Abstract
Cockroaches are surrogate hosts for microbes that cause many human diseases. In spite of their generally destructive nature, cockroaches have recently been found to harbor potentially beneficial and medically useful substances such as drugs and allergens. However, genomic information for the American cockroach (Periplaneta americana) is currently unavailable; therefore, transcriptome and gene expression profiling is needed as an important resource to better understand the fundamental biological mechanisms of this species, which would be particularly useful for the selection of novel antimicrobial peptides. Thus, we performed de novo transcriptome analysis of P. americana that were or were not immunized with Escherichia coli. Using an Illumina HiSeq sequencer, we generated a total of 9.5 Gb of sequences, which were assembled into 85,984 contigs and functionally annotated using Basic Local Alignment Search Tool (BLAST), Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) database terms. Finally, using an in silico antimicrobial peptide prediction method, 86 antimicrobial peptide candidates were predicted from the transcriptome, and 21 of these peptides were experimentally validated for their antimicrobial activity against yeast and gram positive and -negative bacteria by a radial diffusion assay. Notably, 11 peptides showed strong antimicrobial activities against these organisms and displayed little or no cytotoxic effects in the hemolysis and cell viability assay. This work provides prerequisite baseline data for the identification and development of novel antimicrobial peptides, which is expected to provide a better understanding of the phenomenon of innate immunity in similar species.
Introduction
Cockroaches (order: Dictyoptera; suborder: Blattaria) are among the known primitive winged insects, with an extremely high diversity of ~4,000 species worldwide. Thirty of these species are considered as household insects [1]. The American cockroach Periplaneta americana (Linnaeus) is a synanthropic pest that generally inhabits cosmopolitan to urban areas. Cockroaches survive in warm weather with high moisture conditions as well as in unfavorable environments for humans (i.e., sewers and other human-made habitats) [2]. Accordingly, cockroaches physically transmit several human pathogens and allergens from the environment to human habitations [3]. However, the cockroach has also been a beneficial insect for humans, serving as an established model organism for basic research in the fields of neurobiology [4, 5], cardiophysiology [6], blood clotting mechanisms [7], gut microbial diversity [8, 9], and the discovery of allergenic proteins [10].
Innate immunity is the first line of defense of multicellular organisms against invading microbes such as bacteria, fungi, and viruses. Multicellular organisms thus adapt to microbes via their innate immune system through the rapid synthesis and release of various small peptides known as antimicrobial peptides (AMPs) [11].
In insects, AMPs are synthesized from the fat body and various epithelia, which are secreted into the hemolymph. Through the hemolymph, AMPs are directly supplied to the whole body in the context of microbial infection [12]. Moreover, insect autophagy also actively participates along with the innate immunity to evade the microbial infections, and these mechanisms have been extensively studied in the Drosophila model. Furthermore, the signal transaction cascade receptors such as pathogen-associated molecular patterns (PAMPs) and pattern recognition receptors (PRRs) are also activated in response to infection [13, 14]. These combinatorial molecular mechanisms serve to completely protect the insect/host from microbial infection. Since the first insect AMPs were isolated from Hyalophora cecropia in 1980 [15], 259 insect AMPs have been functionally annotated and classified according to their structural and physiochemical properties [16]. Furthermore, the effects of AMPs on innate immunity, and their corresponding molecular and metabolite/peptide synthesis mechanisms differ according to their degrees of evolutionary conservation [17].
AMPs have been exploited and developed into effective antibiotic and antimicrobial drugs from a diversity of insect species [18]. In particular, Lee et al. [19] suggested that cockroaches are a good source of antimicrobial agents. They further found that the cockroach (P. americana) brain tissues showed potent broad-spectrum antimicrobial activities, including against antibiotic-resistant bacteria [20]. AMPs are low-molecular-weight and heat-stable proteins, which are typically cationic and often comprise less than 100 amino acid residues. Despite the large number of AMPs that have been identified from different insect species, little information on their potential applications is available. In general, AMPs are predicted through in silico approaches based on their derived characteristics, i.e., similarity in physiochemical and structural properties to known AMPs [10, 21]. Several reports have indicated that AMPs can be expressed either constitutively or can be induced upon pathogenic challenge [22]. Alternatively, massive developments in high-throughput sequencing technologies have presented a more efficient method for genomic characterization of a species [23]. However, based on the few studies conducted to date, the genetic resources of cockroaches are scarce [1, 21, 24, 25]. Recently, the transcriptome of the German cockroach (Blattella germanica) was reported using next-generation sequencing (NGS) technology, which led to the identification of genes that putatively encode detoxification enzyme systems, insecticide targets, key components in systematic RNA interference, and the immunity and chemoreception pathways [26]. Therefore, identifying new insect AMPs may provide insight into natural interactions between pathogens and proteins. In this present study, we sequenced the P. americana transcriptome using an NGS platform. Libraries representing control and Escherichia coli-immunized P. americana were systematically analyzed for gene expression profiles along with AMP and allergenic protein prediction. This transcriptome data set and AMPs provide a solid baseline for further functional analysis in P. americana.
Materials and Methods
Animals
This experimental design was approved by the Institutional Animal Care and Use Committee (IACUC) of the National Academy of Agricultural Sciences (approval number: NAAS-1114). Adult American cockroaches were obtained from Kosin University, Busan, South Korea. For immunization, each cockroach was injected by log phase E. coli (2 × 106 colony forming units [CFU]) suspended in 10 μL of autoclaved 10 mM sodium phosphate buffer (pH 7.4). Cockroaches were reared at 25 ± 1°C for 18 h before total RNA isolation.
Microorganisms and growth conditions
The bacterial strains E. coli (KACC 13821, ATCC 11775) and Staphylococcus aureus (KACC 10768, ATCC 25923), and the yeast strain Candida albicans (KCTC 7121, ATCC 14053) were purchased from the Korean Agricultural Culture Collection (KACC) and Korean Collection for Type Cultures (KCTC). Both bacteria and the yeast were cultivated overnight in tryptic soy broth (TSB; Difco, USA) at 200 rpm in a 37°C shaking incubator to the stationary phase. Bacteria were cultivated for 3 h in fresh TSB medium under the same condition to the log phase. The strains were stored with 15% glycerol at −70°C until use.
NGS of the cockroach transcriptome
To obtain high-throughput transcriptome data of P. americana, we implemented Illumina-based NGS sequencing. Total RNA was isolated from E. coli-immunized (18 h after injection) and non-immunized (Control) adults. Total RNA was quantitated using a Nanodrop spectrophotometer (Thermo Scientific) and its quality was assessed with the RNA 6000 Nano assay kit (Agilent) and Bioanalyser2100 (Agilent). NGS libraries were generated from 1 μg of total RNA using TruSeq RNA Sample Prep Kit (Illumina), according to the manufacturer’s protocol. In brief, the poly-A-containing RNA molecules were purified using poly-T oligo-attached magnetic beads. After purification, the total poly A+ RNA was fragmented into small pieces using divalent cations under elevated temperature. The cleaved mRNA fragments were reverse-transcribed into first-strand cDNA using random primers. Short fragments were purified with a QiaQuick polymerase chain reaction (PCR) extraction kit and resolved with elution buffer for end repair and addition of poly (A). Subsequently, the short fragments were connected with sequencing adapters. Each library was separated by adjoining distinct MID tags. The resulting cDNA libraries were then paired-end sequenced (2 × 101 bp) with the Illumina HiSeq™ 2000 system. The resulting sequences have been deposited at the NCBI Short Read Archive under submission number SRP067419.
De novo assembly and functional annotations
Complete paired-end sequences were obtained as individual FASTQ files (forward and reverse) from the images by using CASAVA v.1.8.2 base-calling software with an ASCII Q-score offset of 64. Adaptor sequences and low-quality bases with PHRED scores (Q) ≤ 20 were removed. Repeat sequences in raw reads were masked by using RepeatMasker (Ver. 4.0.3) against the human and Drosophila Repbase database (http://www.girinst.org/). Masked sequences were subjected to de novo assembly using CLC Assembly Cell v.4.0 (CLCBio, Inc.; Aarhus, Denmark) with default parameters. Finally, the assembled contigs were subjected to functional annotations with Pendant-ProTM suite (Biomax, Inc.) under default parameters [27].
Digital gene expression (DGE) profiling
To characterize the quantitative expression pattern of individual sequences, the clean sequence reads from the two libraries (Control, E. coli-immunized) were mapped individually to the reference transcriptome by using Bowtie (Ver.0.12.7) with default parameters in TopHat (Ver. 1.3.3). Cufflink (Ver.1.3.0) and Cuffdiff (Ver. 2.0.2) were used to calculate expression profiles with reads per kilobase per million (RPKM) normalization [28]. Genes showing a minimum of 2-fold up- and down-regulation were filtered from the isoforms expression dataset with 1 ≤ log2 (fold-change [FC]) values, and the Gene Ontology (GO) annotations were classified using the WEGO webserver [29].
AMP prediction and classification
The deduced amino acid sequences were subjected to AMP prediction analysis by using a modified bioinformatics strategy. Peptide characteristics of molecular propensity (based on physicochemical properties) and aggregation propensity (in vitro and in vivo) were determined, and AMP prediction was established using a predefined bioinformatics strategy with parameters defined previously [30]. In addition to this previous strategy, the allergenic propensity of the peptides was also determined using Allerdictor software [31]. Finally, the AMPs were mapped with the CAMP database [32] and classified as novel and known AMPs. To classify the predicted AMPs as novel, sequences were matched to the CAMP database by using two programs: PatMatch (no mismatch) for sequences ≤ 20 bp in length [33] and BLASTP (1E-05) for sequences ≥ 20 bp in length. The BLAST results were filtered with a similarity score ≥ 90. Sequences with observed similarity at the given cutoff values were considered as known AMPs, and others were considered as novel AMPs. Finally, the novel and the known AMPs were manually validated for continuous stretches of amino acids to account for the low-complexity regions and assembly artifacts.
Peptide synthesis
All putative and novel peptides were selected based on the various prediction tools used previously [28]. The peptides were synthesized using solid-phase peptide synthesis methods at AnyGen Co. Ltd. (Gwangju, Korea). Then, each peptide was purified to >95% by high-performance liquid chromatography, and the purity was confirmed by mass spectrometry analysis. The peptides were dissolved in acidified distilled water (0.01% acetic acid) and stored at −20°C until used in subsequent experiments.
Antimicrobial activity assay
The radial diffusion assay was performed to test the antimicrobial activities of peptides, as described previously with slight modifications [34]. In brief, bacteria and yeast strains were grown to the mid-logarithmic phase in TSB at 37°C and then washed twice with 10 mM Tris-HCl (containing 5 mM glucose, pH 7.4). A total of 4 × 106 CFU was added to 10 mL of an underlay agarose gel [0.03% (w/v) TSB, 1% (w/v) agarose (Sigma, USA), and 0.02% (v/v) Tween 20 (Sigma, USA) in 10 mM Tris-HCl]. The underlay gel was poured into a 100-mm INTEGRIDTM Petri dish. After agarose solidification, 3-mm-diameter wells were punched and 5 μL of each peptide solution was added to each well. Buffer alone was used as a negative control. Plates were incubated at 37°C for 3 h to allow for diffusion of the peptides. The underlay gel was then covered with 10 mL of nutrient-rich agar overlay (6% TSB and 1% agarose in 10 mM Tris-HCl). The antimicrobial activity of a peptide was measured as the diameter of the cleared zone around each well after 12 h of incubation at 37°C. This experiment was repeated at least 3 times and the same results were obtained.
In addition, antimicrobial activities of the peptides were also tested by broth microdilution assays against E. coli, S. aureus, and C. albicans. Briefly, microbes were grown overnight in Mueller-Hinton Broth (MHB) to the onset of the stationary phase with shaking at 200 rpm. The cultures were diluted in fresh MHB to a final concentration of 2 × 104 CFU/mL. A stock solution of each peptide was prepared to a concentration of 640 μg/mL in 0.01% acetic acid, and was then serially diluted two-fold to reach a concentration of 10 μg/mL. After 90-μL aliquots of the microbial suspension were dispensed into each well of a 96-well polypropylene microtiter plate, 10 μL of the peptide solution was added. The antimicrobial activities of the peptides were assessed by measuring the visible turbidity in each well of the plate after 18 h of incubation at 37°C. Minimum inhibitory concentrations (MICs) are expressed as a specific value that caused complete growth inhibition.
Hemolytic assay
This experiment was approved by the Institutional Animal Care and Use Committee (IACUC) of the National Academy of Agricultural Sciences (approval number: NAAS-1114). The hemolytic activity of the peptides was determined by monitoring the release of hemoglobin from rat erythrocytes at 540 nm. For the hemolytic assay, 20 μL of each peptide solution at a predetermined concentration was added to 180 μL of a 2.5% (v/v) suspension of rat erythrocytes in phosphate-buffered saline (PBS). Melittin (Sigma, USA), a hemolytic and α-helical peptide isolated from bee venom, was used as the positive control. This mixture was incubated for 30 min at 37°C, and 600 μL of PBS was then added to each tube. After 3 min of centrifugation at 10,000 ×g, the supernatant was removed, and the absorbance was measured at 540 nm. Evaluations were made from the results of at least three independent experiments, each carried out in triplicate.
Cell culture and cell viability assay
The human keratinocytes were maintained in Dulbecco’s Modified Eagle Medium supplemented with 10% fetal bovine serum (FBS; Hyclone, USA), penicillin G (100 U/mL), and streptomycin (100 μg/mL) (Invitrogen, USA). Human umbilical vein endothelial cells (HUVECs) were grown on 0.1% gelatin-coated cell culture dishes in M199 medium (Welgene, Korea) supplemented with 20% (v/v) FBS, 3 ng/mL basic fibroblast growth factor (R&D Systems, USA), 5 U/mL of heparin (Sigma, USA), and a penicillin-streptomycin-amphotericin B mixture (100 U/mL potassium penicillin, 100 mg/mL streptomycin sulfate, and 250 ng/mL amphotericin B; Lonza, Belgium) as a complete medium. Cells were cultured at 37°C in a humidified incubator with 5% CO2. Cells were plated in 96-well tissue culture plates (2 × 104 cells per well). After 1 day, they were treated with various concentrations (25, 50, 100, and 200 μg/mL) of peptides. Melittin (Sigma, USA) was used as the positive control. After incubation for 24 h, the viability of the cells was assessed by the Cell Titer 96 AQueous One Solution Cell Proliferation Assay according to the manufacturer’s protocol (Promega, USA). The optical density at 490 nm was measured with a microplate reader (Beckman DTX 8800 Multi Detector, USA).
Results and Discussion
Sequencing and transcriptome assembly
The cDNA library prepared from cockroach samples was sequenced using the Illumina HiSeqTM 2000 sequencer. As a result of sequencing, 4,687,932,060 (52,088,134 reads) and 4,781,794,320 (53,131,048 reads) bases were obtained for E. coli-immunized and non-immunized cockroaches, respectively. We injected live E. coli into the hemocoel of the cockroaches for immunization, although mixtures of bacteria and fungi should have been employed for the possibility of full induction. A comparative study of differential gene expression is required in future studies to determine the effects of various elicitors. In the present study, we focused on the prediction and experimental validation of novel AMP candidates. None of the novel AMPs was identified among known AMPs that are induced by bacteria and fungi, since we excluded known AMPs after comparison with sequences from the UniProtKB database for the novel AMPs selection procedure. In addition, we employed a naïve sample as a non-immunized control to exclude the expression data of overlapping genes for calculating the maximum fold change of differential gene expression. Initially, the total reads were subjected to preprocessing, as described in the Materials and Methods, resulting in 4,302,302,163 (49,317,908 reads) and 4,380,901,481 (50,270,016 reads) bases, for an average of 91% coverage from raw sequences for the immunized and non-immunized samples, respectively (Table 1). Preprocessed sequences were taken for de novo transcriptome assembly by using CLC Assembly Cell v. 4.0. In total, 85,984 contigs were obtained from the assembly, ranging from 200- to 18,078-bp transcripts with an average of 620.8 bp (Fig 1A), which was considered as the draft reference transcriptome for P. americana.
Table 1. Summary of HiSeq and Assembly Statistics from E. coli-immunized and Non-immunized P. americana.
| Description | Samples | Reads | % | Bases | % | Avg. Length (bp) |
|---|---|---|---|---|---|---|
| Raw sequences | 1.TG1110R2526_l1(E. coli-immunized) | 52,088,134 | 100.0 | 4,687,932,060 | 100 | 90.0 |
| 2.TG1110R2527_l1(non-immunized) | 53,131,048 | 100.0 | 4,781,794,320 | 100 | 90.0 | |
| Pre-Processed sequences | 1.TG1110R2526_l1(E. coli-immunized) | 49,317,908 | 94.6 | 4,302,302,163 | 91.77 | 82.5 |
| 2.TG1110R2527_l1(non-immunized) | 50,270,016 | 94.6 | 4,380,901,481 | 91.62 | 82.5 | |
| De novo assembly | Contig | 85,984 | 53,382,468 | 620.8 | ||
| Repeat mask | Contigs_Masked | 85,608 | 44,222,145 | 516.6 |
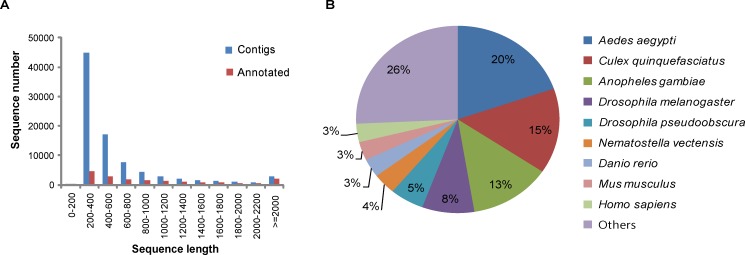
Fig 1. Overview of the P. americana Annotation.
(A) Comparison of sequence length distribution before (blue) and after (red) annotation. (B) Top-hit species distribution of BLAST matches of sequences unique to P. americana.
Functional annotation of unigenes
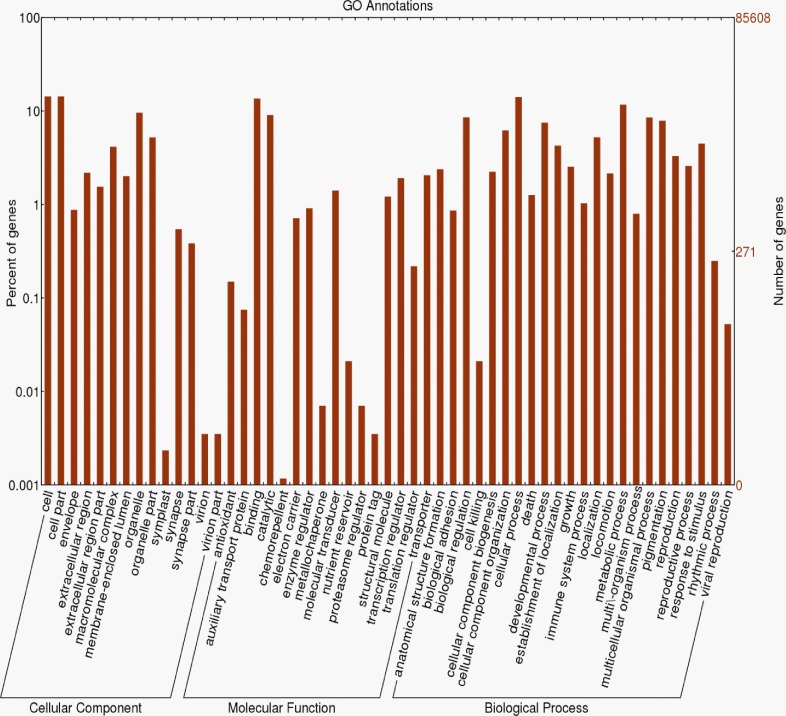
The standardized automated software suite Pendant-Pro (Biomax Informatics) was used to annotate the transcripts. Initially, the assembled transcripts were subjected to repeat masking with a human repeat library, resulting in 44,222,145 reads (85,608 contigs), and masked sequences were subjected to Pendant-Pro with default parameters to obtain the annotations. In total, 17,744 (20.7%) sequences were annotated from 13,726 UniProt protein sequences (Table 2) and the remaining sequences were unannotated (most sequences < 300 bp were not annotated well) (Fig 1A). More than 60% of the annotated sequences were homologous to proteins from mosquitoes (Aedes aegypti, Anopheles gambiae, Culex quinquefasciatus), flies (Drosophila melanogaster, Drosophila pseudoobscura), and mammals (human and mouse) (Fig 1B). Further, the annotated transcripts were grouped into GO subcategories, i.e., biological process (BP), molecular functions (MF), and cellular components (CC), from level-2 GOs. The GO terms cell; cell part; organelle (in CC); binding, catalytic, and transcription regulators (in MF); and cellular process, metabolic process, and pigmentation (in BP) were shown to be the top 3 clusters (Fig 2).
Table 2. Summary Statistics of Gene Ontology Categories.
| Groups | Category | Contigs (%) | |
| A. BLAST | Total contigs | 85,608 (100) | |
| Has UniProt ID | 17,744 (20.7) | ||
| Unique UniProt ID | 13,726 (16.0) | ||
| Category | # Up (%) | # Down (%) | |
| B. Digital Gene Expression | Fold change ≥ 2 | 848 (1.0) | 1,228 (1.4) |
| Known UniProt ID | 232 (0.3) | 448 (0.5) | |
| Unique UniProt ID | 218 (0.3) | 373 (0.4) | |
| Category | # Term | # Contigs (%) | |
| C. Gene Ontology | Biological Process | 4,953 | 11,395 (13.3) |
| Molecular Function | 2,712 | 11,822 (13.8) | |
| Cellular Component | 947 | 10,958 (12.8) | |
Fig 2. Gene Ontology (GO) Classification of the P. americana Transcriptome.
The histogram of the GO annotation was generated automatically using the web histogram tool WEGO (http://wego.genomics.org.cn/cgi-bin/wego/index.pl) based on the most recent GO archive available. The results are summarized into three main GO categories: cellular component, molecular function, and biological process. The right y-axis indicates the number of genes in a category. The left y-axis indicates the percentage of a specific category of genes in that main category. One gene could be annotated into more than one GO term.
DGE profile
To analyze the gene expression profiles of P. americana from the transcriptome data, DGE analysis was performed, as described in the Materials and Methods. In total, 2,076 (2.4%) transcripts were found to be significantly up- and down-regulated with a ≥2-fold change. Among these transcripts, 848 (1.0%) were up-regulated and 1,228 (1.4%) were down-regulated in the immunized condition, which were plotted in a histogram based on the GO categories (S1 Fig).
In silico analysis of allergens and AMPs from P. americana
Isolation of AMPs from insects has been one of most effective and promising strategies in the development of antimicrobial drugs [18]. For the most part, AMPs have been predicted through computational rather than experimental methods. The primary goal of this study was to predict the AMPs from the transcripts of P. americana and validate these predictions experimentally. In total, 86 AMPs were predicted to be novel AMPs (Table 3 and S1 Table), 72 were identified as putative AMPs (S2 Table), as defined in the Materials and Methods, and 180 proteins were predicted as allergens (S3 Table). Both the novel and putative AMPs were identified as non-allergenic peptides and are listed in Table 3. Among the putative AMPs, 54 are known to function as antibacterials, 5 are antifungals, and 3 are antivirals (S1 Table). Three of these transcripts were annotated as being related to the immune response (ISGCock_Contig01_0792, ISGCock_Contig04_0023, and ISGCock_Contig08_4679), 20 transcripts were annotated as being involved in protein binding, and none of the novel AMPs was annotated. The allergenic proteins were grouped into GO subcategories (S2 Fig). Previously known allergen proteins were only predicted from 233 UniProt database sequences of P. americana, and 9 were validated [10]. In our predictions, 57 novel transcripts were predicted as allergens. These novel candidates should be useful for progress in anti-allergen development.
Table 3. In Silico Functional Characterization of Selected Antimicrobial Peptides (AMPs).
| Program | Parameter | Filtered | |
|---|---|---|---|
| PEPSTATS | Length | ≤ 50mer | 18,309 |
| Charge | >0(+) | 64,172 | |
| pI | 8 ≤ pI ≤ 12 | 51,872 | |
| AMPA | Stretch No. | ≥1 | 45,751 |
| Known AMP | BLASTP (E-value) | 1.00E-05 | 83,350 (42) |
| PATMATCH | No mismatch | 30 | |
| CAMP | AMP (Discriminant analysis) | TRUE (score < -0.251) | 30,534 |
| AMP (SVM) | TRUE (no score) | 26,948 | |
| TANGO | AGG | AGG ≤ 500 | 37,092 |
| HELIX | 0 ≤ HELIX ≤ 25 | 38,526 | |
| BETA | 25 ≤ BETA ≤ 100 | 3,711 | |
| AGGRESCAN | Na4vSS | -40 ≤ Na4vSS ≤ 60 | 81,166 |
| EPESTFIND | Protein cleavage site | FALSE | 40,791 |
| Expression change | NC or Increased | -1 < log2 (FC) | 64,725 |
| Final filtered AMPs | 86 | ||
pI, isoelectric point; SVM, support vector machine; NC, no change; FC, fold change.
Experimental validation of putative and novel AMPs
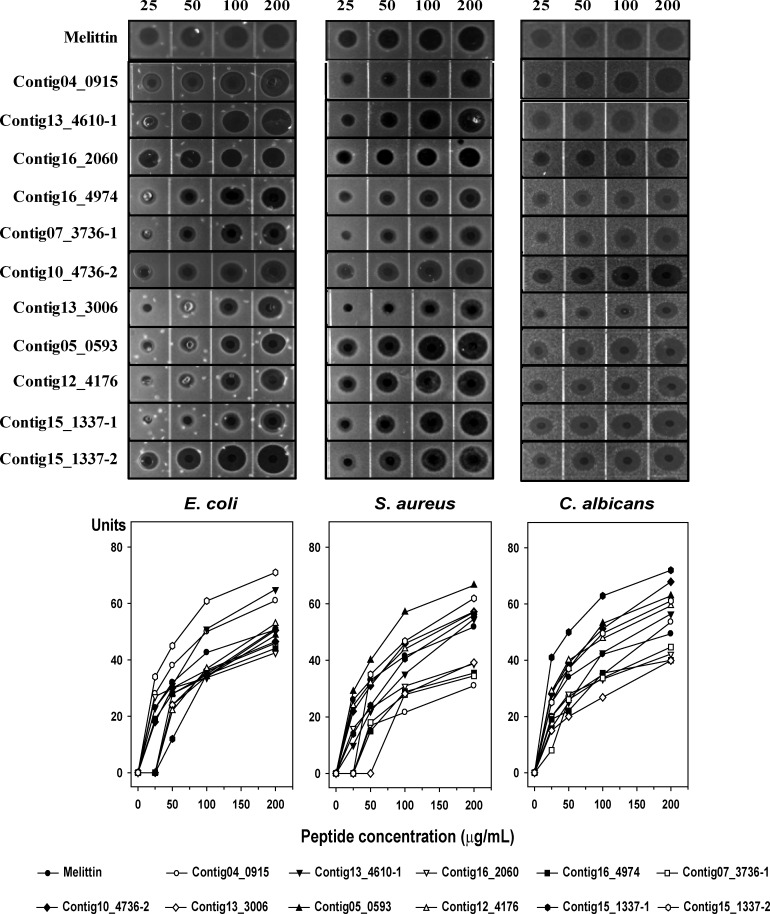
Experimental validation is required to examine the accuracy of any putative and novel AMPs identified. The 86 peptides were sorted according to fold-change in expression, and 21 AMPs with potentially high activity were ultimately selected (Table 4). Twenty-five peptides were synthesized according to results of the AMPA server (http://tcoffee.crg.cat/apps/ampa). We chose the active regions based on AMPA stretch due to efficiency and cost of peptide synthesis for development of antimicrobial agents. We tested their antimicrobial activities against Gram-negative bacteria, Gram-positive bacteria, and yeast using a radial diffusion assay (Fig 3). We found antimicrobial activity in 11 synthetic peptides (ISGCock_Contig04_0915, ISGCock_Contig13_4610–1, ISGCock_Contig16_2060, ISGCock_Contig16_4974, ISGCock_Contig07_3736–1, ISGCock_Contig10_4736–2, ISGCock_Contig13_3006, ISGCock_Contig05_0593, ISGCock_Contig12_4176, ISGCock_Contig15_1337–1, and ISGCock_Contig15_1337–2), which increased in a dose-dependent manner. Remarkably, the peptides ISGCock_Contig04_0915, ISGCock_Contig13_4610–1, and ISGCock_Contig15_1337–2 showed stronger antimicrobial activity than melittin in E. coli. The antimicrobial activities of ISGCock_Contig13_4610–1, ISGCock_Contig10_4736–2, ISGCock_Contig05_0593, ISGCock_Contig12_4176, ISGCock_Contig15_1337–1, and ISGCock_Contig15_1337–2 were greater than that of melittin in S. aureus. Correspondingly, ISGCock_Contig04_0915, ISGCock_Contig13_4610–1, ISGCock_Contig10_4736–2, ISGCock_Contig05_0593, ISGCock_Contig12_4176, ISGCock_Contig15_1337–1, and ISGCock_Contig15_1337–2 showed strong antimicrobial activity against C. albicans.
Table 4. List of Predicted Antimicrobial Peptides (AMPs) from American Cockroach Transcripts.
| Sequence ID | Length | Discriminant | log2(FC) | Sequence | SVM-score | Prediction |
|---|---|---|---|---|---|---|
| ISGCock_Contig04_0915 | 50 | AMP | 3.07 | ALQICTRNMIDDRLPYVADNVRPGTFIKQQRKQKQQRHHTSGTRKRMAKG | -1.01 | non-allergen |
| ISGCock_Contig02_3734 | 44 | AMP | 2.96 | KLHEFKLGYPLATNYACAIARDLILHKIYIIHFLHRLRKKLSHY | -1.04 | non-allergen |
| ISGCock_Contig14_1231 | 41 | AMP | 2.57 | ISYFLFLDFRDIFHSQRRKVNFNAGIHSHKNNNKYKLSSCQ | -1.02 | non-allergen |
| ISGCock_Contig13_3331 | 47 | AMP | 2.55 | IRFGKFKNLRQKQENRCGDIFKQRQGLETCRHRLQFKIDLYISTNDK | -1.00 | non-allergen |
| ISGCock_Contig13_4610–1 | 44 | AMP | 2.55 | HLYPCKLNLKLGKVPFHFLNLNHKGKSIMVNQQTCLYYIICQTR | -0.97 | non-allergen |
| ISGCock_Contig13_4610–2 | 44 | AMP | 2.55 | HLYPCKLNLKLGKVPFHFLNLNHKGKSIMVNQQTCLYYIICQTR | -0.97 | non-allergen |
| ISGCock_Contig01_3774 | 31 | AMP | 2.31 | PPHMQSPLCAPCKIQGRSIVFRTSIVLVNLN | -1.00 | non-allergen |
| ISGCock_Contig12_2253 | 23 | AMP | 2.31 | KQRKEGECGQFLTKVNSGKIITG | -1.00 | non-allergen |
| ISGCock_Contig13_4305 | 36 | AMP | 2.29 | YAHLSNIPIFQVCVCSKVYYIHKHFTNYLRVSKQNC | -1.02 | non-allergen |
| ISGCock_Contig16_2060 | 34 | AMP | 2.29 | ISHNHLTAASITHVKNRGKYIYMHLKFRKTNVLI | -1.04 | non-allergen |
| ISGCock_Contig13_2121 | 31 | AMP | 2.27 | LSPHSSNVKRKEHLLSNCKFNFYRLKLIQIP | -0.99 | non-allergen |
| ISGCock_Contig11_1401 | 41 | AMP | 2.01 | RTSKNYLIITQLKGENLESPKDIRKIIFSNGDRLDCRKSKP | -0.96 | non-allergen |
| ISGCock_Contig16_4974 | 44 | AMP | 1.99 | RKKVWFIFHVCPKLKQRILSDTHAKNKCRLSPLLIKSTKIKNET | -1.00 | non-allergen |
| ISGCock_Contig07_3736–1 | 31 | AMP | 1.88 | CNYISFFRKCKNSQSTMYGCHRMNKCVFSSY | -1.02 | non-allergen |
| ISGCock_Contig07_3736–2 | 31 | AMP | 1.88 | CNYISFFRKCKNSQSTMYGCHRMNKCVFSSY | -1.02 | non-allergen |
| ISGCock_Contig14_0122 | 26 | AMP | 1.70 | SINRFLQHYNISLYTPYNFIIKKTNF | -1.01 | non-allergen |
| ISGCock_Contig05_0163 | 35 | AMP | 1.66 | KSILYLLCRDFRDLHKYAALRIVQSPRRTLYNKLN | -1.00 | non-allergen |
| ISGCock_Contig10_4736–1 | 37 | AMP | 1.57 | LMLCKGFLRHSYKSIHERGTKRGKLCRISRLALSSLP | -0.77 | non-allergen |
| ISGCock_Contig10_4736–2 | 37 | AMP | 1.57 | LMLCKGFLRHSYKSIHERGTKRGKLCRISRLALSSLP | -0.77 | non-allergen |
| ISGCock_Contig07_2123 | 19 | AMP | 1.55 | NIYHFFNINKTQFLLITHN | -1.01 | non-allergen |
| ISGCock_Contig13_3006 | 47 | AMP | 1.49 | ANLLRHKVYGYCVLGPKGSSLGGIHGTWHDHHCSLIQRNPSTSTKGN | -1.00 | non-allergen |
| ISGCock_Contig05_0593 | 18 | AMP | 1.48 | MKTFLRLYRSLINKVLHV | -1.01 | non-allergen |
| ISGCock_Contig12_4176 | 30 | AMP | 1.38 | VVGRKHSILNCIPYLKKKKIMRLVESESIG | -1.06 | non-allergen |
| ISGCock_Contig15_1337–1 | 46 | AMP | 1.31 | KRMKLNAKKLSFCDHLNSYLNLSPTLFIHNSSKQWSHWLWHNGIRI | -1.00 | non-allergen |
| ISGCock_Contig15_1337–2 | 46 | AMP | 1.31 | KRMKLNAKKLSFCDHLNSYLNLSPTLFIHNSSKQWSHWLWHNGIRI | -1.00 | non-allergen |
The underlined peptide sequences were synthesized according to results of the AMPA stretches.
Fig 3. Radial Diffusion Assay.
Antimicrobial activities of 11 selected peptides against E. coli, S. aureus, and C. albicans determined by a radial diffusion assay. Peptide concentration (x-axis) was plotted against the diameter of the microbial growth inhibition zone (y-axis) after incubation for 12 h, and is expressed in units (1 mm = 10 units). Melittin was used as a positive control. Mean values were obtained from tests repeated three times.
Thus, different effects were observed for different strains. ISGCock_Contig15_1337–2 showed the highest antimicrobial activity of the tested peptides in E. coli, ISGCock_Contig05_0593 showed the highest antibacterial effect in S. aureus, and Contig15_1337–1 showed the highest antifungal activity of the tested peptides with C. albicans. Although the antimicrobial activity of AMPs is generally related to the cell membrane components of the microbes known as PAMPs [35], these AMPs showed a broad range (200 μg/mL) of activity toward Gram-negative bacteria, Gram-positive bacteria, and yeast.
We performed additional antimicrobial testing to determine MIC values against E. coli, S. aureus, and C. albicans. Table 5 shows the antimicrobial activities of the selected peptides including melittin as a control peptide. The MICs of melittin for microbes were measured to be between 4 μg/mL and 8 μg/mL. The ISGCock_Contig16_2060, Contig16_4974, ISGCock_Contig10_4736–2, ISGCock_Contig05_0593, and ISGCock_Contig12_4176 peptides showed potent antibacterial activities in E. coli. Most of the peptides were relatively less potent against S. aureus except for ISGCock_Contig16_2060, ISGCock_Contig16_4974, and ISGCock_Contig05_0593 compared to their E. coli-cidal activities. The ISGCock_Contig16_2060, Contig16_4974, ISGCock_Contig05_0593, ISGCock_Contig12_4176, and ISGCock_Contig15_1337–1 peptides showed potent anti-Candida activities and the MIC values were equal to the anti-E. coli activities except for ISGCock_Contig15_1337–1. In contrast, the ISGCock_Contig04_0915, ISGCock_Contig13_4610–1, ISGCock_Contig07_3736–1, ISGCock_Contig13_3006, ISGCock_Contig15_1337–1, and ISGCock_Contig15_1337–2 peptides exhibited higher MIC for most strains, indicating that these peptides may be influenced by the MHB components. Further study is required to elucidate the mechanism and source of the observed antimicrobial activity. Overall, the ISGCock_Contig16_2060, Contig16_4974, ISGCock_Contig05_0593, and ISGCock_Contig12_4176 peptides were prime candidates for development of antimicrobial agents.
Table 5. Minimum Inhibitory Concentration (MIC, μg/mL) for Antimicrobial Activity of the Selected Peptides and Melittin.
| E. coli (ATCC 11775) | S. aureus (ATCC 25923) | C. albicans (ATCC 14053) | |
|---|---|---|---|
| Melittin | 8 | 4 | 8 |
| ISGCock_Contig04_0915 | 64 | >64 | >64 |
| ISGCock_Contig13_4610–1 | 64 | >64 | >64 |
| ISGCock_Contig16_2060 | 16 | 16 | 16 |
| ISGCock_Contig16_4974 | 16 | 16 | 16 |
| ISGCock_Contig07_3736–1 | >64 | >64 | >64 |
| ISGCock_Contig10_4736–2 | 16 | >64 | 32 |
| ISGCock_Contig13_3006 | 64 | >64 | 64 |
| ISGCock_Contig05_0593 | 4 | 16 | 4 |
| ISGCock_Contig12_4176 | 8 | >64 | 8 |
| ISGCock_Contig15_1337–1 | >64 | >64 | 16 |
| ISGCock_Contig15_1337–2 | >64 | >64 | 64 |
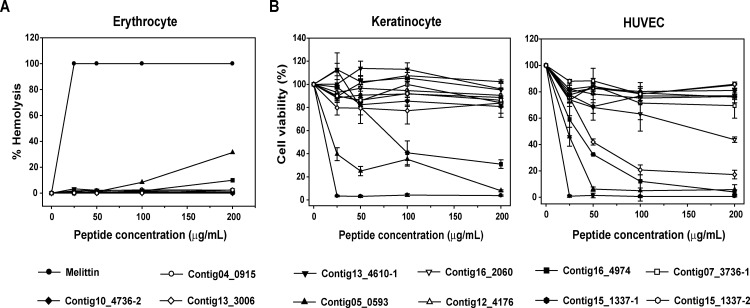
The hemolytic effects of the 11 selected synthetic peptides showing antimicrobial activity in the radial diffusion assay are shown in Fig 4A. Melittin lysed 99% of rat red blood cells at a concentration of 25 μg/mL, whereas no hemolytic activity was observed for the 11 synthetic peptides at this concentration and up to 50 μg/mL, although the ISGCock_Contig05_0593 and Contig16_4974 peptides showed relatively strong hemolytic activity even at a high concentration (200 μg/mL). Nevertheless, the hemolytic activities of the peptides ISGCock_Contig13_4610–1, ISGCock_Contig12_4176, ISGCock_Contig15_1337–1, and ISGCock_Contig15_1337–2 were relatively low compared to that of melittin at a concentration of 100 μg/mL. Therefore, the ISGCock_Contig05_0593 and Contig16 _4974 peptides are thought to be effective at doses less than 100 μg/mL, indicating their potential as therapeutic agents against a vast array of microbial infections (Fig 4A). In addition, we investigated the cell viabilities of normal human cell lines (keratinocytes and HUVECs) after treatment with the selected peptides for 24 h at the same concentration of the hemolysis assay. Most of the peptides did not decrease cell viabilities of the cell lines except for the ISGCock_Contig16_4974 and Contig05_0593 peptides together with the ISGCock_Contig16_2060 and Contig13_3006 peptides in HUVECs (Fig 4B). These two peptides (ISGCock_Contig16_4974 and Contig05_0593) showed hemolytic activity in the hemolysis assay and the data are consistent with the MTS assay results, except for the ISGCock_Contig16_2060 and Contig13_3006 peptides, which suggests that these peptides have a specific cytotoxic effect on eukaryotic cells. In contrast, normal human cells were more susceptible to melittin treatment even at the lowest concentration. Melittin has strong and broad antimicrobial spectrum, but the peptide lacks selectivity in normal cells. The purpose of this experimental study is to find novel peptides, which have potent antimicrobial activities with little or no cytotoxicity. Thus, these data indicate that the selected peptides are useful for the development of novel antimicrobial agents.
Fig 4. Cytotoxic Effects of 11 Selected Peptides.
(A) Hemolytic activity of the peptides. Peptide concentration (x-axis) is plotted against the percentage of hemolysis (y-axis) of rat red blood cells after incubation for 30 min. Melittin was used as the positive control. The percent hemolysis was calculated with the following equation: hemolysis (%) = (A540 of sample − A540 of peptide-free control)/(A540 of 100% control − A540 of peptide-free control) × 100. (B) Cell viability of human keratinocytes and human umbilical vein endothelial cells (HUVECs) after peptides treatment. Cell viability was measured by an MTS assay after a 24-h incubation with the indicated amounts of each peptide. Each symbol represents the mean value estimated from triplicate experiments.
Conclusions
Microbial resistance towards antibiotics threatens the effective prevention and treatment of a wide range of infections caused by bacteria, parasites, viruses, and fungi. In recent years, intensive studies have been undertaken towards the development of more effective antimicrobial drugs. AMPs are vital components of innate immunity that can rapidly respond to diverse microbial pathogens. Insects, as a rich source of AMPs, have attracted considerable research attention with respect to both understanding the insect’s immune system and searching for new molecular models for anti-infective drug design [1, 6, 11].
Here, we have shown the effectiveness of a combination of in silico and in vitro approaches to identify the putative and novel AMPs in P. americana. We performed de novo transcriptome sequencing of E. coli-immunized and non-immunized P. americana and selected 86 AMPs by combining the transcriptome with the successive assembly strategies. We further validated the antimicrobial and hemolytic effects of 11 selected AMPs experimentally, demonstrating broad-range antimicrobial activity.
Reduction in sequencing costs and the availability of high-throughput data facilitated by NGS have provided essential genetic resources to help expand fundamental knowledge of the biology and evolutionary history of an organism. Collectively, the present findings show that the combination of in silico and in vitro approaches could narrow down the identification of potential AMPs, and recent advances in both fields could be used to validate the applications of these 11 candidate AMPs as a template for further development as effective antibiotic therapeutics. Furthermore, these transcriptome sequencing results provide a genetic resource that should facilitate further comprehensive studies on the American cockroach.
Supporting Information
(TIF)
(TIF)
(XLSX)
(XLSX)
(XLSX)
Data Availability
The resulting sequences have been deposited at the NCBI Short Read Archive under submission number SRP067419. All other relevant data are included within the Supporting Information files.
Funding Statement
This work was supported by a grant from the Next-Generation BioGreen 21 Program (Project No. PJ01104304), Rural Development Administration, Republic of Korea. Insilicogen, Inc. provided support in the form of salaries for authors [SS, JP], but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1.Chung HS, Yu TH, Kim BJ, Kim SM, Kim JY, Yu HS, et al. Expressed sequence tags analysis of Blattella germanica. Kor J Parasitiol. 2005;43: 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaramillo-Ramirez GI, Cárdenas-Henao H, González-Obando R, Rosero-Galindo CY. Genetic variability of five Periplaneta americana L. (Dyctioptera: Blattidae) populations in southwestern Colombia using the AFLP molecular marker technique. Neotrop Entomol. 2010;39: 371–378. [DOI] [PubMed] [Google Scholar]
- 3.Kinfu A, Erko B. Cockroaches as carriers of human intestinal parasites in two localities in Ethiopia. Trans R Soc Trop Med Hyg. 2008;102: 1143–1147. 10.1016/j.trstmh.2008.05.009 [DOI] [PubMed] [Google Scholar]
- 4.Stankiewicz M, Browski M, de Lima ME. Nervous system of Periplaneta americana cockroach as a model in toxinological studies: a short historical and actual view. J Toxicol. 2012;2012: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lent DD, Pinter M, Strausfeld NJ. Learning with half a brain. Dev Neurobiol. 2007;67: 740–751. [DOI] [PubMed] [Google Scholar]
- 6.Slama K, Sakai T, Takeda M. Effect of corazonin and crustacean cardioactive peptide on heartbeat in the adult American cockroach (Periplaneta americana). Arch Insect Biochem Physiol. 2006;62: 91–103. [DOI] [PubMed] [Google Scholar]
- 7.Haine ER, Rolff J, Siva-Jothy MT. Functional consequences of blood clotting in insects. Dev Comp Immunol. 2007;31: 456–464. [DOI] [PubMed] [Google Scholar]
- 8.Fang W, Fang Z, Liu Z, Yuan J, Zhang X, Peng H, et al. Phylogenetic analysis of bacterial community in the gut of American cockroach (Periplaneta americana). Acta Microbiologica Sinica 2013;53: 984–994. [PubMed] [Google Scholar]
- 9.Bertino-Grimaldi D, Medeiros MN, Vieira RP, Cardoso AM, Turque AS, Silveira CB, et al. Bacterial community composition shifts in the gut of Periplaneta americana fed on different lignocellulosic materials. SpringerPlus 2013;2: 609 10.1186/2193-1801-2-609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmed A, Minhas K, Namood ES, Aftab O, Khan FS. In silico identification of potential American cockroach (Periplaneta americana) allergens. Iran J Public Health. 2010;39: 109–115. [PMC free article] [PubMed] [Google Scholar]
- 11.Seo MD, Won HS, Kim JH, Mishig-Ochir T, Lee BJ. Antimicrobial peptides for therapeutic applications: a review. Molecules 2012;17: 12276–12286. 10.3390/molecules171012276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bulet P, Stocklin R. Insect antimicrobial peptides: structures, properties and gene regulation. Protein Pept Lett. 2005;12: 3–11. . [DOI] [PubMed] [Google Scholar]
- 13.Moy RH, Cherry S. Antimicrobial autophagy: a conserved innate immune response in Drosophila. J Innate Immun. 2013;5: 444–455. 10.1159/000350326 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomes LC, Dikic I. Autophagy in antimicrobial immunity. Mol Cell. 2014;54: 224–233. 10.1016/j.molcel.2014.03.009 . [DOI] [PubMed] [Google Scholar]
- 15.Yi HY, Chowdhury M, Huang YD, Yu XQ. Insect antimicrobial peptides and their applications. Appl Microbiol Biotechnol. 2014;98: 5807–5822. 10.1007/s00253-014-5792-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang G, Li X, Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44: D1087–D1093. 10.1093/nar/gkv1278 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vilcinskas A. Evolutionary plasticity of insect immunity. J Insect Physiol. 2013;59: 123–129. 10.1016/j.jinsphys.2012.08.018 . [DOI] [PubMed] [Google Scholar]
- 18.Bulet P, Charlet M, Hetru C. Antimicrobial peptides in insect immunity In: Ezekowitz RA, Hoffmann J, editors. Innate immunity. New Jersey: Humana Press; 2003. pp. 89–107. [Google Scholar]
- 19.Lee S, Siddiqui R, Khan NA. Animals living in polluted environments are potential source of antimicrobials against infectious agents. Pathog Glob Health. 2012;106: 218–223. 10.1179/2047773212Y.0000000033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee S, Duce I, Atkins H, Khan NA. Cockroaches and locusts: physicians' answer to infectious diseases. Int J Antimicrob Agents. 2011;37: 279–280. 10.1016/j.ijantimicag.2010.12.005 [DOI] [PubMed] [Google Scholar]
- 21.Fjell CD, Hiss JA, Hancock RE, Schneider G. Designing antimicrobial peptides: form follows function. Nat Rev Drug Discov. 2012;11:37–51. [DOI] [PubMed] [Google Scholar]
- 22.Imler JL, Hoffmann JA. Signaling mechanisms in the antimicrobial host defense of Drosophila. Curr Opin Microb. 2000;3: 16e22. [DOI] [PubMed] [Google Scholar]
- 23.Hornett EA, Wheat CW. Quantitative RNA-Seq analysis in non-model species: assessing transcriptome assemblies as a scaffold and the utility of evolutionary divergent genomic reference species. BMC Genomics. 2012;13: 361 10.1186/1471-2164-13-361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao B, Chen AH, Zhang YY, Jiang GF, Hu CC, Zhu CD. Complete mitochondrial genomes of two cockroaches, Blattella germanica and Periplaneta americana, and the phylogenetic position of termites. Curr Genet. 2012;58: 65–77. 10.1007/s00294-012-0365-7 [DOI] [PubMed] [Google Scholar]
- 25.Chen W, Jiang GF, Sun SH, Lu Y, Ma F, Li B. Identification of differentially expressed genes in American cockroach ovaries and testes by suppression subtractive hybridization and the prediction of its miRNAs. Mol Genet Genomics. 2013;288: 627–638. 10.1007/s00438-013-0777-1 [DOI] [PubMed] [Google Scholar]
- 26.Zhou X, Qian K, Tong Y, Zhu JJ, Qiu X, Zeng X. De novo transcriptome of the hemimetabolous German cockroach (Blattella germanica). PLoS ONE 2014; 9: e106932 10.1371/journal.pone.0106932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walter MC, Rattei T, Arnold R, Güldener U, Münsterkötter M, Nenova K, et al. PEDANT covers all complete RefSeq genomes. Nucleic Acids Res. 2009;37: D408–D411. 10.1093/nar/gkn749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelly DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7: 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34: W293–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoo W, Lee J, Shin Y, Shim J-Y, Jung M, Kang B-C, et al. Antimicrobial peptides in the centipede Scolopendra subspinipes mutilans. Funct Integr Genomics. 2014; 14: 275–283. 10.1007/s10142-014-0366-3 [DOI] [PubMed] [Google Scholar]
- 31.Dang HX, Lawrence CB. Allerdictor: fast allergen prediction using text classification techniques. Bioinformatics 2014;30: 1120–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waghu FH, Gopi L, Barai RS, Ramteke P, Nizami B, Idicula-Thomas S. CAMP: Collection of sequences and structures of antimicrobial peptides. Nucleic Acids Res. 2014;42: D1154–1158. 10.1093/nar/gkt1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan T, Yoo D, Berardini TZ, Mueller LA, Weems DC, Weng S, et al. PatMatch: a program for finding patterns in peptide and nucleotide sequences. Nucleic Acids Res. 2005; 33: W262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torrent M, Pulido D, de la Torre BG, Garcia-Mayoral MF, Nogues MV, Bruix M, et al. Refining the eosinophil cationic protein antibacterial pharmacophore by rational structure minimization. J Med Chem. 2011;54: 5237–5244. 10.1021/jm200701g [DOI] [PubMed] [Google Scholar]
- 35.Müller U, Vogel P, Alber G, Schaub GA. The innate immune system of mammals and insects In: Egesten A, Schmidt A, Herwald H (eds). Trends in innate immunity. Basel: Karger; 2008. pp. 21–44. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
The resulting sequences have been deposited at the NCBI Short Read Archive under submission number SRP067419. All other relevant data are included within the Supporting Information files.