Abstract
Xp11 translocation cancers include Xp11 translocation renal cell carcinoma (RCC), Xp11 translocation perivascular epithelioid cell tumor (PEComa), and melanotic Xp11 translocation renal cancer. In Xp11 translocation cancers, oncogenic activation of TFE3 is driven by the fusion of TFE3 with a number of different gene partners, however, the impact of individual fusion variant on specific clinicopathologic features of Xp11 translocation cancers has not been well defined. In this study, we analyze 60 Xp11 translocation cancers by fluorescence in situ hybridization (FISH) using custom BAC probes to establish their TFE3 fusion gene partner. In 5 cases RNA sequencing (RNA-seq) was also used to further characterize the fusion transcripts. The 60 Xp11 translocation cancers included 47 Xp11 translocation RCC, 8 Xp11 translocation PEComas, and 5 melanotic Xp11 translocation renal cancers. A fusion partner was identified in 53/60 (88%) cases, including 18 SFPQ (PSF), 16 PRCC, 12 ASPSCR1 (ASPL), 6 NONO, and 1 DVL2. We provide the first morphologic description of the NONO-TFE3 RCC, which frequently demonstrates sub-nuclear vacuoles leading to distinctive suprabasal nuclear palisading. Similar sub-nuclear vacuolization was also characteristic of SFPQ-TFE3 RCC, creating overlapping features with clear cell papillary RCC. We also describe the first RCC with a DVL2-TFE3 gene fusion, in addition to an extrarenal pigmented PEComa with a NONO-TFE3 gene fusion. Furthermore, among neoplasms with the SFPQ-TFE3, NONO-TFE3, DVL2-TFE3 and ASPL-TFE3 gene fusions, the RCC are almost always PAX8-positive, cathepsin K-negative by immunohistochemistry, whereas the mesenchymal counterparts (Xp11 translocation PEComas, melanotic Xp11 translocation renal cancers, and alveolar soft part sarcoma) are PAX8-negative, cathepsin K-positive. These findings support the concept that despite an identical gene fusion, the RCCs are distinct from the corresponding mesenchymal neoplasms, perhaps due to the cellular context in which the translocation occurs. We corroborate prior data showing that the PRCC-TFE3 RCC are the only known Xp11 translocation RCC molecular subtype which is consistently cathepsin K positive. In summary, our data expand further the clinicopathologic features of cancers with specific TFE3 gene fusions, and should allow for more meaningful clinicopathologic associations to be drawn.
Keywords: Renal Neoplasm, TFE3, Translocation
INTRODUCTION
Xp11 translocation renal cell carcinomas (RCC) were first officially recognized by the World Health Organization (WHO) in 2004, following the initial detailed morphologic descriptions that were published in 2001 and 2002 (1,2). Xp11 translocation RCC bear chromosome translocations that result in one of a variety of gene fusions that involve the TFE3 transcription factor gene, which maps to the Xp11.2 locus. Reported TFE3 fusion partners include ASPSCR1 (ASPL), PRCC, SFPQ1 (PSF), NONO, CLTC, PARP14, LUC7L3, and KHSRP (3,4,5,6). Xp11 translocation RCC comprise the majority of pediatric RCC and approximately 1–4% adult RCC (7–10). While a variety of morphologic patterns have been described (11), the most common appearance is that of an RCC with papillary architecture, clear cells, and psammoma bodies. By immunohistochemistry, these tumors frequently underexpress cytokeratins, but frequently express melanocytic markers and the cysteine protease cathepsin k, which distinguishes them from more common RCC subtypes (12–14). Overall, outcome is similar to that of clear cell RCC, with increased age and advanced stage being poor prognostic factors (15,16). While immunohistochemistry for overexpressed TFE3 fusion proteins was initially the only method to confirm this diagnosis in formalin-fixed, paraffin embedded archival material (17), break-apart fluorescence in situ hybridization (FISH) demonstrating TFE3 gene rearrangement is now the preferred method (10, 11,18,19). However, TFE3 break-apart FISH does not provide information as to the specific fusion partners of TFE3. In fact, data on the clinicopathologic features of the subtypes of Xp11 translocation RCC associated with specific fusion partners is limited, as demonstration of the fusion partner has typically required fresh tissue for either cytogenetics or reverse transcriptase polymerase chain reaction (RT-PCR) assays.
Two other Xp11 translocation cancers which morphologically overlap with the Xp11 translocation RCC include Xp11 translocation PEComas and melanotic Xp11 translocation renal cancers. Xp11 translocation PEComas differ from typical PEComas in that they typically affect younger patients, have purely or predominantly epithelioid clear cell morphology, are not associated with Tuberous Sclerosis syndrome, do not express muscle markers by immunohistochemistry, and are not associated with TSC2 gene alterations (20–24). The most commonly identified fusion in Xp11 translocation PEComas has been SFPQ-TFE3, with a rare case demonstrating a DVL2-TFE3 gene fusion (22). Melanotic Xp11 translocation renal cancers were initially described before the Xp11 PEComas, and have been thought to overlap most with PEComa, though their renal origin raised the possibility of their representing Xp11 translocation RCC that do not express renal tubular markers (24–26). Significant morphologic overlap between Xp11 translocation RCC, Xp11 translocation PEComa, and melanotic Xp11 translocation renal cancers has been described. In the few cases of melanotic Xp11 translocation renal cancer in which a specific fusion has been identified, the gene fusion has always been SFPQ-TFE3 (24,26).
The development of FISH probes for the commonly identified TFE3 fusion partners allows subtyping of Xp11 translocation neoplasms in archival material, vastly increasing the number of cases that can be analyzed. Subtyping of Xp11 translocation-associated cancers should allow more meaningful clinicopathologic associations to be drawn, such as the differences previously described in a review of the published literature between the ASPSCR1-TFE3 RCC and the PRCC-TFE3 RCC (16). In this study, we apply a large battery of these fusion-partner probes to a cohort of confirmed TFE3-rearranged Xp11 translocation cancers by break-apart FISH, and correlate the subtype with clinicopathologic features. We also corroborate the results in 5 cases using RNA-sequencing.
MATERIALS AND METHODS
Case Selection and FISH analysis
The cases studied included 60 cases collected from the consultation files of 3 of the authors (PA, VER, CRA) and additional cases collected from the institutional files of The Johns Hopkins Medical Institutions and Memorial Sloan Kettering Cancer Center. Twenty of these cases were previously reported but without data as to fusion partner subtype in a prior study demonstrating the efficacy of TFE3 break-apart FISH in renal tumor consultations (11). Two other cases of Xp11 translocation PEComa in this study were included from the original description of this entity (20) and one other case was reviewed by one author (PA) in consultation and subsequently reported by others (27). One case of melanotic Xp11 translocation renal cancer in this study was included in the original description of this entity (25) and two other cases were reviewed by one author (PA) in consultation and subsequently reported by others (28, 29). Immunohistochemistry for PAX8 (which is almost always negative in PEComa)(30) and cathepsin K (which is consistently positive in PEComa) was performed as previously described (12). For RCC, staging was performed using the American Joint Commission on Cancer Manual, 7th edition (31). This study was approved by the Institutional Review Boards at participating Institutions.
FISH on interphase nuclei from paraffin-embedded 4-micron sections was performed applying custom probes using bacterial artificial chromosomes (BAC), covering and flanking genes that were identified as potential fusion partners in the RNA-seq experiment. TFE3 break-apart FISH was performed as previously described (11). BAC clones were chosen according to UCSC genome browser (http://genome.ucsc.edu), see Supplementary Table 1. The BAC clones were obtained from BACPAC sources of Children’s Hospital of Oakland Research Institute (CHORI)(Oakland, CA)(http://bacpac.chori.org). DNA from individual BACs was isolated according to the manufacturer’s instructions, labeled with different fluorochromes in a nick translation reaction, denatured, and hybridized to pretreated slides. Slides were then incubated, washed, and mounted with DAPI in an antifade solution, as previously described (32). The genomic location of each BAC set was verified by hybridizing them to normal metaphase chromosomes. Two hundred successive nuclei were examined using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems, Newton, MA). A positive score was interpreted when at least 20% of the nuclei showed a break-apart signal. Nuclei with incomplete set of signals were omitted from the score.
RNA-sequencing and Data Analysis
To verify FISH results in 5 selected cases with available material, RNA-seq was performed as described previously (5). Briefly, total RNA was extracted after xylene deparaffinization, using the AllPrep DNA/RNA FFPE Kit (Qiagen, Hilden, Germany). The quantity and integrity of the RNA was measured using the Agilent 2100 Bioanalyzer. One hundred nanograms of the RNA was applied for sequencing library preparation using the TruSeq RNA Access Library Prep Kit (Illumina, San Diego, USA) as per the manufacturer’s protocol. Paired-end sequencing (75 bp×2) was performed using the MiSeq Reagent V3 Kit (150 cycles) and the MiSeq sequencing system (Illumina). STAR algorithm was employed for detection of any potential TFE3 fusion. Bowtie2 was employed for alignment and mapping of short sequence reads to the human genome reference hg19 and the fusion transcript SFPQ/PSF-TFE3. Integrative Genomics Viewer (IGV) was employed for data visualization.
RESULTS
A total of 60 cases with known TFE3 gene rearrangements demonstrated by TFE3 break-apart FISH were analyzed for fusion partners. These included 47 Xp11 translocation RCC, 8 Xp11 translocation PEComa (2 renal, 6 non-renal), and 5 melanotic Xp11 translocation renal cancers. A fusion partner was identified in 53 cases (88%), including 18 SFPQ (PSF), 16 PRCC, 12 ASPSCR1 (ASPL), 6 NONO, and 1 DVL2 (Figure 1). In the remaining 7 (12%) cases no gene partner was identified using the available probes, including custom BACs for the above genes and for additional less common TFE3 gene partners reported, such as CLTC, YAP1 (recently shown to be fused to TFE3 in a subset of epithelioid hemangioendotheliomas) (33), or PARP14. The results for specific subtypes are presented according to the specific gene rearrangement identified.
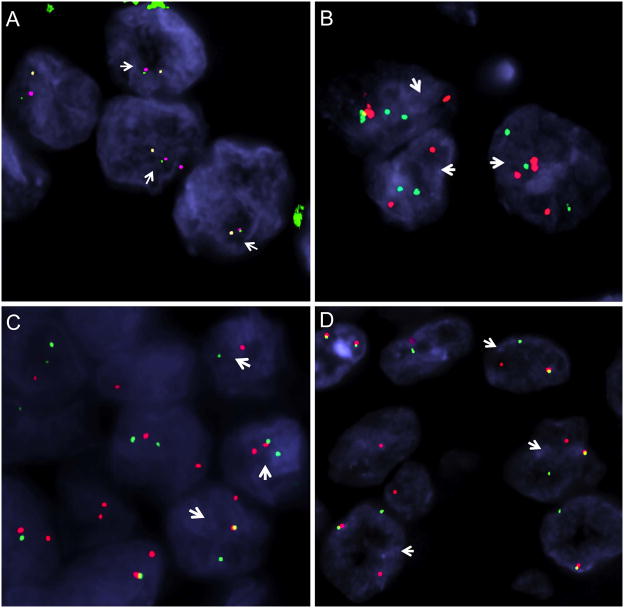
Figure 1.
FISH analysis of TFE3 fusion partner genes. All four neoplasms (A–D) in this composite image demonstrated TFE3 gene rearrangements by FISH. A. RCC with NONO-TFE3 fusion (Table 1, case 3): 3-color FISH fusion assay shows the 5′ NONO (red, centromeric) signal is fused to the 3′TFE3 probes (green, telomeric); while the 5′TFE3 probe (orange, centromeric) is split apart. B. DVL2-TFE3 fusion positive RCC: arrows show 3 cells with DVL2 break-apart signals (red, centromeric; green, telomeric); C. RCC with SFPQ-TFE3 fusion (Table 4, case 4): arrows show 3 cells demonstrating split SFPQ signals (red, centromeric; green, telomeric). D. RCC with PRCC-TFE3 fusion (Table 4, case 4): arrows show 3 cells with break-apart PRCC signals (red, centromeric; green, telomeric).
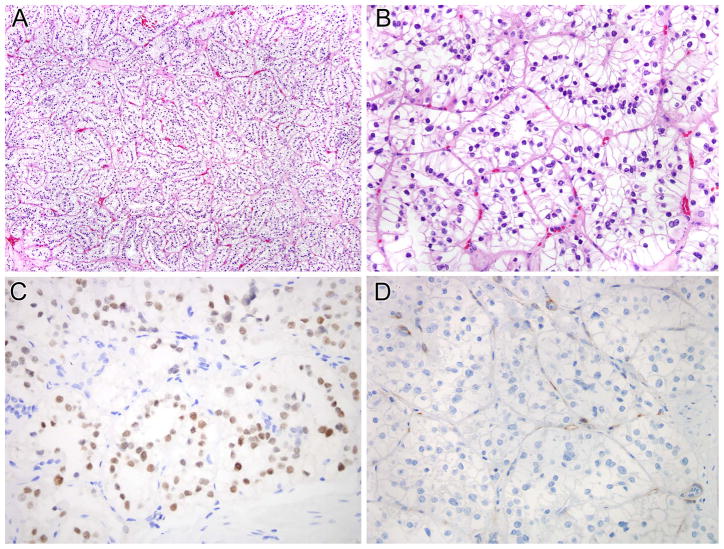
NONO-TFE3 RCC (5 cases)
These five cases affected three males and two females (mean age 38.4 years, median 36 years) (Table 1, cases 1–5). Two cases presented as small localized RCCs, one each presented with regional lymph node and bone metastases, while another presented as localized stage one disease but developed lung metastases after two years. Morphologically, the tumors showed a combination of nested to papillary architecture, and predominantly clear cytoplasm. Four of five cases demonstrated psammomatous calcifications. Four of the five cases demonstrated nuclear palisading with sub-nuclear vacuoles, a pattern which mimics clear cell papillary RCC (34,35) (Figures 2,3). This pattern was focal (present in <50% of the tumor) in two cases and diffuse (present in >50% of the tumor) in the other two. In the fifth case, nuclear palisading resembled a trabecular architecture, leading to an initial impression of a neuroendocrine neoplasm. By immunohistochemistry, all five cases were immunoreactive for PAX8, but none expressed cathepsin K (PAX8+, cathepsin K−). All 5 cases were negative for Carbonic Anhydrase IX (CA-IX). Two of five cases had rare cytokeratin 7 positive cells with the other three cases being completely negative.
Table 1.
NONO-TFE3 Cases
| Case | Age/Sex | Tumor Type | Size and Stage | Immunohistochemistry | Comment | Other publication |
|---|---|---|---|---|---|---|
| 1 | 36/F | RCC | 4.7cm, pT3NXM1 (bone), IV | PAX8+, HMB45 focal+; Cytokeratin 7, CA-IX, EMA, MiTF, Mart1, Cathepsin K− | Presented with back pain | Reference 11, Case 6 |
| 2 | 26/M | RCC | 4cm, pT1NX, I | PAX8+; Cytokeratin focal+; Cytokeratin 7 rare cells +; Cathepsin K, Melan A, HMB45, CA-IX − | Lung metastasis after 2 years | Reference 11, Case 10 |
| 3 | 51/M | RCC | 1.4cm, pT1NX, I (partial nephrectomy) | PAX8+; Racemase focal+; Cytokeratin 7, CA-IX, Cathepsin K, Vimentin, Cytokeratin, EMA− | Reference 11, Case 21 | |
| 4 | 29/M | RCC | 3.5cm, pT1NX, I | PAX8+; Cathepsin K, Cytokeratin 7, CA-IX− | Reference 11, Case 27 | |
| 5 | 50/F | RCC | 9cm, pT3N1, III | PAX8+; EMA+; HMB45 focal +; Cytokeratin 7 rare cells +; Cathepsin K, Vimentin, CA-IX−; | ||
| 6 | 20/M | Orbital Xp11 PEComa | Unknown | Cathepsin K+, HMB45+; PAX8, Desmin, Actin, Cytokeratin, Myogenin, S100− | Melanin Pigment |
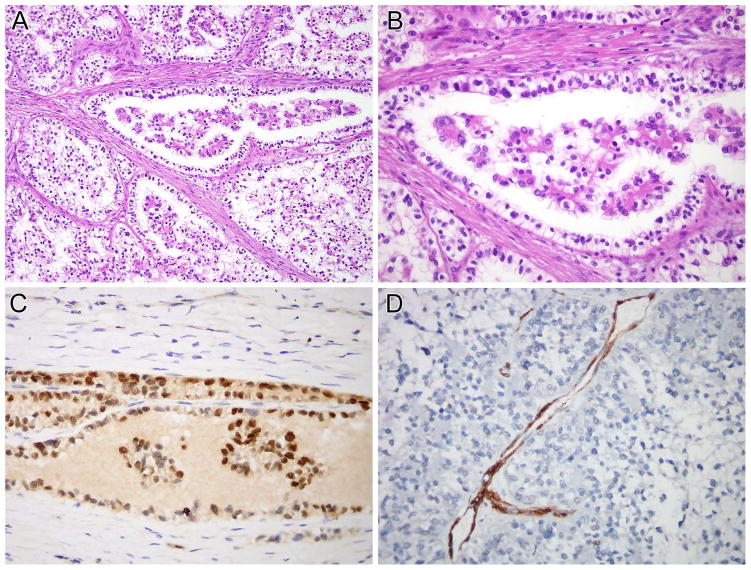
Figure 2.
NONO-TFE3 RCC (Table 1, case 1). A and B, this neoplasm demonstrated nested to papillary architecture. The neoplastic cells have predominantly clear cytoplasm and demonstrate sub-nuclear vacuolization, leading to apical palisading of nuclei. The neoplastic cells demonstrate nuclear labeling for PAX8 (C) but not for cathepsin K (D). Note the intact labeling of endothelial cells as an internal control for cathepsin K labeling.
Figure 3.
NONO-TFE3 RCC (Table 1, case 4). A and B, this neoplasm demonstrated nested to papillary architecture. The neoplastic cells have predominantly clear cytoplasm and demonstrate sub-nuclear vacuolization, leading to apical palisading of nuclei. The neoplastic cells demonstrate nuclear labeling for PAX8 (C) but not for cathepsin K (D). Note the intact labeling of endothelial cells as an internal control for cathepsin K labeling.
NONO-TFE3 PEComa with Melanin Pigment (1 case)
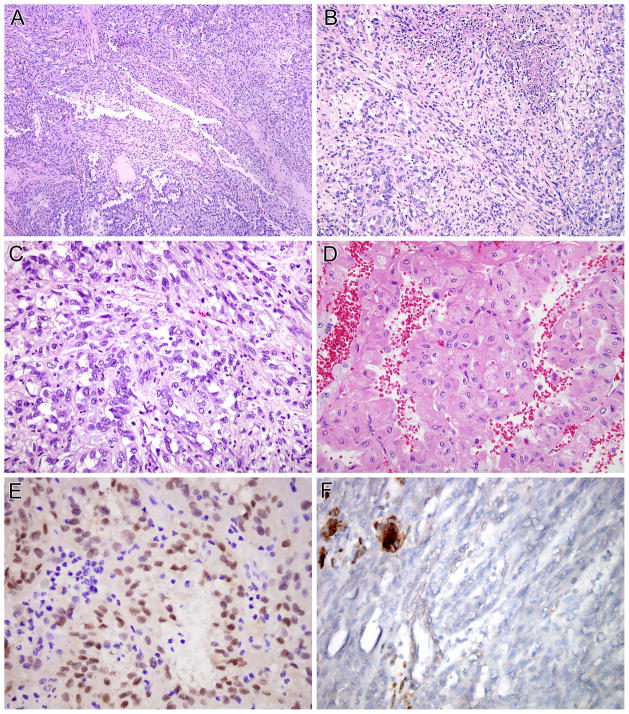
This neoplasm presented as an orbital mass in a 20 year-old male (Table 1, case 6). The morphology was that of a nested epithelioid neoplasm, with clear to finely granular eosinophilic cytoplasm, typical of an Xp11 translocation PEComa, with the exception of abundant melanin pigment (Figure 4). By immunohistochemistry, this neoplasm had the opposite immunoreactivity of the NONO-TFE3 RCC; being positive for cathepsin K, but not for PAX8 (PAX8−, cathepsin K+). Clinical follow-up on this case is not available.
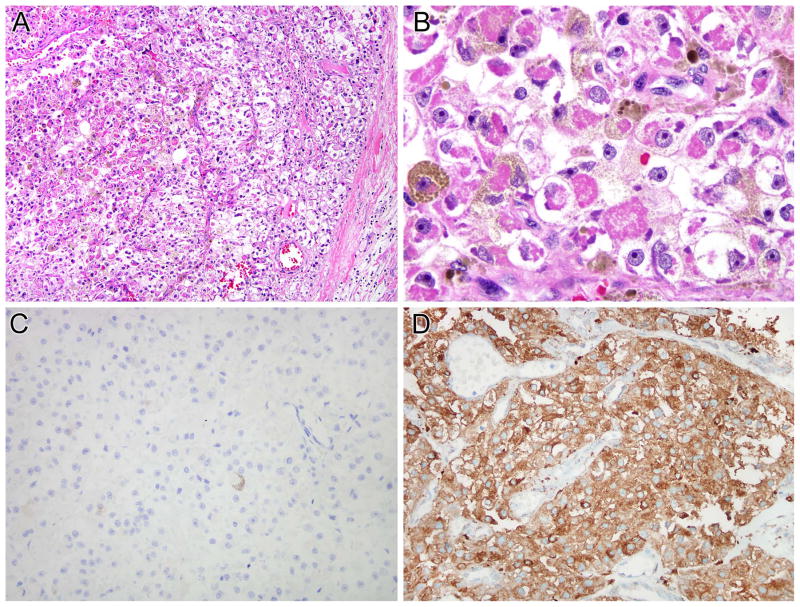
Figure 4.
NONO-TFE3 melanotic PEComa (Table 1, case 6). A and B, this is a neoplasm with nested to alveolar architecture, which features epithelioid cells with clear to finely granular eosinophilic cytoplasm. Fine pigment which proves to be melanin is present in the cytoplasm. The neoplasm was immunoreactive for melan A (not shown). The neoplasm does not label for PAX8 (C) but shows diffuse immunoreactivity for cathepsin K (D).
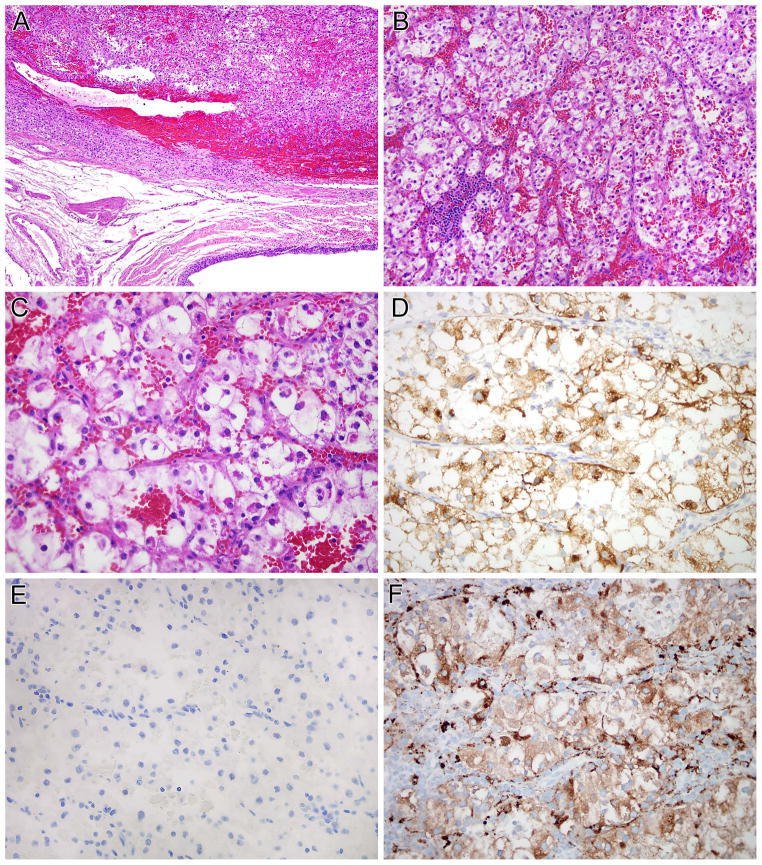
DVL2-TFE3 RCC (1 case)
This neoplasm was a 14.5 cm renal tumor in a 73 year-old male. The tumor demonstrated papillary and solid architecture with focal sarcomatoid areas, had variably eosinophilic cytoplasm, and presented with right perirenal lymph node metastases (pT3N1). By immunohistochemistry, the neoplasm was positive for PAX8, but negative for cathepsin K and melan A (Figure 5). For comparison, we performed PAX8 and cathepsin K immunohistochemistry on a previously reported DVL2-TFE3 PEComa, a 5.5 cm tumor located on the calf (22). The PEComa demonstrated the opposite pattern as the RCC with the same DVL2-TFE3 gene fusion; it was PAX8 negative and cathepsin K positive (not shown).
Figure 5.
DVL2-TFE3 RCC. This neoplasm demonstrates a variety of morphologic patterns. Much of the neoplasm has basophilic to pale cytoplasm, and demonstrates tubular and papillary architecture (A) each merges with sarcomatoid areas (B, C). Other areas on the same slides demonstrated more oncocytic cytoplasm (D). The neoplasm demonstrated nuclear immunoreactivity for PAX8 (E) but did not label for cathepsin K (F). Note the intact labeling of capillaries and associated macrophages as an internal control for cathepsin K labeling.
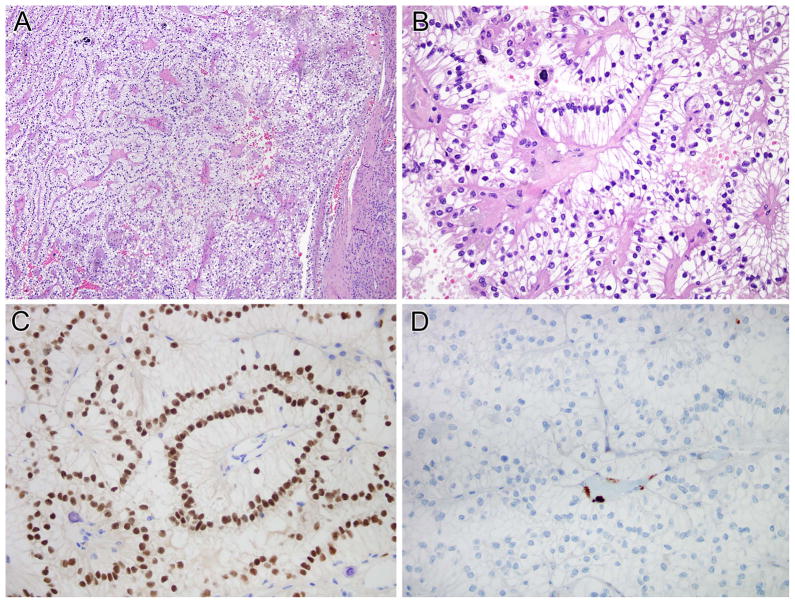
SFPQ-TFE3 RCCs (7 cases)
Seven RCC harbored a SFPQ-TFE3 gene fusion. These cases occurred in patients ranging from 19 – 63 years of age (mean 39, median 36), and 6 of 7 occurred in females (Table 2, cases 1–7). All cases were initially confined to the kidney. Case 1 presented as pT2 disease, had sarcomatoid morphology and later recurred in the retroperitoneum. Morphologically, the tumors showed a combination of nested to papillary architecture, and predominantly clear cytoplasm. All seven cases demonstrated psammomatous calcifications. Five of these seven cases demonstrated striking sub-nuclear vacuoles, similar to those seen in clear cell papillary RCC and (as shown above) in the RCC with NONO-TFE3 gene fusion. In all five cases, this pattern was diffuse (present in >50% of the tumor). In areas in which the sub-nuclear vacuoles were less evident, the morphology was typically that of a nested epithelioid clear cell neoplasm with thin capillary vasculature, closely mimicking clear cell RCC (Figures 6, 7). Six of these 7 RCC were reactive for PAX8 but not for cathepsin K (PAX8+ cathepsin k−). The other case (case 6) was positive for PAX8 and focally positive for cathepsin K in cystic areas of the tumor. All 3 tested cases were negative for cytokeratin 7. Three of 4 cases were completely negative for CA-IX while one case demonstrated rare positive cells.
Table 2.
SFPQ-TFE3 Cases
| Case | Age/Sex | Tumor Type | Size and Stage | Immunohistochemistry | Comment | Other publication |
|---|---|---|---|---|---|---|
| 1 | 36/F | RCC | 7.8cm, pT2NXMX, II | PAX8+; Cathepsin K− | Sarcomatoid, recurred in retroperitoneum after 3 years | Reference 11, Case 2 |
| 2 | 63/F | RCC | 3.5cm, pT1NX, I (partial nephrectomy) | PAX8, Racemase +; Cathepsin K, Cytokeratin 7, CA-IX− | ||
| 3 | 32/F | RCC | 4.7cm, pT1NX, I (partial nephrectomy) | PAX8, Racemase+; Cathepsin K, Cytokeratin 7, CA-IX, EMA− | Ipsilateral partial nephrectomy 4 years later for clear cell RCC | |
| 4 | 24/F | RCC | 3cm, pT1NX, I (partial nephrectomy) | PAX8, CD10, RCC marker, Racemase+; CA-IX rare cells +; Cathepsin K, CK7−, | ||
| 5 | 48/F | RCC | 4cm, pT1NX, I | PAX8+; Cathepsin K, Vimentin, CA-IX− | ||
| 6 | 51/F | RCC | 7.4cm, pT2NX, II (partial nephrectomy) | PAX8, CD10+; Cathepsin K, Cytokeratin AE1/3 focal+; HMB45, Melan A− | ||
| 7 | 19/M | RCC | 1.6cm, pT1NX, I (partial nephrectomy) | PAX8+; Cathepsin K− | ||
| 8 | 35/F | Xp11 PEComa | Kidney, pT3aN1, clinical stage IV | Cathepsin K, HMB45+; PAX8−, Cytokeratin− | Multiple copies of fusion | Reference 11, Case 3 |
| 9 | 4/F | Xp11 PEComa | 1.3cm, Renal sinus | Cathepsin K+, HMB45+; PAX8, Cytokeratin, S100 Melan A, EMA, RCC marker− | ||
| 10 | 27/F | Xp11 PEComa | Bladder | Cathepsin K, HMB45+; PAX8, Melan A, Desmin, Actin, S100− | Developed pelvic lymph node metastases at 6 months | Reference 27 |
| 11 | 9/M | Xp11 PEComa | 7cm, Kidney, pT3N1 clinical stage IV | Cathepsin K, HMB45, Melan A+; PAX8, Cytokeratin, Desmin, S100 protein, CD34− | ||
| 12 | 46/F | Xp11 PEComa | 6.5cm, Thigh | Cathepsin K+; PAX8− | SFPQ exon 9-TFE3 exon 6 fusion | Reference 20, Case 4. |
| 13 | 21/F | Xp11 PEComa | 6cm, Pelvis | Cathepsin K+; HMB45 focal+; PAX8, Melan A, Actin, Desmin, Cytokeratin− | ||
| 14 | 9/F | Xp11 PEComa | 5cm, Uterus. Lymph node metastases at diagnosis. | Cathepsin K, HMB45+; PAX8, Melan A, S100, Desmin, Actin− | SFPQ exon 9- TFE3 exon 6 fusion | Reference 20, Case 2. |
| 15 | 11/M | Melanotic Xp11 Renal Cancer | 21cm, pT3N1M1, IV | Cathepsin K, Melan A, HMB45+; PAX8, Cytokeratin, RCC marker, Actin, Desmin, S100− | Reference 25, case 1 | |
| 16 | 34/F | Melanotic Xp11 Renal Cancer | 4.8cm, pT1NXMX, I (partial nephrectomy) | Cathepsin K, HMB45+; PAX8, CD10, Cytokeratin, EMA, Desmin,− | SFPQ exon 9- TFE3 exon 6 fusion | Reference 28. |
| 17 | 30/F | Melanotic Xp11 Renal Cancer | 12.5cm, pT3NX (clinically stage IV, thrombus in inferior vena cava and pulmonary artery) | Cathepsin K, HMB45, Melan A+; PAX8, Cytokeratin, CD10, Desmin, S100− | SFPQ exon 9- TFE3 exon 6 fusion | Reference 29 |
| 18 | 34/M | Melanotic Xp11 Renal Cancer | 9.7cm pT2NX, II | Cathepsin K, HMB45+; Melan A focal+; PAX8, Cytokeratin, S100− | SFPQ exon 9- TFE3 exon 6 fusion |
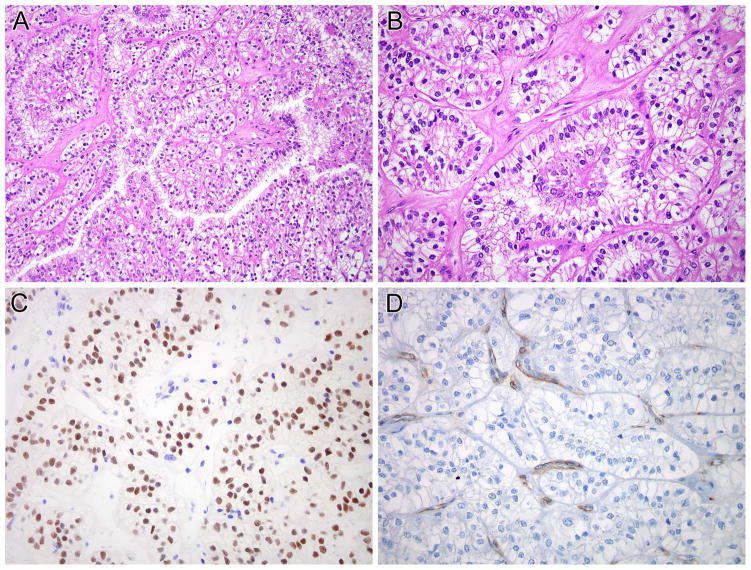
Figure 6.
SFPQ-TFE3 renal cell carcinoma (Table 2, case 5). This neoplasm demonstrated solid nested architecture (A and B), and demonstrated abundant clear cytoplasm with prominent sub-nuclear vacuoles leading to apical palisading of nuclei. This case was originally classified as a clear cell RCC. The neoplasm demonstrated nuclear labeling for PAX8 (C) but was negative for cathepsin K (D). Note the intact staining of endothelial cells as an internal control for cathepsin K labeling.
Figure 7.
SFPQ-TFE3 renal cell carcinoma (Table 2, case 4). A and B, this neoplasm demonstrated solid nested to papillary architecture, and striking sub-nuclear vacuoles leading to apical palisading of nuclei. The neoplasm was diffusely immunoreactive for PAX8 but not for cathepsin K. Note the intact staining of endothelial cells as an internal control for cathepsin K labeling.
SFPQ-TFE3 PEComas (7 cases)
These neoplasms comprised 7 of the 8 Xp11 translocation PEComas available for study (the other case demonstrated a NONO-TFE3 gene fusion described above). Six patients were females and 1 was male. Patient ages ranged from 4 – 46 years (mean 21.6, median 21). Sites of origin included the kidney (2 cases), renal sinus, uterus, bladder, thigh, and pelvis (Table 2, cases 8–14). All cases demonstrated the morphology previously described in Xp11 translocation PEComa; specifically, a solid nested architecture featuring epithelioid cells with predominantly clear to focally eosinophilic cytoplasm. All cases analyzed were diffusely immunoreactive for cathepsin K, but negative for PAX8 (PAX8−, cathepsin k+) (Figure 8). All cases were immunoreactive for HMB45, but only four of six were immunoreactive for melan A. None of these cases demonstrated melanin pigment.
Figure 8.
SPFQ-TFE3 PEComa. (Table 2, case 9). This neoplasm was centered in the renal pelvis (A). The neoplasm was highly vascular, and had a nested to an alveolar architecture (B). The neoplastic cells had abundant clear to finely granular eosinophilic cytoplasm (C). The neoplasm was negative for cytokeratins, but showed labeling for HMB45 (D). The neoplasm was negative for PAX8 (E) but demonstrated diffuse labeling for cathepsin K (F).
Both cases with adequate RNA for RNA seq demonstrated an SFPQ-TFE3 gene fusion, fusing exon 9 of SFPQ with exon 6 of TFE3 (Table 2, cases 12 and 14).
SFPQ-TFE3 Melanotic Xp11 translocation renal cancers (4 cases)
These comprised four of the five tested cases (the other case did not have an identifiable fusion partner). The patients ranged in age from 11 – 34 years (mean 27, median 24) (Table 2, cases 15–18). Two patients presented with disseminated metastatic disease. All of these cases demonstrated a solid nested architecture featuring epithelioid cells with predominantly clear to focally eosinophilic cytoplasm, along with melanin pigment readily identifiable on H&E sections. All of these neoplasms labeled diffusely for cathepsin K, and all were negative for PAX8 (PAX8− cathepsin k+). All 3 cases with adequate RNA for RNA seq demonstrated an SFPQ-TFE3 gene fusion, fusing exon 9 of SFPQ with exon 6 of TFE3 (Table 2, cases 16–18).
ASPSCR1-TFE3 RCC (12 cases)
These patients ranged in age from 15–73 years (mean 33.75, median 27), and nine were females and three males (Table 3). Morphologically, all neoplasms demonstrated the typical morphology of this subtype; specifically, nested to papillary architecture, voluminous clear to eosinophilic cytoplasm, and abundant psammoma bodies. Two of three cases with lymph nodes examined harbored lymph node metastases at diagnosis, and all nine cases tested were negative for cathepsin K.
Table 3.
ASPSCR1-TFE3 Renal Cell Carcinoma
| Case | Age/Sex | Size and Stage | Immunohistochemistry | Comment | Other publication |
|---|---|---|---|---|---|
| 1 | 20/F | 12cm, pT2NXM1, IV | PAX8+; Cathepsin K− | Reference 11, Case 4 | |
| 2 | 67/M | 5.8cm, pT1bNX, I | PAX8+; Cathepsin K, HMB45− | Reference 11, Case 8 | |
| 3 | 23/F | 2cm, pT1aN1MX, III | Cytokeratin 7+, Vimentin focal+; Cathepsin K, CA-IX, HMB45− | Reference 11, Case 14 | |
| 4 | 17/F | 5cm, pT1bNXMX, I | CD10+, RCC+; Cathepsin K− | Reference 11, Case 29 | |
| 5 | 39/F | 14cm, pT3NXM1, IV | RCC marker, CD10+; Cathepsin K− | Diagnosed as urothelial carcinoma | Reference 11, Case 3 |
| 6 | 41/F | 6.5cm, pT1 NXMX, I | PAX8+; Cathepsin K− | Metastasized to spine after 1 year | |
| 7 | 28/F | 8cm, pT2N0, II | PAX8+; Cathepsin K− | Hematuria and sickle cell trait, retroperitoneal recurrence 3 years later | |
| 8 | 33/M | 2.4cm, pT3aN1, III | CD10+ | s/p chemotherapy for lymphoma, also has sarcoma of mediastinum | |
| 9 | 26/M | 7cm, pT3N1, III | PAX8+; Cathepsin K− | ||
| 10. | 23/F | 4cm, pT1NX, I (partial nephrectomy) | Recurred after 1 year→pT3N1 radical nephrectomy (matted lymph nodes) | ||
| 11 | 73/F | 2.2cm, pT1NX, I | PAX8, AE1/3, CD10+; CA-IX, HMB45− | Retroperitoneal lymph node recurrence 7 years later | |
| 12 | 15/F | pTXNXM1, IV | PAX8+; Cathepsin K, CA-IX− | Presented with cervical lymph node metastases |
PRCC-TFE3 RCC (16 cases)
These patients ranged in age from 5 – 63 years (mean 40.3, median 42), with an equal gender distribution, 8 females, 7 males, and one of unknown gender (Table 4). Morphologically, all tumors demonstrated the typical morphology of this subtype; specifically, they demonstrated compact nested to papillary architecture, clear to eosinophilic cytoplasm, and psammoma bodies. One case demonstrated palisading of nuclei and another demonstrated sub-nuclear vacuoles similar to RCC harboring SFPQ-TFE3 and NONO-TFE3 fusions, as described above. Seven of the eleven tested cases were immunoreactive for cathepsin K. Of note, two patients presented with hematogenous metastasis, which has not previously been reported in this subtype of Xp11 translocation RCC.
Table 4.
PRCC-TFE3 Renal Cell Carcinoma
| Case | Age/Sex | Size and Stage | Immunohistochemistry | Comment | Other publication |
|---|---|---|---|---|---|
| 1. | 19/M | 12 cm pT3NXM1 (bone), IV | PAX8+, Cathepsin K+; Desmin, Vimentin focal+; HMB45− | Sarcomatoid | Reference 11, Case 7 |
| 2. | 58/F | 12cm, pT3NXMX, III | Cathepsin K+; CD10, Melan A focal+ | Contralateral clear cell RCC | Reference 11, Case 13 |
| 3. | 29/F | 11.5cm, pT2NX, II | Racemase diffusely+; Cathepsin K, CA-IX, Vimentin focal+ | Hematuria during pregnancy; nephrectomy during delivery | Reference 11, Case 20 |
| 4. | 47/M | 3.5cm, pT3NXMX, III (partial nephrectomy) | Cathepsin K− | Capsular invasion | Reference 11, Case 28 |
| 5. | 21/F | 9cm, pT2N1MX, III | Cathepsin K− | Cystic | Reference 11, Case 30 |
| 6. | 30/M | 3cm, pT1NXMX, I (partial nephrectomy) | Vimentin+; Cathepsin K− | ||
| 7. | Unknown | Unknown | Cathepsin K+ | ||
| 8. | 52/M | 5cm, pT3N0M0 (involved inferior vena cava), III | Cathepsin K+ | Developed bone metastasis 1.5 years later, died of disease | |
| 9. | 5/F | Renal mass with retroperitoneal lymphadenopathy | |||
| 10 | 48/F | 3.7cm, pT1NX, I (partial nephrectomy) | Cathepsin K focal+ | ||
| 11 | 35/F | Renal mass with retroperitoneal lymphadenopathy | Cathepsin K− | Core biopsy | |
| 12 | 50/F | 0.9cm, pT1NX, I (partial nephrectomy) | Cathepsin K, Racemase +; Cytokeratin 7 focal +, Melan A− | Incidental finding on imaging | |
| 13 | 62/M | “Organ Confined” by report | PAX8+ | Bladder recurrence at 12 years→Neck metastasis at 16 years | |
| 14 | 37/M | 8.5cm, pT3NX, III | RCC marker, CD10+; Cam5.2, AE1/3, Cytokeratin 903, HMB45, Melan A− | ||
| 15 | 63/F | 5.3cm, pT1bNX, I | CD10, PAX8+, racemase+; CA-IX− | ||
| 16 | 49/M | 11cm, pT3NXM1, IV (pelvis and intestinal wall) | CD10+; CK7, CK20− |
TFE3-Rearranged Neoplasms with Unknown Fusion Partner (7 cases)
The clinicopathologic features of these cases are summarized in Table 5. These included 6 Xp11 translocation RCC and 1 Melanotic Xp11 translocation cancer. Of note, all of the RCC were cathepsin K positive. Two of the RCC were extensively cystic and 2 had biphasic morphology mimicking t(6;11) RCC as previously described (11). Adequate material for RNA seq was available from the Melanotic Xp11 translocation cancer; however, a TFE3 fusion partner could not be identified
Table 5.
TFE3-Rearranged Cases with Unknown Fusion Partner
| Case | Age/Sex | Tumor Type | Size and Stage | Immunohistochemistry | Comment | Other publication |
|---|---|---|---|---|---|---|
| 1 | 33/M | RCC | 2.2cm, pT1NX, I (partial nephrectomy) | Cathepsin K, PAX8, Melan A+ | Multilocular cystic | Reference 11, case 9 |
| 2 | 28/F | RCC | 3.4cm pT1NXMX, I (partial nephrectomy) | Cathepsin K+ | Reference 11, case 11 | |
| 3 | 16/F | RCC | pT1aNXMX, I (partial nephrectomy) | PAX8, Cathepsin K, Cytokeratin, CD10+; HMB45, Melan A− | Highly cystic | Reference 11, Case 16 |
| 4 | 20/F | RCC | 13.6cm, pT3N2M0, III | Cathepsin K, CD10+ | Went on E2805 (sorafenib), developed pulmonary and paravertebral metastases at 2 years | Reference 11, case 26 |
| 5 | 46/F | RCC | 3.7cm, pT3NX, III (partial nephrectomy) | Cathepsin K+; Cam5.2, Melan A, HMB45 focal+ | Mimic t(6;11) RCC | |
| 6 | 56/F | RCC | 4.5cm, pT1NX, I (partial nephrectomy) | Cathepsin K, PAX8, Melan A, CD117+ | Mimic t(6;11) RCC | |
| 7 | 21/F | Melanotic Xp11 Renal cancer | pT3NX, III | Cathepsin K, HMB45, Melan A+; PAX8, S100, Vimentin, Actin− | Lung nodules 4 years later |
DISCUSSION
In this study, we apply combined molecular methodologies, FISH and RNA seq, to establish a detailed characterization of TFE3 fusion partners in a large cohort of Xp11 translocation positive neoplasms. First, we provide the first morphologic description of the NONO-TFE3 RCC. Previously, a single case of this entity had been reported (3), but a morphologic description and images were not provided. We found that this neoplasm and the SFPQ-TFE3 RCC (see below) frequently demonstrate sub-nuclear vacuoles, leading to palisading of nuclei, similar to the appearance of clear cell papillary RCC. All five cases were immunoreactive for PAX8, but negative for cathepsin K. This immunoprofile contrasts with that of the Xp11 translocation PEComa harboring identical NONO-TFE3 gene fusion (a fusion not previously reported in Xp11 translocation PEComa until now), which was conversely immunoreactive for cathepsin K but not for PAX8. We note that the presence of NONO-TFE3 gene fusion can be suspected based on the pattern of TFE3 rearrangement by FISH, showing constant, small gaps between the telomeric and centromeric TFE3 signals, in keeping with an inversion /intra-chromosomal fusion.
We also report the first RCC with a DVL2-TFE3 gene fusion. This tumor had a variety of morphologic patterns, including oncocytic, tubular and sarcomatoid features. DVL2-TFE3 fusion has been previously reported in an Xp11 translocation PEComa (22). In our study, similar to the NONO-TFE3 neoplasms, we found that the DVL2-TFE3 RCC was PAX8 positive and cathepsin K negative, whereas the DVL2-TFE3 PEComa was cathepsin K positive and PAX8 negative.
We identified 18 neoplasms in this study harboring SFPQ-TFE3 gene fusions. Prior studies have suggested significant overlap among Xp11 translocation RCC, Xp11 translocation PEComa, and melanotic Xp11 translocation renal cancers bearing the SFPQ-TFE3 gene fusion. Some have grouped all neoplasms with the SFPQ-TFE3 gene fusion together (36), and suggested that the distinction may be arbitrary. We have seen cases in which the same renal tumor has been seen by experts at several different major academic centers, and been classified as RCC by some and PEComa by others. Along these lines, we originally classified one of the cases in this study as an RCC based on its nested epithelioid morphology and renal location, but on re-review of the morphology and immunohistochemical profile reclassified it as a PEComa (Table 2, case 8). A complete immunohistochemical profile of tumors classified as RCC with the SFPQ-TFE3 gene fusion has not been reported. We show herein that RCC with the SFPQ-TFE3 gene fusion are immunoreactive for PAX8 and almost always negative for cathepsin K, which distinguishes them from Xp11 translocation PEComa and melanotic Xp11 translocation renal cancer, and frequently have a distinctive morphology featuring sub-nuclear vacuoles and nuclear palisading, similar to that seen in the NONO-TFE3 RCC. The similar morphology of the SPFQ-TFE3 and NONO-TFE3 RCC is intriguing given the highly overlapping functions of SFPQ and NONO in RNA processing, and the fact that they together form protein complexes (37). We note that, in retrospect, subnuclear vacuoles are readily seen in the published images provided in 4 case reports of SFPQ-TFE3 RCC, though not commented on in these publications (38–41). The frequent sub-nuclear vacuolization seen indicates that these Xp11 translocation RCC should also be considered in the differential diagnosis of clear cell papillary RCC. Unlike clear cell papillary RCC, SFPQ-TFE3 and NONO-TFE3 RCC frequently occur in younger patients, frequently have psammomatous calcifications, and show no or minimal immunoreactivity for cytokeratin 7 and CA-IX. We also found that the SFPQ-TFE3 RCC frequently mimics clear cell RCC to a striking degree. In fact, two of the cases in our study were previously diagnosed as clear cell RCC by experienced surgical pathologists with extensive expertise in urologic pathology consultations before they were reviewed in the course of this study. This morphologic mimicry of clear cell RCC also explains the observation that all 5 Xp11 translocation RCC mistakenly included in the cancer genome atlas (TCGA) sequencing study of clear cell RCC contained the SFPQ-TFE3 RCC fusion (42). Based on these findings, we believe that the SFPQ-TFE3 RCC are the Xp11 translocation RCC most likely to mimic clear cell RCC. The frequently younger patient age, focal papillary architecture, psammomatous calcification, and absence of or minimal labeling for CA-IX are clues to the diagnosis of SFPQ-TFE3 RCC.
The usual PAX8-positive cathepsin K-negative immunoprofile of the SFPQ-TFE3 RCC contrasts with those of the Xp11 translocation PEComa and melanotic Xp11 translocation RCC that harbor the same gene fusion, both of which were consistently cathepsin K positive and PAX8 negative. These findings suggest that the SFPQ-TFE3 RCC are a distinctive entity which can be separated in the majority of cases from Xp11 translocation PEComa and melanotic Xp11 translocation cancers. The latter two neoplasms share a similar immunoprofile (PAX8 negative, cathepsin K positive), which supports our view and that of others that melanotic Xp11 translocation renal cancers likely represent a variant of or part of the spectrum of Xp11 translocation PEComa. Further evidence of overlap is the presence of melanin in several extrarenal Xp11 translocation neoplasms, including the NONO-TFE3 PEComa in this study, an SFPQ-TFE3 PEComa of the pancreas, pelvis and cervix reported by Rao et al (24), along with an ovarian melanotic Xp11 translocation neoplasm previously reported in the literature (43). While the phenotype of the Xp11 translocation RCC and Xp11 translocation PEComas is distinct at the immunohistochemical level, it is clear that these lesions are genetically related and can be considered part of a family of cancers driven by TFE3 gene fusions. This suggests the potential for utilization of novel targeted therapies that are effective against one member against the others.
Our data on RCC with the ASPCR1-TFE3 and PRCC-TFE3 RCC adds supporting data to the published findings. We had previously shown that cathepsin K is frequently positive in the PRCC-TFE3 RCC but consistently negative in the ASPCR1-TFE3 RCC, a finding which is confirmed in this study (14). Two of three ASPSCR1-TFE3 RCC in which lymph nodes were resected were associated with lymph node metastasis, which corroborates tendency to lymph node involvement we previously described in this subtype (16). Furthermore, 2 patients with small, seemingly localized ASPCR1-TFE3 RCC who did not initially undergo node sampling at presentation (pT1NX) later recurred with involvement of retroperitoneal lymph nodes, suggesting the possibility that nodal sampling at diagnosis could have impacted outcome. A previous review of the literature showed that PRCC-TFE3 RCC typically present at lower stage than do the ASPCR1-TFE3 RCC (16), but in this study we report the first two cases of PRCC-TFE3 RCC which presented with metastatic disease. We note that neither of the ASPCR1-TFE3 and PRCC-TFE3 gene fusions was or has previously been identified in an Xp11 translocation PEComa. The ASPCR1-TFE3 gene fusion is of course characteristic of the mesenchymal neoplasm alveolar soft part sarcoma (44), but there is no currently identified mesenchymal counterpart to the PRCC-TFE3 RCC. Of note, the PRCC-TFE3 RCC is the one known subtype of Xp11 translocation RCC which does express cathepsin K frequently, whereas our study shows that other Xp11 translocation RCC are typically cathepsin k negative and have a mesenchymal counterpart with the identical gene fusion that is cathepsin k positive (Table 6). It should be noted, however, that all of the Xp11 RCC in this study which do not as of now demonstrate a known fusion partner were positive for cathepsin K, suggesting that the as yet unknown TFE3 fusion partners involved in these cases may function similarly to PRCC.
Table 6.
Specific Recurrent TFE3 Gene Fusions in Epithelial and Mesenchymal Neoplasms
| Gene Fusion | Mesenchymal Neoplasm | Immunohistochemical Profile | Epithelial Neoplasm | Immunohistochemical Profile |
|---|---|---|---|---|
| ASPSCR1- TFE3 | Alveolar Soft Part Sarcoma | PAX8-Cathepsin K+ | ASPSCR1- TFE3 RCC | PAX8+ Cathepsin K− |
| SFPQ-TFE3 | Xp11 PEComa/ Melanotic Xp11 Cancer | PAX8-Cathepsin K+ | SFPQ-TFE3 RCC | PAX8+ Cathepsin K−* |
| NONO-TFE3 | Xp11 PEComa/ Melanotic Xp11 Cancer | PAX8-Cathepsin K+ | NONO-TFE3 RCC | PAX8+ Cathepsin K− |
| DVL2-TFE3 | Xp11 PEComa | PAX8-Cathepsin K+ | DVL2-TFE3 RCC | PAX8+ Cathepsin K− |
| PRCC-TFE3 | ND | ND | PRCC-TFE3 RCC | PAX8+ Cathepsin K +/− |
| YAP1-TFE3 | Epithelioid Hemangioendothelioma Subset | Not studied | ND | ND |
ND=not described; RCC=renal cell carcinoma
One case was focally Cathepsin K positive
Given the variable clinical presentation and morphologic appearances described in this and other manuscripts, one could be tempted to conclude that all RCCs be worked up by molecular techniques to exclude Xp11 translocation RCC. Since the prognosis for Xp11 translocation RCC is similar to that of clear cell RCC, a missed diagnosis of a localized Xp11 translocation RCC as clear cell papillary RCC would result in the patient receiving an inappropriately optimistic report of their prognosis. For cases which present with metastatic disease, the diagnosis of Xp11 translocation RCC would make a patient ineligible for some treatments designed to specifically target clear cell RCC and eligible for treatments that target cancers with TFE3 gene fusions. However, we believe that comprehensive molecular analysis of all RCCs is not cost effective. Our view is that RCCs with classic clinical presentation and morphology that is typical of the specific common subtypes of RCC (such as clear RCC, papillary RCC, and chromophobe RCC) do not require immunohistochemical stains or molecular analysis for diagnosis: the diagnosis can be comfortably reached on routine H&E sections. For other cases in which the morphology or clinical presentation is slightly unusual, we typically perform a limited immunohistochemical profile (such as cytokeratin 7, CA-IX and cathepsin K) to support or refute the suspected diagnosis. If this does not clarify the diagnosis, further workup is indicated. We do not routinely perform TFE3 FISH unless there is a strong clinical suspicion (age less than 30 years), highly suggestive morphology (such as papillary architecture with clear cells and psammoma bodies), or highly suggestive immunohistochemical results (such as diffuse cathepsin K immunoreactivity). We also perform TFE3 FISH for many unclassified RCCs, given the morphologic variability demonstrated by Xp11 translocation RCC including its ability to have a non-descript high grade RCC appearance.
In summary, our study highlights the ability of subset of Xp11 translocation RCC to mimic clear cell papillary RCC and clear cell RCC. Clinical clues such as young age, morphologic clues such as the presence of psammoma bodies, and immunohistochemical clues such as minimal immunoreactivity for CA-IX should suggest the possibility of Xp11 translocation RCC. Our study also highlights the different immunohistochemical phenotypes of Xp11 translocation RCC and PEComas that harbor the same gene fusion. Despite morphologic overlap, PAX8 and cathepsin K can distinguish most cases. We also highlight the strong association of cathepsin K immunoreactivity in Xp11 translocation RCC with the presence of a PRCC-TFE3 gene fusion.
Supplementary Material
Supplemental Figure 1: Visualization of PSF-TFE3 gene fusion. Short sequence reads from RNA-seq were aligned to PSF-TFE3 fusion gene by Bowtie2 and visualized by Integrative Genomics Viewer (IGV), supporting the PSF-TFE3 fusion. Each grey bar represents a short sequence read. The sequence near the PSF-TFE3 junction was demonstrated at the bottom. The junction site is marked with a red arrow.
Acknowledgments
We thank Norman Barker MA, MS, RBP for expert photographic assistance. 1-18-16
Footnotes
Disclosures: Supported in part by: P01CA47179 (CRA), P50 CA 140146-01 (CRA), Cycle for Survival (CRA), Kristin Ann Carr Foundation (CRA), Dahan Translocation Carcinoma Fund (PA)
References
- 1.Argani P, Antonescu CR, Illei PB, Lui MY, Timmons CF, Newbury R, Reuter VE, Garvin AJ, Perez-Atayde AR, Fletcher JA, Beckwith JB, Bridge JA, Ladanyi M. Primary renal neoplasms with the ASPL-TFE3 gene fusion of alveolar soft part sarcoma: A distinctive tumor entity previously included among renal cell carcinomas of children and adolescents. Am J Pathol. 2001;159:179–92. doi: 10.1016/S0002-9440(10)61684-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argani P, Antonescu CR, Couturier J, Fournet J, Sciot R, Debiec-Rychter M, Hutchinson B, Reuter VE, Boccon-Gibod L, Timmons C, Hafez N, Ladanyi M. PRCC-TFE3 renal carcinomas: Morphologic, immunohistochemical, ultrastructural, and molecular analysis of an entity associated with the t(X;1)(p11. 2;q21) Am J Surg Pathol. 2002;26:1553–66. doi: 10.1097/00000478-200212000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Clark J, Lu YJ, Sidhar SK, Parker C, Gill S, Smedley D, Hamoudi R, Linehan WM, Shipley J, Cooper CS. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene. 1997;15:2233–9. doi: 10.1038/sj.onc.1201394. [DOI] [PubMed] [Google Scholar]
- 4.Argani P, Lui MY, Couturier J, Bouvier R, Fournet J, Ladanyi M. A novel CLTC-TFE3 gene fusion in pediatric renal adenocarcinoma with t(X;17)(p11. 2;q23) Oncogene. 2003;22:5374–8. doi: 10.1038/sj.onc.1206686. [DOI] [PubMed] [Google Scholar]
- 5.Huang W, Goldfischer M, Babyeva S, Mao Y, Volyanskyy K, Dimitrova N, Fallon JT, Zhong M. Identification of a novel PARP14-TFE3 gene fusion from 10-year-old FFPE tissue by RNA-seq. Genes Chromosomes Cancer. 2015 May 29; doi: 10.1002/gcc.22261. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 6.Malouf GG, Monzon FA, Couturier J, et al. Genomic heterogeneity of translocation renal cell carcinoma. Clin Cancer Res. 2013;19:4673–84. doi: 10.1158/1078-0432.CCR-12-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altinok G, Kattar MM, Mohamed A, Poulik J, Grignon D, Rabah R. Pediatric renal carcinoma associated with Xp11. 2 translocations/TFE3 gene fusions and clinicopathologic associations. Pediatr Dev Pathol. 2005;8:168–80. doi: 10.1007/s10024-004-9106-3. [DOI] [PubMed] [Google Scholar]
- 8.Ramphal R, Pappo A, Zielenska M, Grant R, Ngan BY. Pediatric renal cell carcinoma: Clinical, pathologic, and molecular abnormalities associated with the members of the mit transcription factor family. Am J Clin Pathol. 2006;126:349–64. doi: 10.1309/98YE9E442AR7LX2X. [DOI] [PubMed] [Google Scholar]
- 9.Komai Y, Fujiwara M, Fujii Y, Mukai H, Yonese J, Kawakami S, Yamamoto S, Migita T, Ishikawa Y, Kurata M, Nakamura T, Fukui I. Adult Xp11 translocation renal cell carcinoma diagnosed by cytogenetics and immunohistochemistry. Clin Cancer Res. 2009;15:1170–6. doi: 10.1158/1078-0432.CCR-08-1183. [DOI] [PubMed] [Google Scholar]
- 10.Zhong M, De Angelo P, Osborne L, Keane-Tarchichi M, Goldfischer M, Edelmann L, Yang Y, Linehan WM, Merino MJ, Aisner S, Hameed M. Dual-color, break-apart FISH assay on paraffin-embedded tissues as an adjunct to diagnosis of Xp11 translocation renal cell carcinoma and alveolar soft part sarcoma. Am J Surg Pathol. 2010;34:757–766. doi: 10.1097/PAS.0b013e3181dd577e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green WM, Yonescu R, Morsberger L, Morris K, Netto GJ, Epstein JI, Illei PB, Allaf M, Griffin C, Argani P. Utilization of a TFE3 break-apart FISH assay in a renal tumor consultation service. Am J Surg Pathol. 2013;37:1150–1163. doi: 10.1097/PAS.0b013e31828a69ae. [DOI] [PubMed] [Google Scholar]
- 12.Argani P, Hicks J, DeMarzo A, Albadine R, Illei P, Ladanyi M, Reuter VE, Netto G. Xp11 Translocation Renal Cell Carcinoma (RCC): Extended Immunohistochemical (IHC) Profile Emphasizing Novel RCC Markers. Am J Surg Pathol. 2010;34:1295–1303. doi: 10.1097/PAS.0b013e3181e8ce5b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martignoni G, Pea M, Gobbo S, Brunelli M, Bonetti F, Segala D, Pan CC, Netto G, Doglioni C, Hes O, Argani P, Chilosi M. Cathepsin-K immunoreactivity distinguishes MiTF/TFE family renal translocation carcinomas from other renal carcinomas. Mod Pathol. 2009;22:1016–22. doi: 10.1038/modpathol.2009.58. [DOI] [PubMed] [Google Scholar]
- 14.Martignoni G, Gobbo S, Camparo P, Brunelli M, Munari E, Segala D, Pea M, Bonetti F, Illei PB, Netto G, Ladanyi M, Chilosi M, Argani P. Differential expression of cathepsin-K in neoplasms harbouring TFE3 gene fusions. Mod Pathol. 2011;24:1313–1319. doi: 10.1038/modpathol.2011.93. [DOI] [PubMed] [Google Scholar]
- 15.Sukov WR, Hodge JC, Lohse CM, et al. TFE3 rearrangements in adult renal cell carcinoma: clinical and pathologic features with outcome in a large series of consecutively treated patients. Am J Surg Pathol. 2012;36:663–670. doi: 10.1097/PAS.0b013e31824dd972. [DOI] [PubMed] [Google Scholar]
- 16.Ellis CL, Eble JN, Subhawong AP, Martignoni G, Zhong M, Ladanyi M, Epstein JI, Netto GJ, Argani P. Clinical heterogeneity of Xp11 translocation renal cell carcinoma: impact of fusion subtype, age and stage. Mod Pathol. 2014;27:875–86. doi: 10.1038/modpathol.2013.208. [DOI] [PubMed] [Google Scholar]
- 17.Argani P, Lal P, Hutchinson B, Lui MY, Reuter VE, Ladanyi M. Aberrant nuclear immunoreactivity for TFE3 in neoplasms with TFE3 gene fusions: A sensitive and specific immunohistochemical assay. Am J Surg Pathol. 2003;27:750–61. doi: 10.1097/00000478-200306000-00005. [DOI] [PubMed] [Google Scholar]
- 18.Rao Q, Williamson SR, Zhang S, Eble JN, Grignon DJ, Wang M, Zhou XJ, Huang W, Tan PH, Maclennan GT, Cheng L. TFE3 break-apart FISH has a higher sensitivity for Xp11. 2 translocation associated renal cell carcinoma compared with TFE3 or cathepsin K immunohistochemical staining alone: expanding the morphologic spectrum. Am J Surg Pathol. 2013;37:804–15. doi: 10.1097/PAS.0b013e31827e17cb. [DOI] [PubMed] [Google Scholar]
- 19.Mosquera JM, Dal Cin P, Mertz KD, Perner S, Davis IJ, Fisher DE, Rubin MA, Hirsch MS. Validation of a TFE3 break-apart FISH assay for Xp11 translocation renal cell carcinomas. Diagn Mol Pathol. 2011;20:129–37. doi: 10.1097/PDM.0b013e31820e9c67. [DOI] [PubMed] [Google Scholar]
- 20.Argani P, Illei P, Netto G, Ro J, Cho HY, Dogan S, Ladanyi M, Martignoni G, Aulmann S, Weiss SW. A Distinctive Subset of PEComas Harbor TFE3 Gene Fusions. Am J Surg Pathol. 2010;34:1395–1406. doi: 10.1097/PAS.0b013e3181f17ac0. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka M, Kato K, Gomi K, Matsumoto M, Kudo H, Shinkai M, Ohama Y, Kigasawa H, Tanaka Y. Perivascular epithelioid cell tumor with SFPQ/PSF-TFE3 gene fusion in a patient with advanced neuroblastoma. Am J Surg Pathol. 2009;33:1416–20. doi: 10.1097/PAS.0b013e3181a9cd6c. [DOI] [PubMed] [Google Scholar]
- 22.Agaram NP, Sung YS, Zhang L, Chen CL, Chen HW, Singer S, Dickson MA, Berger MF, Antonescu CR. Dichotomy of Genetic Abnormalities in PEComas With Therapeutic Implications. Am J Surg Pathol. 2015;39:813–25. doi: 10.1097/PAS.0000000000000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malinowska I, Kwiatkowski DJ, Weiss S, Martignoni G, Netto G, Argani P. Perivascular epithelioid cell tumors (PEComas) harboring TFE3 gene rearrangements lack the TSC2 alterations characteristic of conventional PEComas: further evidence for a biological distinction. Am J Surg Pathol. 2012;36:783–4. doi: 10.1097/PAS.0b013e31824a8a37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao Q, Shen Q, Xia QY, Wang ZY, Liu B, Shi SS, Shi QL, Yin HL, Wu B, Ye SB, Li L, Chen JY, Pan MH, Li Q, Li R, Wang X, Zhang RS, Yu B, Ma HH, Lu ZF, Zhou XJ. PSF/SFPQ Is a Very Common Gene Fusion Partner in TFE3 Rearrangement-associated Perivascular Epithelioid Cell Tumors (PEComas) and Melanotic Xp11 Translocation Renal Cancers: Clinicopathologic, Immunohistochemical, and Molecular Characteristics Suggesting Classification as a Distinct Entity. Am J Surg Pathol. 2015;39:1181–96. doi: 10.1097/PAS.0000000000000502. [DOI] [PubMed] [Google Scholar]
- 25.Argani P, Aulmann S, Karanjawala Z, Fraser RB, Ladanyi M, Rodriguez MM. Melanotic Xp11 translocation renal cancers: A distinctive neoplasm with overlapping features of PEComa, carcinoma, and melanoma. Am J Surg Pathol. 2009;33:609–19. doi: 10.1097/PAS.0b013e31818fbdff. [DOI] [PubMed] [Google Scholar]
- 26.Chang IW, Huang HY, Sung MT. Melanotic Xp11 translocation renal cancer: A case with PSF-TFE3 gene fusion and up-regulation of melanogenetic transcripts. Am J Surg Pathol. 2009;33:1894–901. doi: 10.1097/PAS.0b013e3181ba7a5f. [DOI] [PubMed] [Google Scholar]
- 27.Russell CM, Buethe DD, Dickinson S, Sexton WJ. Perivascular epithelioid cell tumor (PEComa) of the urinary bladder associated with Xp11 translocation. Ann Clin Lab Sci. 2014;44:91–8. [PubMed] [Google Scholar]
- 28.Ritterhouse LL, Cykowski MD, Hassell LA, Slobodov G, Bane BL. Melanotic Xp11 translocation renal cancer: report of a case with a unique intratumoral sarcoid-like reaction. Diagn Pathol. 2014;9:81. doi: 10.1186/1746-1596-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varinot J, Camparo P, Beurtheret S, Barreda E, Compérat E. An adult case of melanotic Xp11 translocation renal cancers: distinct entity or sub-entity? Int J Surg Pathol. 2011;19:285–9. doi: 10.1177/1066896911400736. [DOI] [PubMed] [Google Scholar]
- 30.Argani P, Yonescu R, Morsberger L, Morris K, Netto GJ, Smith N, Gonzalez N, Illei PB, Ladanyi M, Griffin CA. Molecular confirmation of t(6;11)(p21;q12) renal cell carcinoma in archival paraffin-embedded material using a break-apart TFEB FISH assay expands its clinicopathologic spectrum. Am J Surg Pathol. 2012;36:1516–26. doi: 10.1097/PAS.0b013e3182613d8f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edge SB, Byrd DR, Compton C, Fritz AG, Greene FL, Trotti A. American Joint Committee on Cancer Staging Manual. 7. Springer; New York: 2010. pp. 481–491. [Google Scholar]
- 32.Antonescu CR, Zhang L, Chang NE, et al. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer. 2010;49:1114–1124. doi: 10.1002/gcc.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Antonescu CR, Le Loarer F, Mosquera JM, Sboner A, Zhang L, Chen CL, Chen HW, Pathan N, Krausz T, Dickson BC, Weinreb I, Rubin MA, Hameed M, Fletcher CD. Novel YAP1-TFE3 fusion defines a distinct subset of epithelioid hemangioendothelioma. Genes Chromosomes Cancer. 2013;52:775–84. doi: 10.1002/gcc.22073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tickoo SK, dePeralta-Venturina MN, Harik LR, Worcester HD, Salama ME, Young AN, Moch H, Amin MB. Spectrum of epithelial neoplasms in end-stage renal disease: An experience from 66 tumor-bearing kidneys with emphasis on histologic patterns distinct from those in sporadic adult renal neoplasia. Am J Surg Pathol. 2006;30:141–53. doi: 10.1097/01.pas.0000185382.80844.b1. [DOI] [PubMed] [Google Scholar]
- 35.Aydin H, Chen L, Cheng L, et al. Clear cell tubulopapillary renal cell carcinoma: A study of 36 distinctive low-grade epithelial tumors of the kidney. Am J Surg Pathol. 2010;34:1608–1621. doi: 10.1097/PAS.0b013e3181f2ee0b. [DOI] [PubMed] [Google Scholar]
- 36.Zhan HQ, Chen H, Wang CF, Zhu XZ. A case of PSF-TFE3 gene fusion in Xp11.2 renal cell carcinoma with melanotic features. Hum Pathol. 2015;46:476–81. doi: 10.1016/j.humpath.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 37.Yarosh CA, Iacona JR, Lutz CS, Lynch KW. PSF: nuclear busy-body or nuclear facilitator? Wiley Interdiscip Rev RNA. 2015;6:351–67. doi: 10.1002/wrna.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dijkhuizen T, van den Berg E, Wilbrink M, Weterman M, Geurts van Kessel A, Storkel S, Folkers RP, Braam A, de Jong B. Distinct Xp11. 2 breakpoints in two renal cell carcinomas exhibiting X; autosome translocations. Genes Chromosomes Cancer. 1995;14:43–50. doi: 10.1002/gcc.2870140108. [DOI] [PubMed] [Google Scholar]
- 39.Parast MM, Eudy G, Gow KW, Amin M, Shehata B. A unique case of renal carcinoma with Xp11.2 translocations/TFE3 gene fusions in a 3-year-old child, with coexistent von Hippel-Lindau gene mutation. Pediatr Dev Pathol. 2004;7:403–6. doi: 10.1007/s10024-004-1018-8. [DOI] [PubMed] [Google Scholar]
- 40.Zhong M, Weisman P, Zhu B, Brassesco M, Yang Y, Linehan WM, Merino MJ, Zhang D, Rohan S, Cai D, Yang X. Xp11.2 translocation renal cell carcinoma with PSF-TFE3 rearrangement. Diagn Mol Pathol. 2013;22:107–11. doi: 10.1097/PDM.0b013e318278962e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haudebourg J, Hoch B, Fabas T, Burel-Vandenbos F, Carpentier X, Amiel J, Cardot-Leccia N, Michiels JF, Pedeutour F. A novel case of t(X;1)(p11.2;p34) in a renal cell carcinoma with TFE3 rearrangement and favorable outcome in a 57-year-old patient. Cancer Genet Cytogenet. 2010;200:75–8. doi: 10.1016/j.cancergencyto.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 42.Creighton CJ, Morgan M, Gunaratne PH Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–9. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LeGallo RD, Stelow EB, Sukov WR, Duska LR, Alisanski SB, Folpe AL. Melanotic xp11.2 neoplasm of the ovary: report of a unique case. Am J Surg Pathol. 2012;36:1410–4. doi: 10.1097/PAS.0b013e31826277a9. [DOI] [PubMed] [Google Scholar]
- 44.Ladanyi M, Lui MY, Antonescu CR, Krause-Boehm A, Meindl A, Argani P, Healey JH, Ueda T, Yoshikawa H, Meloni-Ehrig A, Sorensen PH, Mertens F, Mandahl N, van den Berghe H, Sciot R, Dal Cin P, Bridge J. The der(17)t(X;17)(p11;q25) of human alveolar soft part sarcoma fuses the TFE3 transcription factor gene to ASPL, a novel gene at 17q25. Oncogene. 2001;20:48–57. doi: 10.1038/sj.onc.1204074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Visualization of PSF-TFE3 gene fusion. Short sequence reads from RNA-seq were aligned to PSF-TFE3 fusion gene by Bowtie2 and visualized by Integrative Genomics Viewer (IGV), supporting the PSF-TFE3 fusion. Each grey bar represents a short sequence read. The sequence near the PSF-TFE3 junction was demonstrated at the bottom. The junction site is marked with a red arrow.