SUMMARY
The ENE (element for nuclear expression) is a cis-acting RNA structure that protects viral or cellular noncoding (nc)RNAs from nuclear decay through triple-helix formation with the poly(A) tail or 3′-terminal A-rich tract. We expanded the roster of 9 known ENEs by bioinformatic identification of ~200 distinct ENEs that reside in transposable elements (TEs) of numerous non-metazoan and one fish species, and in four Dicistrovirus genomes. Despite variation within the ENE core, none of the predicted triple-helical stacks exceeds five base triples. Increased accumulation of reporter transcripts in human cells demonstrated functionality for representative ENEs. Location close to the poly(A) tail argues that ENEs are active in TE transcripts. Their presence in intronless but not intron-containing hAT transposase genes supports the idea that TEs acquired ENEs to counteract the RNA-destabilizing effects of intron loss, a potential evolutionary consequence of TE horizontal transfer in organisms that couple RNA silencing to splicing deficits.
INTRODUCTION
Polyadenylated RNAs, which include messenger (m)RNA and long noncoding (lnc)RNA, are subject to deadenylation-dependent decay in both the nucleus and the cytoplasm (Garneau et al., 2007). Since the process of RNA decay initiates with deadenylation, highly abundant polyadenylated RNAs often harbor cis-acting elements that protect their poly(A) tails (Conrad and Steitz, 2005; Geisberg et al., 2014; Muhlrad and Parker, 2005; Wang et al., 1999). Among them, the element for nuclear expression (ENE) (previously referred to as expression and nuclear retention element) acts primarily in the nucleus to inhibit rapid RNA decay (Conrad et al., 2006). The ENE, initially discovered in an abundant PAN lncRNA from Kaposi’s sarcoma-associated herpesvirus (KSHV) (Conrad and Steitz, 2005), was subsequently found in noncoding and genomic RNAs of diverse viruses (Tycowski et al., 2012). Analogous elements have been recently identified in two mammalian non-polyadenylated nuclear lncRNAs, MALAT1 and MENβ, which is also known as NEAT1_2 (Brown et al., 2012; Wilusz et al., 2012).
Viral ENEs are 40–79 nucleotide (nt)-long, each composed of a stem-loop structure containing an asymmetric internal U-rich loop, which, in conjunction with several adjacent base pairs, constitutes the ENE’s functional core (Conrad et al., 2007; Tycowski et al., 2012). The crystal structure of the KSHV PAN RNA ENE core bound to oligo(A)9 revealed formation of a U•A-U triple helix between the U-rich loop and its oligoadenylate target (Mitton-Fry et al., 2010) (see also Figure 1A). The triple-helical binding interface is further extended by A-minor interactions with three G-C base pairs of the lower stem. Located between the triple helix and the A-minor interactions are two residues that loop out to form a bulge, which we refer to as a linker. Biochemical and genetic analyses suggested that interactions observed in the X-ray structure occur between KSHV PAN RNA’s poly(A) tail and the KSHV PAN RNA ENE (schematized in Figure 1A) (Conrad et al., 2006; Conrad et al., 2007; Mitton-Fry et al., 2010).
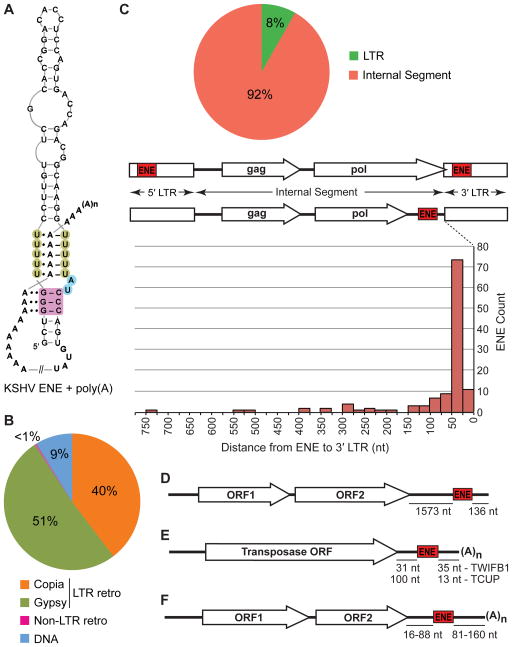
Figure 1. Distribution of the ENEs described in this paper between different TE classes and their positions within TEs and viral genomic RNAs.
(A) Interaction of the KSHV PAN RNA ENE with the poly(A) tail (Conrad et al., 2006; Mitton-Fry et al., 2010). The U residues involved in triple-helix formation are shaded green while the A-minor receptor pairs are colored magenta. Linker residues are shaded blue. Watson-Crick and Hoogsteen base pairings are denoted by dashes and single dots, respectively, while A-minor interactions are indicated by double dots. (B) Distribution of ENEs among different classes of TEs. (C) ENE positions within LTR retrotransposons. The pie chart depicts the partitioning of ENEs between LTRs and Internal Segments. The graph shows distances to the 3′ LTR for ENEs located in Internal Segments. The distances to the poly(A) tail of transcripts cannot be predicted because the sequences around the LTR polyadenylation sites lack conserved motifs. (D) The ENE of the non-LTR retrotransposon Tad1 from N. crassa is predicted to lie ~136 nt upstream of the poly(A) tail of the Tad1 RNA. (E) Positions of the TWIFB1_OSa and TCUP_ZMa ENEs in their respective hAT transposase mRNAs. (F) Location of ENEs in Dicistrovirus genomes. Distance ranges for MCDiV, TSV, MRTV and HiPV are shown (see Figure S3 for details). Not to scale.
A different triplex structure is observed for the MALAT1 ENE. By engaging its 3′-terminal A-rich tract, the MALAT1 element forms a bipartite triple helix containing stacks of five and four U•A-U triples separated by a C+•G-C triplet and a C-G doublet (Brown et al., 2014). As in the KSHV PAN RNA, the triple helix is fortified by A-minor interactions. An analogous, albeit shorter, bipartite triple helix is predicted to form at the 3′ end of MENβ RNA. In vivo decay assays indicate that the MALAT1 blunt-ended triple helix, with the 3′ nucleotide sequestered in a U•A-U triple, inhibits rapid nuclear RNA decay, thus allowing the abundant accumulation of MALAT1 lncRNA in vertebrate cell nuclei.
Transposable elements (TEs) or transposons are mobile genetic elements that contribute significantly to genome evolution (Feschotte and Pritham, 2007; Huang et al., 2012; Kazazian, 2004). TEs are most numerous in plants, constituting up to 80% of total genomic DNA in some species, e.g. maize (Baucom et al., 2009; Hua-Van et al., 2005). Although less abundant, TE sequences also represent a considerable fraction of fungal (3–20%) and metazoan (3–45%) genomes (Hua-Van et al., 2005).
TEs are divided into two classes based on their transposition intermediates: class I or retrotransposons propagate via RNA intermediates and class II or DNA transposons move without intermediary RNAs (Wicker et al., 2007). Retrotransposons can be further divided into long terminal repeat (LTR)-possessing TEs, which resemble animal retroviruses, and non-LTR TEs comprising LINEs and SINEs.
Transcription of a canonical LTR retrotransposon begins in the 5′ LTR and terminates within the 3′ LTR producing a 3′-polyadenylated RNA, which typically serves both as an mRNA for translation of TE proteins and as a template for reverse transcription to produce a transposition intermediate (Schulman, 2013). The major 3′-polyadenylated transcript of a typical non-LTR retrotransposon, which also serves both functions, is largely collinear with the TE DNA. DNA transposons produce at least one mRNA, that for the transposase protein, which catalyzes the transposition process (Feschotte and Pritham, 2007; Wicker et al., 2007).
hAT DNA transposons are “cut-and-paste” elements found in almost all eukaryotic genomes (Atkinson, 2015; Kempken and Windhofer, 2001). Autonomous hAT TEs are typically 2.5–5.0 kb long and carry a single gene, which encodes the transposase protein. Transposase mRNAs from many active plant and fungal hAT TEs have been well characterized (see Table S1 for references).
Here, we performed a genome-wide bioinformatics search for ENEs in eukaryotic organisms. We found ~200 distinct types of ENE-like structures, mostly in plant and fungal, and less frequently, in slime mold TEs. We also found analogous structures in two endogenous retroviruses (ERVs) from the stickleback fish Gasterosteus aculeatus and in four dicistroviral genomes. These findings establish that ENEs are present not only in lncRNAs but also in mRNAs. This large collection of ENEs provides a wealth of information on the ENE sequence and secondary structure variation, modes of interaction with the poly(A) tail and the length of RNA major groove triple helices. We tested one dicistroviral and three plant elements and showed that they can increase the accumulation of an intronless reporter transcript, demonstrating their functionality. Inspection of the published transposition-competent plant and fungal hAT TEs revealed the presence of ENEs in intronless, but not in intron-containing, transposase genes. We consider the possibility that ENEs compensate for intron loss by TE genes, which occurs evolutionarily because of the coupling of TE silencing to RNA splicing in lower eukaryotes (Dumesic and Madhani, 2013; Dumesic et al., 2013).
RESULTS
To identify cellular ENEs, we first searched genomic RefSeq databases of all available eukaryotic genomes using the Infernal tools (Nawrocki and Eddy, 2013). This exercise revealed multiple ENE-like hits in plant and fungal genomes, many occurring in multiple identical copies in a given organism. This finding indicated that ENEs are abundant in repetitive elements. To overcome extensive redundancy and the lack of repetitive element annotation when exploring entire genomes, we instead searched Repbase, a comprehensive database of eukaryotic repetitive DNA elements, which contains only one consensus sequence for each element family from a given organism (Bao et al., 2015). The Repbase search revealed ~200 distinct ENE-like structures in numerous plant, fungal and slime mold TEs and in two fish ERVs (Figures 1–3). For instance, it identified 12 unique ENE sequences in Arabidopsis thaliana TEs, even though the A. thaliana genome harbors much larger number of ENEs (~150) because each appears in multiple identical or very similar copies. In addition, probing the RefSeq viral genomic database yielded 4 novel ENE-like structures in 4 out of 18 sequenced Dicistroviruses (Figure S3). Another dicistroviral ENE was identified in our previous work (Tycowski et al., 2012).
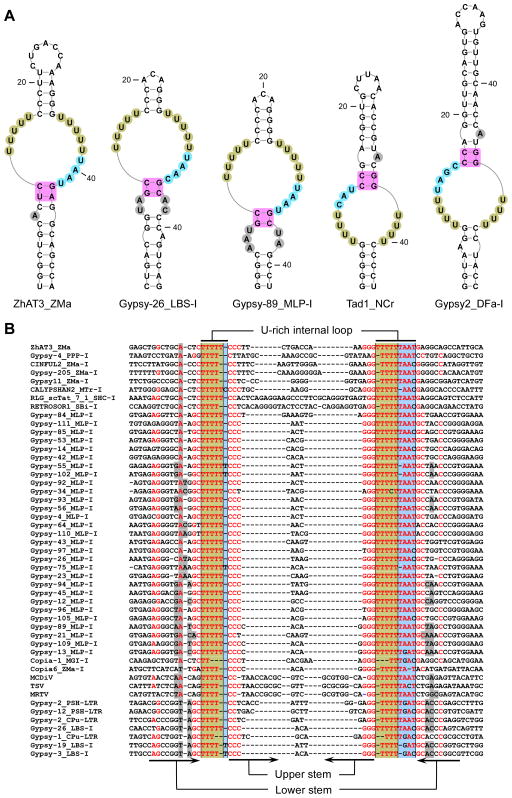
Figure 3. Single-domain ENEs.
(A) Predicted secondary structures of selected ENEs. Note that the Tad1_NCr and Gypsy2_DFa-I ENEs are in inverted orientation. (B) Alignment of TE DNA and Dicistrovirus cDNA sequences corresponding to 50 ENEs and their flanking regions. Only standard-orientation ENEs are included. For clarity, sequences with ≥95% redundancy were discarded. Nucleotide coloring and shading are as in Figure 2. The organism name abbreviations are as follows: Dictyostelium fasciculatum - DFa, Polysphondylium pallidum PN500 – PPP, Saccharum hybrid cultivar –SHC, Melampsora larici-populina –MLP, Mycosphaerella graminicola IPO323 – MGI, Punctularia strigosozonata HHB-11173 –PSH, Coniophora puteana- CPu, Laccaria bicolor S238N – LBS. Other organism name abbreviations are as in Figure 2. MCDiV, TSV and MRTV stand for mud crab dicistrovirus, Taura syndrome virus and Macrobrachium rosenbergii Taihu virus, respectively. See also Figure S3.
Distribution and location of ENEs in TEs and dicistroviral genomes
The TE ENEs are most common in LTR retrotransposons, with 51% residing in the members of the gypsy and 40% in those of the copia superfamily, respectively (Figure 1B). The vast majority (92%) of the LTR retrotransposon ENEs are located close to the 3′LTR within so-called Internal Segments (Figure 1C). The rest (8%) are within LTRs. Only one non-LTR retrotransposon was found to possess an ENE: the Tad1 TE from N. crassa (Figure 1D) and related species (data not shown). Here, the ENE is positioned about 136 nt from the 3′ terminus of the TE.
Nine percent of the TE ENEs were identified in DNA transposons, i.e. in cut-and-paste plant elements belonging to the hAT superfamily. Here, the 3′UTRs of the transposase mRNA harbor the ENEs, which in the rice TWIFB1 and the maize TCUP TEs are located 35 and 13 nt upstream of the poly(A) tail, respectively (Figure 1E).
In Dicistroviruses, each ENE is located in the region of the genomic RNA that corresponds to the 3′UTR when the genome serves as a message for the synthesis of viral proteins. A genetically encoded poly(A) tail appears 81–160 nt downstream of the ENE (Figures 1F and S3).
Organization of the functional cores of the TE and Dicistrovirus ENEs
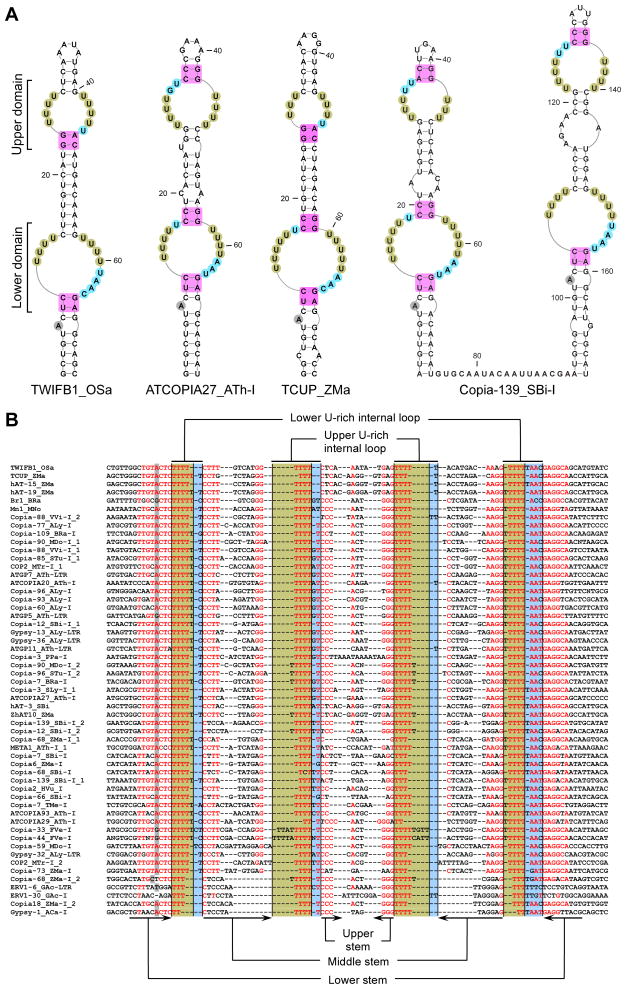
The majority of the plant ENE-like structures, as well as both fish elements, are composed of two functional cores, which we call domains: a highly conserved lower and a less conserved upper domain (Figures 2 and S1). The right-side U tracts (shaded green) of the lower domain are invariably abutted by a 3- or 4-nt long linker (shaded blue), while the left-side U tracts (shaded green) are sometimes abutted by a 1- or 2-nt linker (shaded blue). The A-minor receptor (shaded magenta) of the lower stem is usually composed of Y-R pairs (Y=pyrimidine, R=purine) rather than the G-C, which is invariably found in the ENEs of herpesviral PAN RNAs (Conrad et al., 2007; Mitton-Fry et al., 2010; Tycowski et al., 2012) and of cellular MALAT1 and MENβ (Brown et al., 2014; Brown et al., 2012) lncRNAs. Note that the lower stem is interrupted by a conserved bulge (shaded gray), which most frequently is represented by an A residue, but G substitutions can occur.
Figure 2. Double-domain ENEs.
(A) Predicted secondary structures for selected ENEs. Note the two adjacent double-domain ENEs in the Copia_139 TE from Sorghum bicolor. The U residues predicted to form triple helices with the poly(A) tail are shaded green while the A-minor receptor residues are colored magenta. Linker residues are shaded blue. The bulged nucleotide at a conserved position within the lower stem is shaded gray. (B) Alignment of transposon sequences corresponding to selected ENEs and their flanking regions. For clarity, sequences with ≥95% redundancy were discarded. Nucleotide shading is as in A. Red residues are at least 60% conserved. Each ENE is designated beginning with the name of its host TE, followed by the three-letter code of the host organism, and then, for ENEs from LTR retrotransposons, followed by the location either in the Internal Segment (I) or the Long Terminal Repeat (LTR). If a TE carries two ENEs, the name ends with the copy identifier. Organism names are abbreviated as follows: Ajellomyces capsulatus - ACa, Arabidopsis lyrata - ALy, Arabidopsis thaliana - ATh, Brassica rapa - BRa, Fragaria vesca - FVe, Hordeum vulgare - HVu, Gasterosteus aculeatus – GAc, Malus x domestica - MDo, Medicago truncatula - MTr, Morus notabilis – MNo, Oryza sativa - OSa, Physcomitrella patens - PPa Sorghum bicolor - SBi, Solanum lycopersicum - SLy, Solanum tuberosum - STu, Tuber melanosporum - TMe, Vitis vinifera - VVi and Zea mays - ZMa. The TE nomenclature was taken from Repbase18.04. See also Figures S1 and S2.
The upper domains are more variable and closely resemble the functional core of the KSHV PAN RNA ENE (see Figure 1A) in that the A-minor receptors are almost always G-C pairs (shaded magenta) and the linker is always 1- or 2-nt long (shaded blue). Note that the U tracts in Copia-33_FVe-I and Copia-44_FVe-I are interrupted by non-U residues (Figures 2B and S2); thus the upper domains in these ENEs may form bipartite triple helices akin to those found in MALAT1 and MENβ lncRNAs (Brown et al., 2014; Brown et al., 2012).
Single-domain ENEs are found predominantly in fungal and slime mold TEs and in several Dicistroviral genomes (Figures 3 and S3). They are most closely related to the lower domain of the double-domain ENEs (see Figure 2) in that i) they possess a 3 or 4 nt-long linker (shaded blue) and ii) the A-minor stem is invariably interrupted at a conserved position by a bulge or a small internal loop (shaded gray).
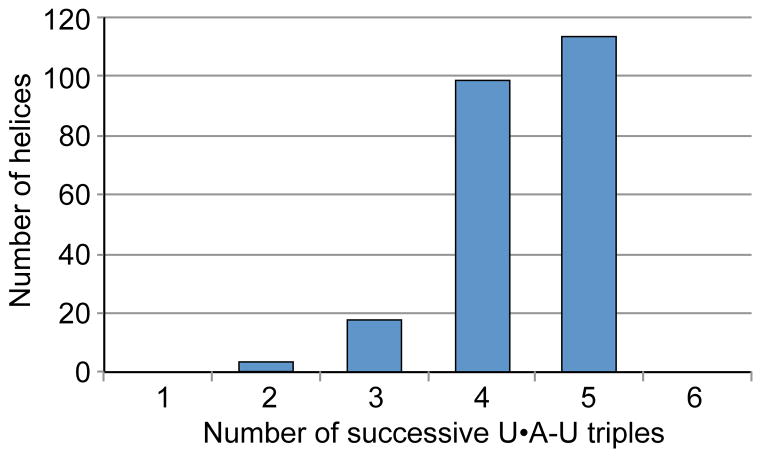
Except for the upper domain in two Fregaria ENEs (Figure S2), which is predicted to form an interrupted U•A-U helix, all domains in other ENEs reported here exhibit the potential to form contiguous U•A-U triple helices, most frequently composed of four or five triples, although domains that may form only three or two triples also occur (Figures 2, 3 and 4). The upper domains of the double-domain ENEs tend to possess shorter stacks than the lower domains. For example, three-triple-long helices are ~3 times more frequent in the upper domains. Whether this also holds for two-triple-long stacks cannot be assessed because only 2 such stacks were found in double-domain ENEs, 1 each in a lower and upper domain. Taken together, our collection reveals unexpected variability in the composition of the functional cores of ENEs.
Figure 4.
Length distribution of the predicted major groove U•A-U triple helices formed between TE or Dicistrovirus ENEs and downstream poly(A) tails.
Transcript stabilization by the ENEs
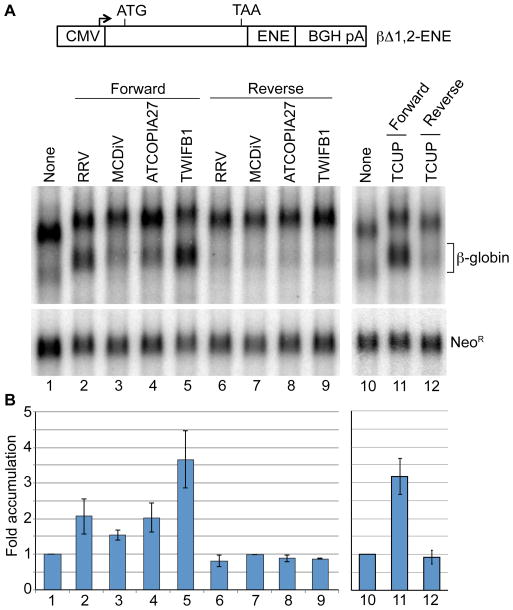
To address whether the newly-identified ENEs function in RNA stabilization, we tested four structures for their ability to increase accumulation of an intronless β-globin mRNA reporter (Conrad and Steitz, 2005). The MCDiV, ATCOPIA27_ATh-I, TWIFB1_OSa and TCUP_ZMa ENEs were each inserted into the β-globin 3′UTR 167 nt upstream of the poly(A) tail and the resulting chimeras transiently expressed in HEK293T cells. The rhesus rhadinovirus (RRV) ENE served as a positive control. Figure 5 shows that, relative to the no insert control (lane 1), the RRV ENE increased β-globin mRNA levels 2.1-fold (lane 2), while the MCDiV, ATCOPIA27_ATh-I and TWIFB1_OSa ENEs showed 1.5-, 2.0- and 3.7-fold stabilization, respectively (lanes 3–5). The TCUP_ZMa ENE increased β-globin mRNA levels 3.2-fold (lane 11). None of the ENE inserts in antisense orientation increased the levels of β-globin mRNA, but either lowered its accumulation slightly or did not alter its levels (compare lanes 6–9 with 1 and lane 12 with 10). In summary, all of the ENEs tested increased accumulation of the intronless β-globin reporter transcript, but to varying degrees.
Figure 5. ENEs increase the levels of a heterologous intronless transcript.
β-globin constructs (βΔ1,2-ENE) containing the RRV, MCDiV, ATCOPIA27_ATh-I, TWIFB1_OSa or TCUP_ZMa ENE were transiently expressed in HEK293T cells. The ENE was inserted in either forward or reverse orientation, as indicated. (A) Northern blot analysis of β-globin transcript expression using a full-size β-globin RNA probe. The same blot was probed with a mixture of Neo1, Neo2 and Neo3 oligonucleotides for NeoR mRNA expressed from the same βΔ1,2-ENE plasmid. The unmarked band above β-globin may represent either an unrelated RNA or a non-polyadenylated β-globin precursor. (B) Quantification of Northern blot signals. To control for expression and loading, the levels of β-globin transcripts were normalized to those of NeoR mRNA. The βΔ1,2 signal (lanes 1 and 10) was set to 1. The bars show the average values from three experiments ±SD.
Conserved features of the newly-discovered ENEs contribute to RNA stabilization
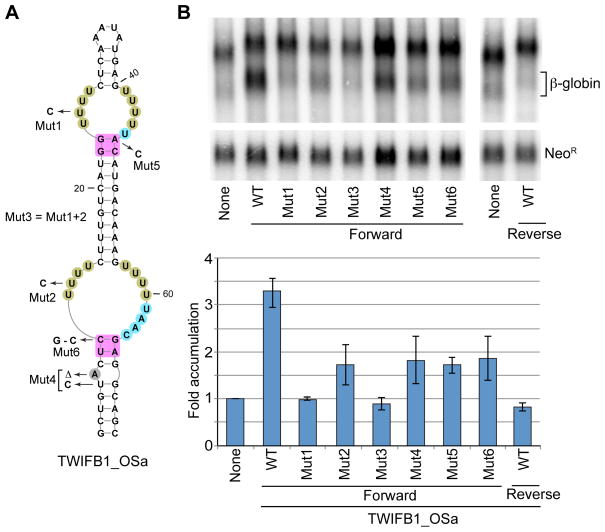
The majority of the ENEs described here contain two domains (Figure 2 and data not shown). To assess the contribution of individual domains to the stabilization of a reporter RNA, we mutated one U to C in each domain or in both domains of the TWIFB1_OSa ENE. The resultant construct was inserted into the intronless β-globin reporter and assayed for β-globin mRNA accumulation (Figure 6). Mutant 1 (Mut1), predicted to disrupt the triple helix in the upper domain only, abolished stabilization activity. Surprisingly, disruption of the triple helix in the lower domain (Mut2) reduced the accumulation of β-globin mRNA, but only by ~45%. Disruption of the triple helices in both domains (Mut3) resulted in only slight, not statistically significant, decrease in levels of β-globin transcript when compared to Mut1. We conclude that both ENE domains contribute to the accumulation of the intronless β-globin mRNA, but to varying degrees; the upper domain exhibits more stabilization activity.
Figure 6. Contribution of conserved elements in double-domain ENEs to RNA stabilization.
(A) TWIFB1_OSa ENE. Nucleotide shading is as in Figure 2. Mutants designated Mut1-Mut6 were used in the experiments shown in (B). (B) β-globin constructs (βΔ1,2) containing either the WT or a mutant TWIFB1_OSa ENE were transiently expressed in HEK293T cells and analyzed as described in Figure 5. The bars depict the average values from three experiments ±SD.
Many of the ENEs described here possess new features, e.g. a bulge or a small internal loop at a conserved position in the A-minor stems adjacent to the 3 or 4 nt-long linkers (see Figures 2 and 3). To test whether this interruption of the A-minor stem is important, we deleted the bulged A and strengthened the stem by substituting the adjacent U-G pair with C-G (Mut4, Figure 6A). Indeed, these changes decreased the accumulation of β-globin mRNA by ~40% (Figure 6B). Thus, relaxation of the A-minor receptor stem increases the stabilizing activity of a double-domain ENE.
Our large collection of ENEs also revealed considerable variability in the composition of A-minor receptors (see Figures 2, 3, S3, S2 and S3). For instance, the TWIFB1_OSa harbors a non-canonical G-A receptor in the upper domain (Figure 6A, shaded magenta). We mutated the G-A to G-C (Mut5) and observed ~45% reduction in reporter β-globin mRNA accumulation (Figure 6B). Thus, a G-A receptor confers greater stabilizing activity than the G-C invariably found in all previously characterized ENEs (Brown et al., 2012; Conrad et al., 2007). However, this appears to hold for domains with 1 nt-long linkers only (data not shown).
Another common feature is the frequent occurrence of C-G as the first A-minor receptor pair (counting from the U-rich loop) in the lower stem of the lower domain of the double-domain ENEs (see Figure 2). This feature is also frequent in the single-domain ENEs (see Figure 3). In both cases, this non-canonical A-minor receptor correlates with a longer (3 or 4 nt) linker. Changing the C-G to the canonical G-C in TWIFB1_OSa (Mut6, Figure 6A) decreased the accumulation of β-globin mRNA by ~40% (Figure 6B), although both base pairs conferred equal stabilization activity on the MCDiV ENE (data not shown). This difference may be, at least in part, caused by the formation of an alternative structure in the TWIFB1_OSa Mut6 ENE that may contribute to inhibition of the stabilization activity. Indeed, because of the UAAC rather than UAAU linker, the A-minor receptor stem could conceivably be extended by 3 base pairs into the U-rich loop in the TWIFB1_OSa Mut6, looping out C65. We conclude that a longer linker apparently permits more variability in the composition of the A-minor receptor.
Intronless hAT transposase mRNAs accumulate ENEs
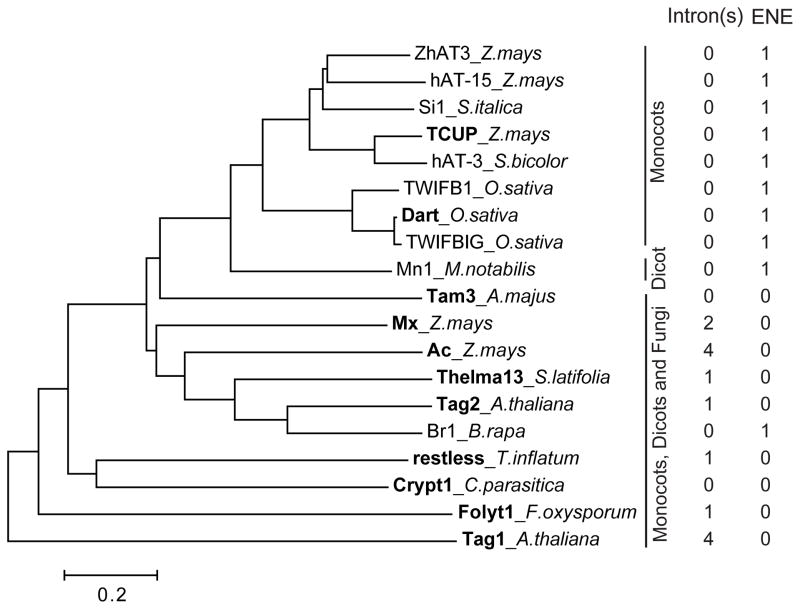
We inspected the reported transposition-competent hAT TEs from plants and fungi (compiled in Table S1) for the presence of introns in the transposase gene and subsequently searched each TE sequence for the presence of an ENE. Interestingly, this well-characterized transposase gene in the rice Dart and maize TCUP TEs lacks introns (see Table S1 for references). Yet, we found both TEs to possess an ENE. In contrast, none of the hAT TEs with an intron-containing transposase gene was found to possess an ENE (Figure 7).
Figure 7. The absence of an intron(s) in the hAT tranposase gene correlates with the presence of an ENE.
The hAT transposase sequences were aligned with MUSCLE (Edgar, 2004), and the tree was constructed using the neighbor joining option from MEGA6 (Tamura et al., 2013). TEs with experimentally-demonstrated transposition activity (see Table S1 for references) are shown in bold.
All the ENE-containing hAT TEs from our collection belong to the Dart/TCUP clade (Figure 7 and data not shown) except for the Br1 TE, which constitutes an independent branch. As in Dart and TCUP, the transposase gene in all other ENE-possessing TEs is bioinformatically predicted to lack introns. We conclude that the presence of an ENE correlates with the absence of intron(s) in the hAT transposase gene.
DISCUSSION
We identified a large collection of ENEs in plant, fungal and slime mold TEs (Figures 1–3). These ENEs are found predominantly in LTR retrotransposons, but they also occur in plant DNA transposons of the hAT superfamily (Figure 1B). They are rare in non-LTR retrotransposons, where we found only one TE, the fungal Tad1, to possess an ENE (Figures 1 and 3). Interestingly, two ERVs from the stickleback fish also possess ENE-like structures (Figure S1). Since only two stickleback ERVs are present in the Repbase18.04 and both harbor such structures, ENEs may be more numerous in the stickleback genome.
We previously identified an ENE in the RNA genome of a Dicistrovirus, PSTV (Tycowski et al., 2012). Here, we report ENEs in four additional Dicistroviruses (Figure S3), bringing the total number to 5 out of 18 sequenced genomes. Since Dicistroviruses replicate in the cytoplasm (Bonning and Miller, 2010), this finding predicts a cytoplasmic function for the ENE, most likely to increase mRNA levels.
The TE ENEs fall into two classes. Whereas the double-domain ENEs are found predominantly in plants, the single-domain elements occur mostly in fungi and slime molds. Whether an organism possesses single or multiple domain ENEs may be dictated by the length of poly(A) tails of its transcripts. Whereas short poly(A) tails in fungal and slime mold transcripts [60–80 nt (Adams et al., 1980; Brown and Sachs, 1998)] may be able to associate with only one ENE domain, the two-fold longer tails in plants (Subtelny et al., 2014) may confer the ability to interact simultaneously with two domains. The latter type of interaction is also expected for the β-globin reporter mRNA in HEK293T cells, which is predicted to possess an ~200 nt long poly(A) tail, based on previous measurements of the mRNA poly(A) size in mammalian cells (Eckmann et al., 2011). This prediction is also in agreement with the recent demonstration that the median length of mRNA poly(A) tails in HEK293T cells is ~30% greater than that in Arabidopsis (Subtelny et al., 2014).
The upper domains of the double-domain ENEs appear to exhibit greater stabilizing activity than their lower-domain counterparts. For instance, the bulk of stabilizing activity in the TWIFB1_OSa ENE resides in the upper domain (Figure 6). This might be partly due to better accessibility of upper domains for interaction with the poly(A) tail. Once such an interaction is initiated, the lower domains also contribute. Note that the lower domains often possess linkers on both sides of the U-rich loops, perhaps allowing such domains to engage the poly(A) tail in both orientations, thus maximizing the chance of triple-helix formation. More experiments will be required to verify the above assumptions.
The crystal structure of the MALAT1 ENE complexed with the A-rich tract revealed a restriction on the length of an U•A-U helix, which is imposed by steric clash between groups on the Hoogsteen and Watson (A-rich) strands of helices longer than 6 triples (Brown et al., 2014). Complete absence of continuous U•A-U triple helices predicted to be longer than 5 triples in our collection of more than 200 ENE domains (Figure 4) supports this observation. Interestingly, helices longer than 5 triples are predicted to be interrupted by non-canonical triples and/or bulged nucleotides in two strawberry ENEs (Figure S2). These non-canonical triples and/or bulged nucleotides might constitute helical reset points for extended helices, as proposed for the C•G-C triple/C-G doublet that interrupts the MALAT1 triple helix (Brown et al., 2014).
Conservation of sequence, structure and poly(A)-proximal location suggests that the newly-discovered ENEs are functional
All TE ENEs reside in regions predicted to specify the 3′UTRs of TE transcripts (Figure 1C–E). Among LTR retrotransposons, each ENE is positioned either in the LTR itself or close to the 3′LTR, within the Internal Segment (Figure 1C). Thus, TE transcripts terminating somewhere in the 3′LTR position the ENE in proximity to the poly(A) tail. The precise distance from the ENE to the poly(A) tail cannot be predicted because the sequences around polyadenylation sites in non-metazoan LTR retrotransposon RNAs lack conserved motifs (Benachenhou et al., 2013). Conversely, in the stickleback’s ERV1-6, an AATAAA abuts the ENE (Figure S1), predicting that a poly(A) tail in the ERV1-6 transcript appears ~20 nt downstream of the ENE. Likewise, the ENE in the Tad1 non-LTR retrotransposon from N. crassa and related fungal species lies ~136 nt from its 3′ terminus. Although the N. crassa Tad1 was demonstrated to be an active TE (Cambareri et al., 1994), its transcript(s) has not been characterized. However, since non-LTR retrotransposon transcripts are largely co-linear with their DNAs, the Tad1 transcript would carry the ENE ~136 nt upstream of the poly(A) tail (Figure 1D), a distance that is also favorable for engaging the poly(A) tail in triple helix formation. In hAT TEs, which contain only a single ORF that encodes the hAT transposase, the ENE always lies downstream of this ORF (Figure 1E). In the rice TWIFB1 and maize TCUP TEs, where the 3′ ends of the transposase mRNAs have been mapped (Itoh et al., 2007; Smith et al., 2012), the ENE is located very close (i.e. 35 and 13 nt upstream, respectively) to the poly(A) tail.
Their poly(A)-proximal location (Figure 1), coupled with the remarkable conservation of their functional cores (Figures 2 and 3), suggest that the ENEs described here are functional in their natural contexts. Note that conservation applies not only to the U residues themselves but also to the symmetry of the U tracts within the U-rich loops, which is also required for triple helix formation. Stabilization of the β-globin intronless reporter RNA by the tested ENEs (Figure 5) further supports this conclusion.
Why are ENEs abundant in TE transcripts but not in other cellular RNAs?
We hypothesize that the accumulation of ENEs by plant and fungal TEs is the result of TE silencing via siRNA-based mechanisms (Castel and Martienssen, 2013). This idea has been inspired by the recent findings of Dumesic and Madhani (Dumesic and Madhani, 2013; Dumesic et al., 2013) that in Cryptococcus yeast, the production of siRNAs is mechanistically linked to mRNA splicing. A connection between siRNA-mediated gene silencing and RNA splicing has been reported also for other organisms, including fission yeast and plants (Bayne et al., 2008; Lee et al., 2013; Tabach et al., 2013; Zhang et al., 2013). During splicing-dependent siRNA production in Cryptococcus, a spliceosome-associated complex called SCANR carries out the synthesis — from stalled splicing intermediates — of double-stranded (ds)RNAs, which are subsequently processed into siRNAs. Because of frequent horizontal transfer (HT) between organisms with distinct, and therefore often suboptimal, cis-acting splicing signals, we expect that TE transcripts might well generate such stalled intermediates, unless they are intronless. Indeed, among the spliceosome-stalled RNA processing intermediates in Cryptococcus, TE transcripts were observed to be significantly enriched (Dumesic et al., 2013). Such coupling between siRNA production and intron splicing would undoubtedly lead to evolutionary pressure on HT-prone TEs to lose introns in order to avoid silencing.
The loss of introns, however, poses a challenge for mRNA transcripts. Since splicing facilitates mRNA export, intronless transcripts are retained in the nucleus where they are exposed to powerful deadenylation-dependent exosome-based decay machineries (Conrad et al., 2006; Le Hir et al., 2003). The ENE, which is known to inhibit this nuclear RNA decay (Conrad et al., 2006), is well suited to rescue nucleus-retained intronless TE transcripts. Accordingly, we find that LTR retrotransposons and DNA TEs, which are both more prone to HT than non-LTR retrotransposons ((Hua-Van et al., 2005; Schaack et al., 2010) and references therein) and are therefore predicted to lose introns more frequently, preferentially accumulate ENEs (Figure 1B). Most importantly, we observe for the hAT transposase gene that the presence of an ENE indeed correlates with the lack of introns (Figure 7). Such a correlation analysis cannot be currently performed for retrotransposons because information concerning the exon/intron structure of transcripts derived from these elements is not available.
Are there TE-derived ENEs in non-TE cellular mRNAs and lncRNAs? It is well established that TEs constitute a rich source of regulatory elements, which are often co-opted over evolutionary time by host genomes (Feschotte, 2008; Rebollo et al., 2012). Searching plant and fungal transcriptomes uncovered several ENEs in plant non-TE RNAs (data not shown). Since these ENEs are flanked by TE sequences, they most likely represent recent transposition events. Whether they have been undergoing purifying selection remains to be established.
In conclusion, we have uncovered the widespread occurrence of triple-helix-forming RNA stability elements in plant and fungal TE transcripts. Since these elements preferentially accumulate in intronless RNAs, our findings add to a growing list of examples of significant connections between RNA processing and small RNA-mediated gene silencing.
EXPERIMENTAL PROCEDURES
Bioinformatics
Cellular genomic RefSeq (NCBI, release 65), viral genomic RefSeq (NCBI, release 70) and Repbase (GIRI, release 18.04) databases were iteratively queried for ENE structures using the Infernal 1.1 tools (Nawrocki and Eddy, 2013). The initial covariance models (CMs) for either standard or inverted ENEs were built from the alignments of viral ENE sequences (Tycowski et al., 2012). For the next iteration, the initial alignments were supplemented with hits satisfying inclusion thresholds from the first search and new CMs were constructed.
Sequenced dicistroviral genomes were also queried for ENEs by a mfold-aided folding of their 3′UTRs followed by manual inspection.
To remove identical or similar (95% or higher redundancy) sequences from our ENE set, the Skipredundant algorithm (EMBOSS) was used. Multiple sequence alignments were prepared with the help of the LocARNA software (Smith et al., 2010). Phylogenetic trees were drawn using MEGA6 tools (Tamura et al., 2013).
To determine the exon/intron structure of the hAT transposase genes, DNA sequences were translated in three frames and the resulting peptides aligned to known hAT transposases using the tblastn algorithm. Inserts were then manually inspected for splicing signals.
Oligonucleotide probes
Neo1 - GCA TCA GAG CAG CCG ATT GTC TGT TG
Neo2 - GCA TCA GCC ATG ATG GAT ACT TTC TCG G
Neo3 - CGG CCA TTT TCC ACC ATG ATA TTC GGC AAG C
Plasmids and Mutagenesis
The βΔ1,2 plasmid was previously described (Conrad and Steitz, 2005) as were the βΔ1,2-RRV_F and βΔ1,2-RRV_R (Tycowski et al., 2012).
The βΔ1,2-ATCOPIA27-F and βΔ1,2-ATCOPIA27-R constructs were generated by inserting into the ApaI site of the βΔ1,2 plasmid, in either forward (F) or reverse (R) orientation, a synthetic DNA fragment composed of ATCOPIA27_ATh-I ENE plus the 5′ and 3′ flanking sequences, 5 nt each. The βΔ1,2-TWIFB1-F and βΔ1,2-TWIFB1-R plasmids were constructed as above except that the insert contained the TWIFB1_OSa ENE. The MCDiV ENE-containing βΔ1,2 constructs were prepared as above except that the 5′ and 3′ ENE-flanking sequences were 6 and 5 nt, respectively. The 5′ and 3′ ENE-flanking sequences in the βΔ1,2-TCUP constructs were 3 and 4 nt, respectively.
The βΔ1,2-TWIFB1 mutant constructs were produced using the QuikChange kit (Stratagene) according to the manufacturer’s protocol.
Cell culture, transfections and RNA analyses
HEK293T cells were grown in DMEM medium supplemented with 10% fetal bovine serum and transfections were performed using the TransIT-293 reagent (Mirus) according to the manufacturer’s protocol. For Northern blot analyses, RNA was isolated 48 hours post-transfection using Trizol reagent (Life Technologies). Ten micrograms of total RNA were resolved on a 1.2% agarose/6.5% formaldehyde gel and capillary transferred to a Zeta-Probe GT membrane (Bio_Rad). To detect β-globin transcripts, a uniformly-32P-labeled full-size β-globin RNA probe, prepared by in vitro transcription, was used. To detect NeoR mRNA, 5′-32P-labeled DNA oligonucleotides were used as specified in the figure legends. RNA bands were quantified on a Storm PhosphorImager. To calculate the fold accumulation of β-globin mRNA, β-globin signal in each lane was divided by that of NeoR and the normalized values for each construct were then divided by the normalized value of βΔ1,2 without an ENE insert. Transfection experiments were repeated 3 times and are considered to be biological replicates.
Supplementary Material
Acknowledgments
We thank Walter Moss for bioinformatics advice, Jess Brown, Seyed Torabi, Johanna Withers, Ming Xie and Angie Miccinello for critical reading of the manuscript and the entire Steitz lab for stimulating discussions. This work was supported by grants GM026154 and CA16038 from the NIH. J.A.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTION
K.T.T. performed the bioinformatic searches and analyses; K.T.T. and M.-D.S. performed the experiments; all authors analyzed the data; K.T.T. and J.A.S. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams DS, Noonan D, Jeffery WR. The poly(adenylic acid)-protein complex is restricted to the nonpolysomal messenger ribonucleoprotein of Physarum polycephalum. Biochemistry. 1980;19:1965–1970. doi: 10.1021/bi00550a036. [DOI] [PubMed] [Google Scholar]
- Atkinson PW. hAT Transposable Elements. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0054-2014. [DOI] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucom RS, Estill JC, Chaparro C, Upshaw N, Jogi A, Deragon JM, Westerman RP, Sanmiguel PJ, Bennetzen JL. Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet. 2009;5:e1000732. doi: 10.1371/journal.pgen.1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne EH, Portoso M, Kagansky A, Kos-Braun IC, Urano T, Ekwall K, Alves F, Rappsilber J, Allshire RC. Splicing factors facilitate RNAi-directed silencing in fission yeast. Science. 2008;322:602–606. doi: 10.1126/science.1164029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benachenhou F, Sperber GO, Bongcam-Rudloff E, Andersson G, Boeke JD, Blomberg J. Conserved structure and inferred evolutionary history of long terminal repeats (LTRs) Mob DNA. 2013;4:5. doi: 10.1186/1759-8753-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonning BC, Miller WA. Dicistroviruses. Annu Rev Entomol. 2010;55:129–150. doi: 10.1146/annurev-ento-112408-085457. [DOI] [PubMed] [Google Scholar]
- Brown CE, Sachs AB. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol Cell Biol. 1998;18:6548–6559. doi: 10.1128/mcb.18.11.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Bulkley D, Wang J, Valenstein ML, Yario TA, Steitz TA, Steitz JA. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat Struct Mol Biol. 2014;21:633–640. doi: 10.1038/nsmb.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENbeta noncoding RNAs. Proc Natl Acad Sci U S A. 2012;109:19202–19207. doi: 10.1073/pnas.1217338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambareri EB, Helber J, Kinsey JA. Tad1-1, an active LINE-like element of Neurospora crassa. Mol Gen Genet. 1994;242:658–665. doi: 10.1007/BF00283420. [DOI] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad NK, Mili S, Marshall EL, Shu MD, Steitz JA. Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol Cell. 2006;24:943–953. doi: 10.1016/j.molcel.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Conrad NK, Shu MD, Uyhazi KE, Steitz JA. Mutational analysis of a viral RNA element that counteracts rapid RNA decay by interaction with the polyadenylate tail. Proc Natl Acad Sci USA. 2007;104:10412–10417. doi: 10.1073/pnas.0704187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad NK, Steitz JA. A Kaposi’s sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J. 2005;24:1831–1841. doi: 10.1038/sj.emboj.7600662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumesic PA, Madhani HD. The spliceosome as a transposon sensor. RNA Biol. 2013;10:1653–1660. doi: 10.4161/rna.26800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumesic PA, Natarajan P, Chen C, Drinnenberg IA, Schiller BJ, Thompson J, Moresco JJ, Yates JR, 3rd, Bartel DP, Madhani HD. Stalled spliceosomes are a signal for RNAi-mediated genome defense. Cell. 2013;152:957–968. doi: 10.1016/j.cell.2013.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann CR, Rammelt C, Wahle E. Control of poly(A) tail length. Wiley Interdiscip Rev RNA. 2011;2:348–361. doi: 10.1002/wrna.56. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Geisberg JV, Moqtaderi Z, Fan X, Ozsolak F, Struhl K. Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell. 2014;156:812–824. doi: 10.1016/j.cell.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua-Van A, Le Rouzic A, Maisonhaute C, Capy P. Abundance, distribution and dynamics of retrotransposable elements and transposons: similarities and differences. Cytogenet Genome Res. 2005;110:426–440. doi: 10.1159/000084975. [DOI] [PubMed] [Google Scholar]
- Huang CR, Burns KH, Boeke JD. Active transposition in genomes. Annu Rev Genet. 2012;46:651–675. doi: 10.1146/annurev-genet-110711-155616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Tanaka T, Barrero RA, Yamasaki C, Fujii Y, Hilton PB, Antonio BA, Aono H, Apweiler R, Bruskiewich R, Bureau T, Burr F, Costa de Oliveira A, Fuks G, Habara T, et al. Curated genome annotation of Oryza sativa ssp. japonica and comparative genome analysis with Arabidopsis thaliana. Genome Res. 2007;17:175–183. doi: 10.1101/gr.5509507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Kempken F, Windhofer F. The hAT family: a versatile transposon group common to plants, fungi, animals, and man. Chromosoma. 2001;110:1–9. doi: 10.1007/s004120000118. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Nott A, Moore MJ. How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci. 2003;28:215–220. doi: 10.1016/S0968-0004(03)00052-5. [DOI] [PubMed] [Google Scholar]
- Lee NN, Chalamcharla VR, Reyes-Turcu F, Mehta S, Zofall M, Balachandran V, Dhakshnamoorthy J, Taneja N, Yamanaka S, Zhou M, Grewal SI. Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance. Cell. 2013;155:1061–1074. doi: 10.1016/j.cell.2013.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitton-Fry RM, DeGregorio SJ, Wang J, Steitz TA, Steitz JA. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science. 2010;330:1244–1247. doi: 10.1126/science.1195858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Parker R. The yeast EDC1 mRNA undergoes deadenylation-independent decapping stimulated by Not2p, Not4p, and Not5p. EMBO J. 2005;24:1033–1045. doi: 10.1038/sj.emboj.7600560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Romanish MT, Mager DL. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet. 2012;46:21–42. doi: 10.1146/annurev-genet-110711-155621. [DOI] [PubMed] [Google Scholar]
- Schaack S, Gilbert C, Feschotte C. Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol Evol. 2010;25:537–546. doi: 10.1016/j.tree.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman AH. Retrotransposon replication in plants. Curr Opin Virol. 2013;3:604–614. doi: 10.1016/j.coviro.2013.08.009. [DOI] [PubMed] [Google Scholar]
- Smith AM, Hansey CN, Kaeppler SM. TCUP: a novel hAT transposon active in maize tissue culture. Front Plant Sci. 2012;3:6. doi: 10.3389/fpls.2012.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Heyne S, Richter AS, Will S, Backofen R. Freiburg RNA Tools: a web server integrating INTARNA, EXPARNA and LOCARNA. Nucleic Acids Res. 2010;38:W373–377. doi: 10.1093/nar/gkq316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabach Y, Billi AC, Hayes GD, Newman MA, Zuk O, Gabel H, Kamath R, Yacoby K, Chapman B, Garcia SM, Borowsky M, Kim JK, Ruvkun G. Identification of small RNA pathway genes using patterns of phylogenetic conservation and divergence. Nature. 2013;493:694–698. doi: 10.1038/nature11779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Borah S, Shi M, Steitz JA. Conservation of a triple-helix-forming RNA stability element in noncoding and genomic RNAs of diverse viruses. Cell Rep. 2012;2:26–32. doi: 10.1016/j.celrep.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Day N, Trifillis P, Kiledjian M. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol Cell Biol. 1999;19:4552–4560. doi: 10.1128/mcb.19.7.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26:2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CJ, Zhou JX, Liu J, Ma ZY, Zhang SW, Dou K, Huang HW, Cai T, Liu R, Zhu JK, He XJ. The splicing machinery promotes RNA-directed DNA methylation and transcriptional silencing in Arabidopsis. EMBO J. 2013;32:1128–1140. doi: 10.1038/emboj.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.