and H3K27me3 is classically associated with repression. We identified peaks of H3K4me3 and H3K27me3 in 12 ChIP libraries (six SLE, six healthy controls) and using 4 libraries of DNA input samples as background controls. As expected, H3K4me3 was predominantly enriched around TSSs whereas H3K27me3 had broad peaks with slight enrichment around TSSs (Supplementary Figures 1 & 2). High H3K4me3 was associated with high RNA abundance and high H3K27me3 was associated with low RNA abundance (Figure 1A). H3K4me3 and H3K27me3 at TSSs were negatively correlated (correlation coefficient = -0.47), as expected). The levels of both modifications were not evenly distributed and formed three distinctive classes (Figure 1B & C). Class I TSSs had low levels of both histone modifications and a moderate expression level of transcripts downstream of these TSSs; class II TSSs had high H3K27me3, low-to-medium H3K4me3 and repressed expression of downstream transcripts; class III TSSs had low H3K27me3 with high H3K4me3 and high expression of downstream transcripts. Over 90% of all RefSeq TSSs could be classified into one of these classes. Although not forming a distinct set, genes with both high H3K4me3 and high H3K27me3 are considered poised. Therefore, our ChIP-seq results appeared to conform with the previously identified characteristics associated with H3K4me3 and H3K27me3 [29].
Figure 1. . The correlation between H3K4me3 and H3K27me3 around transcription start sites and transcription level of the genes associated with the transcription start sites.
(A) The correlation coefficients were computed with nonparametric Spearman's method. The strength of the correlation was tracked along the relative location toward the transcription start sites (TSSs). Levels of H3K4me3 were positively associated with transcript abundance and levels of H3K27me3 were negatively associated with transcript abundance. (B) Classification of TSSs based on their H3K4me3 and H3K27me3. Average H3K4me3 and H3K27me3 between -250bp and +250bp of each TSS were computed and plotted after adjusting to background signals and log10-transformation. The bimodal and trimodal distributions of H3K4me3 and H3K27me3 were also plotted along the sides of the scatterplot. The circled dashed lines represent the borders of clustered TSSs. TSSs of these three clusters were used as seeds of an SVM model to classify other TSSs (Supplementary methods) and extra TSSs of each class were identified by the model. TSSs labeled in gray were ones that could not be definitively classified into any class, which accounted for about 8% of the total TSSs. (C) The barplot shows the average transcription level of genes associated with each class. Genes associated with class III TSSs had higher transcription level while class II TSSs were associated with transcription level lower than global average (the dashed line).
TSS: Transcription start site.
Differential H3K4me3 & H3K27me3 in SLE
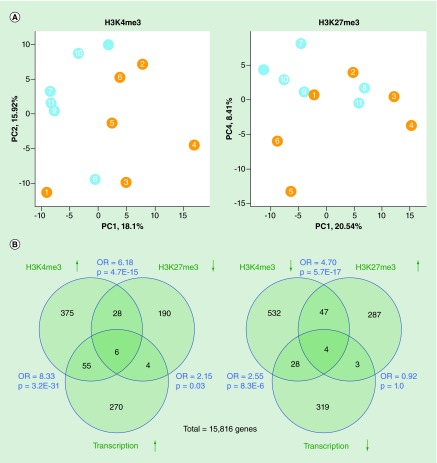
To understand whether H3 methylation was affected in SLE monocytes, we first evaluated overall patterns. Principal component analysis of H3K4me3 and H3K27me3 at all TSSs showed that SLE patients had modified patterns of both histone modifications (Figure 2A). More TSSs had significant changes of H3K4me3 in SLE than H3K27me3 changes (1788 vs 815; p < 0.01). Changes in the two histone modifications were negatively correlated (Pearson's r = 0.091; p = 2.3E-52). TSSs not belonging to any of the three classes identified in Figure 1 were 3.3-times more likely to have significant changes of either histone modification (p = 3.0E-18), suggesting that these poised sites were susceptible to inflammatory influence. The strongest associations were between increased transcript abundance and increased H3K4me3 at the promoter. Strong associations were also seen between increased H3K4me3 and decreased H3K27me3 and vice versa (Figure 2B). These are expected relationships and give confidence in the validity of the data.
Figure 2. . H3K4me3 and H3K27me3.
(A) Principal components analysis of H3K4me3 and H3K27me3 at transcription start sites. Both data sets included six control samples (blue) and six systemic lupus erythematosus patients (orange). Systemic lupus erythematosus and control samples are distinct. (B) Overlap between genes with differential expression and differential H3K4me3 or H3K4me3 at their transcription start sites. The total genes include ones with detectable transcription and both histone modifications. The OR and p-values were calculated by Fisher's exact test.
OR: Odds ratio.
To gain insights into the types of genes impacted by H3 methylation changes in SLE, we used DAVID to identify gene sets [30]. Genes with increased TSS H3K4me3 in SLE were enriched with the immunoglobulin superfamily of genes (n = 15; p = 1.1E-7). Genes with decreased TSS H3K4me3 in SLE were enriched with immune response genes, particularly MHC class II genes (n = 7; p = 2.5E-10). Therefore, TSS with altered H3K4me3 were enriched in immunologically active genes.
More than 80% of the sites with a significant change in H3K4me3 were intronic or intergenic (Supplementary Figure 3). We hypothesized that differential H3K4me3 and H3K27me3 in SLE occurs in other regulatory regions, such as enhancers, insulators and intragenic promoters. The ENCODE project identified 136,573 DNase hypersensitivity sites (DHS) in CD14+ monocytes. DHS sites typically mark regulatory regions. 3705 of these regions were generic to all 125 cell types included in the ENCODE data set whereas 5202 were unique to monocytes. In the following analyses, the 136,573 DHS regions found in monocytes will be referred to as the monocyte DHS regions. Monocyte DHS regions were more likely to have significant H3K4me3 changes in SLE than generic DHS (Supplementary Figure 4). This strongly supports the hypothesis that SLE-specific H3K4me3 changes preferentially occurred within regions with regulatory functions. We therefore examined regulatory regions in greater detail.
Differential H3K4me3 & H3K27me3 at promoters
The histone modifications associated with active, repressed, poised and intermediate transcription are now well understood. We used this framework to understand the changes seen in SLE. Of the monocyte DHS regions, 16,194 had a promoter-like histone signature in ENCODE (high H3K4me3, low H3K4me1 and low CCCTC binding factor [CTCF] binding). About 90% of these regions were intragenic or located within proximal upstream regions of genes. 4583 of these promoter sites (28%) had SLE-specific alterations in H3K4me3 and 1714 sites (11%) had SLE-specific alterations in H3K27me3. Promoters defined by annotation within 1 kb of a TSS were 2.5-fold more likely have increased H3K4me3 than decreased H3K4me3. However, promoters more distal to the TSS were more likely to have decreased H3K4me3 than increased H3K4me3 in SLE. This suggested a complexity to the architecture at each locus. Promoters with decreased H3K4me3 and H3K27me3 were significantly overlapped with each other, possibly signifying nucleosome remodeling (n = 486; odds ratio = 3.28; p = 8.8E-77). These data suggested that SLE monocytes had a promoter architecture that had been substantially altered.
Differential H3K4me3 & H3K27me3 at enhancers
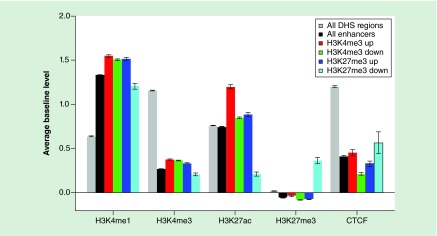
To contrast with the promoter findings, we separately examined differential H3K4 methylation at putative enhancers. We identified 39,930 monocyte DHS regions that were at least 1 kb away from any known TSSs and had an enhancer-like histone signature, based on the ENCODE data. The median distance of these regions to the nearest TSSs was about 25 kb. 12,109 (30%) and 3046 (8%) of these enhancers had a significant change (p < 0.01) in H3K4me3 and H3K27me3 (respectively) in SLE. H3K4me3 at enhancers was about 2.5-times more likely to be decreased than to be increased in SLE (8634 vs 3475) whereas H3K27me3 at enhancers was over seven-times more likely to be increased in SLE (2681 vs 365). Enhancers with decreased H3K4me3 and increased H3K27me3 significantly overlapped with each other (n = 1038; odds ratio = 2.47; p = 1.8E-96). Enhancers with increased H3K4me3 in SLE had higher baseline levels of H3K27ac in controls, a mark of active enhancers (Figure 3). Enhancers with decreased H3K4me3 or increased H3K27me3 in SLE had lower H3K27ac in controls, suggesting that they had low or intermediate activity. These data suggest that SLE is associated with changes that restrict the set of active genes in SLE but augment activity at previously active enhancers.
Figure 3. . Histone methylation at enhancers.
Enhancers were grouped based on whether they had H3K4me3 or H3K27me3 change in systemic lupus erythematosus (SLE) and the change direction. The averages of four histone modifications and CTCF in CD14+ monocytes were compared across regions exhibiting changes in H3K4me3 or H3K27me3 in SLE. Enhancers generally had higher H3K4me1 and lower H3K4me3 and CTCF than other DHS regions (gray bars). Enhancers with increased H3K4me3 in SLE (red) had the highest level of H3K27ac, a mark of active enhancers.
CTCF: CCCTC-binding factor; DHS: DNase hypersensitivity site.
Transcription factor binding sequences within differential H3K4me3 sites
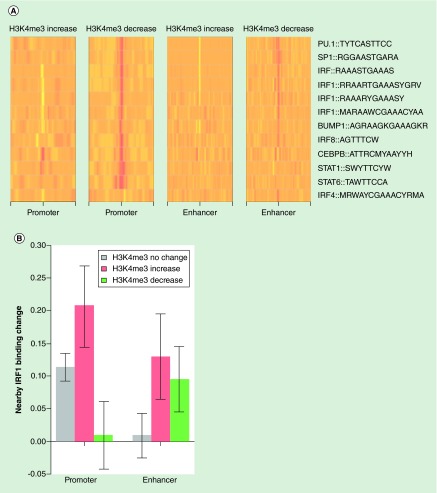
We now understood that SLE monocytes had acquired an epigenome that exhibited significant changes and further that the changes impacted immunologically relevant genes. To understand the forces driving changes to histone methylation in SLE, we derived conserved transcription factor motifs. We collected 2414 known position weight matrices (PWMs) of DNA-binding proteins from databases including ENCODE, TRANSFAC, Jasper and UniProbe [29,31–34]. These PWMs were searched against the human genome to identify potential binding sites of transcription factors. By comparing the frequency of PWMs between sites with and without H3K4me3 change in SLE, we identified a subset of PWMs that were significantly more likely to be found at promoters and/or enhancers with altered H3K4me3 in SLE (Figure 4B). These transcription factor motifs include those for known pioneer transcription factors (PU.1 and CEBPB) [35,36], and monocyte development (BLIMP1) [37]. SLE is associated with responses to type I interferons, and we also found transcription factor motifs related to interferon responses (STAT1, STAT6, IRF1, IRF4, IRF8) [5–6,38]. Most of these PWMs were over-represented within sites with decreased H3K4me3 compared with sites without significant H3K4me3 change. IRF4 sites were notable for being over-represented only among enhancers with decreased H3K4me3. CEBPB and STAT1 sites were enriched at promoters with both increased and decreased H3K4me3. In fact, CEBPB sites were enriched at all four sets of regulatory regions, befitting its role as a pioneer factor. These data implicate a relatively small number of signaling pathways in the altered epigenome of SLE. The pioneer transcription factors define monocyte-specific regulatory regions and the interferon-responsive transcription factors nucleate chromatin modifying complexes that set the level of gene expression at each locus. The same set of PWMs did not have any association with H3K27me3 changes in SLE.
Figure 4. . Position weight matrice analysis of promoters and enhancers impacted by systemic lupus erythematosus.
(A) Enrichment of selected position weight matrices (PWMs) around subsets of promoters and enhancers having significant H3K4me3 change in systemic lupus erythematosus (SLE). The relative enrichment was computed using the promoters or enhancers without significant H3K4me3 change as background. Most of the PWMs were enriched (red) around promoters and enhancers with decreased H3K4me3 while some PWMs were less likely to be found (pale yellow) around promoters and enhancers with increased H3K4me3. (B) Differential H3K4me3 in SLE was associated with differential IRF1 binding at nearby sites. While IRF1 binding was increased around monocyte promoters in general, there was a positive correlation between IRF1 binding change and H3K4me3 change in SLE. At enhancers, increased IRF1 binding was associated with H3K4me3 change in either direction.
IRF1 binding activity around differential H3K4me3 sites
This in silico analysis of transcription factor binding sites showed enrichment of interferon-related sites where H3K4me3 was decreased. We had previously published ChIP-seq data for IRF1 [39] and therefore we examined actual IRF1 binding in these same patients and controls. Our previous study identified over 3000 IRF1 binding sites with increased binding in SLE [27]. Consistent with the PWM matching analysis above, these sites were highly enriched within 500 bp of the promoter-like DHS regions, especially the ones with decreased H3K4me3 in SLE (Supplementary Figure 5).
To further clarify the relationship of IRF1 binding and H3K4me3, we examined the relationship between changes in IRF1 binding in SLE and H3K4me3 changes in the same samples. The direction of differential H3K4me3 at promoters was positively correlated with the direction of differential IRF1 binding (Figure 4B). IRF1 binding activity at sites proximal to promoters with increased H3K4me3 was increased by about 20% in SLE. On the other hand, differential H3K4me3, toward either direction, within enhancers, was associated with increased IRF1 binding. This result suggests that IRF1 functions as a regulator of histone modifications at both promoters and enhancers. The association with increased H3K4me3 at promoters and any change in H3K4me3 at enhancers highlights the distinct mechanisms. The finding of actual IRF1 binding at sites with decreased H3K4me3 suggests nucleosome remodeling could have occurred. Nevertheless, at a subset of sites with increased IRF1 binding, H3K4me3 also increased in SLE, suggesting a mechanistic link.
Impact of promoter/enhancer differential H3K4me3 on gene transcription
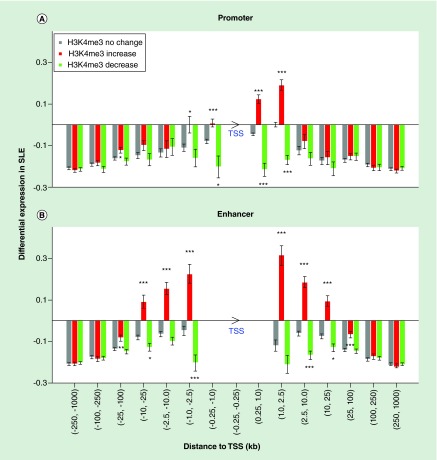
To investigate how differential H3K4me3 at promoters and enhancers affects gene expression, we mapped all monocyte promoters and enhancers to known gene TSSs and obtained over 1.3 million TSS - promoter/enhancer pairs. The differential H3K4me3 was strongly associated with differential expression of nearby genes in SLE, based on our RNA-seq data from the same patients and controls (Figure 5). Decreased H3K4me3 at promoters immediately upstream of TSSs was associated with decreased expression while increased H3K4me3 immediately downstream of the TSS was associated with increased expression. Differential H3K4me3 at intragenic promoters was associated with differential transcription from about 2.5 kb downstream of TSSs.
Figure 5. . Impact of differential H3K4me3 on differential gene expression in systemic lupus erythematosus had a distance-dependent pattern.
All promoter/enhancer-TSS pairs were grouped based on their distance. Promoters (A) and enhancers (B) were split into subsets based on their differential H3K4me3. The number of stars represents the significance of difference compared with promoters or enhancers without H3K4me3 change (p = 0.05, 0.01 and 0.001). The arrowhead of TSS indicates the direction of transcription.
SLE: Systemic lupus erythematosus; TSS: Transcription start site.
Differential H3K4me3 at enhancers, on the other hand, was associated with differential transcription from 25 kb or even longer distance both upstream and downstream. The positive correlation of differential H3K4me3 with differential expression suggests that increasing H3K4me3 at enhancers is associated with their activation. About 92% of the genes with significantly increased transcription in SLE had at least one monocyte enhancer within 100 kb of their TSSs and over 50% of these genes had an enhancer with significantly increased H3K4me3. These data highlight the profound impact of the altered enhancer landscape. Genes with increased transcription in SLE had more enhancers on average than other genes (12.8 vs 8.7). Therefore, differential histone modifications at enhancers plays an important role in differential gene expression in SLE, likely molding the character of the cell.
Histone methyltransferase & demethylase expression
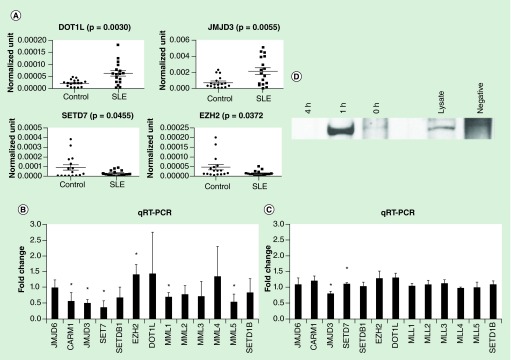
We hypothesized that the changes in histone methylation in SLE monocytes could be due to dysregulated expression of histone methyltransferases and demethylase enzymes. Using new cohorts of 17 SLE patients and 17 controls and examining transcript abundance by qRT-PCR, we found that JMJD3 and DOT1L had increased expression in the SLE cohort. Conversely, SETD7 and EZH2 had decreased expression in the new SLE cohort compared with controls (Figure 6A). Increased JMJD3 and decreased EZH2 would be expected to favor H3K27 demethylation, consistent with our ChIP-seq data. Thus, there may be altered levels of chromatin modification enzymes that favor some of the changes observed in SLE.
Figure 6. . Gene expression.
(A) Histone methylation enzyme expression in SLE patients. New cohorts of 17 SLE patients and 17 controls were used to examine the expression pattern of some histone methyltransferases and demethylase enzymes. qRT-PCR was used to quantitate the abundance of RNA. JMJD6, CARM1, JMJD3, SET7, SETDB1, EZH2, DOT1L, MML1, MMl2, MML3, MML4, MMl5, SETD1B were examined. Only the four genes shown were significantly different between SLE and control groups. (B) Interferon treatment of THP-1 cells induces changes in the expression of SETD7 and JMJD3. Cells were treated with 1000U/ml IFN-α2 for 18 h. qRT-PCR was used to quantitate the RNA abundance of histone methylation enzymes. In the bar graph, the results are expressed as a fold change from the vehicle-treated sample to normalize across the broad expression levels seen. The error bars represent SEM. Asterisks indicate p < 0.05. n = 3. (C) Interferon treatment of female primary monocytes (as above) changes the expression of several histone methylation enzymes. Asterisks indicate p < 0.05. n = 4. (D) IRF1 was immunoprecipitated from D54MG cells and the blot was probed with anti-JMJD3. Treatments are as indicated.
SLE: Systemic lupus erythematosus.
Interferon effects
Type I interferons are a recognized contributor to the development of lupus [4,6]. We examined whether IFN-α2 could mimic some of the effects we observed. Initially, we treated THP-1 cells with IFN-α2 or vehicle for 18 h. We measured levels of methyltransferases and demethylase enzymes by qRT-PCR. The expression of JMJD3 was decreased and SETD7 was increased (Figure 6B). To further investigate, we utilized primary human monocytes in the same assay, recognizing that primary cells behave differently than cell lines. The expression of EZH2 was significantly increased and the expression levels of CARM1, JMJD3, SETD7, MML1 and MML5 were all decreased in interferon-treated cells (Figure 6C). We further validated these effects in the THP-1 cell line, a monocyte line. We found a similar but distinct set of histone methyltransferases regulated by IFN-α2. Thus, interferon may contribute to the decreased expression of JMJD3 seen in the SLE patients but IFN-α2 does not seem to globally mimic the altered expression levels of histone methyltransferase and demethylase enzymes we observed in SLE patients, at least within the time frame used experimentally. We were intrigued by the regulation of JMJD3 by interferon. This histone demethylase typically targets H3K27me3 or H3K27me2 for demethylation [40–42]. It has been implicated in inflammatory responses but not in interferon responses [43,44]. We performed a co-immunoprecipitation to determine whether it could interact with IRF1. We found an interferon-dependent interaction between IRF1 and JMJD3 (Figure 6D).
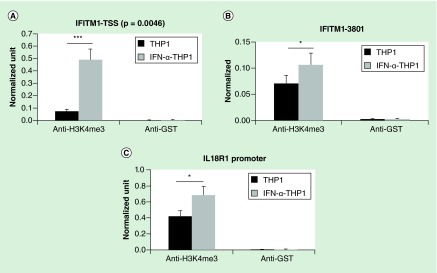
We had previously observed that type I interferons could drive a pattern of histone acetylation that was similar to that seen in SLE patients [22] and had demonstrated that type I interferons could drive protein–protein interactions related to histone acetylation [45]. We therefore selected two known IRF1 target genes that had exhibited increased H3K4me3 in SLE monocytes and examined H3K4me3 in the THP1 monocyte cell line with and without interferon treatment (Figure 7). ChIP assays for H3K4me3 were performed and qRT-PCR used to detect DNA. IFITM1 had two peaks of H3K4me3, both of which were increased in SLE and IL18R1 had a single peak of H3K4me3 in the distal promoter that was increased in SLE. THP1 cells were treated for 18 h. For all three peaks, IFN-α2 led to increased H3K4me3. These data suggest that type I interferons can drive histone modifications directly.
Figure 7. . Interferon treatment of THP1 cells changes the H3K4me3 at two IRF1 target genes.
Using the peaks identified by ChIP-seq, primers were designed to examine the changes in H3K4me3 at the major promoter peaks seen in systemic lupus erythematosus. There were two IFITM1 peaks at the TSS (A), and upstream (B) regions. There was a single promoter peak at IL18R1 (C). THP1 cells were treated with 1000 U/ml IFN-α2 for 18 h and ChIP assays performed. Interferon treatment induced H3K4me3 at the same peaks seen in the systemic lupus erythematosus patients for these two genes. The error bars represent standard error of the mean. GST is shown as a negative control antibody. The differences at each peak are significant with p < 0.05; n = 4.
GST: Glutathione-S-transferase; TSS: Transcription start site.
Discussion
SLE is a disease with diverse effects on organ systems and profound effects on all cells within the immune system. We examined monocytes as a cell that is a key participant in inflammation and as a cell type that influences cells in its surroundings. Monocyte function and survival have both been found to be abnormal in SLE patients and monocyte infiltration into renal parenchyma has been associated with worse renal function in SLE [46,47]. We found that H3K4me3 was significantly altered across the genome. We found regulatory regions in general exhibited high levels of change with 28% of promoters and 40% of monocyte enhancers exhibiting change in H3K4me3 in SLE. Proximal promoters with increased H3K4me3 enriched with DAVID categories of phosphoprotein, acetylation, immune response, Toll-like receptor and chromatin DNA binding (p < 10-3). Proximal promoters with decreased H3K4me3 were enriched with DAVID categories of regulation of leukocyte activation, regulation of cell activation, immune response, regulation of cytokine production, negative regulation of cell apoptosis and positive regulation immune system process (p < 10-5). These changes are likely to have substantial effects on cell behavior. Although functions of monocytes have been described as aberrant in SLE, the magnitude of the changes in the epigenome in SLE was surprising. This level of change exceeds that seen in the polarization of primary monocytes and macrophages [16,48–50]. These data suggest that disease effects on cells can significantly change behavior.
The overall findings of a transcriptome and epigenome that favor increased transcription are concordant with the global DNA demethylation that has been described in lymphocytes in SLE [8,51–55], in the sense that both favor increased gene expression. About 1000 genes with detectable expression in monocytes had no active promoter or monocyte enhancers within 100 kb of their TSSs. While this subset of genes already had low expression in normal monocytes, expression levels were further lowered by about one-third in SLE. Thus, the activation of genes in SLE was not uniform, suggesting an epigenome molded by specific signals not wholesale changes. Increased expression in SLE was associated with a higher number of enhancers. Additionally, the increased H3K4me3 was associated specifically with enhancers active in monocytes. These data suggest that SLE is associated with activation of genes with monocyte intrinsic functions. Indeed, our transcriptome analysis found increased expression of genes involved in cytokine production and immune responses [24]. Enhancer status gives powerful insights into both the history of the cell and its specific pattern of gene expression [15–16,56–57]. This study is the first to examine enhancer status in SLE and our findings were remarkable for the extent of change observed. One interpretation is that the monocytes have been ‘polarized’ by the diseases and this drives aberrant function. This is a subtly different interpretation of longstanding data demonstrating that most hematopoietic cells exhibit aberrant function in SLE. In the past, these data were interpreted as representing dysfunction that predisposed to disease or dysfunction imposed by the disease acutely. Our data support a model where the cells become ‘polarized’ in the disease and become integrated into the disease process, both impacted by the disease and contributing to disease.
To gain insights into what cellular pathways could drive the altered epigenetic landscape, we used an analysis of PWMs. The identification of potential transcription factor binding sites can be a powerful approach because it is unbiased and it directly maps to the site of the histone modification. At the promoter, among sites with increased H3K4me3 in SLE, only CEBPB and STAT1 binding sites were enriched. CEBPB is a monocyte pioneer transcription factor and STAT1 is a downstream effector of interferon signaling. Among promoters with decreased H3K4me3 in SLE, we saw enrichment of STAT1 and CEBPB as well as enrichment of PU.1, SP1, IRF1, BLIMP1, IRF8 and STAT6. PU.1 is another pioneer transcription factor and SP1 is associated with chromatin looping [58]. BLIMP1, a monocyte differentiation factor, was previously identified by us as associated with increased H4 acetylation peaks in SLE [23,37]. IRF8 induces β-interferon. Consistently, IRF4 binding sites were under-represented with increased or decreased H3K4me3 at promoters and IRF4 acts to oppose IRF1 [59,60]. A similar finding was seen at enhancers, although the enrichment was less substantial. Thus, these potential transcription factors meet the test of biological plausibility because they all represent pioneer transcription factors or transcription factors involved in type I IFN responses. We also noted that at ˜10% of sites, with decreased H3K4me3, H3K27me3 at promoters.was also decreased. We hypothesize that these cases represent nucleosome remodeling, a common feature where transcription factors bind [61–64]. Unfortunately, that important question cannot be answered with the current data set where the primary cells were exhausted. Decreased gene expression in SLE was strongly associated with decreased H3K4me3, suggesting that nucleosome remodeling to accommodate transcription factor binding may occur but is not the major mechanism of gene activation. Indeed, only about 10% of TSSs and enhancers exhibited concordantly decreased H3K4me3 and H3K27me3 in SLE, the expected finding at sites at nucleosome remodeling.
This study is unique because it utilized matched samples from the same patients and controls and because these same samples had previously been examined by RNA-seq and IRF1 ChIP-seq [24,39]. This allowed us great power to define associations. This study fills an important knowledge gap by providing critical information on histone modifications in SLE. While DNA methylation has been examined and appears to mold the expression pattern of critical T cell genes in SLE [65,66], there has been only one prior study of histone methylation in SLE which examined total peripheral blood mononuclear cells [67]. Epigenetic changes in cells act in a combinatorial pattern and additional studies will be required to define the critical changes arising in SLE and understand the effects in multiple cell types, the role of the transcription factors in driving these changes and how durable the changes are in this chronic disease. This study was also limited by a small sample size, imposed by the large blood volume requirement. Additional limitations include data from a small number of samples. We utilized input and GST immunoprecipitation DNA as controls and did not have sufficient material to do additional marks such as total H3 as a control or marks of enhancers such as H3K4me1. Nevertheless, the findings from this study were statistically robust. It will be important to examine other histone marks, other cell types and additional patients. We selected patients with low disease activity to minimize the effects of medications. Moving forward, it will be important to examine different phenotypes of SLE patients.
In summary, this study demonstrated substantial differences in histone methylation patterns in SLE. The finding of interferon-related transcription factor binding sites embedded in the peaks with differential height in SLE supported an important role for type I interferons in remodeling the epigenome in SLE. We found that loci with increased H3K4me3 peak height in SLE also had increased IRF1 binding by ChIP-seq and had increased expression, supporting a mechanistic role.
Conclusion
The epigenome of SLE patients is highly altered and may contribute to pathologic cell behavior. We found influences of type I interferons and also found a dysregulated landscape of histone methyltransferases and demethylases in SLE. Regulatory regions were consistently altered in SLE with 30% of monocyte enhancers impacted by SLE.
Future perspective
Targeting histone modifications has been a major pharmaceutical effort recently and resetting the epigenome may be required to interrupt the pathologic pattern of gene expression in SLE. This study suggests that targeting histone modifications may be beneficial. Intensive efforts to direct chromatin modifying enzymes to specific subsets of genes remain preclinical but represent a new horizon for therapeutics.
Executive summary.
Systemic lupus erythematosus (SLE) is an autoimmune disease with protean manifestations.
Current therapies are unsatisfactory.
Methods
ChIP-seq was used to identify SLE-specific changes in H3K4me3 and H3K27me3.
Results
H3K4me3 was both increased and decreased at regulatory regions in SLE.
Monocyte enhancers were more likely to have decreased H3K4me3 than increased H3K4me3 in SLE whereas promoters were more likely to have increased H3K4me3 in SLE.
Type I IFN was able to mimic some effects found in SLE.
Conclusion
Type I IFN and pioneer transcription factors represent powerful forces molding H3K4me3 in SLE monocytes.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge the contributions of patients, nurses and physicians and thank Eric Rappaport, Stephen Mahoney and Kristen Hunter from the nucleic acid core facility at CHOP for SOLiD sample preparation as well as Juan Perin for alignments.
Footnotes
Financial & competing interests disclosure
This study was supported in part by the Wallace Chair of Pediatrics, and NIH grants R01 ES017627 and AR43727. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Clarke A. Economic impact of lupus. Am. J. Manag. Care. 2001;7(16 Suppl.):S496–S501. [PubMed] [Google Scholar]
- 2.Garris C, Jhingran P, Bass D, Engel-Nitz NM, Riedel A, Dennis G. Healthcare utilization and cost of systemic lupus erythematosus in a US managed care health plan. J. Med. Econ. 2013;16(5):667–677. doi: 10.3111/13696998.2013.778270. [DOI] [PubMed] [Google Scholar]
- 3.Garris C, Oglesby A, Sulcs E, Lee M. Impact of systemic lupus erythematosus on burden of illness and work productivity in the United States. Lupus. 2013;22(10):1077–1086. doi: 10.1177/0961203313498795. [DOI] [PubMed] [Google Scholar]
- 4.Baechler EC, Batliwalla FM, Karypis G, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc. Natl Acad. Sci. USA. 2003;100(5):2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banchereau J, Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25(3):383–392. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Kirou KA, Lee C, George S, et al. Coordinate overexpression of interferon-alpha-induced genes in systemic lupus erythematosus. Arthritis Rheum. 2004;50(12):3958–3967. doi: 10.1002/art.20798. [DOI] [PubMed] [Google Scholar]
- 7.Absher DM, Li X, Waite LL, et al. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T-cell populations. PLoS Genet. 2013;9(8):e1003678. doi: 10.1371/journal.pgen.1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coit P, Jeffries M, Altorok N, et al. Genome-wide DNA methylation study suggests epigenetic accessibility and transcriptional poising of interferon-regulated genes in naive CD4+ T cells from lupus patients. J. Autoimmun. 2013;43:78–84. doi: 10.1016/j.jaut.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renauer PA, Coit P, Sawalha AH. The DNA methylation signature of human TCRalphabeta+CD4-CD8- double negative T cells reveals CG demethylation and a unique epigenetic architecture permissive to a broad stimulatory immune response. Clin. Immunol. 2015;156(1):19–27. doi: 10.1016/j.clim.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao M, Liu S, Luo S, et al. DNA methylation and mRNA and microRNA expression of SLE CD4+ T cells correlate with disease phenotype. J. Autoimmun. 2014;54:127–136. doi: 10.1016/j.jaut.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Strickland FM, Hewagama A, Wu A, et al. Diet influences expression of autoimmune-associated genes and disease severity by epigenetic mechanisms in a transgenic mouse model of lupus. Arthritis Rheum. 2013;65(7):1872–1881. doi: 10.1002/art.37967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedrich CM, Rauen T, Apostolidis SA, et al. Stat3 promotes IL-10 expression in lupus T cells through trans-activation and chromatin remodeling. Proc. Natl Acad. Sci. USA. 2014;111(37):13457–13462. doi: 10.1073/pnas.1408023111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu N, Qiu X, Luo Y, et al. Abnormal histone modification patterns in lupus CD4+ T cells. J. Rheumatol. 2008;35(5):804–810. [PubMed] [Google Scholar]
- 14.Pham TH, Benner C, Lichtinger M, et al. Dynamic epigenetic enhancer signatures reveal key transcription factors associated with monocytic differentiation states. Blood. 2012;119(24):e161–171. doi: 10.1182/blood-2012-01-402453. [DOI] [PubMed] [Google Scholar]
- 15.Pham TH, Langmann S, Schwarzfischer L, et al. CCAAT enhancer-binding protein beta regulates constitutive gene expression during late stages of monocyte to macrophage differentiation. J. Biol. Chem. 2007;282(30):21924–21933. doi: 10.1074/jbc.M611618200. [DOI] [PubMed] [Google Scholar]
- 16.Lavin Y, Winter D, Blecher-Gonen R, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159(6):1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roadmap Epigenomics C. Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••Identifies the major chromatin features across the genome.
- 18.Farh KK, Marson A, Zhu J, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518(7539):337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Many genetic variants assocaited with disease fall into regulatory regions such as promoters and enhancers. This manuscript reveals the strength of the association and the difficulty in attributing causality.
- 19.Garrett S, Dietzmann-Maurer K, Song L, Sullivan KE. Polarization of primary human monocytes by IFN-gamma induces chromatin changes and recruits RNA Pol II to the TNF-alpha promoter. J. Immunol. 2008;180(8):5257–5266. doi: 10.4049/jimmunol.180.8.5257. [DOI] [PubMed] [Google Scholar]
- 20.Lee JY, Kim NA, Sanford A, Sullivan KE. Histone acetylation and chromatin conformation are regulated separately at the TNF alpha promoter in monocytes and macrophages. J. Leuk. Biol. 2003;73:862–871. doi: 10.1189/jlb.1202618. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Z, Maurer K, Perin JC, Song L, Sullivan KE. Cytokine-induced monocyte characteristics in SLE. J. Biomed. Biotechnol. 2010;2010:507475. doi: 10.1155/2010/507475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z, Song L, Maurer K, Bagashev A, Sullivan KE. Monocyte polarization: the relationship of genome-wide changes in H4 acetylation with polarization. Genes Immun. 2011;12(6):445–456. doi: 10.1038/gene.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z, Song L, Maurer K, Petri MA, Sullivan KE. Global H4 acetylation analysis by ChIP-chip in systemic lupus erythematosus monocytes. Genes Immun. 2010;11(2):124–133. doi: 10.1038/gene.2009.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi L, Zhang Z, Yu AM, et al. The SLE transcriptome exhibits evidence of chronic endotoxin exposure and has widespread dysregulation of non-coding and coding RNAs. PLoS ONE. 2014;9(5):e93846. doi: 10.1371/journal.pone.0093846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garrett S, Fitzgerald MC, Sullivan KE. LPS and poly I:C induce chromatin modifications at a novel upstream region of the IL-23 p19 promoter. Inflammation. 2008;31(4):235–246. doi: 10.1007/s10753-008-9070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sullivan KE, Reddy AB, Dietzmann K, et al. Epigenetic regulation of tumor necrosis factor alpha. Mol. Cell. Biol. 2007;27(14):5147–5160. doi: 10.1128/MCB.02429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi L, Perin JC, Leipzig J, Zhang Z, Sullivan KE. Genome-wide analysis of interferon regulatory factor I binding in primary human monocytes. Gene. 2011;487(1):21–28. doi: 10.1016/j.gene.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 31.Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000;16(1):16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 32.Matys V, Kel-Margoulis OV, Fricke E, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34(Database issue):D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wingender E, Chen X, Fricke E, et al. The TRANSFAC system on gene expression regulation. Nucleic Acids Res. 2001;29(1):281–283. doi: 10.1093/nar/29.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hume MA, Barrera LA, Gisselbrecht SS, Bulyk ML. UniPROBE, update 2015: new tools and content for the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2015;43(Database issue):D117–122. doi: 10.1093/nar/gku1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garber M, Yosef N, Goren A, et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol. Cell. 2012;47(5):810–822. doi: 10.1016/j.molcel.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25(21):2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This manuscript was the first to define pioneer transcription factors.
- 37.Chang DH, Angelin-Duclos C, Calame K. BLIMP-1: trigger for differentiation of myeloid lineage. Nat. Immunol. 2000;1(2):169–176. doi: 10.1038/77861. [DOI] [PubMed] [Google Scholar]
- 38.Baechler EC, Bauer JW, Slattery CA, et al. An interferon signature in the peripheral blood of dermatomyositis patients is associated with disease activity. Mol. Med. 2007;13(1–2):59–68. doi: 10.2119/2006-00085.Baechler. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Z, Shi L, Song L, Ephrem E, Petri M, Sullivan K. IRF1 marks activated genes in SLE and can induce target gene expression. Arthritis Rheum. 2015;67(3):785–796. doi: 10.1002/art.38964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen S, Ma J, Wu F, et al. The histone H3 Lys 27 demethylase JMJD3 regulates gene expression by impacting transcriptional elongation. Genes Dev. 2012;26(12):1364–1375. doi: 10.1101/gad.186056.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130(6):1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 42.Xiang Y, Zhu Z, Han G, Lin H, Xu L, Chen CD. JMJD3 is a histone H3K27 demethylase. Cell Res. 2007;17(10):850–857. doi: 10.1038/cr.2007.83. [DOI] [PubMed] [Google Scholar]
- 43.Satoh T, Takeuchi O, Vandenbon A, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat. Immunol. 2010;11(10):936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- 44.De Santa F, Narang V, Yap ZH, et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 2009;28(21):3341–3352. doi: 10.1038/emboj.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leung YT, Shi L, Maurer K, et al. Interferon regulatory factor 1 and histone H4 acetylation in systemic lupus erythematosus. Epigenetics. 2015;10(3):191–199. doi: 10.1080/15592294.2015.1009764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dai C, Liu Z, Zhou H, Li L. Monocyte chemoattractant protein-1 expression in renal tissue is associated with monocyte recruitment and tubulo-interstitial lesions in patients with lupus nephritis. Chin. Med. J. (Engl.) 2001;114(8):864–868. [PubMed] [Google Scholar]
- 47.Hill GS, Delahousse M, Nochy D, et al. Predictive power of the second renal biopsy in lupus nephritis: significance of macrophages. Kidney Int. 2001;59(1):304–316. doi: 10.1046/j.1523-1755.2001.00492.x. [DOI] [PubMed] [Google Scholar]
- 48.Kittan NA, Allen RM, Dhaliwal A, et al. Cytokine induced phenotypic and epigenetic signatures are key to establishing specific macrophage phenotypes. PLoS ONE. 2013;8(10):e78045. doi: 10.1371/journal.pone.0078045. [DOI] [PMC free article] [PubMed] [Google Scholar]; • One of the first papers to identify the chromatin changes associated with macrophage subtypes, it strongly attributed function to epigenetic changes.
- 49.Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaikkonen MU, Spann NJ, Heinz S, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol. Cell. 2013;51(3):310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gorelik G, Richardson B. Aberrant T cell ERK pathway signaling and chromatin structure in lupus. Autoimmun. Rev. 2009;8(3):196–198. doi: 10.1016/j.autrev.2008.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richardson B, Ray D, Yung R. Murine models of lupus induced by hypomethylated T cells. Methods Mol. Med. 2004;102:285–294. doi: 10.1385/1-59259-805-6:285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richardson B, Sawalha AH, Ray D, Yung R. Murine models of lupus induced by hypomethylated T cells (DNA hypomethylation and lupus…) Methods Mol. Biol. 2012;900:169–180. doi: 10.1007/978-1-60761-720-4_8. [DOI] [PubMed] [Google Scholar]
- 54.Zhu X, Liang J, Li F, Yang Y, Xiang L, Xu J. Analysis of associations between the patterns of global DNA hypomethylation and expression of DNA methyltransferase in patients with systemic lupus erythematosus. Int. J. Dermatol. 2011;50(6):697–704. doi: 10.1111/j.1365-4632.2010.04804.x. [DOI] [PubMed] [Google Scholar]
- 55.Cornacchia E, Golbus J, Maybaum J, Strahler J, Hanash S, Richardson B. Hydralazine and procainamide inhibit T cell DNA methylation and induce autoreactivity. J. Immunol. 1988;140(7):2197–2200. [PubMed] [Google Scholar]; • This is one of the first papers to identify DNA demethylation as a common feature in systemic lupus erythematosus.
- 56.Vahedi G, A CP, Hand TW, et al. Helper T-cell identity and evolution of differential transcriptomes and epigenomes. Immunol. Rev. 2013;252(1):24–40. doi: 10.1111/imr.12037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vahedi G, Takahashi H, Nakayamada S, et al. STATs shape the active enhancer landscape of T cell populations. Cell. 2012;151(5):981–993. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This is an early paper that identifies transcription factors and chromatin interactions in immunologically competent cell differentiation.
- 58.Deshane J, Kim J, Bolisetty S, Hock TD, Hill-Kapturczak N, Agarwal A. Sp1 regulates chromatin looping between an intronic enhancer and distal promoter of the human heme oxygenase-1 gene in renal cells. J. Biol. Chem. 2010;285(22):16476–16486. doi: 10.1074/jbc.M109.058586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ahyi AN, Chang HC, Dent AL, Nutt SL, Kaplan MH. IFN regulatory factor 4 regulates the expression of a subset of Th2 cytokines. J. Immunol. 2009;183(3):1598–1606. doi: 10.4049/jimmunol.0803302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Negishi H, Ohba Y, Yanai H, et al. Negative regulation of Toll-like-receptor signaling by IRF-4. Proc. Natl Acad. Sci. USA. 2005;102(44):15989–15994. doi: 10.1073/pnas.0508327102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Owen-Hughes T, Workman JL. Remodeling the chromatin structure of a nucleosome array by transcription factor-targeted trans-displacement of histones. EMBO J. 1996;15(17):4702–4712. [PMC free article] [PubMed] [Google Scholar]
- 62.Ballare C, Castellano G, Gaveglia L, et al. Nucleosome-driven transcription factor binding and gene regulation. Mol. Cell. 2013;49(1):67–79. doi: 10.1016/j.molcel.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 63.Ballare C, Zaurin R, Vicent GP, Beato M. More help than hindrance: nucleosomes aid transcriptional regulation. Nucleus. 2013;4(3):189–194. doi: 10.4161/nucl.25108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Le Dily F, Bau D, Pohl A, et al. Distinct structural transitions of chromatin topological domains correlate with coordinated hormone-induced gene regulation. Genes Dev. 2014;28(19):2151–2162. doi: 10.1101/gad.241422.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sawalha AH, Jeffries M. Defective DNA methylation and CD70 overexpression in CD4+ T cells in MRL/lpr lupus-prone mice. Eur. J. Immunol. 2007;37(5):1407–1413. doi: 10.1002/eji.200636872. [DOI] [PubMed] [Google Scholar]
- 66.Zhao M, Sun Y, Gao F, et al. Epigenetics and SLE: RFX1 downregulation causes CD11a and CD70 overexpression by altering epigenetic modifications in lupus CD4+ T cells. J. Autoimmun. 2010;35(1):58–69. doi: 10.1016/j.jaut.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 67.Dai Y, Zhang L, Hu C, Zhang Y. Genome-wide analysis of histone H3 lysine 4 trimethylation by ChIP-chip in peripheral blood mononuclear cells of systemic lupus erythematosus patients. Clin. Exp. Rheumatol. 2010;28(2):158–168. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.