Abstract
Aim
We investigated the pathway of autophagy signaling linked to sarcopenia of mice.
Methods
Young adult (3‐month) and aged (24‐ month) C57BL/6J mice were used. Using real‐time PCR, Western blotting, and immunohistochemical microscopy, we evaluated the amounts of p62/SQSTM1, LC3, and Beclin‐1 in the quadriceps muscle change with aging in mice.
Results
Marked fiber atrophy (30%) and many fibers with central nuclei were observed in the aged mice. Western blotting using homogenate of the cytosolic fraction clearly showed that the amounts of p62/SQSTM1 and Beclin‐1 proteins were significantly increased in the aged skeletal muscle. The amounts of these proteins in both nuclear and membrane fractions did not change significantly with age. Immunofluorescence labeling indicated that aged mice more frequently possessed p62/SQSTM1‐positive fibers in the cytosol in quadriceps muscle than young ones (aged: 14% vs. young: 1%). In aged muscle, p62/SQSTM1‐positive fibers were significantly smaller than the surrounding p62/SQSTM1‐negative fibers. Aging did not elicit significant changes in the mRNA levels of p62/SQSTM1 and Beclin‐1, but decreased LC3 mRNA level. In aged muscle, the location of p62/SQSTM1 immunoreactivity was similar to that of Beclin‐1 protein, but not LC3.
Conclusion
Sarcopenia in mice appears to include a marked defect of autophagy signaling.
Keywords: Sarcopenia; p62/SQSTM1; LC3, Beclin‐1; Skeletal muscle; Atrophy
Introduction
Sarcopenia is a term that refers to a loss of skeletal muscle mass associated with a decrease in muscle strength and increased fatigability. Sarcopenia is the common denominator of the ageing process, responsible for a general and substantial decline in physical performance, which leads ultimately to physical disability. The aetiology of sarcopenia is complex and characterized by the contribution of multiple factors, such as physical activity, nutritional intake, oxidative stress, and hormonal changes.1, 2, 3
Age‐related muscle loss is a result of reductions in the size and number of muscle fibres possibly due to a prominent role of accelerated apoptosis.4 A most important factor in the development of muscle loss is the imbalance between protein synthesis and protein degradation.5 Skeletal muscle contains various proteolytic systems regulated by lysosome, calpain, and proteasome. Although evidence suggests that lysosomal degradation contributes to accelerated muscle proteolysis, such as in muscle disuse and denervation, the major pathway responsible for the degradation of contractile protein in skeletal muscle is orchestrated through the ubiquitin‐proteasome system (UPS).5, 6 In a variety of conditions, such as cancer, diabetes, denervation, uremia, sepsis, disuse, and fasting, skeletal muscle undergoes atrophy through the degradation of myofibrillar proteins via the UPS.7, 8, 9 Recent advances assert that muscle atrophy in these conditions shares a common mechanism in the induction of the muscle‐specific E3 ubiquitin ligase atrophy gene‐1 (atrogin‐1)/muscle atrophy F‐box and muscle ring‐finger protein 1 (MuRF1).6, 8, 10, 11 Therefore, it is believed that these atrogenes also contribute to the fibre degradation under normal ageing, in spite of the lack of direct evidence demonstrating this.12, 13, 14
There are contradictory data and varying interpretations in the literature on the effect of ageing on proteasome‐mediated protein degradation in skeletal muscle. Attaix, Combaret, and colleagues concluded in their reviews of the literature that it currently remains unclear where proteasome activity and proteasome function are increased or decreased in aged skeletal muscle.5, 7 Interestingly, recent findings indicate that atrogin‐1‐knockout mice are short‐lived and experience higher loss of muscle mass during ageing than control mice,15 indicating that the activity of E3 ubiquitin ligase is required to preserve muscle mass during ageing in mice. As indicated by Sandri et al.,15 chronic inhibition of these atrogenes should not be considered a therapeutic target to counteract sarcopenia because this does not prevent muscle loss but instead exacerbates weakness. Moreover, MuRF1 null mice experience higher decay of muscle strength during ageing than controls, although muscle mass is at least in part preserved in these mice.16 Collectively, these findings seem to indicate that the induction of UPS does not occur in mammalian sarcopenic muscle, and the attenuation of UPS by several interventions is not effective for preventing this symptom.
Autophagy is a vital cellular process by which intracellular components are degraded within lysosomes.17, 18 Three major mechanisms of autophagy have been described as follows: (i) microautophagy, in which lysosomes directly take up cytosol, inclusions, and organelles for degradation; (ii) chaperone‐mediated autophagy, in which soluble proteins with a particular pentapeptide motif are recognized and transported across the lysosomal membrane for degradation; and (iii) macroautophagy, in which a portion of cytoplasm, including subcellular organelles, is sequestered within a double membrane‐bound vacuole that ultimately fuses with a lysosome.19
A decline in autophagy during normal ageing has been described for invertebrates and higher organisms.20 Inefficient autophagy has been attributed a major role in the apparent age‐related accumulation of damaged cellular components, such as undegradable lysosome‐bound lipofuscin, protein aggregates, and damaged mitochondria.21 Interestingly, the degree of age‐related changes in autophagy appears to be organ‐specific. Some researchers demonstrated the upregulation of LC3‐II (active form) and p62/SQSTM1 in the sarcopenic muscles of mice.22 Demontis and Perrimon23 showed that the function of the autophagy/lysosome system of protein degradation declined during ageing in the skeletal muscle of Drosophila. This results in the progressive accumulation of polyubiquitin and p62/SQSTM1 protein aggregates in senescent Drosophila muscle. Importantly, to the best of our knowledge, changes of autophagy with age have been examined in mammalian skeletal muscle only by biochemical approaches (western blot using crude homogenates) but not morphological (e.g. immunofluorescence) ones. Therefore, it has remained to be elucidated where autophagy‐linked molecules are upregulated (i.e. cytosol, nucleus, or cell membrane) in sarcopenic muscle.
The purpose of this study was to determine whether autophagy‐linked molecules are upregulated in sarcopenic mouse muscle and where they are localized in muscle fibres.
Materials and methods
Experimental animals
Young adult (3 months; n = 10) and aged (24 months; n = 10) C57BL/6J mice were used in this study. The animals were housed in a temperature (22 ± 2°C) and humidity (60 ± 5%)‐controlled room regulated to provide alternating 12‐h periods of light and darkness and were allowed to feed (commercial chow) and drink ad libitum. Using excess pentobarbital, mice were killed and the quadriceps and triceps brachii muscles dissected. The muscles were quickly weighed, frozen in liquid nitrogen, and then stored at −80°C. This experimental procedure was approved by the Committee for Animal Research in Toyohashi University of Technology.
Primary antibodies
The antibodies employed in the present study were as follows: affinity‐purified mouse monoclonal antibody to pan‐cadherin (1:1500, Cat. No. ab6528‐100, Abcam Ltd, UK) and to HSP70 (1:2400, Cat. No., ab2787, Abcam Ltd, UK); affinity‐purified rabbit polyclonal antibody to p62/SQSTM1 (1:100–1:1000, Cat. No. M‐225, MBL, Nagoya, Japan), to Beclin‐1 (1:1000, Cat. No. #9315, Cell Signaling, Beverly, USA), to LC3 (1:100–1:1000, Cat. No. PM036, MBL, Nagoya, Japan), to ubiquitin (linkage‐specific K63, 1:2000, Cat. No., ab179434, Abcam Ltd, UK), and to lamin A/C (1:1000, Cat. No., #2032, Cell Signaling, Beverly, USA); and affinity‐purified goat polyclonal antibody to MAP‐LC3 (1:50, Cat. No. SC‐16756, Santa Cruz Biotechnology, Santa Cruz, CA, USA).
Tissue preparation, gel electrophoresis, and immunoblotting
The quadriceps muscle of both mice was homogenized in 10–20 volumes of 50 mM of Tris‐HCl pH 7.4, 5 mM of EDTA, 10 µg/mL of phenylmethylsulfonyl fluoride, 0.5 µg/mL of leupeptin, 0.2 µg/mL of aprotinin, 0.2% NP‐40, 0.1% Triton X‐100, and 1 mM of Na3VO4 in a polytron (DIAX 900, Heidolph‐Instruments, Schwabach, Germany) for 30 s. The homogenized tissues were then centrifuged at 15 000 × g for 25 min at 4°C, and the protein concentration of the supernatant was colorimetrically determined (Bio‐Rad protein determination kit, Bio‐Rad Laboratories, Richmond, CA, USA). Sodium dodecylsulfate‐polyacrylamide gel electrophoresis (10–15% acrylamide for detection of p62/SQSTM1, Beclin‐1, and LC3) and western blotting were performed next as described previously.24 Proteins separated by Sodium dodecylsulfate‐polyacrylamide gel electrophoresis were electrophoretically transferred onto nitrocellulose membranes (Hybond‐ECL Western, Amersham, Arlington Heights, IL, USA). The blots were then incubated with blocking buffer composed of 0.1% Tween‐20 and 1% gelatin in 10 mM of Tris‐buffered saline (10 mM of Tris, 135 mM of NaCl, 1 mM of KCl, and 0.02% NaN3, pH 7.4) for 10 min. The blots were next incubated with primary antibodies overnight, and with goat anti‐rabbit IgG ‐conjugated AP (1:10 000, Promega Co., Madison, WI, USA) for l h, and were visualized with Western blue, a stabilized substrate for alkaline phosphatase (Promega Co.). A densitometric analysis of each blot was performed with a computerized image processing system (NIH Image 1.63).
Subcellular fractionation
To compare the amount of LC3, p62/SQSTM1, and Beclin‐1 protein between young and aged quadriceps muscles in the cytosol, nucleus, and membrane, rapidly frozen muscle tissue (25–50 mg) was fractionated into cytosol, membrane, nucleus, and cytoskeleton using a Subcellular ProteoExtract proteome kit from Calbiochem (EMD Biosciences, La Jolla, CA, USA) according to the manufacturer's directions. We identified pure subcellular fractions using antibodies for heat shock protein 70 (HSP70), lamin A, and cadherin, each of which was selectively detected in cytosol, nucleus, and membrane fractions, respectively.
Immunofluorescence
Mouse quadriceps muscle was isolated and frozen in isopentane. Serial 7‐µm transverse sections made with a cryostat (Microm Coldtome, HM‐520, Germany) were mounted on silanized slides (Dako Japan, Tokyo). Cryosections were fixed with methanol (3 min) and incubated in blocking solution (2.5–10 % normal horse serum in phosphate buffered saline (PBS)) for 20 min at room temperature. Sections were incubated with polyclonal anti‐p62 and anti‐LC3 antibodies at 4°C overnight. After being washed in PBS, sections were incubated with anti‐rabbit fluorescein isothiocyanate (FITC)‐conjugated (1:100 final dilution; Rockland Immunochemicals, Inc., USA) and anti‐goat Rhodamine‐conjugated (1:100 final dilution; Chemicon International Inc., USA) secondary antibodies. The muscle sections were mounted in a slowfade antifade kit with 4′‐6‐diamidino‐2‐phenylindole (Vector Laboratories, Inc., Burlingame, CA, USA). Images were acquired on an Olympus BX50 inverted microscope with a fluorescent attachment (Olympus Japan, Inc.) and Photonic Science CCD camera (DP70, Olympus Japan, Inc.). As described previously,25 we calculated in percentages of myofibres possessing p62/SQSTM1 immunoreactivity in cytosol by analysing 500–700 fibres from 10–15 different parts of cross‐sections using six animals. We conducted the analysis of cross‐sectional area (CSA)‐showing p62‐positive and p62‐negative fibres in aged quadriceps muscles of mice by analyzing all measurable p62‐positive fibres and the surrounding p62‐negative fibres (100–120 numbers).
Real‐time PCR
Reverse transcription‐PCR was performed using total RNA samples obtained from muscle tissues as described previously.26 Real‐time PCR using the DNA‐binding dye SYBR Green was employed for the detection of PCR products for RNA sample obtained from cells after synthesized cDNA. The following PCR primers (Sigma‐Aldrich Japan, Hokkaido, Japan) were used: p62/SQSTM1, 5′‐TGTGGAACATGGAGGGAAGAG (forward) and 5′‐TGTGCCTGTGCTGGAACTTTC (reverse); MAP1lc3b (LC3B), 5′‐GACGGCTTCCTGTACATGGTTT (forward) and 5′‐TGGAGTCTTACACAGCCATTGC (reverse); Beclin‐1, 5′‐GTGCGCTACGCCCAGATC (forward) and 5′‐GATGTGGAAGGTGGCATTGAA (reverse); and GAPDH, 5′‐TGTGTCCGTCGTGGATCTGA (forward) and 5′‐CCTGCTTCACCACCTTCTTGA (reverse). The ratio of the other signals to that of GAPDH was calculated for every sample.
Statistical analysis
All values were expressed as means ± standard deviation. A one‐way analysis of variance was used to evaluate the significance of differences in morphometric parameter (the percentage of p62/SQSTM1‐positive fibres) and in the mRNA and protein levels of autophagy‐linking molecules in muscle. A Scheffés post hoc test was conducted if the analysis of variance indicated a significant difference. P < 0.05 was considered to be statistically significant.
Results
The body weight of the aged mice (31.9 ± 0.6 g) was 18.6% higher than that of the young adult mice (26.9 ± 0.3 g). The levels of ageing‐associated sarcopenia were similar in each of the skeletal muscles from aged mice compared with that of the young adults, as shown by the ~ 27% reduction in muscle wet weight with age. The degree of muscle loss was even more striking when muscle mass was normalized to body weight (milligram per gram body wt), which was 33.2–38.4% lower in aged mice than in young adult ones. As previously reported,27 the muscle of aged mice contained several atrophic, centrally nucleated fibres. The percentage (19.1 ± 1.5%) of muscle fibres with central nuclei was significantly increased in the aged quadriceps muscle. Compared with that of young mice, the mean fibre cross‐sectional area was significantly lower in the quadriceps muscle of aged mice.27
The expression of autophagy‐related genes was assessed in young (3 months old) and old (24 months old) mice (Figure 1). No significant differences were observed for p62/SQSTM1 or LC3B expression between young and old mice. We observed a significant age‐dependent decrease in Beclin‐1 mRNA in quadriceps muscle (P < 0.05).
Figure 1.
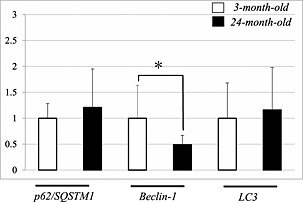
The mRNA expression levels of autophagy‐related genes in the quadriceps muscle of 3‐month‐old and 24‐month‐old mice. Significant age‐related decrease was observed in the amount of Beclin‐1 mRNA, but not in the other mRNAs. Values are means ± standard deviation (n = 8/group). The values are expressed as fold change relative to that in the 3‐month‐old mice, which was taken as 1.0.
We conducted control experiments with subcellular pure fractions using selective markers (HSP70, lamin A, and cadherin). We observed selective immunoblots of HSP70 in cytosol, lamin A in the nucleus, and cadherin in the membrane fraction in spite of minor HSP70 bands also being present in the membrane fraction (Figure 2A). We observed a prominent band of p62/SQSTM1 at 62 kDa using all of the fractions in mouse quadriceps muscles (Figure 2B). The cytosol fraction of aged mice possessed more abundant p62/SQSTM1 protein than that of young mice (Figure 2B). In contrast, we recognized similar p62/SQSTM1 bands in nuclear and membrane fractions between young and aged mouse muscles. Densitometric analysis showed a significant increase in the cytosol fraction in quadriceps muscle of aged mice, but not in the other two fractions, compared with young mice (Figure 2C).
Figure 2.
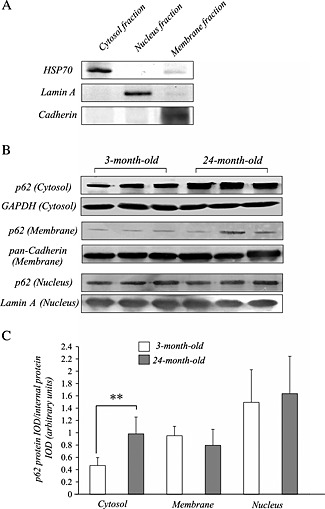
Control experiments with subcellular pure fractions using selective markers indicate selective immunoblots of HSP70 in cytosol, lamin A in the nucleus, and cadherin in the membrane fraction (A). HSP70 was also detected at a low level in the membrane fraction. Western blot analysis showed that p62/SQSTM1 protein was more abundantly expressed in the cytosolic fraction of the quadriceps muscle of aged mice than in that of young mice (B and C). No significant difference in the amount of p62/SQSTM1 in the nuclear and membrane fractions was observed in the quadriceps muscle between 3‐month‐old and 24‐month‐old mice (B and C). The integrated optical density of p62/SQSTM1 protein was normalized to the integrated optical density of each internal control (GAPDH for cytosol fraction, pan‐cadherin for membrane fraction, or lamin A protein for nucleus fraction) (arbitrary units). Values are means ± standard deviation (n = 6/group).
To obtain the reliability of using antibodies to LC3, we conducted the western blot to detect the LC3‐II bands. We succeeded to observe the apparent LC3‐I and LC3‐II bands of quadriceps muscle crude extracts (homogenates) from a 24‐h fasted mice (Figure 3A), in which autophagy‐dependent signalling is highly activated. In both young and aged mouse muscles, western blot analysis showed a prominent band of LC3 at 22 kDa (LC3‐I) but not 18 kDa (LC3‐II) in both cytosol and membrane fractions (Figure 3B). We did not observe any bands of LC3 using nuclear fractions of both muscle samples. The densitometric analysis showed no significant changes in the amount of LC3‐I protein with age (Figure 3C). Our western blot detects apparent bands of Beclin‐1. Similar to p62/SQSTM1, we observed an age‐related increase in Beclin‐1 protein in the cytosol fraction, but not in the two other fractions (Figure 3D and 3E).
Figure 3.
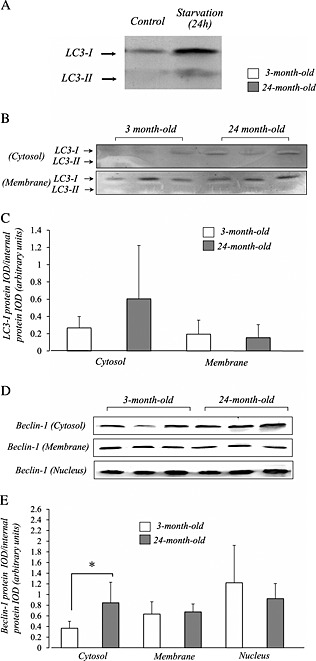
The LC3‐I and LC3‐II bands were detected in the quadriceps muscle crude extracts (homogenates) from a 24‐h fasted mice (A). Compared with 3‐month‐old mice, aged mice did not show a significant increase in LC3‐I protein content (B and C). We could not detect the active form (LC3‐II) in quadriceps in both 3‐month‐old and 24‐month‐old mice. Similar to p62/SQSTM1, a selective increase of Beclin‐1 protein was observed in the cytosol fraction of aged mice (C and D). The integrated optical density of LC3‐I and Beclin‐1 protein was normalized to the integrated optical density of each internal control (arbitrary units) (E). Values are means ± standard deviation (n = 6/group).
In both young and aged mouse muscles, western blot analysis showed several prominent bands of Lys63 polyubiquitinated proteins in both cytosol and membrane fractions (Figure 4A). We did not observe any bands of these proteins using nuclear fractions of both muscle samples. The densitometric analysis showed no significant changes in the amount of Lys63 polyubiquitinated proteins with age (Figure 4B).
Figure 4.
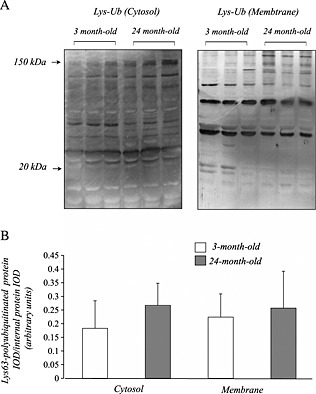
In both young and aged muscles of mice, several prominent bands of Lys63 polyubiquitinated proteins were detected in both cytosol and membrane fractions (A). The densitometric analysis showed no significant changes in the amount of Lys63 polyubiquitinated proteins in quadriceps muscle with age (B). The integrated optical density of Lys63 polyubiquitinated protein was normalized to the integrated optical density of each internal control (arbitrary units). Values are means ± standard deviation (n = 6/group).
To obtain a clear understanding of the cellular localization of p62/SQSTM1 and LC3 proteins in young and aged quadriceps muscles of mice, we performed immunofluorescence staining of single cryosections. p62/SQSTM1 and LC3 immunoreactivity was visualized using FITC‐conjugated or Rhodamine‐conjugated antibodies. In young quadriceps muscle, immunolabelling of p62/SQSTM1 was detected in the membrane of several muscle fibres, but not in the cytosol (Figure 5A). Apparent p62/SQSTM1 immunoreactivity was also observed in the membrane and cytosol of aged muscle fibres (Figure 5D). In contrast, the immunolabelling pattern of LC3 did not significantly differ between young and aged muscles (Figure 5B and 5E). We did not apparent p62/SQSTM1 and LC3 immunoreactivity in both young and aged muscles in the case of without primary antibodies. We conducted the analysis of CSA showing p62‐positive and p62‐negative fibres in aged quadriceps muscle of mice. We observed a significant smaller CSA of p62‐positive muscle fibres in aged muscle (Figure 5H).
Figure 5.
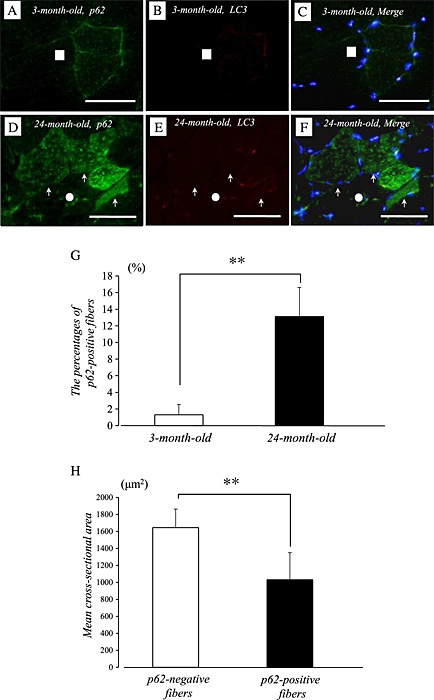
Serial cryosections of the quadriceps muscle of 3‐month‐old and 24‐month‐old mice. p62/SQSTM1 and LC3 immunoreactivity was visualized using FITC‐conjugated or Rhodamine‐conjugated antibodies. In young quadriceps muscle, immunofluorescence labelling showed that p62/SQSTM1 was present in the membrane and at a low level in the cytosol of several muscle fibres (A and C). Marked increases of p62/SQSTM1 immunoreactivity were observed in the membrane and the cytosol of aged muscle fibres (D). No apparent difference in LC3 immunoreactivity was observed in the muscle between 3‐month‐old and 24‐month‐old mice (B and E). p62/SQSTM1‐positive muscle fibres exhibit significant smaller cross‐sectional area than p62/SQSTM1‐negative fibres in the muscle of aged mice (H). White circles and squares indicate the same fibres on different immuno images (A–F). White arrows denote the muscle fibres possessing p62/SQSTM1. Bar = 50 µm.
Discussion
Two main conclusions can be derived from the findings of the present study. First, there was a marked increase in the amount of several autophagy‐regulated molecules (p62/SQSTM1 and Beclin‐1) in the quadriceps muscles of mice with ageing. Second, our immunofluorescence and quantitative analyses showed that many muscle fibres possess p62/SQSTM1 immunoreactivity in the cytosol of aged mice.
It is proposed that the autophagy pathway is of potential importance for protein degradation by lysosome.28, 29 Autophagy is a highly regulated process for the bulk degradation of cytoplasmic components including protein aggregates and organelles. Autophagy has been implicated in cell death simultaneously or independently of apoptosis. Skeletal muscle is one of the tissues where autophagy has been detected in specific disorders such as Danon and Pompe disease as well as X‐linked congenital autophagic vacuolar myopathy.30 More recently, autophagy‐linked molecules such as LC3 and Beclin‐1 have been shown to be upregulated in skeletal muscle under denervation,31 starvation,32 and cachexia.33 In addition, one recent review has proposed a possible linkage of autophagy signalling to the fibre atrophy of sarcopenia.34 Although some researchers found the upregulation of LC3 protein22 in aged muscle by western blot using crude homogenates, no descriptive evidence using several analyses has shown the enhanced expression of autophagy‐linked molecules with age. To our knowledge, this is the first study to involve a systematic investigation of autophagy signalling of skeletal muscle in aged animals using several different approaches (real‐time PCR, western blot for fractionated homogenates, and immunofluorescence). Our present study demonstrated that the localization pattern of p62/SQSTM1 markedly differs from that of LC3 in aged quadriceps muscle. One interpretation of the selective increase of p62/SQSTM1 protein in fibre cytosol is an autophagic defect in muscle during ageing. Indeed, cytosol accumulation of p62/SQSTM1 is observed in muscle fibres of sporadic inclusion‐body myositis,35 in knockout mice of Vps15, class III phosphatidylinositol 3‐kinase,36 and in mdx mice,37 a Duchenne muscular dystrophy (DMD) model. All of these mice possess apparent autophagic defect.
We report here that the protein expression of Beclin‐1 was increased in the aged animals, which is in agreement with a previous study conducted in rat heart.38 The roles of Beclin‐1 are manifold,39 and it was first described as a tumour suppressor. With regard to autophagy in muscle adaptation, O'Leary and Hood31, 40 demonstrated that the amounts of Beclin‐1 and LC3‐II proteins were markedly elevated in rat hindlimb muscle following 7 days of denervation. Mouse tibialis anterior muscle has also been shown to have elevated mRNA expression of autophagy‐related genes including LC3, Gabarapl1, PI3KIII, Ulk2, Atg12I, Atg4b, and Beclin‐1 after 3 or 7 days of denervation.41 Ogata et al. 32 have reported increases in LC3‐II and p62/SQSTM1 in soleus and plantaris muscles of rats following 1–3 days of fasting. Some controversy exists as to the adaptive changes in mRNA expression of autophagy‐related molecules with ageing. The upregulation of p62/SQSTM1 and Beclin‐1 mRNA could not be detected by our real‐time PCR analysis. Although rapid induction of these molecules has been observed in skeletal muscle subjected to denervation,31 and starvation,32 protein degradation under sarcopenia occurs more slowly. In addition, our analysis using subcellular‐fractionated homogenates clearly demonstrated that the abundance of these autophagy‐related molecules occurred only in the cytosol fraction, not in the membrane and nuclear fractions, of sarcopenic muscle.
We failed to detect marked increases in LC3‐I and LC3‐II (active form) proteins in aged quadriceps muscle. This result clearly differs from the finding of Wenz et al., who demonstrated marked upregulation of LC3‐I and LC3‐II proteins in biceps femoris muscle.22 They utilized C57BL6 mice similar to our group, but these mice were a little younger (22 months old) than those of utilized in our study (24 months old). Although we cannot provide an accurate mechanism explaining these results, there may be several reasons for them. One possibility is the difference of the analysed muscle (biceps femoris vs. quadriceps). Recently, Iida et al. 42 demonstrated different manners of adaptation for LC3‐I and LC3‐II proteins among several muscles using klotho mutant mice, a type of short‐lived mouse models displaying several ageing‐related phenotypes. They indicated marked increases of the ratios of LC3‐II to LC3‐I in masseter and tongue muscles but not hindlimb gastrocnemius muscle in klotho mice. A second possibility is the difference in the manner of analysis. We used fractionated homogenate, whereas Wenz et al. 22 used crude homogenate of total muscle. Because we did not determine the amount of autophagy‐related proteins in the cytoskeletal fraction, LC3‐I and LC3‐II proteins may exist in this fraction in sarcopenic muscle.
Other candidates may regulate the localization of p62/SQSTM1 and Beclin‐1 in the quadriceps muscle of aged mice. At the M‐band, the mechanically modulated kinase of titin interacts with a complex of NBR1, p62/SQSTM1, and MuRFs.43, 44 This complex dissociates under mechanical arrest, and MuRF1 and MuRF2 translocate to the nucleus.43, 45 p62/SQSTM1 was also shown to translocate together with MuRF1 and MuRF2 to the nucleus in mechanically arrested skeletal muscles.45 However, it is not considered that MuRF1 and MuRF2 accumulate in both cell cytosol and/or myonucleus in sarcopenic muscle, because several researchers15, 16 have recently indicated little possibility of age‐related upregulation of E3‐ubiquitin ligases (atrogin‐1, MuRF1, etc.) in skeletal muscle. Intriguingly, both p62/SQSTM1 and NBR1 are similarly and prominently accumulated in cell cytosol in muscle fibres of sporadic inclusion‐body myositis.35, 46 In sarcopenic muscle fibre, the localization of MuRF may not be regulated by the MuRF‐linked molecules (p62/SQSTM1 and/or NBR1).
It is believed that E3 ubiquitin ligases (atrogin‐1 and MuRF1) contribute to the fibre degradation under normal ageing, in spite of the lack of direct evidence demonstrating this.12, 13, 14 However, recent findings indicate that atrogin‐1‐knockout mice are short‐lived and experience higher loss of muscle mass during ageing than control mice.15 In addition, MuRF1 null mice experience higher decay of muscle strength during ageing than controls.16 As indicated by Sandri et al.,15 chronic inhibition of these atrogenes should not be considered a therapeutic target to counteract sarcopenia. Demontis and Perrimon23 showed that the function of the autophagy/lysosome system of protein degradation declined during ageing in the skeletal muscle of Drosophila. This results in the progressive accumulation of polyubiquitin protein aggregates in senescent Drosophila muscle, although our analysis by western blot did not exhibit significant increase in the Lys63 polyubiquitinated proteins in both cytosol and membrane fractions in aged mice muscles (Figure 4). Our present study demonstrated the selective induction of p62/SQSTM1 and Beclin‐1 but not LC3 in the cytosol of sarcopenic muscle fibres. The selective accumulation of p62/SQSTM1 in skeletal muscle seems to indicate an autophagic defect. Indeed, more recent study47 using biopsy samples of young and aged human volunteers clearly showed the age‐dependent autophagic defect such as the decrease in the amount of Atg7 protein and in the ratio of LC3‐II/LC3‐I proteins. Collectively, sarcopenia appears to include a partial defect of autophagy signalling.
The results presented in this paper highlight the adaptive changes in autophagy‐linked molecules in sarcopenic muscle of mice. Autophagy signalling may regulate protein degradation in aged muscle. More work is required to define the functional role of upregulated p62/SQSTM1 and Beclin‐1 protein in aged muscle.
Funding
This work was supported by a research Grant‐in‐Aid for Scientific Research C (No. 26350815) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest
None declared.
Acknowledgements
All authors of this manuscript have complied with the guidelines of ethical authorship and publishing as stated in the Journal of Cachexia, Sarcopenia, and Muscle 2010; 1:7–8 (von Haehling S, Morley JE, Coats AJ, and Anker SD).
Sakuma, K. , Kinoshita, M. , Ito, Y. , Aizawa, M. , Aoi, W. , and Yamaguchi, A. (2016) p62/SQSTM1 but not LC3 is accumulated in sarcopenic muscle of mice. Journal of Cachexia, Sarcopenia and Muscle, 7: 204–212. doi: 10.1002/jcsm.12045.
References
- 1. Aoi W, Sakuma K. Oxidative stress and skeletal muscle dysfunction with aging. Curr Aging Sci. 2011; 4: 101–109. [DOI] [PubMed] [Google Scholar]
- 2. Roubenoff R, Hughes VA. Sarcopenia: current concepts. J Gerontol A Biol Sci Med Sci 2000; 55: M716–M724. [DOI] [PubMed] [Google Scholar]
- 3. Sakuma K, Yamaguchi A. Molecular mechanisms in aging and current strategies to counteract sarcopenia. Curr Aging Sci 2010; 3: 90–101. [DOI] [PubMed] [Google Scholar]
- 4. Marzetti E, Calvani R, Cesari M, Buford TW, Lorenzi M, Behnke BJ, et al Mitochondrial dysfunction and sarcopenia of aging: from signaling pathways to clinical trials. Int J Biochem Cell Biol 2013; 45: 2288–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Combaret L, Dardevet D, Béchet D, Taillandier D, Mosoni L, Attaix D. Skeletal muscle proteolysis in aging. Curr Opin Clin Nutr Metab Care 2009; 12: 37–41. [DOI] [PubMed] [Google Scholar]
- 6. Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, et al Foxo transcription factors induce the atrophy‐related ubiquitin ligase atrogin‐1 and cause skeletal muscle atrophy. Cell 2004; 117: 399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Attaix D, Aurousseau E, Combaret L, Kee A, Larbaud D, Rallière C, et al Ubiquitin‐proteasome‐dependent proteolysis in skeletal muscle. Reprod Nut Dev 1998; 38: 153–165. [DOI] [PubMed] [Google Scholar]
- 8. Itakura E, Mizushima N. p62 targeting to the autophagosome formation site requires self‐oligomerization but not LC3 binding. J Cell Biol 2011; 192: 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mitch WE, Goldberg AL. Mechanisms of muscle wasting: the role of ubiquitin‐proteasome pathway. N Engl J Med 1996; 335: 1897–1905. [DOI] [PubMed] [Google Scholar]
- 10. Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, et al Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 2001; 294: 1704–1708. [DOI] [PubMed] [Google Scholar]
- 11. Cai D, Frantz JD, Tawa NE Jr, Melendez PA, Oh BC, Lidov HG, et al IKKbeta/NF‐kappaB activation causes severe muscle wasting in mice. Cell 2004; 119: 285–298. [DOI] [PubMed] [Google Scholar]
- 12. Sakuma K, Yamaguchi A. Sarcopenia: molecular mechanisms and current therapeutic strategy In Cell Aging. NY: Nova Science Publisher; 2011. p93–152. [Google Scholar]
- 13. Sakuma K, Aoi W, Yamaguchi A. Current understanding of sarcopenia: possible candidates modulating muscle mass. Pflugers Arch 2015; 467: 213–229. [DOI] [PubMed] [Google Scholar]
- 14. Sakuma K, Aoi W, Yamaguchi A. The intriguing regulators of muscle mass in sarcopenia and muscular dystrophy. Front Aging Neurosci 2014; 6: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sandri M, Barberi L, Bijlsma AY, Blaauw B, Dyar KA, Milan G, et al Signalling pathways regulating muscle mass in ageing skeletal muscle: the role of the IGF1‐Akt‐mTOR‐FoxO pathway. Biogerontology 2013; 14: 303–323. [DOI] [PubMed] [Google Scholar]
- 16. Hwee DT, Baehr LM, Philp A, Baar K, Bodine SC. Maintenance of muscle mass and load‐induced growth in Muscle RING Finger 1 null mice with age. Aging Cell 2014; 13: 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meijer AJ, Codogno P. Regulation and role of autophagy in mammalian cell. Int J Biochem Cell Biol 2004; 36: 2445–2462. [DOI] [PubMed] [Google Scholar]
- 18. Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol 2007; 9: 1102–1109. [DOI] [PubMed] [Google Scholar]
- 19. Wang CW, Klionsky DJ. The molecular mechanism of autophagy. Mol Med 2003; 9: 65–76. [PMC free article] [PubMed] [Google Scholar]
- 20. Cuervo AM, Bergamini E, Brunk UT, Dröge W, Ffrench M, Terman A. Autophagy and aging: the importance of maintaining “clean” cells. Autophagy 2005; 1: 131–140. [DOI] [PubMed] [Google Scholar]
- 21. Terman A, Brunk UT. Oxidative stress, accumulation of biological ‘garbage’, and aging. Antioxid Redox Signal 2006; 8: 197–204. [DOI] [PubMed] [Google Scholar]
- 22. Wenz T, Rossi SG, Rotundo RL, Spiegelman BM, Moraes CT. Increased muscle PGC‐1α expression protects from sarcopenia and metabolic disease during aging. Proc Natl Acad Sci U S A 2009; 106: 20405–20410. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23. Demontis F, Perrimon N. FOXO/4E‐BP signaling in Drosophila muscles regulates organism‐wide proteostasis during aging. Cell 2010; 143: 813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sakuma K, Akiho M, Nakashima H, Nakao R, Hirata M, Inashima S, et al Cyclosporin A modulates cellular localization of MEF2C protein and blocks fiber hypertrophy in the overloaded soleus muscle of mice. Acta Neuropathol (Berl) 2008; 115: 663–674. [DOI] [PubMed] [Google Scholar]
- 25. Akiho M, Nakashima H, Sakata M, Yamasa Y, Yamaguchi A, Sakuma K. Expression profile of Notch‐1 in mechanically overloaded plantaris muscle of mice. Life Sci 2010; 86: 59–65. [DOI] [PubMed] [Google Scholar]
- 26. Aoi W, Naito Y, Mizushima K, Takanami Y, Kawai Y, Ichikawa H, et al The microRNA regulates PGC‐1α in mouse skeletal muscle in response to physical activity. Am J Physiol Endocrinol Metab 2010; 298: E799–E806. [DOI] [PubMed] [Google Scholar]
- 27. Sakuma K, Akiho M, Nakashima H, Akima H, Yasuhara M. Age‐related reductions in expression of serum response factor and myocardin‐related transcription factor A in mouse skeletal muscles. Biochim Biophys Acta 2008; 1782: 453–461. [DOI] [PubMed] [Google Scholar]
- 28. Sandri M. Autophagy in skeletal muscle. FEBS Lett 2010; 584: 1411–1416. [DOI] [PubMed] [Google Scholar]
- 29. Sandri M. Autophagy in health and disease. 3. Involvement of autophagy in muscle atrophy. Am J Physiol Cell Physiol 2010; 298: C1291–C1297. [DOI] [PubMed] [Google Scholar]
- 30. Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 2008; 132: 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. O'Leary MF, Hood DA. Denervation‐induced oxidative stress and autophagy signaling in muscle. Autophagy 2009; 5: 230–231. [DOI] [PubMed] [Google Scholar]
- 32. Ogata T, Oishi Y, Higuchi M, Muraoka I. Fasting‐related autophagic response in slow‐ and fast‐twitch skeletal muscle. Biochem Biophys Res Commun 2010; 394: 136–140. [DOI] [PubMed] [Google Scholar]
- 33. McClung JM, Judge AR, Powers SK, Yan Z. p38 MAPK links oxidative stress to autophagy‐related gene expression in cachectic muscle wasting. Am J Physiol Cell Physiol 2010; 298: C542–C549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rajawat YS, Hilioti Z, Bossis I. Aging: central role for autophagy and the lysosomal degradative system. Ageing Res Rev 2009; 8: 199–213. [DOI] [PubMed] [Google Scholar]
- 35. Nogalska A, Terracciano C, D'Agostino C, Engel WK, Askanas V. p62/SQSTM1 is overexpressed and prominently accumulated in inclusions of sporadic inclusion‐body myositis muscle fibers, and can help differentiating it from polymyositis and dermatomyositis. Acta Neuropathol 2009; 118: 407–413. [DOI] [PubMed] [Google Scholar]
- 36. Nemazanyy I, Blaauw B, Paolini C, Caillaud C, Protasi F, Mueller A, et al Defects of Vps15 in skeletal muscles lead to autophagic vacuolar myopathy and lysosomal disease. EMBO Mol Med 2013; 5: 870–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. De Palma C, Morisi F, Cheli S, Pambianco S, Cappello V, Vezzoli M, et al Autophagy as a new therapeutic target in Duchenne muscular dystrophy. Cell Death Dis 2012; 3: e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wohlgemuth SE, Seo AY, Marzetti E, Lees HA, Leeuwenburgh C. Skeletal muscle autophagy and apoptosis during aging: effects of calorie restriction and life‐long exercise. Exp Gerontol 2010; 45: 138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cao Y, Klionsky DJ. Physiological functions of Atg6/Beclin 1: a unique autophagy‐related protein. Cell Res 2007; 17: 839–849. [DOI] [PubMed] [Google Scholar]
- 40. O'Leary MF, Hood DA. Effect of prior chronic contractile activity on mitochondrial function and apoptotic protein expression in denervated muscle. J Appl Physiol 2008; 105: 114–120. [DOI] [PubMed] [Google Scholar]
- 41. Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, et al FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab 2007; 6: 472–483. [DOI] [PubMed] [Google Scholar]
- 42. Iida R, Kanko S, Suga T, Morito M, Yamane A. Autophagic‐lysosomal pathway functions in the masseter and tongue muscles in the klotho mouse, a mouse model for aging. Mol Cell Biochem 2011; 348: 89–98. [DOI] [PubMed] [Google Scholar]
- 43. Lange S, Xiang F, Yakovenko A, Vihola A, Hackman P, Rostkova E, et al The kinase domain of titin controls muscle gene expression and protein turnover. Science 2005; 308: 1599–1603. [DOI] [PubMed] [Google Scholar]
- 44. Puchner EM, Alexandrovich A, Kho AL, Hensen U, Schäfer LV, Brandmeier B, et al Mechanoenzymatics of titin kinase. Proc Natl Acad Sci U S A 2008; 105: 13385–13390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ochala J, Gustafson AM, Diez ML, Renaud G, Li M, Aare S, et al Preferential skeletal muscle myosin loss in response to mechanical silencing in a novel rat intensive care unit model: underlying mechanisms. J Physiol 2011; 589: 2007–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. D'Agostino C, Nogalska A, Cacciottolo M, Engel WK, Askanas V. Abnormalities of NBR1, a novel autophagy‐associated protein, in muscle fibers of sporadic inclusion‐body myositis. Acta Neuropahol (Berl) 2011; 122: 627–636. [DOI] [PubMed] [Google Scholar]
- 47. Carnio S, LoVerso F, Baraibar MA, Longa E, Khan MM, Maffei M, et al Autophagy impairment in muscle induces neuromuscular junction degeneration and precocious aging. Cell Reports 2014; 8: 1509–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]


