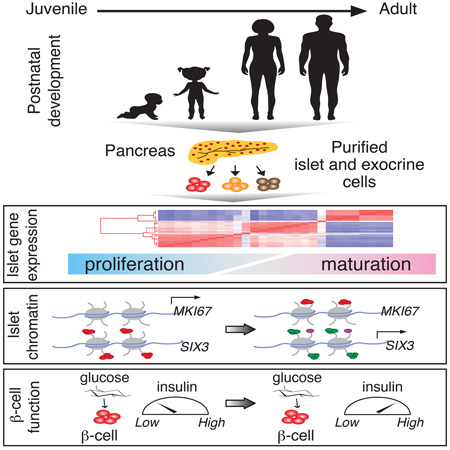
SUMMARY
Intensive efforts are focused on identifying regulators of human pancreatic islet cell growth and maturation to accelerate development of therapies for diabetes. After birth, islet cell growth and function are dynamically regulated; however establishing these age-dependent changes in humans has been challenging. Here we describe a multimodal strategy for isolating pancreatic endocrine and exocrine cells from children and adults to identify age-dependent gene expression and chromatin changes on a genomic scale. These profiles revealed distinct proliferative and functional states of islet α-cells or β-cells, and histone modifications underlying age-dependent gene expression changes. Expression of SIX2 and SIX3, transcription factors without prior known functions in the pancreas and linked to fasting hyperglycemia risk, increased with age specifically in human islet β-cells. SIX2 and SIX3 were sufficient to enhance insulin content or secretion in immature β-cells. Our work provides a unique resource to study human-specific regulators of islet cell maturation and function.
Graphical abstract
INTRODUCTION
The pancreas is a vital compound organ with an exocrine component comprised of acinar and duct cells that aid with digestion of nutrients, and an endocrine component which has hormone producing cells organized into structures called Islet of Langerhans, that regulate metabolism of glucose and other macromolecules (Arda et al., 2013; Benitez et al., 2012). Pancreatic islet β-cells are the sole source of insulin in the body; loss or impairment of pancreatic islet β-cells is a common cause of diabetes mellitus. Thus, the goals of human β-cell replacement therapies for type 1 diabetes (T1DM) or β-cell functional restoration in type 2 diabetes (T2DM) motivate global efforts to identify factors that regulate and stimulate β-cell insulin production and secretion.
After birth, pancreatic islet cells proliferate, expand and mature (Chen et al., 2011; Goodyer et al., 2012; Gregg et al., 2012; Meier et al., 2008), providing a physiological setting to study the genetic mechanisms underlying these crucial changes. Postnatal β-cell proliferation drives the expansion of β-cell mass, a feature that may influence T2DM risk (Butler et al., 2007). Although several cell cycle genes promoting or inhibiting human β-cell proliferation have been identified, the genetic programs controlling postnatal β-cell expansion remain poorly understood (Chen et al., 2011; Fiaschi-Taesch et al., 2009; Goodyer et al., 2012).
Glucose-stimulated insulin secretion is a hallmark feature of functional β-cells that may also change after birth (reviewed in Benitez et al., 2012). Prior work in rodents showed that, compared to adult islets, neonatal islets have blunted insulin secretion in response to glucose or other secretagogues (Aguayo-Mazzucato et al., 2011; Avrahami et al., 2015). Likewise, studies with human fetal β-cells demonstrated that glucose-stimulated insulin secretion was impaired (Otonkoski et al., 1988; Rorsman et al., 1989). While molecular analysis suggested involvement of different mechanisms including expression levels of specific transcription factors, Glucokinase and KATP channel activity (Jermendy et al., 2011; Rorsman et al., 1989; Taniguchi et al., 2000), systematic analysis of gene expression on a genomic scale during human islet cell maturation has not been reported. Moreover, assessment of functions like insulin secretion has not been systematically compared in islets from children and adults.
Regulation of gene expression is governed by several levels of molecular control, including chromatin structure and histone modifications. Studies by the ENCODE consortium and others have shown that chromatin modifications can modulate the activity or silencing of gene transcripts (The ENCODE Project, 2012). Elucidation of genome-wide distribution of histone marks can provide mechanistic insights into the regulation of gene expression including the pancreas (Bramswig et al., 2013; Pasquali et al., 2014; Zhou et al., 2013). However, the delineation of chromatin states in the context of human postnatal islet development from purified cells has not been achieved.
Here we describe a systems approach to identify age-dependent gene expression programs in human islet cells that includes (1) procurement of pancreatic tissue from children and adults, (2) developing robust and reliable cell purification methods, (3) generation of comprehensive transcriptome and histone modification maps, and (4) systematic assays of islet physiology and function. Our work generated datasets identifying several hundred genes whose expression changed with age in human islet cells, including regulators of human postnatal islet development and function.
RESULTS
Purification of pancreatic cells from juvenile and adult donors
To measure pancreatic gene expression changes with age in humans, we established an infrastructure involving a network of collaborators, which includes organ procurement organizations and islet isolation centers to obtain and isolate islets from human donors in a timely manner, including donors of young age. For this study, we targeted a rare set of juvenile donors under 9 years of age, in whom islet β-cells are expanding, permitting studies of human β-cell proliferation and function. In addition, we procured both juvenile and adult human pancreata with uniform, strict criteria, crucial for optimizing the quality and reliability of gene expression analysis (see Experimental Procedures).
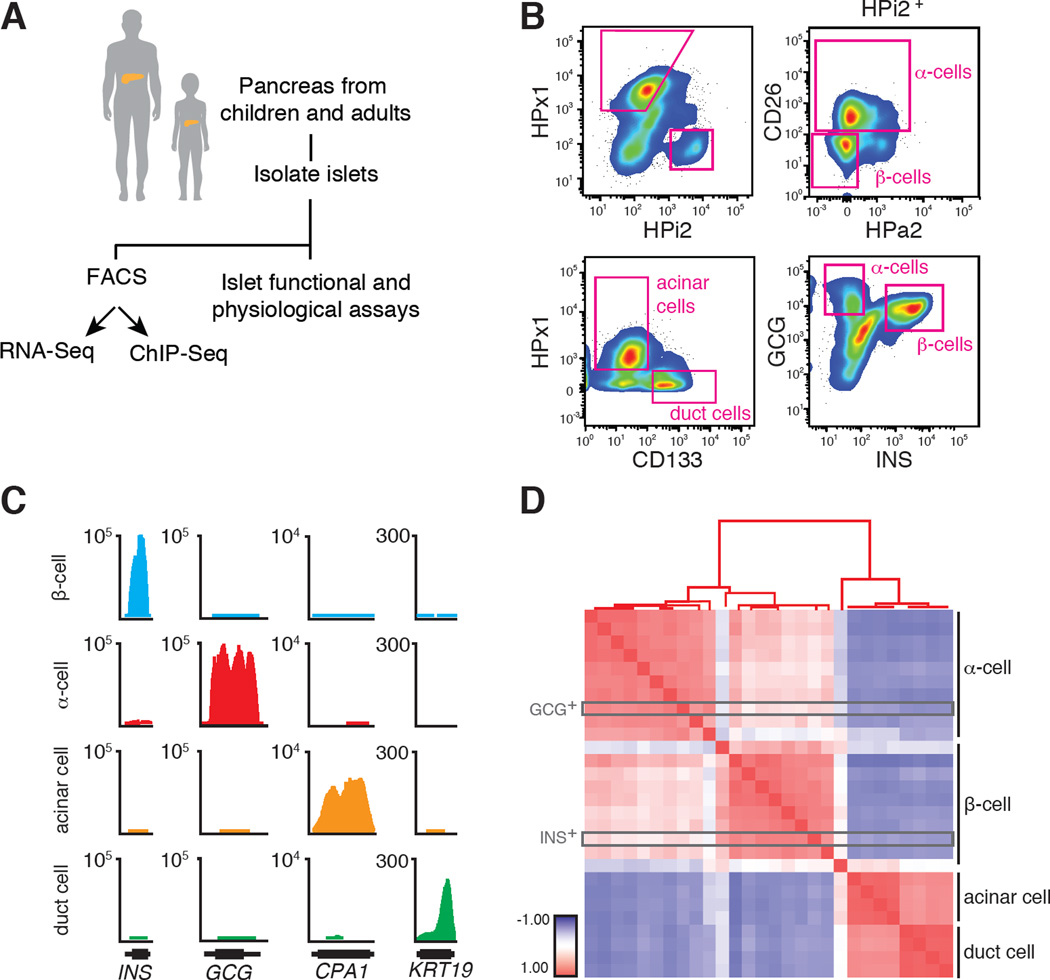
Pancreatic cells were initially separated by fluorescence activated cell sorting (FACS) into endocrine (HPi2) and exocrine (HPx1) populations (Dorrell et al., 2008, 2011) (Figures 1A–B and S1A). Unlike sorting of adult islet α-cells, however, juvenile α-cells were not separated from β-cells by existing FACS strategies using the HPa2 antibody (Bramswig et al., 2013; Dorrell et al., 2008) (Figure S1B). Prior work suggested that cluster of differentiation antigen 26 (CD26, also known as DPP4) is expressed in human α-cells (Dorrell et al., 2011), and FACS analysis revealed that juvenile or adult HPi2pos islet cells from multiple donors can be successfully separated into α-cell-enriched and β-cell-enriched fractions using anti-CD26 antibody (Figures 1B and S1A). In addition, we purified CD133pos duct cells and HPx1pos acinar cells (Dorrell et al., 2008; Sugiyama et al., 2007) (Figures 1B and S1A). We used quantitative RT-PCR (qPCR; Figure S1C) to confirm enrichment or depletion of expected marker genes for β-cells (INS), α-cells (GCG), δ-cells (SST), acinar (CPA1) and duct cells (KRT19). We also performed intracellular sorting with GLUCAGON- and INSULIN-specific antibodies (Hrvatin et al., 2014) to isolate juvenile α- and β-cells (Figure 1B), and found that the molecular signatures of GCGpos and INSpos cells matched those of cells purified with surface antigens (Figure 1D, also see below). Thus, our FACS strategies generated highly enriched endocrine and exocrine pancreatic cell subsets from children and adult donors.
Figure 1. Cell purification approach to isolate native juvenile and adult islet cell subpopulations.
(A) Schematic summary of this study design. (B) FACS plots showing the distribution of distinct pancreatic cell populations. (C) RNA-Seq tracks representing the enrichment of transcript reads in GCG, INS, CPA1 and KRT19 loci. Gene model (black, 5’ to 3’) excludes introns. (D) Pearson Correlation Coefficient matrix of all RNA-Seq samples used in this study. +1 indicates perfect correlation (red), 0 indicates no correlation (white), −1 indicates anti-correlation (blue). The positions of intracellular sorted juvenile samples are boxed in grey.
High-depth transcriptome maps of human juvenile and adult pancreatic cells
To obtain comprehensive gene expression profiles, we performed high-throughput RNA sequencing (RNA-Seq), yielding 28 expression libraries (Table S1). An average of approximately 130 million paired-end sequences per sample uniquely mapped to the human genome, with a total of >3.5 billion sequence reads (Table S1). Consistent with our initial qPCR assessment (Figure S1C), RNA-Seq transcript counts and abundance matched the sorted cell type (Figure 1C). Pearson Correlation Analysis (de Hoon et al., 2004), which measures overall similarity between samples, followed by unsupervised hierarchical clustering of these 28 RNA-Seq samples showed high correlation within each cell type. This included appropriate clustering of GCGpos-cells with HPi2pos HPa2pos CD26pos α-cells and INSpos-cells with HPi2pos HPa2neg CD26neg β-cells, as well as distinct clustering of acinar cells and primary pancreatic duct cells (Figure 1D). A separate correlation analysis demonstrated that our RNA-Seq data were well-matched to recent transcriptome profiling of adult human acinar and β-cells (Morán et al., 2012) (Figure S1D). Thus, our cell purification strategy generated high-quality, age-dependent gene expression profiles of human pancreatic endocrine and exocrine cells.
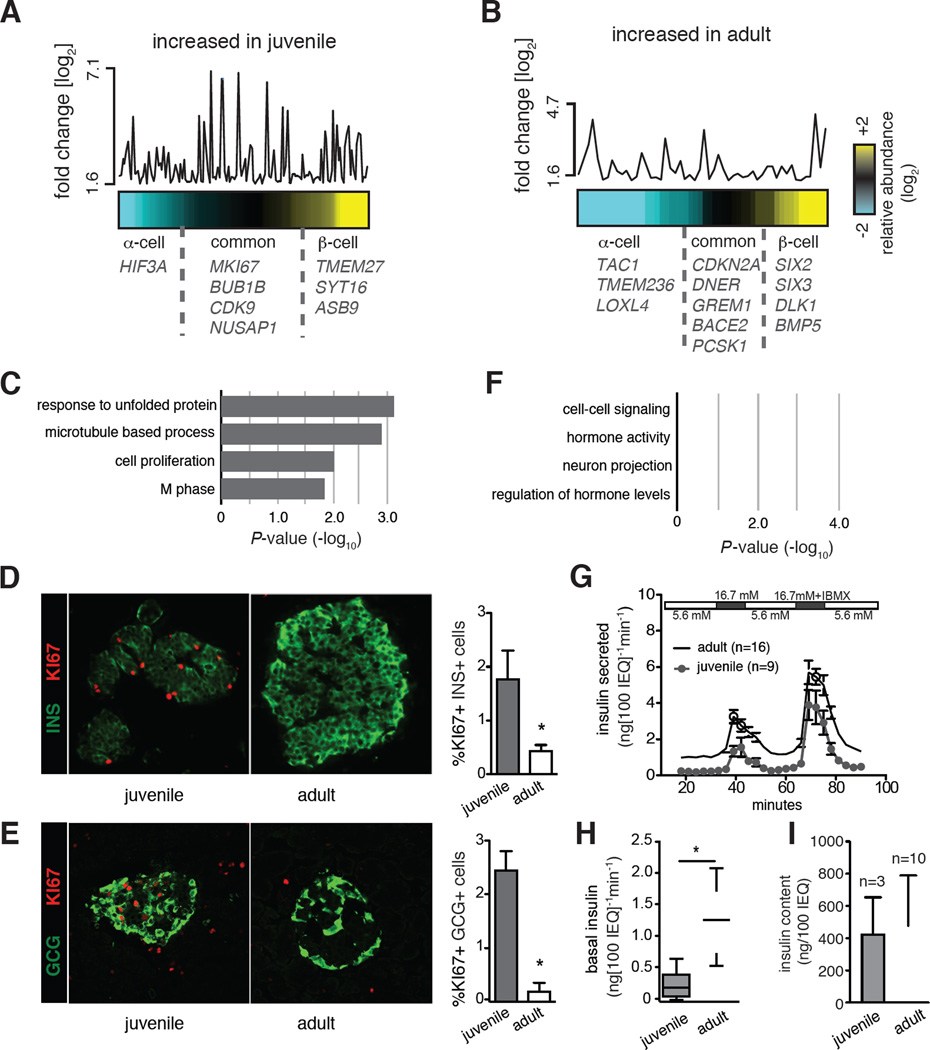
Age-dependent changes of cell growth, fate and function in humans are determined by intrinsic and extrinsic factors that vary greatly between individuals. Thus, as expected, we found the dispersion (a statistical measure to estimate variance; Anders and Huber, 2010) of transcript counts by age to be high. To gain additional statistical power, we combined α- and β-cell data sets into ‘juvenile’ (< 9 years) or ‘adult’ (> 28 years) age groups. Using the DE-Seq algorithm (Anders and Huber, 2010) we identified more than 500 genes whose expression changed significantly in α- or β-cells with age (fold change > 1.5, FDR 0.2, Table S2). We computed the abundance of age-dependent transcripts in α- and β-cells for each age category and represented this as heat maps (Figures 2A and 2B). We observed a subset of genes increased in juveniles (Figure 2A) or adults (Figure 2B) that were enriched in either α- or β-cells. However, the majority of genes differentially expressed with age were shared between α- and β-cells, consistent with the view that genetic programs common to both cells regulate postnatal development (Bramswig et al., 2013). For instance, we found that CDKN2A (p16) and CDKN2B (p15), cyclin dependent kinase inhibitors with functions in cellular senescence and organismal aging (López-Otín et al., 2013), were increased in both adult α- and β-cells. Thus, our bioinformatics analysis successfully identified age-dependent gene expression changes in human islet cells.
Figure 2. Age-dependent gene expression and functional changes in human islet cells.
Heat maps representing the relative abundance of age-dependent genes increased either in juvenile (A) or adult (B) samples. The y-axis shows the fold change of expression between different age groups. Each point corresponds to an age-dependent gene and its position is aligned with the column position in the heat map. Age-dependent genes more abundant in β-cells are colored yellow, in α-cells colored cyan, equal abundance is represented in black. Names of select genes are indicated on the plots. For the full list, see Table S2. (c) GO Term enrichment analysis of genes increased in juvenile (C) or adult (F) samples. Top scoring biological process terms are graphed. Immunostaining for KI67 and (D) Insulin (INS) or (E) Glucagon (GCG) in juvenile (2-year-old) and adult (31-year-old) pancreatic sections. Quantifications on the right (* t-test P < 0.05). Error bars indicate S.D. (G) Dynamic GSIS results of perifused human juvenile (n=9) and adult (n=16) islets, IEQ: islet equivalent. (H) Box plots show secreted insulin levels of juvenile (n=9) and adult (n=16) islets in the last fraction exposed to 5.6 mM glucose (basal) before a step increase to 16.7 mM glucose. (I) Insulin content of equivalent numbers of juvenile (n=3) and adult islets (n=10). Also see Experimental Procedures (* t-test P<0.05; Error bars indicate S.E.).
Age-dependent gene expression in human islet cells indicates distinct functional states
Gene ontology (GO) analysis suggested molecular functions of the enriched gene sets in α- and β-cells. Genes with greater expression in juvenile α-cells and β-cells were enriched for terms related to cell cycle, proliferation and nucleotide biosynthesis, consistent with a neonatal proliferative state (Figure 2C). For example, we found genes including MKI67, BUB1B, NUSAP1 and CDK9, which encode products known to regulate mitosis (M phase) and cell cycle progression. In addition, HSPA1 and HSPH1 are involved in the unfolded protein responses, and were also found to be significantly more abundant in juvenile islets. Consistent with these findings, immunohistology showed that proliferation marker Ki67 (encoded by MKI67) was increased both in juvenile α-cells and β-cells, compared to those cells in adult islets (Figures 2D and 2E).
By contrast, genes with transcripts increased in adulthood were enriched for GO terms like hormone activity, and regulation of hormone levels, consistent with physiological functions of mature endocrine islet cells (Figure 2F). When we applied the same analysis to pancreatic acinar and duct cell samples, unexpectedly, we found fewer than 20 genes whose expression significantly changed with age, and there was no enrichment for a specific GO term (Table S3, Figure S2A). Thus, our findings reveal specific gene expression programs that control endocrine cell function as humans age. Prior work in rodents has shown that insulin secretion changes with age (Aguayo-Mazzucato et al., 2011; Blum et al., 2012; Rorsman et al., 1989; Avrahami et al., 2015); however, systematic studies of human islet insulin secretion with advancing age have not been previously reported. We used in vitro perifusion to measure insulin secretion by juvenile and adult human islets used for gene expression studies (see Experimental Procedures). Compared to equivalent numbers of juvenile islets, adult human islets had higher insulin secretion when exposed to basal glucose (5.6 mM, Figures 2G–H, S2B) and high glucose (16.7 mM, Figures 2G and S2C). By contrast, total insulin content was indistinguishable in equivalent numbers of juvenile or adult islets (Figure 2I), consistent with our observation that insulin mRNA did not change detectably with age (Table S2). Together, these studies demonstrate functional differences in islet proliferation and insulin secretion between children and adults, consistent with our analysis of age-dependent gene expression in endocrine cells.
Analysis of histone marks reveals age-dependent chromatin signatures underlying gene expression changes in human islet cells
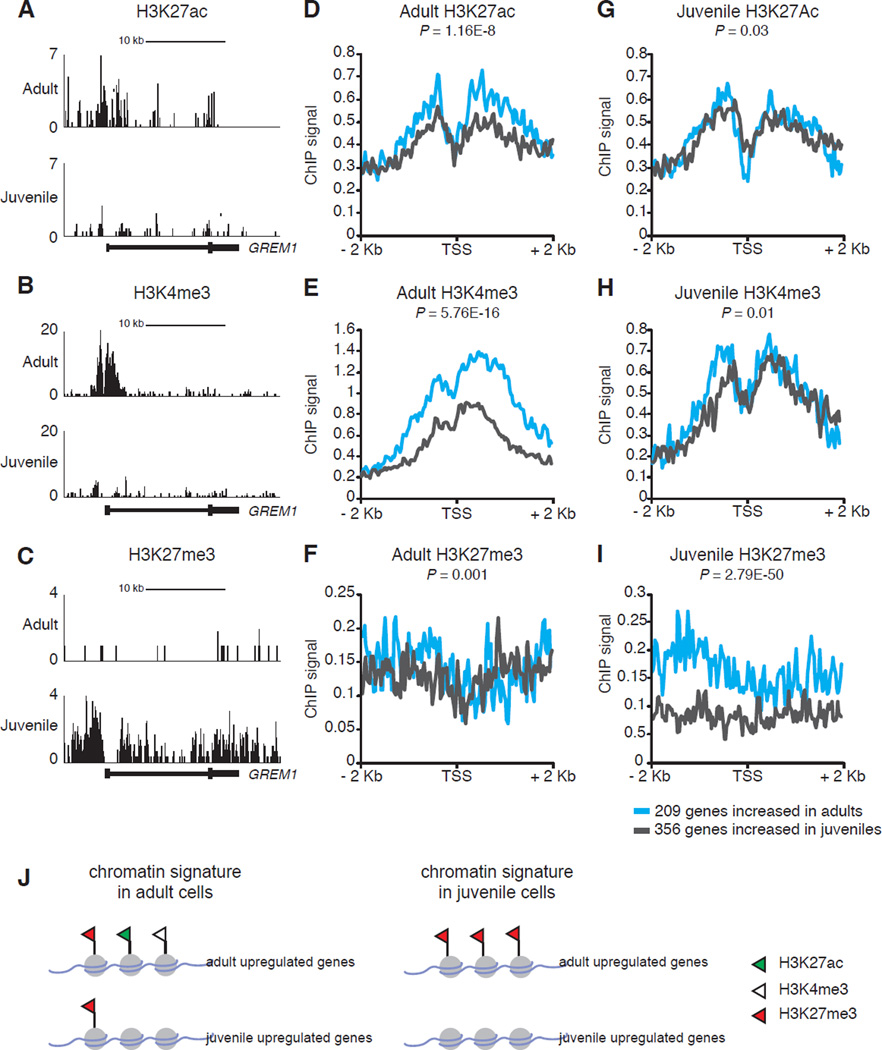
To elucidate mechanisms underlying age-dependent gene expression changes in human islets, we generated genome-wide occupancy maps of histone marks using ChIP-Seq from purified juvenile or adult pancreatic cells (Table S4). We studied the abundance of three well-defined histone modifications: H3K4me3 and H3K27ac, modifications that are known to correlate with active transcription, and H3K27me3 known to be present at inactive or silenced genomic loci (Heintzman et al., 2009; The ENCODE Project, 2012).
To evaluate the chromatin dynamics involving age-dependent gene expression in human islets, we focused on the promoter regions of genes we identified as differentially expressed with age in human α- or β-cells (Table S2). We calculated the mean ChIP-Seq signal of the three histone marks 2 kb upstream or downstream of the transcription start site (TSS) of age-dependent genes. For the 209 genes whose expression is increased in adult α- or β-cells (adult upregulated, Table S2), the ChIP-Seq signal from adult islet samples showed a significant enrichment of H3K27ac and H3K4me3 marks (Figures 3A–B, 3D–E, and S3) when compared to 356 genes whose expression is higher in juvenile samples (juvenile upregulated, Table S2). However, we did not detect substantial changes in levels of H3K27me3, a repressive modification, at the TSS of all age-dependent genes in adult samples (Figure 3F, J). Surprisingly, the ChIP-Seq results from juvenile samples did not show a similar pattern: H3K27ac and H3K4me3 levels were not detectably increased at juvenile-upregulated genes (Figures 3G-I, and S3). Rather, in juvenile islet samples we observed relative absence of the repressive H3K27me3 mark at juvenile-upregulated genes and significant enrichment of the repressive H3K27me3 mark at genes whose expression increases later in adult α- or β-cells (Figures 3C, I, J). As a further control, we randomly selected from our RNA-Seq datasets a subset of genes that are expressed in both α- or β-cells (normalized read counts >100) whose expression does not change with age (fold change < 0.5) and examined the histone ChIP-Seq signal at their TSS (Figure S3, red lines). We found that in juvenile samples the control gene set has substantially higher H3K4me3, H3K27ac and low H3K27me3 signal, correlating with their gene expression status. Thus, our genome-wide histone maps revealed evidence for distinct modes of histone-mediated regulation of age-dependent genes in juvenile and adult human islet cells (Figure 3J).
Figure 3. Histone ChIP-seq maps reveal distinct modes of chromatin modifications associated with age-dependent genes in human islets.
UCSC browser tracks showing (A) H3K27ac, (B) H3K4me3, (C) H3K27me3 normalized ChIP-Seq signal at the GREM1 locus. (D–I) Plots represent mean aggregate signal 2 kb upstream or downstream around the transcriptional start site (TSS) of age-dependent genes that are increased in adult (cyan lines, total of 209 genes) or juvenile samples (grey lines, total of 356 genes). For a complete list of genes see Table S2. Histone ChIP-Seq signals obtained from (D–F) 48-year-old adult donor, (G) 0.8-year-old and (H–I) 0.5-year-old juvenile donors. Wilcoxon rank sum test was used to calculate the P values. (J) Schematic depicting histone modifications found at genes expressed in an age-dependent manner in juvenile and adult islet samples.
Regulators of age-dependent islet gene expression programs and their linkage to diabetes
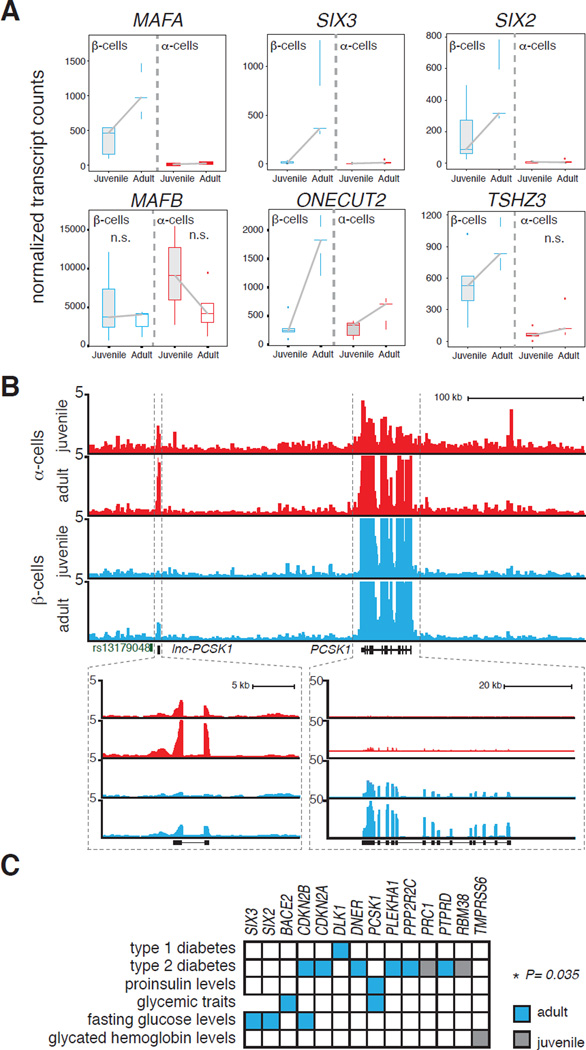
Transcription factors (TFs) are crucial regulators of islet development and disease (Conrad et al., 2014). For instance, mutations in 9 of 15 genes that have been linked with maturity-onset diabetes of the young (Sellick et al., 2004) (MODY) or pancreatic agenesis (Lango Allen et al., 2012) encode TFs, like PDX1, HNF1β, NEUROD1, PTF1A and GATA6. Comparison of age-regulated genes found here with a human TF compendium (Weirauch et al., 2014) revealed 25 TFs, most of which have not been yet linked to diabetes or studied in the context of islet development and function (Table S2). We analyzed the mean expression levels of each TF in juvenile or adult α- or β-cells, and identified several TFs whose expression increased with age specifically in β-cells, such as ONECUT2, MAFA, TSHZ3, SIX2 and SIX3 (Figure 4A). Prior work in rodents has shown that Mafa and Mafb regulate β-cell maturation by controlling genes that function in insulin synthesis, secretion and glucose-sensing (Aguayo-Mazzucato et al., 2011; Artner et al., 2007, 2010). Our analysis reveals age-dependent expression of MAFA specifically in human β-cells; by contrast, MAFB is expressed both in α- and β-cells at levels that do not detectably vary with age (Figure 4A).
Figure 4. Age-dependent islet genes are enriched for genes linked to diabetes and related metabolic traits.
(A) Box plots displaying normalized transcript counts of TFs whose expression changes significantly with age (except for MAFB) in human α- or β-cells, (n=5 juvenile, n=5 adult). (B) RNA-Seq profiles of α- and β-cells from juvenile and adult samples highlighting the PCSK1 and lncPCSK1-1 locus. (C) Matrix indicating the overlap of age-dependent genes and GWAS genes linked to diabetes and associated metabolic traits (* Chi-square test with Yates' correction).
In addition to mRNAs encoding proteins, we identified over 50 non-coding RNAs whose expression changed with age in α- and/or β-cells (Table S2). In six cases, the lncRNA is adjacent to a protein-coding gene whose expression also changes with age. For example, the long non-coding RNA lnc-PCSK1-1 neighbors PCSK1 which encodes a proprotein convertase essential for proinsulin processing (Figure 4B). While PCSK1 mRNA increased with age in β-cells, lnc-PCSK1-1 was most abundant and increased with age in α-cells. A single nucleotide polymorphism (SNP, rs13179048) was previously associated with PCSK1 in a genome-wide association study (GWAS) (Manning et al., 2012) examining fasting glucose levels and we found that this SNP is substantially closer to lnc-PCSK1-1 (6 kb) in the genome than to PCSK1 (160 kb). Thus, our findings suggest that lnc-PCSK1-1 deserves consideration in evaluating the role of this locus to disease risk.
GWAS reports have identified potential causal genetic variants associated with diabetes or related metabolic traits, such as fasting glucose impairment or altered fasting insulin levels. We compared genes in reported GWAS loci linked to these traits to genes whose expression changed with age in α-cells and β-cells; our analysis revealed significant enrichment of genes increased in adult β-cells or α-cells associated with risk for pre-diabetes phenotypes or diabetes (Figure 4C). Notably, our analysis revealed the TFs SIX2 and SIX3, which are encoded at loci previously linked by GWAS to impaired fasting glucose (Kim et al., 2011), but whose roles in β-cells has not been reported. Thus, we next analyzed SIX2 and SIX3 function in human β-cells.
Sine Oculis homeobox (SIX) family transcription factors are human β-cell regulators
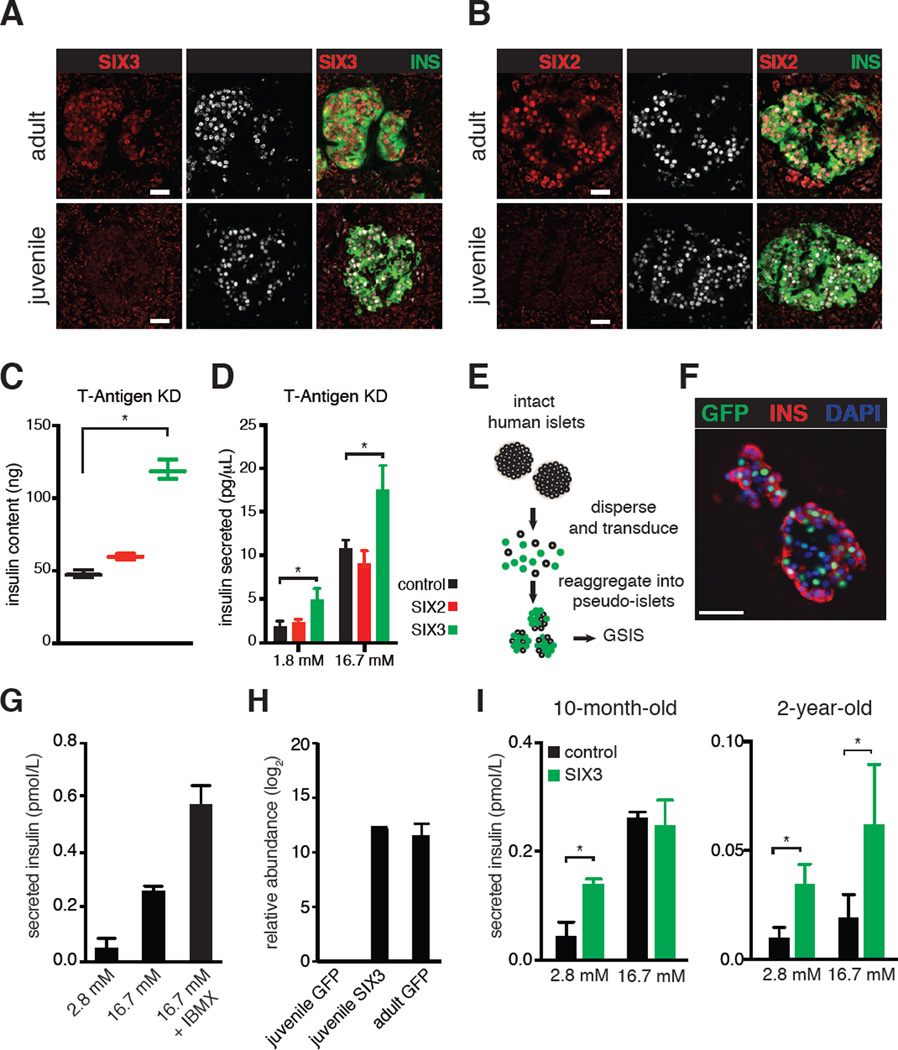
Based on their increased expression in adult β-cells, we postulated that SIX2 and SIX3 could regulate key age-dependent features of β-cells, such as insulin production or secretion. SIX2 and SIX3 belong to the Sine Oculis family of homeodomain TFs and have functions in kidney, forebrain and eye development (Kumar, 2009). We developed immunohistology methods to detect SIX2 and SIX3 protein (Figure S4A), and demonstrated that SIX2 and SIX3 localized to the nucleus of adult β-cells (Figures 5A–B, identified by co-expression of INS and PDX1). By contrast, we did not detect SIX2 or SIX3 in juvenile β-cells (Figures 5A–B), consistent with our RNA-Seq results. To test the function of SIX2 and SIX3, initially we used a human β-cell line, EndoC-βH1, derived from human fetal pancreas by simian virus 40 large T-Antigen transformation (Ravassard et al., 2011). EndoC-βH1 cells proliferate, have modest insulin content and secretion, and do not express detectable SIX2 or SIX3 mRNA (Figure S4B), features characteristic of immature β-cells. Using a lentivirus system, we generated EndoC-βH1 cells that stably express SIX2, SIX3, or GFP alone (Figure S4A). Compared to controls, SIX3-producing cells had increased insulin content (Figure S4C). Upon glucose challenge, both SIX2- and SIX3-producing cells secreted significantly more insulin than controls (Figure S4D). We assessed the expression of PDX1, NKX6.1 and MAFA mRNA but found no significant increase of these by SIX2- and SIX3-producing cells (Figure S4E). These findings indicate that SIX2 and SIX3 are β-cell specific factors sufficient to enhance insulin production and secretion, possibly in parallel to other TFs known to regulate β-cell functional maturation.
Figure 5. SIX3 and SIX2 increase with age specifically in human β-cells and enhance β-cell maturation.
SIX3 (A) and SIX2 (B) immunostaining of adult (31-year-old) and juvenile (4-year-old) human pancreas sections, scale bar: 100 µm. Insulin content (C) and secreted insulin levels (D) of EndoC-βH1 cells expressing GFP, SIX2 or SIX3 without T-Antigen (T-Antigen KD). Asterisks indicate statistically significant results reproduced in multiple experimental replicates (P < 0.05, two-way ANOVA followed by Fisher’s Least Significant Difference test). Bars indicate S.D. (E) Schematic detailing pseudo-islet techniques. Human islet cells are enzymatically dispersed, and transduced with lentivirus which co-expresses a GFP transgene. 5–7 days after dispersion, islet cells spontaneously re-aggregate into pseudo-islet clusters can be assayed using glucose-stimulated insulin secretion (GSIS) in vitro. (F) Representative immunohistologic images of human pseudo-islets stained with Insulin, GFP or DAPI, scale bar: 50 µm. (G) Bar graphs show secreted insulin levels of cultured human pseudo-islets exposed to glucose or glucose +IBMX. Secreted insulin is normalized to total insulin content. (H) Bar graphs indicate relative abundance of SIX3 transcript detected in pseudo-islets after transduction with GFP or SIX3 lentiviral vectors. (I) Graphs show secreted insulin levels from cultured pseudo-islets obtained from juvenile donors, expressing GFP alone (control) or SIX3. Secreted insulin is normalized to total insulin content (* P < 0.05, t-test). Error bars indicate S.D.
Recent studies showed that conditional deletion of T-Antigen reduced proliferation in the EndoC-βH2 cell line (Scharfmann et al., 2014), a change accompanied by enhanced insulin production and glucose-stimulated insulin secretion (GSIS). Using specific siRNAs, we knocked down T-Antigen efficiently in EndoC-βH1 cells (hereafter EndoC-βH1TKD), and observed markedly reduced Ki67 production (Experimental Procedures; see also Figures S4F–G), enhanced insulin production and glucose-stimulated insulin secretion (Figure 5C–D). Thus, EndoC-βH1TKD cells permitted assessment of SIX2 or SIX3 effects in non-proliferating β-cells after T-Antigen knock down. We found insulin content and secretion were not detectably altered by SIX2 expression in EndoC-βH1TKD cells (Figure 5C). By contrast, SIX3 expression increased insulin content and secretion of EndoC-βH1TKD cells (Figure 5C–D).
To gain further insights into β-cell gene regulation by SIX2 and SIX3, we performed RNA-Seq experiments in EndoC-βH1TKD cells stably expressing these factors. Comparison of EndoC-βH1TKD cells expressing SIX3 to control cells expressing GFP revealed over 200 differentially expressed genes (log2 fold change > 0.5, FDR 0.2, Table S5). A significant number of these overlapped with human islet age-dependent genes (P < 0.001, Chi-square test with Yates’ correction, Figure S5A), with enrichment of GO terms such as cell-cell signaling, growth factor and calcium binding (Figure S5B). Similar analysis of SIX2pos EndoC-βH1TKD cells revealed a much smaller set of genes, and there was no significant overlap with the islet age-dependent genes. This suggests that the gene expression differences we observe in SIX3pos EndoC-βH1TKD cells likely reflect SIX3 activity.
SIX3 functional studies in primary juvenile human islets
To assess the function of SIX3 in primary human islets, we developed lentiviral methods for expressing transgenes in human primary islet cells (Figure 5E, see Experimental Procedures). Pancreatic islets can be dispersed to single cell suspensions for efficient viral transduction; these cells spontaneously re-aggregate into ‘pseudo-islet’ clusters (Hopcroft et al., 1985; Zaldumbide et al., 2013). Using this approach we detected marker gene production (GFP, Figure 5F) in 70–80% of islet cells after dispersion, lentiviral transduction and re-aggregation. Insulin secretion stimulated by a step-increase of glucose or glucose + IBMX was robust in transgenic GFPpos pseudo-islets (Figure 5G), demonstrating that fundamental mechanisms regulating insulin secretion remained intact in pseudo-islets.
To test the prediction that SIX3 expression could enhance insulin secretion in juvenile islet cells, we obtained islets from juvenile donors and generated pseudo-islets that were transduced with lentivirus carrying either a control GFP or SIX3 expression construct. Using qPCR we found that juvenile pseudo-islets transduced with GFP virus, as expected, did not have detectable SIX3 mRNA, whereas juvenile pseudo-islets transduced with SIX3 virus produced SIX3 mRNA at levels comparable to adult pseudo-islets transduced with GFP virus (Figure 5H). We assessed insulin secretion by juvenile pseudo-islets and found that compared to GFP controls, SIX3 mis-expression significantly enhanced insulin release at basal glucose levels (2.8 mM), while stimulated secretion levels remained similar or improved (Figure 5I). These findings are consistent with our perifusion studies of primary islets from children and adults (Figure 2D). Taken together, our gain-of-function experiments using either EndoC-βH1 cells or primary juvenile human islets provide evidence that expression of SIX2 or SIX3 were sufficient to enhance cardinal functions of human β-cells.
DISCUSSION
Here we present the transcription and chromatin landscape of age-dependent human pancreatic cell maturation, based on organ procurement and flow cytometry methods to isolate primary cells from adult and rare child donors. Our study generated a unique gene expression atlas and maps of major histone modifications during postnatal development and maturation of human pancreatic endocrine cells. Thus, our study provides several resources that should impact understanding of islet genetics and signaling relevant to metabolism, diabetes and regenerative biology.
Prior studies have described the purification of pancreatic cells using either cell surface antibodies, non-specific zinc-chelating dye, or cell fixation and intracellular labeling (Blodgett et al., 2015; Dorrell et al., 2011; Hrvatin et al., 2014; Morán et al., 2012). However, none of these studies enriched for cells obtained from juvenile islets. Here, we used surface marker-based purification of islet cell subtypes from juvenile as well as adult donors, with appropriate enrichment of cells in each isolated fraction. Our method complements existing protocols for islet cell sorting and will be particularly useful for future applications that require live, native cells, such as open chromatin analysis (Buenrostro et al., 2013) or derivation of cell lines (Scharfmann et al., 2014).
Our data identified several hundred genes whose expression changes with age in human islet cells, most without known functions in the pancreas. We showed that the age-dependent gene expression programs are dynamic in endocrine α-cells and β-cells, and do not represent a general feature of aging cells. Genes with age-dependent expression cluster in classes with different functions, showing that postnatal development and maturation of islet endocrine cells are highly dynamic and involve multiple distinct molecular and signaling pathways. Our observation that genes like SIX2 and SIX3 may regulate hallmark features of β-cells such as insulin content or secretion, but are not expressed for at least several years after birth, suggests that the duration of human β-cell postnatal development may be longer than previously considered (McKnight et al. 2011; Arda et al., 2013). Our studies suggest that insulin secretion increases with age in human β-cells. However, definitive evidence establishing increased β-cell insulin secretion with advancing age, and when this occurs in postnatal life, will require data from a larger set of human donors than achieved here.
Our comparative analysis of histone modifications from human juvenile and adult islet cells provides insights about the chromatin dynamics of age-dependent genes. Chromatin marks associated with active gene expression, like H3K4me3 and H3K27ac, are significantly enriched in adult samples for adult-upregulated genes, but not in juvenile samples for juvenile-upregulated genes. Conversely, the H3K27me3 repressor mark is enriched at the adult-upregulated genes in juvenile samples but not in adult samples. These findings suggest that the Polycomb repressor complex (PRC2) establishes and maintains the H3K27me3 modification in juvenile islet cells to suppress expression of adult-specific genes. Thus, PRC2 might regulate the maturation of human β-cell function observed in our studies. During adulthood, the abundance of the repressive H3K27me3 mark at specific genes is reduced and the expression of adult-upregulated genes coincides with increased levels of H3K4me3 and H3K27ac at those loci. Trithorax Group (TrxG) proteins regulate H3K4me3 methylation in β-cells (Zhou et al., 2013); thus our findings further suggest that the TrxG controls age-dependent gene expression and maturation in human α- and β-cells.
One aspect of our analysis included TFs, a class of genes prior studies have shown are crucial regulators of pancreas and islet development (reviewed in Benitez et al 2012; Arda et al 2013). Comparatively less is known about TF regulation of postnatal human islet cell growth and maturation. Our analysis revealed age-dependent expression of more than a score of TFs, and some factors, like MAFA, SIX2 and SIX3, showed age-dependent induction specifically in β-cells. SIX2 and SIX3 were sufficient to enhance cardinal features of human β-cells like insulin content or secretion. Our gain-of-function studies also suggest that SIX2 and SIX3 may have distinct roles and are not functionally redundant, consistent with findings in other settings (reviewed by Kumar, 2009).
From GWAS studies of T1DM, T2DM and prediabetes, over 100 risk loci have emerged, but the function of candidate risk genes identified through these efforts has been elusive (Rutter, 2014). Unexpectedly we show here that genes whose expression in islet cells is age-dependent, like SIX2, SIX3, BACE2 and DLK1, are significantly enriched in gene sets linked by GWAS to T1DM and T2DM risk. DLK1 encodes a transmembrane protein containing Epidermal Growth Factor motifs with possible roles in Notch and MAP Kinase signaling (Wang et al., 2015). Similar to SIX2, SIX3 and BMP5 mRNA, DLK1 mRNA was highly enriched in β-cells and levels were increased in adult islets. In transgenic mice, mis-expression of Dlk1 in β-cells increased insulin production, β-cell mass, and insulin secretion (Wang et al., 2015). Findings and resources generated in this study, including stable SIX3-expressing human β-cell lines, should advance further studies to delineate the molecular mechanisms linking SIX3 and DLK1 functions to human β-cells. Expression of the mouse homologues Six2, Six3 and Dlk1 was not detected in adult mouse β-cells (Benitez et al., 2014; Benner et al., 2014; Kalousova et al., 2010; Ku et al., 2012; Martens et al., 2014), highlighting the importance of the human-specific approaches reported here.
Generating replacement islet β-cells from renewable sources such as embryonic stem cells, or non-islet cells, including exocrine duct cells is an intensive area of research (Lee et al., 2013; Pagliuca et al., 2014; Rezania et al., 2014; Russ et al., 2015). These approaches have been largely guided by findings from non-human experimental systems, especially mouse pancreas developmental biology. While recent progress toward this goal has been made, progeny from these produce insulin at levels far below those measured in native β-cells, and fail to recapitulate regulated insulin secretion characterized in islet β-cells. Our work provides a crucial genetic framework describing innate human-specific gene expression changes that control functional maturation of human islet cells. We speculate that integration of findings from this study into future β-cell engineering approaches could enhance development of insulin-producing cells from stem cell sources, including improvements in the quality and levels of insulin production and regulated insulin secretion.
EXPERIMENTAL PROCEDURES
Human Pancreas and Islet Procurement
All studies involving human pancreas or islets were conducted in accordance with Stanford University Institutional Review Board guidelines. De-identified human pancreata, islets or acinar tissues were obtained from healthy, non-diabetic organ donors with BMI<30, less than 15 hours of cold ischemia time, and deceased due to acute trauma or anoxia. Organs and islets were procured through Integrated Islet Distribution Network (IIDP), National Diabetes Research Institute (NDRI), and International Institute for the Advancement of Medicine (IIAM). For FACS, RNA-Seq and ChIP-Seq studies, islets from eight adult donors (ages 28, 33, 40, 42, 48, 54, 66 years) and seven juvenile donors (ages 0.5, 0.8, 1.4, 1.5, 5, 5, 6 years) were used (also see Tables S1 and S4). For perifusion assays, islets from 16 adult donors (ages 19, 20, 20, 23, 29, 38, 39, 40, 43, 43, 43, 48, 50, 51, 60, 60 years) and nine juvenile donors (ages 0.5, 1.2, 1.6, 3, 5, 5, 6, 9 years) were used. For total insulin content and corresponding basal secretion, islets from ten adult donors (ages 20, 20, 24, 28, 31, 37, 51, 53, 55, 65 years) and three juvenile donors (ages 4, 6, 9 years) were used. For immunohistologic analysis of proliferation, pancreata from four adult (ages 27, 30, 31, 37 years) and four juvenile donors (ages 1, 2, 3, 4 years) were used. For pseudo-islet studies islets from 0.8-, 2-, 42-year-old donors were used.
Islet Perifusion Assays, Insulin Content Analysis
For each assay 60 size-matched islets (approximately 100 islet equivalents [IEQ]) from juvenile (n=9) or adult (n=16) donors were perifused in an islet perifusion apparatus using two secretagogues. Insulin secretion normalized to IEQ using the method described (Dai et al., 2012; Kayton et al., 2015). To eliminate confounding effects that may stem from islet viability or fitness, only islet samples that matched a defined ‘normal profile’ were considered for this study (Kayton et al., 2015). Basal insulin was defined by the insulin concentration of the last fraction before introduction of 16.7 mM glucose. For insulin content analysis, 60 adult or juvenile islets of 180 µm diameter were processed (100 IEQs) as detailed in Kayton et al., 2015. Insulin was quantified using a human insulin ELISA kit following the manufacturer's protocol (10-1113-10, Mercodia, Inc).
Pseudo-islet generation and GSIS Assays
Juvenile islets were dispersed into monolayer cell suspension using enzymatic digestion (Accumax, Invitrogen). For each experimental group, approximately 1×106 cells were transduced with lentivirus corresponding to 1×109 viral copies in 1 mL as determined by the Lenti-X qPCR titration kit (Clontech, 631235). Transduced islet cells were seeded in ultra-low attachment 96-well plates (Corning) and cultured at 37°C with 5% CO2 in medium containing 10% fetal bovine serum (Hyclone), RPMI 1640 (Gibco), 2.25 g/dL glucose, 1% penicillin/streptomycin (v/v, Gibco). After 24 hours, the islet cells were pooled into 6-well ultra-low attachment plates and maintained under the same culture conditions for seven days. At the day of GSIS, 30–40 pseudo-islets were handpicked and pre-incubated in KRBB without glucose for one hour, which was repeated three times, for a total of three hours. After the initial equilibration, stimulated insulin secretion was performed by static incubation of pseudo-islets for one hour in KRBB containing 2.8 mM (basal) followed by 16.7 mM (high) glucose, and 16.7 mM (high) glucose +IBMX. Pseudo-islets were then lysed in acid-ethanol solution to extract the total insulin content. Human insulin from supernatants and pseudo-islet lysates were quantified using a human insulin ELISA kit (Mercodia Inc., Sweden).
Supplementary Material
Acknowledgments
We gratefully acknowledge organ donors and their families for tissue procurement. We thank R. Banerjee, W. Goodyer, S. Kundu, H. Chen for advice or help with tissue procurement, G. Oliver (St. Jude Children’s Research Hospital, TN USA) for advice on SIX3 antibody, D. Cohen (Harvard Medical School, MA USA), D. Blodgett and D. Harlan (U. Mass Medical School, MA USA) for protocols and help with intracellular sorting, C. Dorrell (Oregon Health Sciences University) for FACS advice, and K. Lee for assistance with confocal microscopy in the Stanford Cell Sciences Imaging Facility (supported by NIH grant 1S10OD01058001A1). We thank P. Batista, D. Simsek-Buck and members of the Kim lab for comments on the manuscript. H.E.A. was supported by a postdoctoral fellowship from the JDRF and a training grant to the Endocrinology Division, Department of Medicine, Stanford (5T32DK007217-39, NIDDK). R.C.S. was supported by the A.P. Giannini Foundation. M.S.S. is a cofounder and member of the scientific advisory board of Personalis. He is a member of the scientific advisory board (SAB) of Axiomx, Genapsys and SensOmics. H.Y.C. is a founder of Epinomics and served on the SAB of RaNA Therapeutics. Joint work in the Chang, Snyder, Scharfmann, Powers and Kim groups was supported by the NIH Beta Cell Biology Consortium (UO1DK089532 to S.K.K.). Work in the Powers group was also supported by grants from the NIH (DK72473, DK89572, DK104211), the JDRF, the Department of Veterans Affairs, the Vanderbilt Diabetes Research and Training Center (DK20593), and in the Kim group by the Helmsley Charitable Trust, the H.L. Snyder Foundation, the Elser Foundation, the Doolittle Trust, the JDRF and the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Accession Numbers
The data presented in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002), under the accession number GEO SuperSeries GSE79469.
AUTHOR CONTRIBUTIONS
H.E.A., H.Y.C., and S.K.K. conceived and supervised the project, H.E.A., L.L., E.A.T., Y.R., R.C.S., C.D., X.G., H.P. and P.W. performed the experiments, M.S.S. supervised sequencing, M.G. and R.S. shared materials, R.B. and J.W. isolated human islets, J.T. and K.Q. performed statistical analysis, H.E.A., J.T., C.D., A.C.P., S.K.K. analyzed the data and wrote the manuscript.
REFERENCES
- Aguayo-Mazzucato C, Koh A, El Khattabi I, Li W-C, Toschi E, Jermendy A, Juhl K, Mao K, Weir GC, Sharma A, et al. Mafa expression enhances glucose-responsive insulin secretion in neonatal rat beta cells. Diabetologia. 2011;54:583–593. doi: 10.1007/s00125-010-2026-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arda HE, Benitez CM, Kim SK. Gene Regulatory Networks Governing Pancreas Development. Dev. Cell. 2013;25:5–13. doi: 10.1016/j.devcel.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artner I, Blanchi B, Raum JC, Guo M, Kaneko T, Cordes S, Sieweke M, Stein R. MafB is required for islet β cell maturation. Proc. Natl. Acad. Sci. 2007;104:3853–3858. doi: 10.1073/pnas.0700013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artner I, Hang Y, Mazur M, Yamamoto T, Guo M, Lindner J, Magnuson MA, Stein R. MafA and MafB regulate genes critical to beta-cells in a unique temporal manner. Diabetes. 2010;59:2530–2539. doi: 10.2337/db10-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avrahami D, Li C, Zhang J, Schug J, Avrahami R, Rao S, Stadler MB, Burger L, Schübeler D, Glaser B, et al. Aging-Dependent Demethylation of Regulatory Elements Correlates with Chromatin State and Improved β Cell Function. Cell Metab. 2015;22:619–632. doi: 10.1016/j.cmet.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bene F, Tessmar-Raible K, Wittbrodt J. Direct interaction of geminin and Six3 in eye development. Nature. 2004;427:745–749. doi: 10.1038/nature02292. [DOI] [PubMed] [Google Scholar]
- Benitez CM, Goodyer WR, Kim SK. Deconstructing pancreas developmental biology. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a012401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez CM, Qu K, Sugiyama T, Pauerstein PT, Liu Y, Tsai J, Gu X, Ghodasara A, Arda HE, Zhang J, et al. An Integrated Cell Purification and Genomics Strategy Reveals Multiple Regulators of Pancreas Development. PLoS Genet. 2014;10:e1004645. doi: 10.1371/journal.pgen.1004645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner C, Meulen T, van der Cacéres E, Tigyi K, Donaldson CJ, Huising MO. The transcriptional landscape of mouse beta cells compared to human beta cells reveals notable species differences in long non-coding RNA and protein-coding gene expression. BMC Genomics. 2014;15:620. doi: 10.1186/1471-2164-15-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blodgett DM, Nowosielska A, Afik S, Pechhold S, Cura AJ, Kennedy NJ, Kim S, Kucukural A, Davis RJ, Kent SC, et al. Novel Observations From Next-Generation RNA Sequencing of Highly Purified Human Adult and Fetal Islet Cell Subsets. Diabetes. 2015;64:3172–3181. doi: 10.2337/db15-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum B, Hrvatin S, Schuetz C, Bonal C, Rezania A, Melton DA. Functional beta-cell maturation is marked by an increased glucose threshold and by expression of urocortin 3. Nat. Biotechnol. 2012;30:261–264. doi: 10.1038/nbt.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramswig NC, Everett LJ, Schug J, Dorrell C, Liu C, Luo Y, Streeter PR, Naji A, Grompe M, Kaestner KH. Epigenomic plasticity enables human pancreatic alpha to beta cell reprogramming. J. Clin. Invest. 2013;123:1275–1284. doi: 10.1172/JCI66514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler PC, Meier JJ, Butler AE, Bhushan A. The replication of β cells in normal physiology, in disease and for therapy. Nat. Clin. Pract. Endocrinol. Metab. 2007;3:758–768. doi: 10.1038/ncpendmet0647. [DOI] [PubMed] [Google Scholar]
- Chen H, Gu X, Liu Y, Wang J, Wirt SE, Bottino R, Schorle H, Sage J, Kim SK. PDGF signalling controls age-dependent proliferation in pancreatic [bgr]-cells. Nature. 2011;478:349–355. doi: 10.1038/nature10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad E, Stein R, Hunter CS. Revealing transcription factors during human pancreatic βcell development. Trends Endocrinol. Metab. 2014;25:407–414. doi: 10.1016/j.tem.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Brissova M, Hang Y, Thompson C, Poffenberger G, Shostak A, Chen Z, Stein R, Powers AC. Islet-enriched gene expression and glucose-induced insulin secretion in human and mouse islets. Diabetologia. 2012;55:707–718. doi: 10.1007/s00125-011-2369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrell C, Abraham SL, Lanxon-Cookson KM, Canaday PS, Streeter PR, Grompe M. Isolation of major pancreatic cell types and long-term culture-initiating cells using novel human surface markers. Stem Cell Res. 2008;1:183–194. doi: 10.1016/j.scr.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Dorrell C, Schug J, Lin CF, Canaday PS, Fox AJ, Smirnova O, Bonnah R, Streeter PR, Stoeckert CJ, Jr, Kaestner KH, et al. Transcriptomes of the major human pancreatic cell types. Diabetologia. 2011 doi: 10.1007/s00125-011-2283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterházy D, Stützer I, Wang H, Rechsteiner MP, Beauchamp J, Döbeli H, Hilpert H, Matile H, Prummer M, Schmidt A, et al. Bace2 Is a β Cell-Enriched Protease that Regulates Pancreatic β Cell Function and Mass. Cell Metab. 2011;14:365–377. doi: 10.1016/j.cmet.2011.06.018. [DOI] [PubMed] [Google Scholar]
- Fiaschi-Taesch N, Bigatel TA, Sicari B, Takane KK, Salim F, Velazquez-Garcia S, Harb G, Selk K, Cozar-Castellano I, Stewart AF. Survey of the Human Pancreatic β-Cell G1/S Proteome Reveals a Potential Therapeutic Role for Cdk-6 and Cyclin D1 in Enhancing Human β-Cell Replication and Function In Vivo. Diabetes. 2009;58:882–893. doi: 10.2337/db08-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodyer WR, Gu X, Liu Y, Bottino R, Crabtree GR, Kim SK. Neonatal β Cell Development in Mice and Humans Is Regulated by Calcineurin/NFAT. Dev. Cell. 2012;23:21–34. doi: 10.1016/j.devcel.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg BE, Moore PC, Demozay D, Hall BA, Li M, Husain A, Wright AJ, Atkinson MA, Rhodes CJ. Formation of a human β-cell population within pancreatic islets is set early in life. J. Clin. Endocrinol. Metab. 2012;97:3197–3206. doi: 10.1210/jc.2012-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes MG, Urbanek M, Hivert M-F, Armstrong LL, Morrison J, Guo C, Lowe LP, Scheftner DA, Pluzhnikov A, Levine DM, et al. Identification of HKDC1 and BACE2 as Genes Influencing Glycemic Traits During Pregnancy Through Genome-Wide Association Studies. Diabetes. 2013;62:3282–3291. doi: 10.2337/db12-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinforma. Oxf. Engl. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Hopcroft DW, Mason DR, Scott RS. Structure-function relationships in pancreatic islets: support for intraislet modulation of insulin secretion. Endocrinology. 1985;117:2073–2080. doi: 10.1210/endo-117-5-2073. [DOI] [PubMed] [Google Scholar]
- Hrvatin S, Deng F, O’Donnell CW, Gifford DK, Melton DA. MARIS: Method for Analyzing RNA following Intracellular Sorting. PLoS ONE. 2014;9:e89459. doi: 10.1371/journal.pone.0089459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jermendy A, Toschi E, Aye T, Koh A, Aguayo-Mazzucato C, Sharma A, Weir GC, Sgroi D, Bonner-Weir S. Rat neonatal beta cells lack the specialised metabolic phenotype of mature beta cells. Diabetologia. 2011;54:594–604. doi: 10.1007/s00125-010-2036-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalousova A, Mavropoulos A, Adams BA, Nekrep N, Li Z, Krauss S, Stainier DY, German MS. Dachshund homologues play a conserved role in islet cell development. Dev. Biol. 2010;348:143–152. doi: 10.1016/j.ydbio.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayton NS, Poffenberger G, Henske J, Dai C, Thompson C, Aramandla R, Shostak A, Nicholson W, Brissova M, Bush WS, et al. Human Islet Preparations Distributed for Research Exhibit a Variety of Insulin Secretory Profiles. Am. J. Physiol. - Endocrinol. Metab. ajpendo.00437.2014. 2015 doi: 10.1152/ajpendo.00437.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Go MJ, Hu C, Hong CB, Kim YK, Lee JY, Hwang J-Y, Oh JH, Kim D-J, Kim NH, et al. Large-scale genome-wide association studies in East Asians identify new genetic loci influencing metabolic traits. Nat. Genet. 2011;43:990–995. doi: 10.1038/ng.939. [DOI] [PubMed] [Google Scholar]
- Ku GM, Kim H, Vaughn IW, Hangauer MJ, Myung Oh C, German MS, McManus MT. Research resource: RNA-Seq reveals unique features of the pancreatic β-cell transcriptome. Mol. Endocrinol. Baltim. Md. 2012;26:1783–1792. doi: 10.1210/me.2012-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP. The sine oculis homeobox (SIX) family of transcription factors as regulators of development and disease. Cell. Mol. Life Sci. 2009;66:565–583. doi: 10.1007/s00018-008-8335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lango Allen H, Flanagan SE, Shaw-Smith C, De Franco E, Akerman I, Caswell R, Ferrer J, Hattersley AT, Ellard S. GATA6 haplo insufficiency causes pancreatic agenesis in humans. Nat. Genet. 2012;44:20–22. doi: 10.1038/ng.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Sugiyama T, Liu Y, Wang J, Gu X, Lei J, Markmann JF, Miyazaki S, Miyazaki J, Szot GL, et al. Expansion and conversion of human pancreatic ductal cells into insulin-secreting endocrine cells. eLife. 2013;2 doi: 10.7554/eLife.00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The Hallmarks of Aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AK, Hivert M-F, Scott RA, Grimsby JL, Bouatia-Naji N, Chen H, Rybin D, Liu C-T, Bielak LF, Prokopenko I, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens GA, Motté E, Kramer G, Stangé G, Gaarn LW, Hellemans K, Nielsen JH, Aerts JM, Ling Z, Pipeleers D. Functional characteristics of neonatal rat βcells with distinct markers. J. Mol. Endocrinol. 2014;52:11–28. doi: 10.1530/JME-13-0106. [DOI] [PubMed] [Google Scholar]
- Meier JJ, Butler AE, Saisho Y, Monchamp T, Galasso R, Bhushan A, Rizza RA, Butler PC. β-Cell Replication Is the Primary Mechanism Subserving the Postnatal Expansion of β-Cell Mass in Humans. Diabetes. 2008;57:1584–1594. doi: 10.2337/db07-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morán I, Akerman İ, van de Bunt M, Xie R, Benazra M, Nammo T, Arnes L, Nakić N, García-Hurtado J, Rodríguez-Seguí S, et al. Human β Cell Transcriptome Analysis Uncovers lncRNAs That Are Tissue-Specific, Dynamically Regulated, and Abnormally Expressed in Type 2 Diabetes. Cell Metab. 2012;16:435–448. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otonkoski T, Andersson S, Knip M, Simell O. Maturation of insulin response to glucose during human fetal and neonatal development. Studies with perifusion of pancreatic islet like cell clusters. Diabetes. 1988;37:286–291. doi: 10.2337/diab.37.3.286. [DOI] [PubMed] [Google Scholar]
- Pagliuca FW, Millman JR, Gürtler M, Segel M, Van Dervort A, Ryu JH, Peterson QP, Greiner D, Melton DA. Generation of functional human pancreatic β cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali L, Gaulton KJ, Rodríguez-Seguí SA, Mularoni L, Miguel-Escalada I, Akerman İ, Tena JJ, Morán I, Gómez-Marín C, van de Bunt M, et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet. 2014;46:136–143. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravassard P, Hazhouz Y, Pechberty S, Bricout-Neveu E, Armanet M, Czernichow P, Scharfmann R. A genetically engineered human pancreatic βcell line exhibiting glucose-inducible insulin secretion. J. Clin. Invest. 2011;121:3589–3597. doi: 10.1172/JCI58447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezania A, Bruin JE, Arora P, Rubin A, Batushansky I, Asadi A, O’Dwyer S, Quiskamp N, Mojibian M, Albrecht T, et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 2014;32:1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- Rorsman P, Arkhammar P, Bokvist K, Hellerström C, Nilsson T, Welsh M, Welsh N, Berggren PO. Failure of glucose to elicit a normal secretory response in fetal pancreatic beta cells results from glucose insensitivity of the ATP-regulated K+ channels. Proc. Natl. Acad. Sci. U. S. A. 1989;86:4505–4509. doi: 10.1073/pnas.86.12.4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ HA, Parent AV, Ringler JJ, Hennings TG, Nair GG, Shveygert M, Guo T, Puri S, Haataja L, Cirulli V, et al. Controlled induction of human pancreatic progenitors produces functional beta- like cells in vitro. EMBO J. 2015 doi: 10.15252/embj.201591058. e201591058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter GA. Dorothy Hodgkin Lecture 2014 Understanding genes identified by genome-wide association studies for Type 2 diabetes. Diabet. Med. 2014;31:1480–1487. doi: 10.1111/dme.12579. [DOI] [PubMed] [Google Scholar]
- Scharfmann R, Pechberty S, Hazhouz Y, von Bülow M, Bricout-Neveu E, Grenier-Godard M, Guez F, Rachdi L, Lohmann M, Czernichow P, et al. Development of a conditionally immortalized human pancreatic β cell line. J. Clin. Invest. 2014;124:2087–2098. doi: 10.1172/JCI72674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellick GS, Barker KT, Stolte-Dijkstra I, Fleischmann C, Coleman RJ, Garrett C, Gloyn AL, Edghill EL, Hattersley AT, Wellauer PK, et al. Mutations in PTF1A cause pancreatic and cerebellar agenesis. Nat. Genet. 2004;36:1301–1305. doi: 10.1038/ng1475. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Rodriguez RT, McLean GW, Kim SK. Conserved markers of fetal pancreatic epithelium permit prospective isolation of islet progenitor cells by FACS. Proc. Natl. Acad. Sci. 2007;104:175–180. doi: 10.1073/pnas.0609490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi S, Tanigawa K, Miwa I. Immaturity of glucose-induced insulin secretion in fetal rat islets is due to low glucokinase activity. Horm. Metab. Res. Horm. Stoffwechselforschung Horm. Métabolisme. 2000;32:97–102. doi: 10.1055/s-2007-978598. [DOI] [PubMed] [Google Scholar]
- Wang Y, Lee K, Moon YS, Ahmadian M, Kim K-H, Roder K, Kang C, Sul HS. Overexpression of Pref-1 in pancreatic islet β-cells in mice causes hyperinsulinemia with increased islet mass and insulin secretion. Biochem. Biophys. Res. Commun. 2015;461:630–635. doi: 10.1016/j.bbrc.2015.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaldumbide A, Alkemade G, Carlotti F, Nikolic T, Abreu JR, Engelse MA, Skowera A, de Koning EJ, Peakman M, Roep BO, et al. Genetically engineered human islets protected from CD8-mediated autoimmune destruction in vivo. Mol. Ther. 2013;21:1592–1601. doi: 10.1038/mt.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JX, Dhawan S, Fu H, Snyder E, Bottino R, Kundu S, Kim SK, Bhushan A. Combined modulation of polycomb and trithorax genes rejuvenates β cell replication. J. Clin. Invest. 2013;123:4849–4858. doi: 10.1172/JCI69468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.