Abstract
Background
Skeletal muscle protein loss is an adaptive response to various patho‐physiological situations, and the ubiquitin proteasome system (UPS) is responsible for the degradation of the bulk of muscle proteins. The role of E2 ubiquitin‐conjugating enzymes is still poorly understood in skeletal muscle.
Methods
We screened for E2s expression levels in C2C12 myotubes submitted to the catabolic glucocorticoid dexamethasone (Dex).
Results
One micromolar Dex induced an accumulation of proteasome substrates (polyUb conjugates) and an overexpression of the muscle‐specific E3 ligase MuRF1 and of six E2 enzymes, UBE2A, UBE2B, UBE2D1, UBE2D2, UBE2G1, and UBE2J1. However, only MuRF1 and UBE2B were sensitive to mild catabolic conditions (0.16 μM Dex). UBE2B knockdown induced a sharp decrease of total (−18%) and K48 (−28%) Ub conjugates, that is, proteasome substrates, indicating an important role of UBE2B in the overall protein breakdown in catabolic myotubes.
Conclusions
Interestingly, these results indicate an important role of UBE2B on muscle protein homeostasis during catabolic conditions.
Keywords: Ubiquitin, E2 Ubiquitin‐conjugating enzyme, E3 ligase, Ubiquitin proteasome system, C2C12 myotubes
Introduction
Ubiquitination is a widespread post‐translational modification of proteins that regulates cell metabolism within eukaryotes.1 Using a single protein, ubiquitin (Ub), several signals can be created that will give rise to different fates for each modified protein. Proteins targeted with Ub chains linked through K48 represent an important part of Ub signals and are bona fide proteasome substrates. Other protein modifications include Ub chains with other Ub lysine residues (K6, K11, K27, K29, K33, or K63), forked (e.g. K6–K11), heterologous (Ub‐SUMO) or simply monoUb.2, 3 Besides K48 and K11 chains, other protein modifications with Ub generally do not involve protein degradation through the 26S proteasome, which highlight the complexity of Ub signalling in eukaryotic cells. Modification of proteins by Ub signal involves several hundreds of enzymes that combine together for achieving high‐specificity targeting and fine‐tuning of metabolic pathways and cell growth. This is in particular the case for skeletal muscle, a tissue with high plasticity that can atrophy and recover very rapidly.4, 5 Indeed, it is now assumed that the ubiquitin proteasome system (UPS) is mainly responsible for controlling muscle mass in nearly any catabolic situations. For these reasons, identifying the enzymes that physiologically target muscle proteins for breakdown by the 26S proteasome is of considerable interest and represents a promising way for the development of future therapeutical strategies.
Ubiquitination of a protein requires the sequential action of three classes of proteins. Ubiquitin is first activated by a single E1 (Ub‐activating enzyme) that transfers high‐energy Ub to one of the 35 mammalian E2s (Ub‐conjugating enzymes). The E2s transfer Ub on target proteins in conjunction with the third class of enzymes (>600), namely, E3 ubiquitin ligases. An E2 is able to cooperate with different E3s and vice versa, which enables the specific targeting of virtually any cellular protein. The E3s recognize the target protein to be degraded and thus bring specificity to the ubiquitination machinery, but most E3s lack enzymatic activity so that only the E2–E3 couple is functionally relevant. Whereas most studies focused on the role of E3 ligases, it turned out that E2s possess an important role and influence the fate of the substrate. Indeed, E2s are directly responsible for determining the type of chain built on the substrate together with the identity of the residue modified by Ub, which means E2s are central players in the ubiquitination machinery.6, 7, 8 The UPS is highly activated during skeletal muscle atrophy as witnessed by increased expression of several components of this pathway and is believed to be the main actor of muscle wasting in several catabolic conditions.9, 10, 11, 12, 13, 14, 15 The muscle‐specific E3 ligases MuRF1 and MAFbx are particularly sensitive markers of the atrophying programme, but other E3s have also been linked to the development of muscle wasting and different targets have been identified.4 However, the identity of the E2s catalyzing Ub transfer remains elusive, as only in vitro ubiquitination assays have been performed (see Polge et al.16 for a recent review). Indeed, multiple E2–E3 combinations may be possible for a substrate but with different signals that are linked or not to 26S proteasome‐dependent degradation.6, 8, 17, 18 Overexpression of different E2s was reported in several catabolic models, but only a limited number of E2 enzymes were addressed in previous studies.12, 14, 15, 19, 20, 21, 22, 23, 24 UBE2B is probably the only E2 enzyme that responded to most catabolic situations tested. In addition, this enzyme is highly expressed in skeletal muscle, represents nearly half of Ub conjugation when combined with its isoform UBE2A, and expression levels of UBE2B are tightly linked to atrophy and recovery situations.13, 19, 25 However, no muscular target has yet been attributed to UBE2B, and its role during atrophying situations is still unknown.
In this work, we screened the expression levels of E2s that are highly expressed using C2C12 myotubes that were treated or not with dexamethasone (Dex). We found that only a limited number of E2s were responsive to Dex and that only the expression of UBE2B and the muscle‐specific E3 ligase MuRF1 was sensitive to moderate levels of Dex. Interestingly, UBE2B knockdown induced an accumulation of actin and myosin heavy chain (MHCI), indicating that UBE2B is an important factor of myofibrillar protein degradation. Surprisingly, this was restricted to UBE2B, as its isoform UBE2A did not exhibit any role on the bulk of muscle proteins. Altogether, we report that a single E2 can profoundly alter protein balance in muscle cells and that close isoforms may have non‐overlapping roles in cells.
Materials and methods
Antibodies and materials
Anti‐flag (clone M2), anti‐actin, and anti‐MHCI (skeletal myosin heavy chain I) antibodies were from Sigma; anti‐polyUb (FK1), anti‐Ub K48‐specific, and anti‐Ub K63‐specific antibodies were from Millipore; anti‐UBE2B antibody was a generous gift from Pr. S. S. Wing (McGill University, Montreal, Canada), and the anti‐MuRF1 antibody was from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Immunoblots were revealed using the Li‐COR Odyssey (ScienceTec, Courtaboeuf, France) procedures. Blots were then stained using Blot‐FastStain (G‐Biosciences, MO, USA), and densitometry was performed using the ImageJ software v.1.34s (NIH, Bethesda, MA, USA) to ensure even loading. C2C12 muscle cells were purchased from LGC Promochem (Teddington, UK). The fa‐C2C12 clone expressing stably flag α‐actin and GST‐MuRF1 were already described.26 Anti‐protease cocktail, water‐soluble Dex, the primers, and siRNAs were from Sigma‐Aldrich (Saint‐Quentin Fallavier, France). All primers and siRNAs were designed using the PrimerBlast (http://www.ncbi.nlm.nih.gov/tools/primer‐blast/) and siRNA Wizard v3.1 (http://www.sirnawizard.com) software, respectively, and are listed in the Supporting Information section.
Constructions
The original sequence of rat UBE2B was provided by Pr. S. S. Wing (McGill University, Montreal, Canada). Full‐length‐UBE2B cDNA was amplified by PCR using the Platinum Pfx DNA polymerase (Invitrogen, Paislay, UK) and subcloned in pAED4 expression vector.27 GST‐MuRF1 construction was already described.26
Cell culture, treatments, and fractionation of cellular proteins
C2C12 and fa‐C2C12 myoblasts were grown under 5% CO2 in Dulbecco's modified Eagle medium (DMEM, Sigma‐Aldrich, Saint‐Quentin Fallavier, France) containing 10% foetal bovine serum (FBS, Gibco, Invitrogen, Paislay, UK) and supplemented with l‐glutamine, non essential amino acids, and gentamycin (Gibco, growth medium, GM). For myoblast differentiation, cells were grown to near confluence and then shifted to DMEM containing 2% horse serum (differentiation medium) for 6 days.
A catabolic state was induced in myotubes by adding Dex up to 48 h before harvesting the cells. Briefly, myotubes were washed and scraped off the plate into 1X phosphate‐buffered saline and then sonicated for 30 s at maximum power using a UP50H sonicator (Hielscher, Teltow, Germany) in lysis buffer (5 mM Tris, pH 7.5, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 10 mM NEM, 1% Triton X‐100/anti‐proteases (Protease Inhibitor Mixture/Sigma)) as previously described.28 Cell lysate was then centrifuged at 10 000 g for 10 min at 4 °C, and the supernatant (soluble fraction) was kept at −80 °C until use. The pellets enriched in myofibrillar proteins were resuspended in homogenization buffer and sonicated to re‐suspend the proteins. Protein content was determined using the BCA protein assay kit (Pierce, Rockford, IL, USA).
GST pulldown assays
GST‐MuRF1 and UBE2B were expressed in Escherichia coli BL21(DE3) (Merck, Darmstadt, Germany) using 100 μM IPTG. Bacteria were then pelleted, resuspended in 1X PBS buffer, 100 μM PMSF, 1% Triton X100, and lyzed using a French press. The lysate was then centrifuged at 16 000 g, and the supernatant was used to carry out the following experiments. GST‐MuRF1 were then purified with Glutathione‐Sepharose 4B beads (GE Healthcare) according to the manufacturers' instructions and used as a 50% slurry for subsequent GST‐pulldown experiments.
qRT–PCR and knockdown experiments
Total RNA was purified from C2C12 myotubes using Trizol (Fisher Gibco). qRT–PCR was performed using the Quantitect Reverse transcription kit (Qiagen) and the SYBR Green supermix (Bio‐Rad) according to the manufacturer's instructions using a cfx96 real‐time system (BioRad). Calculations were made using the comparative ∆Ct method (Pfaffl) with YWHAZ, HPRT1, and GusB Housekeeping genes (see Supporting Information for primers).
Knockdown experiments were carried out on fa‐C2C12 myoyubes grown in multiwell cell culture plates and treated or not with Dex. Myotubes were transfected or not at day 4 of differentiation with a couple of siRNAs targeting either UBE2A (si‐UBE2A), UBE2B (si‐UBE2B), or no known protein sequence (sc‐siRNA, negative control) using the CADY peptide as previously described.12 Two different siRNAs were tested for each target (UBE2A and UBE2B). They gave similar results in preliminary experiments, so that we mixed them for the experiments presented in this manuscript. Another control group was systematically performed where only 1X PBS was present (mock transfection). A couple of siRNAs was used for each transfection. Briefly, siRNAs were diluted in 1X PBS (20 μM). The CADY peptide was resuspended in water (20 μM) and diluted (4 μM final) in 1X PBS containing either no siRNA or siRNA (50 nM final) targeting UBE2A, UBE2B or no known coding sequence. Myotubes were washed with 1X PBS and the CADY‐siRNA mixture (200 µl) was added to the cells for 5 min at 37 °C. Differentiation medium lacking the antibiotics and the serum was added (400 µl), and myotubes were incubated for 2 h at 37 °C. Complete differentiation medium containing or not Dex (1 μM) was then added (2.4 ml) in each well, and cells were harvested upon 2 days of Dex treatment as described earlier. The experiments were repeated three times for UBE2B and twice for UBE2A knockdown and gave reproducible results.
Statistical analysis and data processing
Densitometric analyses were performed on immunoblots using the ImageJ software v.1.34s (NIH, Bethesda, MA, USA). Data are presented as means ± SEM for qRT–PCR and immunoblotting. Statistical significance was tested using the Mann‐Whitney U test (Figures 1, 2(C), and 4), one‐way analysis of variance (ANOVA) (Figures 2(A) and (B) and 3(A) and (B)) or two‐way ANOVA.
Figure 1.
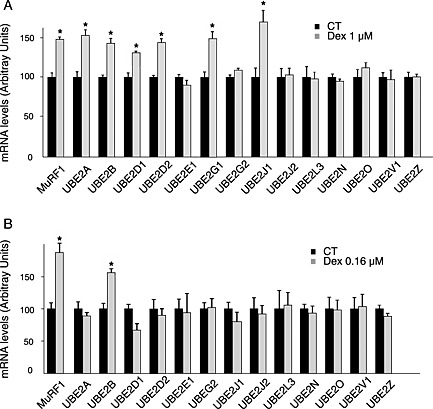
A limited number of E2 enzymes are up regulated upon Dex treatment. C2C12 myotubes were untreated (CT) or treated with either 1 μM (A) or 0.16 μM (B) Dex. mRNA levels for E2s highly expressed in skeletal muscle were determined by qRT–PCR. The E3 ligase MuRF1 was used as a control of catabolic conditions. Only UBE2B was up regulated upon low levels of Dex. * P < 0.01, n = 6.
Figure 2.
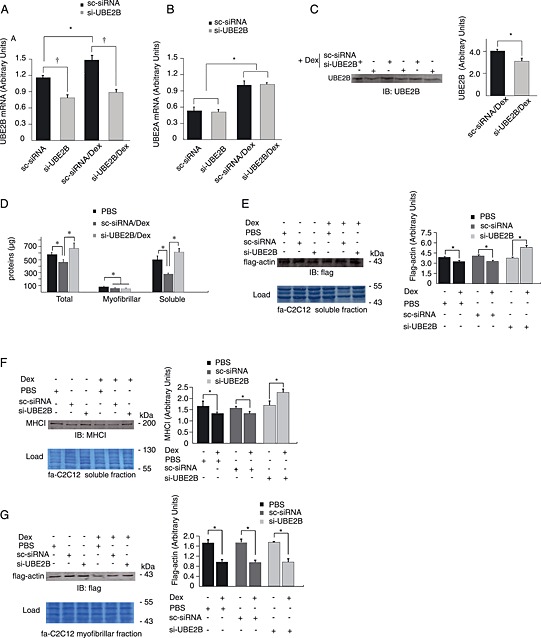
UBE2B knockdown induced an accumulation of soluble proteins. UBE2B was partially invalidated in C2C12 myotubes using cell penetrating peptides for siRNA transfection. C2C12 myotubes were transfected with mock (sc‐siRNA) or a mixture of siRNA directed against UoBE2B mRNA (si‐UBE2B) and treated or not with Dex (1 μM). (A) UBE2B mRNA levels decreased by 30% and 43% in control and Dex‐treated C2C12 myotubes, respectively. Dex treatment induced an upregulation of UBE2B mRNA in mock‐treated myotubes, while UBE2B knockdown suppressed this increase. (B) UBE2A expression was not affected by UBE2B knockdown and Dex treatment increased similarly UBE2A mRNA levels in both sc‐siRNA and si‐UBE2B in C2C12 myotubes. (C) UBE2B protein levels were depressed by UBE2B knockdown (−24%), n = 6, fa‐C2C12 myotubes stably expressing flag‐actin (Polge et al. 12) were treated or not with Dex (1 μM) and transfected with either sc‐siRNA (mock control) or si‐UBE2B siRNAs. Untreated myotubes received identical amount of PBS 1X instead of Dex. (D) Proteins levels were determined, and untreated sc‐siRNA transfected myotubes were used as reference (PBS). Total, myofibrillar, and soluble‐enriched fractions were similarly depressed upon Dex treatment in sc‐siRNA‐transfected (mock) C2C12 myotubes when compared with the PBS group (P < 0.05). UBE2B transfection induced an accumulation of total and soluble proteins in Dex‐treated fa‐C2C12 myotubes. (E) flag‐actin was depressed in the soluble fraction upon Dex treatment in both untransfected and sc‐siRNA (mock) transfected fa‐C2C12 myotubes. In contrast, UBE2B knockdown induced an accumulation of flag‐actin. (F) Myosin heavy chain (MHCI) was depressed in the soluble fraction upon Dex treatment in fa‐C2C12 myotubes and, like α‐actin, accumulated when UBE2B was partially invalidated by knockdown. (G) flag‐actin levels decreased upon Dex treatment in the myofibrillar fraction of fa‐C2C12 myotubes and UBE2B knockdown did not affect α‐actin loss. Load: proteins were stained using Blot‐FastStain reagent. Portions of the analysed membranes are shown and densitometry was performed on large areas. * P < 0.01 for transfection‐linked modification of mRNA levels. † P < 0.01 for Dex treatment, n = 6.
Figure 4.
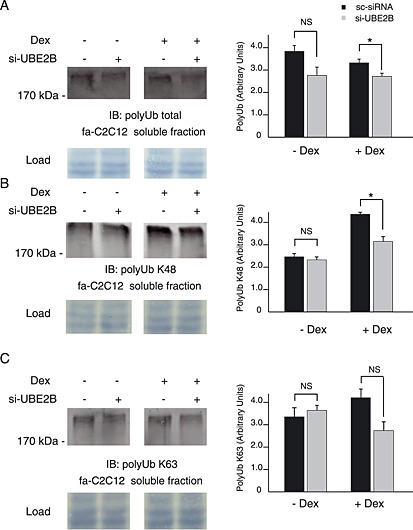
UBE2B knockdown decreased polyUb conjugates levels in fa‐C2C12 myotubes. Immunoblotting was performed on soluble proteins from untreated and Dex‐treated fa‐C2C12 myotubes using antibodies directed against polyUb (all linkages), K63, and K48 conjugates. (A) UBE2B knockdown significantly decreased total polyUb conjugates levels in Dex‐treated myotubes. (B) UBE2B knockdown significantly reduced K48‐linked polyUb conjugates in Dex‐treated myotubes (−28%) but not in untreated cells. (C) PolyUb conjugates via K63 linkages were not significantly modified in either Dex‐treated or untreated fa‐C2C12 myotubes. Load: membranes were stained using Blot‐FastStain dye and densitometric analysis was used to correct for uneven loading (a portion of the membrane is shown). * P < 0.05, n = 6.
Figure 3.
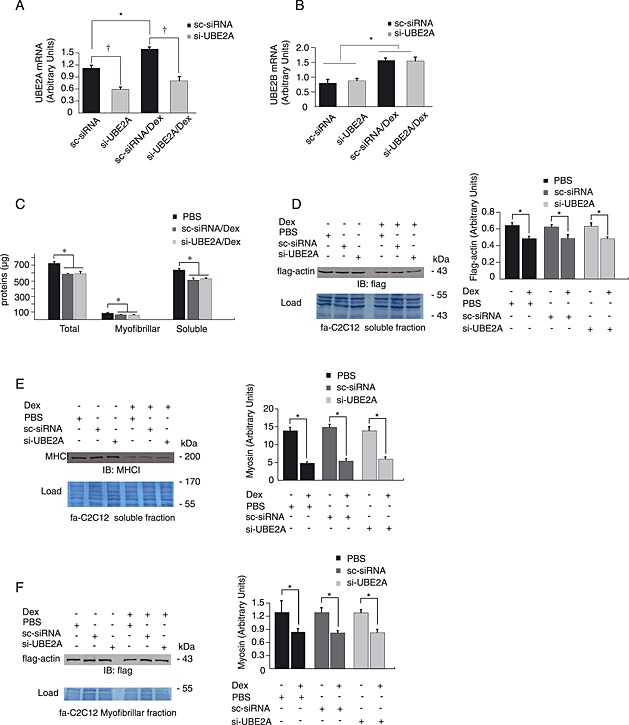
UBE2A knockdown did not modify myofibrillar protein losses in catabolic myotubes. Transfection was performed as described earlier. (A) UBE2A mRNA levels were reduced by 48% and 50% in untreated and Dex‐treated fa‐C212 myotubes, respectively, while no effect was observed on UBE2B (B). n = 6, * P < 0.01 for transfection‐linked modification of mRNA levels. † P < 0.01 for Dex treatment. fa‐C2C12 myotubes were treated or not with Dex (1 μM) and transfected with either sc‐siRNA (mock control) or si‐UBE2B siRNAs. Untreated myotubes received identical amounts of PBS 1X instead of Dex. (C) Proteins levels were determined and untreated sc‐siRNA transfected myotubes were used as reference (PBS). Total, myofibrillar, and soluble‐enriched fractions were similarly depressed upon Dex treatment in both mock (sc‐siRNA) and si‐UBE2A transfected myotubes (P < 0.05). (D) and (E) flag‐actin and MHCI were not protected by UBE2A knockdown in the soluble fraction of fa‐C2C12 myotubes treated with Dex. (F) No effect of UBE2A knockdown was observed on α‐actin levels in the myofibrillar fraction of fa‐C2C12 myotubes. Load: membranes were stained using Blot‐FastStain dye and densitometric analysis was used to correct for uneven loading (a portion of the membrane is shown). * P < 0.05, n = 6.
Results
Expression of E2 and E3 enzymes in catabolic fa‐C2C12 myotubes
The mRNA levels of several enzymes implicated in the ubiquitination machinery were determined in myotubes subjected to Dex treatment. In accordance with previous reports, the muscle‐specific E3 ligase MuRF1 and MAFbx mRNA levels were elevated with Dex treatment (1 μM) in fa‐C2C12 myotubes (Figure 1(A) and Supporting Information Figure 1(A)). Several E2 enzymes were selected based on their mRNA abundance in skeletal muscle (NextBio body Atlas database, https://www.nextbio.com) and also on previous measurements using other catabolic situations.14, 15, 24, 29, 30, 31, 32 Using 1 μM Dex, 6 out of the 14 selected E2 enzymes exhibited enhanced mRNA levels, namely, UBE2A, UBE2B, UBE2D1, UBE2D2, UBE2G1, and UBE2J1 (Figure 1(A)). We next repeated this experiment using a milder Dex treatment in order to detect targets more sensitive to a catabolic treatment. Using 0.16 μM Dex, only MuRF1 and UBE2B mRNA levels were elevated when compared with controls (Figure 1(B)), suggesting that these enzymes were particularly sensitive and important for the establishment of a catabolic situation.
UBE2B knockdown stabilizes α‐actin and myosin heavy chain
We used the amphipathic peptide CADY for performing siRNA transfection to specifically knockdown UBE2B in fa‐C2C12 myotubes treated with Dex (1 μM). When transfection of a siRNA encoding for no known target sequence was performed (sc‐siRNA), UBE2B mRNA levels were elevated upon Dex treatment (+30%, P < 0.05, Figure 2(A)). In contrast, transfection with UBE2B‐specific siRNAs (si‐UBE2B) decreased UBE2B mRNA levels in both untreated and Dex‐treated fa‐C2C12 myotubes (−30%/−43%, P < 0.05, Figure 2(A)). Murine UBE2B and UBE2A share 73% sequence similarity at the nucleotide level and the selection of siRNAs specific for each isoform was thus cautiously performed. Furthermore, redundant function may be shared by so closed isoforms. Consequently, we tested whether UBE2B knockdown could have an effect on UBE2A mRNA levels. In our conditions, UBE2B knockdown had no effect on UBE2A mRNA levels in both control and Dex‐treated fa‐C2C12 myotubes (Figure 2(B)). We then addressed the efficiency of UBE2B knockdown on UBE2B protein levels and observed a significant decrease (−24%, Figure 2(C)). Available antibodies are not able to distinguish between UBE2A and UBE2B, so that we underestimated the actual effect of the knockdown experiment on UBE2B protein levels.
Others showed that UBE2B and the isoform UBE2A account for roughly half of the ubiquitinating activity in skeletal muscles.19 Thus, we addressed the impact of UBE2B knockdown on fa‐C2C12 protein levels. Dexamethasone treatment (1 μM) decreased similarly soluble and myofibrillar protein levels in fa‐C2C12 myotubes transfected with mock sc‐siRNA when compared with untreated controls (≈20%, P < 0.05, Figure 2(D)). Interestingly, UBE2B knockdown increased total protein levels, and this increase was only due to elevated soluble proteins. It should be noticed that in our experimental conditions (intense sonication), a fair amount of contractile proteins are present in the soluble fraction. We next addressed the identity of proteins that were spared by UBE2B knockdown. In the soluble fraction flag‐actin was depressed upon Dex treatment and, as for total proteins, transfection with the control siRNA (sc‐siRNA) did not improve flag‐actin content in the myofibrillar fraction (Figure 2(E)). However, UBE2B knockdown increased flag‐actin levels in the soluble fraction (Figure 2(E)). Similarly, MHCI levels decreased upon Dex treatment in the soluble fraction in both PBS and sc‐siRNA groups but were enhanced in fa‐C2C12 myotubes knocked down for UBE2B (Figure 2(F)). In the myofibrillar fraction, we also observed decreased flag‐actin levels (Figure 2(G)). In contrast with the soluble fraction, this decrease was not abolished by UBE2B knockdown, which is in accordance with the lack of effect of UBE2B knockdown on the myofibrillar protein fraction (Figure 2(D) and (G)).
UBE2A knockdown
As UBE2B shares 96% identity with UBE2A protein sequence, we next addressed whether UBE2A knockdown could have similar effects on protein levels. Upon UBE2A‐siRNA transfection, UBE2A mRNA levels were no longer increased in Dex‐treated fa‐C2C12 myotubes, while UBE2B expression was still elevated by Dex treatment (Figure 3(A) and (B)). Thus, UBE2A knockdown specifically reduced UBE2A mRNA levels without affecting UBE2B mRNA in both Dex‐treated and untreated fa‐C2C12 myotubes (Figure 3(A) and (B)). However, total myofibrillar and soluble protein loss was similar in control sc‐siRNA transfected cells and in UBE2A knocked‐down myotubes (Figure 3(C)). Similarly, flag‐actin and MHCI levels were not improved by siUBE2A transfection. Indeed, Dex treatment still induced decreased contractile protein levels in both the myofibrillar and soluble fractions (Figure 3(D–F)).
K48‐Ub conjugates are depressed upon UBE2B knockdown in Dex‐treated C2C12 myotubes
UBE2A and UBE2B were previously described as important contributors of polyUb conjugates in skeletal muscle.33, 34 Thus, UBE2B knockdown should have an impact on polyUb levels. In the presence of siUBE2B, we found that polyUb conjugates were depressed by 26% (NS) and 17% (P < 0.05) in the soluble fraction from PBS and Dex‐treated C2C12 myotubes, respectively (Figure 4(A)). In contrast, no significant variation was observed in the myofibrillar fraction (Supporting Information Figure 2(A)). We then addressed the impact of UBE2B knockdown on specific Ub chain linkages. Interestingly, K48 Ub conjugates were not altered by UBE2B knockdown in CT myotubes, while these conjugates were depressed by 28% in Dex‐treated myotubes (P < 0.02, Figure 4(B)). K63 Ub conjugates tended to decrease upon UBE2B knockdown in the soluble fraction from Dex‐treated myotubes (Figure 4(C), P = 0.11). No variation was observed in the myofibrillar fraction for both K48 and K63 polyUb conjugates (Supporting Information Figure 2(B) and (C)).
UBE2B and MuRF1 do not interact
Previous work showed that MuRF1 is implicated in the targeting of actin and myosin heavy chains.20, 26, 35 We thus investigated whether MuRF1 could interact with UBE2B for subsequently targeting contractile proteins. Previous studies showed that recombinant UBE2B and the E3 ligase Rad18 forms stable dimers in vitro, indicating that UBE2B‐E3 interactions may be strong enough for pulldown experiments.36 We co‐produced recombinant GST‐MuRF1 and UBE2B in E. coli and purified potential complex using the GST moiety. Co‐production in the same clone ended up with the presence of both GST‐MuRF1 and UBE2B in the supernatant (Figure 5 upper panel). Purification of GST‐MuRF1 on commercial beads was efficient, but no UBE2B was present in the eluate (Figure 5 lower panel), suggesting that the two proteins did not interact in these conditions or that the interaction was labile and/or transient. E3s generally interact with higher affinity with E2 charged with ubiquitin than with uncharged E2.37 Therefore, we added E1, ubiquitin and ATP to the bacterial lysate to charge the E2 before performing the GST‐MuRF1 pull down (see Supporting Information Figure 3 legend for details). Again, we were unable to detect an interaction between GST‐MuRF1 and UBE2B (Supporting Information Figure 3 ).
Figure 5.
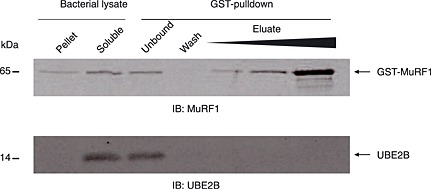
UBE2B does not exhibit affinity for the muscle‐specific E3 ligase MuRF1. Recombinant GST‐MuRF1 and UBE2B were co‐produced in BL21(DE3) Escherichia coli and GST pulldown was performed (three independent experiments). Immunoblotting (IB) revealed that GST‐MuRF1 (upper panel) and UBE2B (lower panel) were mainly present as soluble proteins (bacterial lysate, soluble). GST pulldown efficiently purified GST‐MuRF1 as witnessed by the presence of the recombinant protein in the eluate (upper panel). In contrast, UBE2B did not appear in the blot despite increasing amounts of eluate fraction, thus suggesting that UBE2B did not interact with GST‐MuRF1 in our conditions.
Discussion
We recently reported that α‐actin is a UPS substrate and that the E3 ligase MuRF1 controls the levels of α‐actin in the myofibrillar fraction during catabolic situations.26 In the past decade, most efforts were put on identifying the E3 enzymes responsible for the targeting of myofibrillar proteins, whereas few studies addressed the role of E2 enzymes in muscle wasting. In this work, we provide evidence that a limited subset of E2 enzymes is upregulated at the mRNA levels following Dex treatment. In addition, we found that actin and MHCI accumulated in the soluble fraction when UBE2B was knocked down, thus suggesting an important role of UBE2B in contractile protein homeostasis in catabolic conditions. Finally, MuRF1 did not interact with UBE2B despite the fact that these proteins negatively impact the same targets (actin and myosin), which suggests that different E2–E3 couples may act sequentially and/or concomitantly on myofibrillar protein turnover.
Specificity of protein targeting by the UPS involves two classes of enzymes, E2 ubiquitin carrier proteins and E3 ligases. Several hundreds of E3 ligases ensure the specific recognition step of the targeted proteins, while at least 35 E2 are necessary for bringing Ub to the substrates. Amongst E3s, Ring‐finger and Ring‐finger‐like E3s do not generally possess any catalytic activity and act as a platform for Ub and the substrate. Depending on the E2–E3 couple formed, the ubiquitination signal is different (mono‐Ub, poly‐Ub with different possible linkages, heterogeneous Ub‐SUMO chains, etc.) and sustains various roles, from basic degradation by the UPS to histone rearrangement during DNA repair (see Kravtsova‐Ivantsiv and Ciechanover38 for a review). Interestingly, recent reports hypothesized that it is the E2 enzyme that defines the chain type and thus the future of the substrate.6, 7, 8, 17, 39 This implicates an underestimated role of E2 enzymes in the targeting of UPS substrates and another step of complexity in the fine‐tuning of cellular proteins by this proteolytic system.
The 14‐kDa E2/Rad6B now called UBE2B was one of the first studied E2s in catabolic skeletal muscles. Together with other components of the UPS, UBE2B is highly upregulated in several models of skeletal muscle atrophy including sepsis, cancer, hindlimb suspension, fasting, and myostatin treatment.10, 11, 14, 15, 19, 21, 22, 23, 40, 41 Using catabolic cultured myotubes (1 μM Dex treatment), we found here that UBE2B was upregulated at the mRNA level together with five other E2 enzymes including its isoform UBE2A (Figure 1(A)). Interestingly, lowering Dex concentration still induced components of the UPS, but only MuRF1 (but not MAFbx, Supporting Information Figure 1 ) and UBE2B were upregulated, suggesting that UBE2B and MuRF1 are crucial actors during an atrophying situation (Figure 1(B)). Accordingly, we previously reported that UBE2B is tightly regulated during both atrophy and recovery in skeletal muscles.13, 14 In addition, the combined Ub binding capacity of UBE2A and UBE2B represent half of the total capacity of skeletal muscles, further suggesting a potential important role in skeletal muscle homeostasis.19, 33 Surprisingly, knock‐out mice for UBE2B did not exhibit any marked phenotype but a slightly smaller muscle mass and the authors hypothesized that UBE2A and UBE2B might have redundant functions in skeletal muscles.19 However, a common problem with knock out is the compensatory effect that often occurs and masks the deletion effects. We therefore addressed the role of UBE2B in atrophying myotubes using a transient knockdown approach. Using Dex treatment, total protein loss was equally distributed between soluble and myofibrillar enriched fractions in mock‐transfected myotubes, but this was completely abolished in UBE2B knockdown myotubes (Figure 2(D)). Surprisingly, soluble and myofibrillar‐enriched fractions behaved differently in knockdown myotubes, as proteins were repeatedly (i.e. in three independent experiments) spared in the soluble fraction but not in the myofibrillar enriched fraction (Figure 2(D)). Amongst the proteins that accumulated upon UBE2B knockdown, flag‐actin and MHCI were significantly spared. It should be noticed that the soluble fraction was obtained following intense sonication of myotube homogenates and that all the loosely attached myofibrillar proteins, for example, the easily releasable myofilaments42 were probably present in this fraction. Easily releasable myofilaments may be either degradation intermediates or recently translated proteins rapidly degraded. This means that we cannot distinguish between monomeric and partially unstructured myofibrillar components in our assays. In addition, UBE2B knockdown mainly affected K48‐linked Ub chains (−28%) that represent bona fide proteasome substrates. Altogether, this suggests that UBE2B possesses an important role in the processing of the myofibrillar apparatus upon Dex treatment. UBE2B shares 96% identity at the protein level with the isoform UBE2A, and redundant function was hypothesized. In our assays, UBE2A mRNAs were unaffected by UBE2B knockdown, suggesting that the two isoforms were differentially regulated. This was confirmed by UBE2A knockdown that was unable to accumulate total protein, flag‐actin, or MHCI in both control and Dex‐treated myotubes (Figure 3(C)–(F)). For the first time in skeletal muscle cells, we found that a given E2 has an impact on a peculiar subset of proteins and that this E2 acts preferentially (at least quantitatively) in the soluble fraction.
UBE2B and the E3 ligase UBR2 are known to mediate transcriptional silencing via histone H2A ubiquitination in testis,43 and when combined with the UBR1 E3 ligase, UBE2B is implicated in the ubiquitination of myc proteins.44 In Hela cells, UBE2B and Mdm2 target p53 for degradation by the proteasome,45 while UBE2B interacts with the E3 ligase Rad18 and is implicated in meiotic functions in testis46 and DNA repair through PCNA ubiquitylation.36 Last, but not least, UBE2B‐mediated histone (H2B) ubiquitylation in conjunction with hBRE1 E3 ligase is responsible for Histone H3 methylation and subsequent increased transcriptional activity in Hela cells.47 Thus, UBE2B is a good example of the multiplicity of action of a single E2 depending on the cellular context. However, and surprisingly, no physiological target has been identified so far in skeletal muscle for UBE2B despite the high mRNA abundance and the important upregulation of UBE2B observed in this organ during catabolic situations. The role of UBE2B on actin and MHCI levels might be either direct or indirect, but these targets represent the first muscle proteins being impacted by UBE2B activity.
As an E3 serves as a platform for bringing E2s and substrates in the vicinity, interaction between E2s and substrates are rather limited to a couple of residues, so that reconstitution of the trio E2–E3–substrate is the only way for deciphering protein degradation mechanisms. UBE2B collaborates at least with two members of the UBR E3 family (UBR1 and UBR2). In our conditions, we found that both UBR1 and UBR2 were overexpressed with high levels of Dex (1 μM) but were not responsive to mild catabolic conditions using 0.16 μM Dex (Supporting Information Figure 1(B)). In addition, the role of UBR1 and UBR2 during skeletal muscle atrophy is still controversial as they are mildly upregulated at the mRNA levels in different catabolic situations (diabetes, cancer, and sepsis) but do not seem to be involved in the atrophying programme of catabolic skeletal muscles.9, 48, 49, 50, 51 We thus focused on MuRF1 that was previously demonstrated to target actin and MHCs in myotubes.12, 20, 35 In our conditions, MuRF1 and UBE2B were equally responsive to low doses of Dex (Figure 1) but these UPS enzymes did not interact (Figure 5). Phosphorylation of UBE2B impacts its affinity for E3 ligases,25 so that we cannot rule out that phosphorylation may be needed for efficient MuRF1–UBE2B binding. However, phospho‐mimic UBE2B induced a decrease in binding affinity towards UBR1/E3α ligase but not a total blockade,25 which obviously could not explain the lack of affinity between UBE2B and MuRF1 in our assays. Moreover, this lack of interaction between UBE2B and MuRF1 is in agreement with former data showing that MuRF1 was implicated in the targeting of α‐actin present in the myofibrillar‐enriched fraction,12 while present data indicate a role of UBE2B on the soluble α‐actin content (Figure 2). More work is clearly needed for identifying the E3 ligase that recognizes actin for subsequent ubiquitination by UBE2B.
E2s and E3s act in concert for specifically targeting proteins for degradation. This means that identifying physiologically relevant E2–E3–substrates combinations may provide new therapeutical strategies for fighting against diseases. We provide here evidence that UBE2B is one of the six main E2s recruited during highly catabolic situations and that UBE2B affects a major fraction of cellular proteins including actin and myosin heavy chain. However, this action is probably independent from MuRF1 and restricted to the soluble compartment. Future work will have to fully identify UBE2B‐E3 and E2‐MuRF1 couples responsible in vivo for myofibrillar protein targeting.
Conflict of interest
Cécile Polge, Roza Leulmi, Marianne Jarzaguet, Agnes Claustre, Lydie Combaret, Daniel Béchet, Anne‐Elisabeth Heng, Didier Attaix, and Daniel Taillandier declare that they have no conflict of interest.
Abbreviations
- Ub
ubiquitin
- E1
ubiquitin‐activating enzyme
- E2
ubiquitin‐conjugating enzyme
- E3
ubiquitin‐protein ligase
- UPS
ubiquitin proteasome system
Supporting information
Supporting info item
Supporting info item
Supporting info item
Acknowledgements
The authors are grateful to Dr. May Morris for providing the CADY peptide used for the knockdown experiments. This work was supported by grants to D. T., C. P., and/or D. A. from the Institut National de la Recherche Agronomique and the Association Française contre les Myopathies.
The authors certify that they comply with the ethical guidelines for authorship and publishing of the Journal of Cachexia, Sarcopenia and Muscle (von Haehling S, Morley JE, Coats AJS, Anker SD. Ethical guidelines for authorship and publishing in the Journal of Cachexia, Sarcopenia and Muscle. J Cachexia Sarcopenia Muscle. 2010;1:7–8.)
Polge, C. , Leulmi, R. , Jarzaguet, M. , Claustre, A. , Combaret, L. , Béchet, D. , Heng, A. ‐E. , Attaix, D. , and Taillandier, D. (2016) UBE2B is implicated in myofibrillar protein loss in catabolic C2C12 myotubes. Journal of Cachexia, Sarcopenia and Muscle, 7: 377–387. doi: 10.1002/jcsm.12060.
References
- 1. Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin‐proteasome system. Nat Rev Mol Cell Biol 2008;9:679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ciechanover A, Stanhill A. The complexity of recognition of ubiquitinated substrates by the 26S proteasome. Biochim Biophys Acta 2014;1843:86–96. [DOI] [PubMed] [Google Scholar]
- 3. Polge C, Uttenweiler‐Joseph S, Leulmi R, Heng AE, Burlet‐Schiltz O, Attaix D, et al. Deciphering the ubiquitin proteome: limits and advantages of high throughput global affinity purification‐mass spectrometry approaches. Int J Biochem Cell Biol 2013;45:2136–2146. [DOI] [PubMed] [Google Scholar]
- 4. Sandri M. Protein breakdown in muscle wasting: role of autophagy‐lysosome and ubiquitin‐proteasome. Int J Biochem Cell Biol 2013;45:2121–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Polge C, Heng AE, Combaret L, Bechet D, Taillandier D, Attaix D. Recent progress in elucidating signalling proteolytic pathways in muscle wasting: potential clinical implications. Nutrition, metabolism, and cardiovascular diseases: NMCD 2013;23:S1–S5. [DOI] [PubMed] [Google Scholar]
- 6. David Y, Ziv T, Admon A, Navon A. The E2 ubiquitin‐conjugating enzymes direct polyubiquitination to preferred lysines. J Biol Chem 2010;285:8595–8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Napolitano LM, Jaffray EG, Hay RT, Meroni G. Functional interactions between ubiquitin E2 enzymes and TRIM proteins. Biochem J 2011;434:309–319. [DOI] [PubMed] [Google Scholar]
- 8. van Wijk SJ, Timmers HT. The family of ubiquitin‐conjugating enzymes (E2s): deciding between life and death of proteins. Faseb J 2010;24:981–993. [DOI] [PubMed] [Google Scholar]
- 9. Lecker SH, Solomon V, Price SR, Kwon YT, Mitch WE, Goldberg AL. Ubiquitin conjugation by the N‐end rule pathway and mRNAs for its components increase in muscles of diabetic rats. J Clin Invest 1999;104:1411–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lin SY, Chen WY, Lee FY, Huang CJ, Sheu WH. Activation of ubiquitin‐proteasome pathway is involved in skeletal muscle wasting in a rat model with biliary cirrhosis: potential role of TNF‐alpha. Am J Physiol Endocrinol Metab 2005;288:E493–501. [DOI] [PubMed] [Google Scholar]
- 11. McFarlane C, Plummer E, Thomas M, Hennebry A, Ashby M, Ling N, et al. Myostatin induces cachexia by activating the ubiquitin proteolytic system through an NF‐kappaB‐independent, FoxO1‐dependent mechanism. J Cell Physiol 2006;209:501–514. [DOI] [PubMed] [Google Scholar]
- 12. Polge C, Heng AE, Jarzaguet M, Ventadour S, Claustre A, Combaret L, et al. Muscle actin is polyubiquitinylated in vitro and in vivo and targeted for breakdown by the E3 ligase MuRF1. Faseb J 2011;25:3790–3802. [DOI] [PubMed] [Google Scholar]
- 13. Taillandier D, Aurousseau E, Combaret L, Guezennec CY, Attaix D. Regulation of proteolysis during reloading of the unweighted soleus muscle. Int J Biochem Cell B 2003;35:665–675. [DOI] [PubMed] [Google Scholar]
- 14. Taillandier D, Aurousseau E, MeynialDenis D, Bechet D, Ferrara M, Cottin P, et al. Coordinate activation of lysosomal, Ca2+‐activated and ATP‐ubiquitin‐dependent proteinases in the unweighted rat soleus muscle. Biochem J 1996;316:65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Voisin L, Breuille D, Combaret L, Pouyet C, Taillandier D, Aurousseau E, et al. Muscle wasting in a rat model of long‐lasting sepsis results from the activation of lysosomal, Ca2+‐activated, and ubiquitin‐proteasome proteolytic pathways. J Clin Invest 1996;97:1610–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Polge C, Attaix D, Taillandier D. Role of E2‐Ub‐conjugating enzymes during skeletal muscle atrophy. Front Physiol 2015;6:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 2009;10:755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, et al. Certain pairs of ubiquitin‐conjugating enzymes (E2s) and ubiquitin‐protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem 2007;282:17375–17386. [DOI] [PubMed] [Google Scholar]
- 19. Adegoke OAJ, Bedard N, Roest HP, Wing SS. Ubiquitin‐conjugating enzyme E2(14 k)/HR6B is dispensable for increased protein catabolism in muscle of fasted mice. Am J Physiol‐Endoc M 2002;283:E482–E489. [DOI] [PubMed] [Google Scholar]
- 20. Clarke BA, Drujan D, Willis MS, Murphy LO, Corpina RA, Burova E, et al. The E3 Ligase MuRF1 degrades myosin heavy chain protein in dexamethasone‐treated skeletal muscle. Cell Metab 2007;6:376–385. [DOI] [PubMed] [Google Scholar]
- 21. Combaret L, Tilignac T, Claustre A, Voisin L, Taillandier D, Obled C, et al. Torbafylline (HWA 448) inhibits enhanced skeletal muscle ubiquitin‐proteasome‐dependent proteolysis in cancer and septic rats. Biochem J 2002;361:185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mansoor O, Beaufrere B, Boirie Y, Ralliere C, Taillandier D, Aurousseau E, et al. Increased mRNA levels for components of the lysosomal, Ca2+‐activated, and ATP‐ubiquitin‐dependent proteolytic pathways in skeletal muscle from head trauma patients. Proc Natl Acad Sci U S A 1996;93:2714–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Temparis S, Asensi M, Taillandier D, Aurousseau E, Larbaud D, Obled A, et al. Increased ATP‐ubiquitin‐dependent proteolysis in skeletal‐muscles of tumor‐bearing rats. Cancer Res 1994;54:5568–5573. [PubMed] [Google Scholar]
- 24. Wing SS, Banville D. 14‐kDa ubiquitin‐conjugating enzyme: structure of the rat gene and regulation upon fasting and by insulin. Am J Physiol 1994;267:E39–E48. [DOI] [PubMed] [Google Scholar]
- 25. Kumar B, Lecompte KG, Klein JM, Haas AL. Ser(120) of Ubc2/Rad6 regulates ubiquitin‐dependent N‐end rule targeting by E3{alpha}/Ubr1. J Biol Chem 2010;285:41300–41309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Polge C, Heng AE, Jarzaguet M, Ventadour S, Claustre A, Combaret L, et al. Muscle actin is polyubiquitinylated in vitro and in vivo and targeted for breakdown by the E3 ligase MuRF1. Faseb J 2011;25:3790–3802. [DOI] [PubMed] [Google Scholar]
- 27. Wyckoff EE, Phillips JD, Sowa AM, Franklin MR, Kushner JP. Mutational analysis of human uroporphyrinogen decarboxylase. Biochim Biophys Acta 1996;1298:294–304. [DOI] [PubMed] [Google Scholar]
- 28. Ventadour S, Jarzaguet M, Wing SS, Chambon C, Combaret L, Bechet D, et al. A new method of purification of proteasome substrates reveals polyubiquitination of 20S proteasome subunits. J Biol Chem 2007;282:5302–5309. [DOI] [PubMed] [Google Scholar]
- 29. Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, et al. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. Faseb J 2004;18:39–51. [DOI] [PubMed] [Google Scholar]
- 30. Banduseela VC, Ochala J, Chen YW, Goransson H, Norman H, Radell P, et al. Gene expression and muscle fiber function in a porcine ICU model. Physiol Genomics 2009;39:141–159. [DOI] [PubMed] [Google Scholar]
- 31. Wittwer M, Fluck M, Hoppeler H, Muller S, Desplanches D, Billeter R. Prolonged unloading of rat soleus muscle causes distinct adaptations of the gene profile. Faseb J 2002;16:884. [DOI] [PubMed] [Google Scholar]
- 32. Pattison JS, Folk LC, Madsen RW, Childs TE, Booth FW. Transcriptional profiling identifies extensive downregulation of extracellular matrix gene expression in sarcopenic rat soleus muscle. Physiol Genomics 2003;15:34–43. [DOI] [PubMed] [Google Scholar]
- 33. Rajapurohitam V, Bedard N, Wing SS. Control of ubiquitination of proteins in rat tissues by ubiquitin conjugating enzymes and isopeptidases. Am J Physiol‐Endoc M 2002;282:E739–E745. [DOI] [PubMed] [Google Scholar]
- 34. Solomon V, Lecker SH, Goldberg AL. The N‐end rule pathway catalyzes a major fraction of the protein degradation in skeletal muscle. J Biol Chem 1998;273:25216–25222. [DOI] [PubMed] [Google Scholar]
- 35. Fielitz J, Kim MS, Shelton JM, Latif S, Spencer JA, Glass DJ, et al. Myosin accumulation and striated muscle myopathy result from the loss of muscle RING finger 1 and 3. J Clin Invest 2007;117:2486–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Notenboom V, Hibbert RG, van Rossum‐Fikkert SE, Olsen JV, Mann M, Sixma TK. Functional characterization of Rad18 domains for Rad6, ubiquitin, DNA binding and PCNA modification. Nucleic Acids Res 2007;35:5819–5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Siepmann TJ, Bohnsack RN, Tokgoz Z, Baboshina OV, Haas AL. Protein interactions within the N‐end rule ubiquitin ligation pathway. J Biol Chem 2003;278:9448–9457. [DOI] [PubMed] [Google Scholar]
- 38. Kravtsova‐Ivantsiv Y, Ciechanover A. Non‐canonical ubiquitin‐based signals for proteasomal degradation. J Cell Sci 2012;125:539–548. [DOI] [PubMed] [Google Scholar]
- 39. Wenzel DM, Stoll KE, Klevit RE. E2s: structurally economical and functionally replete. Biochem J 2011;433:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Samuels SE, Taillandier D, Aurousseau E, Cherel Y, LeMaho Y, Arnal M, et al. Gastrointestinal tract protein synthesis and mRNA levels for proteolytic systems in adult fasted rats. Am J Physiol‐Endoc M 1996;271:E232–E238. [DOI] [PubMed] [Google Scholar]
- 41. Lecker SH, Solomon V, Price SR, Kwon YT, Mitch WE, Goldberg AL. Ubiquitin conjugation by the N‐end rule pathway and mRNAs for its components increase in muscles of diabetic rats. J Clin Invest 1999;104:1411–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Neti G, Novak SM, Thompson VF, Goll DE. Properties of easily releasable myofilaments: are they the first step in myofibrillar protein turnover? Am J Physiol‐Cell Ph 2009;296:C1383–C90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. An JY, Kim EA, Jiang Y, Zakrzewska A, Kim DE, Lee MJ, et al. UBR2 mediates transcriptional silencing during spermatogenesis via histone ubiquitination. Proc Natl Acad Sci U S A 2010;107:1912–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gross‐Mesilaty S, Reinstein E, Bercovich B, Tobias KE, Schwartz AL, Kahana C, et al. Basal and human papillomavirus E6 oncoprotein‐induced degradation of Myc proteins by the ubiquitin pathway. Proc Natl Acad Sci U S A 1998;95:8058–8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen S, Wang DL, Liu Y, Zhao L, Sun FL. RAD6 regulates the dosage of p53 by a combination of transcriptional and posttranscriptional mechanisms. Mol Cell Biol 2012;32:576–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Inagaki A, Sleddens‐Linkels E, Wassenaar E, Ooms M, van Cappellen WA, Hoeijmakers JH, et al. Meiotic functions of RAD18. J Cell Sci 2011;124:2837–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kim J, Guermah M, McGinty RK, Lee JS, Tang ZY, Milne TA, et al. RAD6‐mediated transcription‐coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 2009;137:459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Krawiec BJ, Nystrom GJ, Frost RA, Jefferson LS, Lang CH. AMP‐activated protein kinase agonists increase mRNA content of the muscle‐specific ubiquitin ligases MAFbx and MuRF1 in C2C12 cells. Am J Physiol Endocrinol Metab 2007;292:E1555–E1567. [DOI] [PubMed] [Google Scholar]
- 49. An JY, Seo JW, Tasaki T, Lee MJ, Varshavsky A, Kwon YT. Impaired neurogenesis and cardiovascular development in mice lacking the E3 ubiquitin ligases UBR1 and UBR2 of the N‐end rule pathway. Proc Natl Acad Sci U S A 2006;103:6212–62127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wray CJ, Sun X, Gang GI, Hasselgren PO. Dantrolene downregulates the gene expression and activity of the ubiquitin‐proteasome proteolytic pathway in septic skeletal muscle. J Surg Res 2002;104:82–87. [DOI] [PubMed] [Google Scholar]
- 51. Zhang G, Lin RK, Kwon YT, Li YP. Signaling mechanism of tumor cell‐induced up‐regulation of E3 ubiquitin ligase UBR2. Faseb J 2013;27:2893–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting info item
Supporting info item
Supporting info item


