Abstract
As a neutrophilic bacterium, Helicobacter pylori is growth deficient under extreme acidic conditions. The gastric pathogen is equipped with an acid survival kit, regulating urease activity by a pH-gated urea channel, opening below pH 6.5. After overcoming acid stress, the bacterium’s multiplication site is situated at the gastric mucosa with near neutral pH. The pathogen exhibits exceptional genetic variability, mainly due to its capability of natural transformation, termed competence. Using single cell analysis, we show here that competence is highly regulated in H. pylori. DNA uptake complex activity was reversibly shut down below pH 6.5. pH values above 6.5 opened a competence window, in which competence development was triggered by the combination of pH increase and oxidative stress. In contrast, addition of sublethal concentrations of the DNA-damaging agents ciprofloxacin or mitomycin C did not trigger competence development under our conditions. An oxygen-sensitive mutant lacking superoxide dismutase (sodB) displayed a higher competent fraction of cells than the wild type under comparable conditions. In addition, the sodB mutant was dependent on adenine for growth in broth and turned into non-cultivable coccoid forms in its absence, indicating that adenine had radical quenching capacity. Quantification of periplasmically located DNA in competent wild type cells revealed outstanding median imported DNA amounts of around 350 kb per cell within 10 min of import, with maximally a chromosomal equivalent (1.6 Mb) in individual cells, far exceeding previous amounts detected in other Gram-negative bacteria. We conclude that the pathogen’s high genetic diversity is a consequence of its enormous DNA uptake capacity, triggered by intrinsic and extrinsic oxidative stress once a neutral pH at the site of chronic host colonization allows competence development.
Author Summary
Natural transformation, i.e. the capacity to take up DNA from the environment, is one of the crucial means for horizontal gene transfer and genetic diversity in bacteria. The human gastric pathogen Helicobacter pylori is confronted with acid stress before entering its multiplication site, the gastric mucosa. The bacterium causes lifelong chronic gastritis and is perfectly adapted to the human host, crucially by displaying unusual genetic diversity. Using a single cell approach and well-controlled conditions, we show here that the amount of imported DNA in competent H. pylori is outstanding, far exceeding previous measurement with other Gram-negative bacteria. Furthermore, DNA uptake activity was tightly regulated and limited to pH above 6.5, conditions thought to be met in close contact with the gastric mucosa. In addition, we show that within this pH competence window, competence development was triggered by an increase in pH in combination with the level of oxidative stress. Our data provide explanations for the extraordinary high genetic diversity, often referred to as genome plasticity of this unusual microaerobic pathogen.
Introduction
The human stomach is a hostile niche. After overcoming the acid barrier, bacterial colonization at the gastric mucosa represents a constant battle with the host immune system. In particular, the pathogen is confronted with ROS-mediated oxidative stress produced by NADPH oxidases that release superoxide (O2 -) [1]. Progression of disease towards carcinogenesis is also accompanied by DNA damage of host cells [2, 3]. Helicobacter pylori are genetically highly variable, microaerobic Gram-negative bacteria that successfully colonize the human gastric mucosa of half the world’s population, with variable prevalence in different human populations [4]. The bacterium causes lifelong gastritis and is also associated with mucosal-associated lymphoma and gastric adenocarcinoma [5]. H. pylori is a neutrophilic organism, but highly adapted to acid survival. For this purpose, the pathogen has a potent urease enzyme, which is tightly regulated via substrate accessibility by a pH-regulated channel [6, 7]. The microaerobic pathogen is also equipped with a set of oxidative stress enzymes, including catalase (KatA), alkyl hydroperoxide reductase (AhpC) and superoxide dismutase (SodB), which are important for host colonization [8–10]. H. pylori is not only exceptional regarding its colonization capacity of a harsh environment, but also regarding its enormous genetic diversity, crucially based on horizontal gene transfer and high recombination frequency [11]. In H. pylori horizontal gene transfer can be accomplished by natural transformation [12], i. e. the capacity to take up naked DNA from the environment. However, also direct exchange of DNA between H. pylori cells by conjugation-like mechanisms were shown to occur [13–15] and were discussed to play a yet underestimated role in genetic diversity of the gastric pathogen [16]. In contrast to all other known bacteria, DNA uptake in H. pylori during natural transformation is established by a type IV secretion system that is encoded by two separate operons, comB2-B4 and comB6-B10 [12, 17]. By tracking fluorescent DNA during uptake, we previously showed that DNA uptake in H. pylori is a two-step process [18]. The ComB system mediated transport of external dsDNA over the outer membrane into the periplasm. Our data also indicated that the conserved inner membrane localized ComEC channel, previously described by [19, 20] is involved in subsequent transport of probably ssDNA into the cytoplasm of H. pylori [18]. Monitoring fluorescent DNA during natural transformation was subsequently applied to other Gram-negative bacteria, like Vibrio and Neisseria [21, 22]. A maximum of 40 kb of imported DNA was detected in the bacterial periplasmic space of Neisseria, limited by the binding capacity of the periplasmic DNA-binding protein ComE (ComEA homologue) [21], which is absent in H. pylori [23].
In most bacteria, the capacity for natural transformation is tightly regulated, by timely restricting competence phase and/or by preferential uptake of DNA from related species. Quorum sensing for induction of competent state enables Bacillus subtilis, Streptococcus pneumoniae and Vibrio cholerae to increase the chance of import of DNA from siblings [24–26]. In Streptococcus pneumoniae, natural transformation was additionally triggered by antibiotics or DNA-damaging agents [27]. In Vibrio competence is also regulated by the presence of the natural habitat marker chitin and regulatory circuits appeared to be interconnected [28]. In Neisseria species-specific DNA uptake sequences [29] recognized by an outer membrane receptor [30] select for DNA of related bacteria. A DNA uptake sequence also facilitated DNA uptake in Haemophilus influenza [31], with variants of this sequence and core bases recently detailed by a next generation sequencing approach [32]. Furthermore, H. influenzae was suggested to restrict competence state to conditions of starvation [33].
A previous study implicated also sequence bias to occur in H. pylori natural transformation [34]–probably at the level of recombination. However, single cell analysis clearly showed that H. pylori did not discriminate between own and foreign DNA at the level of DNA uptake [18], which confirmed previous predictions of absence of redundant sequence motifs in the gastric pathogen [35]. In addition, natural transformation was long thought to occur constitutively in H. pylori. However, multiple competence phases and strain-dependent differences in natural transformation were described [36]. A study suggested that DNA damage exerted by the fluoroquinolone ciprofloxacin triggered natural transformation [37]. Interestingly, increased transformation rate was observed for H. pylori grown on agar plates incubated at slightly increased pH or different atmospheric composition [38].
Using a single cell approach, tracking fluorescent DNA in competent bacteria, we show here that H. pylori tightly regulates its competence state. A pH above 6.5 opened a window of opportunity, in which competence development was triggered by further increase in pH in combination with oxidative stress. Furthermore, single cell quantification of imported DNA provided evidence of a stunning DNA uptake activity, providing an explanation for the pathogen’s outstanding genetic diversity.
Results
Natural transformation is dynamically regulated in H. pylori
In previous studies H. pylori competence was usually quantified by monitoring the expression of a resistance marker after contact with external DNA [36, 39]. This assay is not only dependent on the competent state but also on downstream effects like timely expression of the cassette and survival of the microaerobic bacterium under selection conditions. In addition to resistance marker expression, the transcription of a subset of genes essential for natural transformation was characterized in other studies [37, 38]. Here we directly monitor active DNA uptake by covalently labelling λ DNA with a Cy3 fluorophore and microscopically tracking DNA import in single cells. We showed previously that active DNA uptake of covalently modified DNA, such as Cy3-labelled λ DNA, occurred over the outer membrane at distinct locations and that DNA was trapped in the periplasm [18]. In this study, we wanted to use the assay for direct visualization of the competent state of H. pylori. Therefore, we first checked that the fraction of cells with incorporated fluorescent DNA, i. e. with at least one DNA focus, corresponding to active DNA uptake activity over the outer membrane, correlated with transformation rate. Thus, in parallel to the fluorescent assay counting cells with active DNA uptake activity, a second aliquot of the same cell suspension was incubated with a 1,022 kb DNA fragment harboring the rpsL(A128G) mutation for streptomycin resistance in order to measure natural transformation rate. We took cells suspensions from different growth phases before, during and after competence development as described below. As expected, the number of cells with one or more DNA foci correlated exponentially with the log number of transformants. Transformation rate ranged between 2 x 10−6 and 2 x 10-1 relative to total CFU (Fig 1). The lower limit of counted bacteria with active DNA transport was 0.2% (n ≥ 500 cells). When 10% cells with DNA foci were detected, transformation rate was around 1% (10-2). At a competent cell fraction of approximately 30% we observed a saturation of transformation rate of around 10−1 in H. pylori N6. As control and previously published for H. pylori strain J99 [18], N6 mutants with a defect in one of the essential proteins of the ComB system (comB4 or comB6, [12, 17]) were impaired for natural transformation and displayed absence of active DNA uptake. These data reveal an enormous range of competence status in H. pylori spanning an order of log 5. The dynamics were controlled by variation of the amount of cells expressing active DNA uptake complexes. Hence, our assay of direct measurement of active DNA uptake was proven suitable for monitoring competence development.
Fig 1. Transformation rate in H. pylori correlated with the fraction of cells with active outer membrane DNA transport.
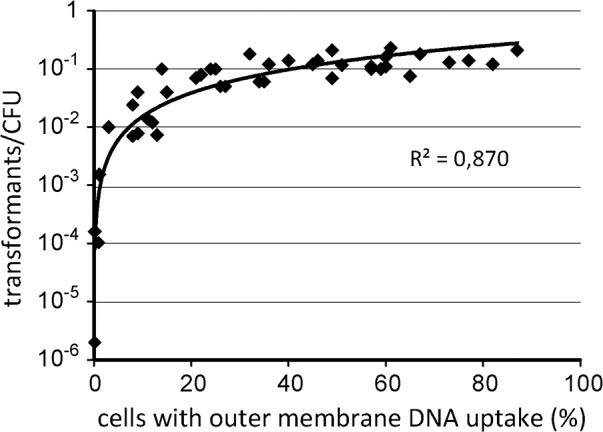
Cell suspensions were derived from the experiments illustrated in Fig 4. Bacteria with different competent fractions of cells were taken during or after competence development; non-competent cells were taken before competence development. H. pylori N6 were incubated in parallel with either Cy3-labelled λ DNA or a PCR fragment of rpsL(A128G), conferring streptomycin resistance. The data show the principle suitability of measuring the fraction of cells with active outer membrane DNA transport for the evaluation of competence development. Cumulative data from 13 independent experiments (n = 42).
Please note that highly transformable bacteria (transformation rate 10−1 to 10−2 tested using the streptomycin resistance point mutation marker) showed reduced transformation rate, when other resistance markers were used. When these cells were transformed in parallel with a chloramphenicol resistance cassette flanked by approximately 500 bp of homologous up- and downstream regions, 7 to 170-fold (median of 13-fold, n = 8) reduced transformation rates were observed. As expected, the transformation rate of the replicative vector pILL2157 [40] was log 3.8–6.4 less (median of log 5.9, n = 6) compared to the point mutation marker. Homologous recombination can occur in very short homologous regions [39]. Therefore, the chance for integrating a point mutation marker as compared to a complete resistance cassette is more likely, considering genetic barriers, in particular, restriction modification systems. A replicative plasmid has to reconstitute completely in the cytoplasm after single strand DNA uptake. Thus, the likeliness for full reconstitution of a plasmid is even less, compared to successful recombination of a resistance cassette with homology regions. Therefore, the data indicate that these highly transformable bacteria had substantial and to some extent variable genetic barriers that reduced the integrity of incoming DNA. These comprise, in particular, restriction modification systems, which were previously studied [41]. It also confirmed that the point mutation marker most faithfully detected DNA uptake.
H. pylori competent cells import huge amounts of DNA within short time
For H. pylori the term “panmictic population structure” was used to express its enormous genetic variability [42]. We wondered if DNA import activity was also unusually high. For this purpose, we exploited the fact that H. pylori imports DNA independent of its source [18] and used DNA of the bacteriophage λ as standard for a 48.5 kb molecule. As mentioned above, we covalently labelled λ DNA with Cy3. In order to visualize the DNA molecules in a spread out way, a drop of DNA was added on an agarose pad and spread over the surface under concurrent air flow. Subsequently, the pad was sealed by a cover slip and mounted for microscopy. Under our experimental conditions, we expected that the chance for fully spread out λ DNA molecules is low. Hence, total fluorescence intensity of single Cy3 λ DNA molecules with a minimal length of ½ λ DNA (equals 8.6 μm, [43], Fig 2A) was determined from at least n = 120 molecules. This cut-off minimized the risk to quantify eventual breakage products and can maximally lead to a theoretical error of a factor two. However, the standard deviation of the measured fluorescence intensities was between 20.2% and 21.3%, confirming that we indeed measured mainly complete single molecules. Bleaching controls and linearity tests with different exposure times revealed high stability of the Cy3 fluorophore and guaranteed that bleaching was negligible under the experimental conditions (S1 Appendix, Fig A). Microaerobically grown competent H. pylori were challenged with the same lot of Cy3 λ DNA (1 μg/ml) in TSB-FBS for 10 min under microaerobic conditions at 37°C before DNaseI treatment for 5 min was performed (Fig 2B).
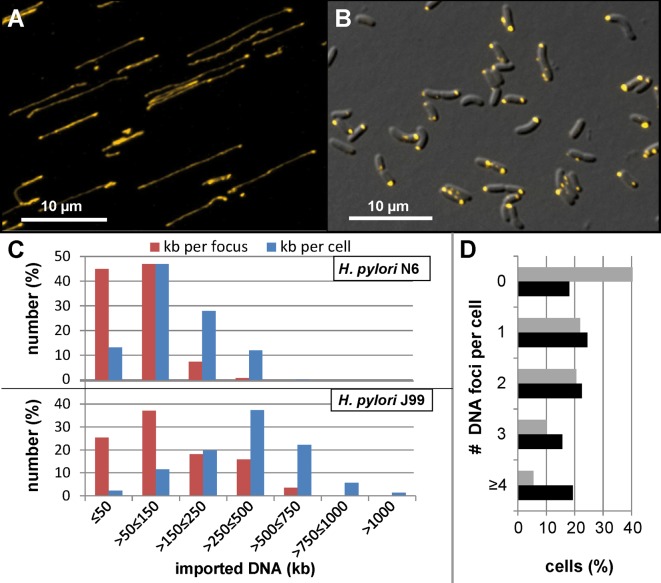
Fig 2. Quantification of imported DNA into H. pylori cells.
A, Cy3 image of single Cy3-labelled λ DNA molecules, stained in yellow; B, DIC/Cy3 overlay image of H. pylori J99 after 10 min of DNA uptake under microaerobic conditions; C, results of the quantitative analysis of DNA uptake. For H. pylori, fluorescence intensity in single DNA foci (red) and in whole cells (blue) was determined in two different wild type strains (N6 and J99) after microaerobic growth in TSB-FBS into competent phase. D, histogram of the number of foci per cell. Grey bars, strain N6; black bars, strain J99. Fluorescence intensity was compared to that of single λ DNA molecules and expressed as imported kb. The images were acquired using a Zeiss Axio Observer Z1 and ZEN software, blue edition, version 2.0.0.0. ncells = 928 or 1483 and nfoci = 1903 or 1740 were analyzed for J99 or N6, respectively.
The amount of imported DNA was extraordinarily high in H. pylori cells. For strain J99, most of the cells had imported more than 250 kb (Fig 2C). We calculated a median value of 108 kb per DNA focus and 351 kb per cell. The maximum DNA amount per focus was 758 kb and 1.6 Mb per cell. Nearly 20% of all cells harbored 4 or more locally distinct DNA foci (Fig 2D). Hence, individual cells were capable of import of a chromosomal equivalent within short periods of time. The values for strain N6 were somewhat lower but still impressive. Here, most of the cells imported more than 50 kb, with a median of 124 kb. The median DNA amount in single foci was 55 kb. Maximum import of 392 kb per focus and 507 kb per cell were observed in strain N6. Thus, in contrast to other Gram-negative bacteria [21], the periplasm of H. pylori had a manifold higher maximal capacity for imported DNA. We analysed whether the DNA packaging density within single foci was also outstanding. Assuming that the fluorescence intensity in the analyzed regions of interest (ROIs) roughly correlated with the localization of DNA, we estimated the packaging density of periplasmic DNA by measuring DNA focus area. As a result, most of the imported DNA was packed in an area of 0.1–0.5 μm2 per λ molecule (S1 Appendix, Fig B). Minimally, we detected an area of 0.04 μm2 in J99. Assuming a spherical dimension of a DNA focus, this maximum package density corresponded to 0.006 μm3 per λ molecule, a DNA density of around 100-fold less than established in the icosahedron capsid of bacteriophage λ with an inner diameter of 55 nm [44].
Imported DNA cannot complement purine deficiency
Since in other bacteria, DNA uptake was suggested to putatively play a role as nutrient supply, in particular in purine acquisition [45], we checked whether H. pylori competent bacteria can use external DNA as a purine source. For this purpose, H. pylori was grown in TSB-FBS and it was checked that at least 50% of the cells exhibited competence before the medium was exchanged for minimal medium without any purine source but with a supplementation of 10 μg/ml of DNA (H. pylori N6 genomic DNA or salmon sperm DNA). As control, growth in minimal medium was restored by addition of at least 5 μg/ml adenine (corresponding to 37 μM) and higher adenine levels (50 μg/ml) did not lead to enhanced growth (S1 Appendix, Fig C). However, when we added genomic DNA of H. pylori or of salmon sperm, the competent bacteria were not able to grow in minimal medium lacking adenine. A substantial fraction of bacteria was still capable to import DNA after 18–24 h of incubation in minimal medium as checked by transient detection of periplasmic DNA that was non-covalently stained by YOYO-1 and loss of signal within 60 min of incubation in the presence of external DNase (S1 Appendix, Fig D). Using a mutant capable of DNA import into the periplasm but deficient of DNA import into the cytoplasm (ΔcomEC), the YOYO-1 signal had been shown to be rather stable in the periplasm of this mutant, thus, suggesting that complete loss of YOYO-1 signal in the wild type was due to import of DNA into the cytoplasm [18]. In addition, import of DNA into the cytoplasm was indirectly shown by transformability of the cells after incubation in minimal medium for 18–24 h using the rpsL(A128G) point mutation marker (transformation rate between 2 x 10−2 and 1.4 x 10−1). The results indicate that DNA was imported but could not complement for purine deficiency.
Activity of DNA uptake complexes is reversibly shut down at slightly acidic pH
H. pylori encounters different pH values in its habitat, the human stomach. We tested the pH dependency of DNA uptake. The pH of BB-FBS was titrated at 37°C with HCl or NaOH to values between 5.0 and 7.5 in 0.5 unit steps and pH stability of these media was confirmed to be within ± 0.1 pH units. In order to keep the pH constant, the 10 min incubation was performed at 37°C under aerobic atmosphere, since higher CO2 concentrations used for microaerobic conditions led to pH decrease (see below). We started from a competent cell suspension exhibiting 63% ± 7.4% competent cells, which we defined as 100% (Fig 3). These values were obtained by incubation of the cells under our standard DNA uptake conditions in TBS-FBS at 37°C, except that the incubation was performed under aerobic conditions (when TSB-FBS exhibits pH 7.5). Changing the medium to BB-FBS with a pH of 7.5 demonstrated that medium did not influence the fraction of competent cells. As illustrated in Fig 3, the activity of DNA uptake was highly sensitive towards slightly acidic pH values. Below pH 6.5, DNA uptake complexes shut down activity and were practically inactive at pH 5.5. Interestingly, preincubation at pH 5.0 for 10 min and subsequent shift to pH 7.5 reactivated DNA uptake in H. pylori (Fig 3). Hence, we conclude that a neutral pH is a prerequisite of DNA uptake activity in H. pylori. In addition, the pH dependent regulation was tight, impairing DNA uptake at slight acidic conditions. Therefore, DNA uptake activity most likely occurs under neutral conditions, e.g. when H. pylori is in direct contact with the gastric epithelial cells.
Fig 3. DNA uptake complexes are reversibly shut down at acidic pH values.
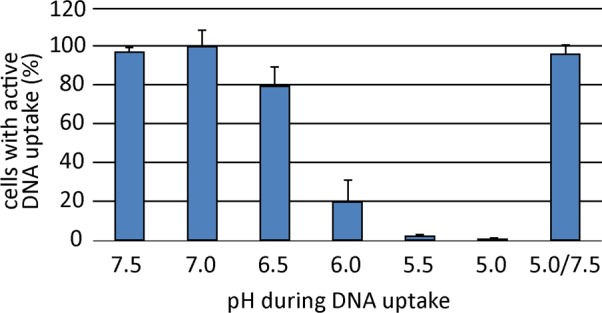
Competent bacteria were incubated in BB-FBS that had been titrated with HCl or NaOH to the indicated pH values. Uptake of Cy3-labelled λ DNA occurred for 10 min under aerobic conditions. Cells were washed once in the respective pH medium before DNase treatment for 5 min in TSB-FBS at pH 7.5. The fraction of competent cells relative to the control condition incubated in TSB-FBS at pH 7.5 are depicted. Values stem from at least three experiments; error bars, standard deviation.
Neutral pH is a prerequisite of competence development triggered by oxidative stress
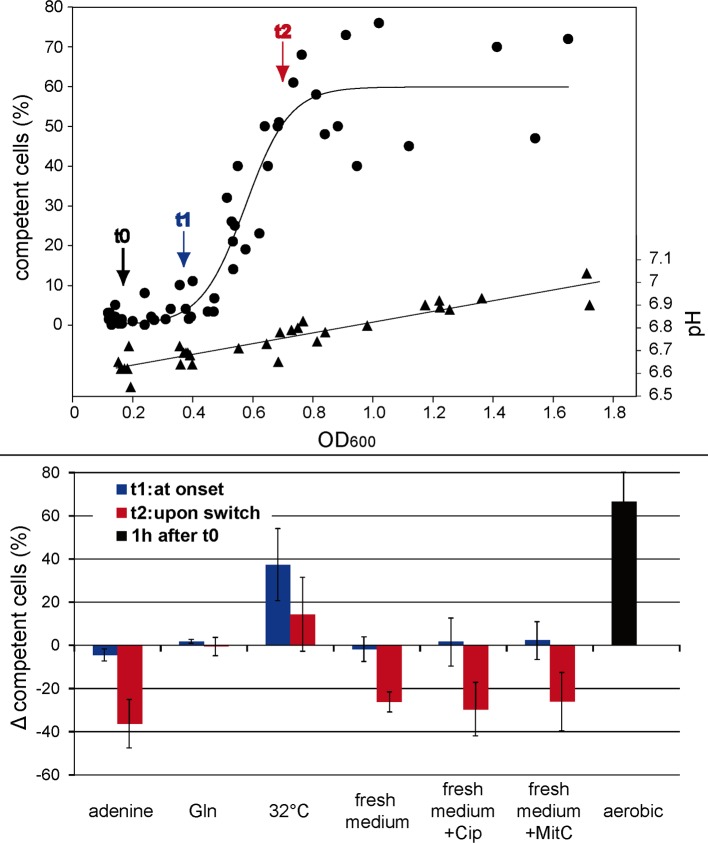
We next monitored competence development, defined by the fraction of cells with active DNA uptake. Cells were grown microaerobically in BB-FBS overnight to an optical density of OD600 0.19 ± 0.05 (t0), i. e. conditions of exponential growth phase. At this low OD600 H. pylori hardly exhibited any active DNA uptake complex (2.2 ± 2.86%). Between an optical density of OD600 0.39 ± 0.05 (t1) and 0.7 ± 0.11 (t2), around 50% of the cells switched into the competent state (Fig 4). Concomitantly, the pH of the medium rose by 0.2 units during this incubation period (Fig 4).
Fig 4. Competence development during microaerobic growth.
Competence development was monitored by the fraction of cells with active outer membrane DNA uptake at distinct growth phases under microaerobic atmosphere at 37°C (n = 16; large graph, circles, with left y-axis, sigmoidal curve fit using Sigma Plot 11.0). pH was monitored during growth (large graph, triangles). H. pylori N6 was grown overnight in BB-FBS to an OD600 of 0.19 ± 0.05 (t0) at which only a minor fraction of cells displayed competence (2.2 ± 2.86%). The onset of competence development was defined at t1 at which cells exhibited a mean OD600 of 0.39 ± 0.05 and 4 ± 2.8% of competent cells (~ t0 + 3–4 hours). At t2 (~ t0 + 6–8 hours) upon switch into competent state (50.6 ± 15.6% of cells with active outer membrane DNA uptake) cells exhibited a mean OD600 of 0.7 ± 0.11. At t0, either effectors (0.5 mM adenine or 0.5 mM glutamine) were added or the medium was exchanged by fresh BB-FBS medium with our without supplementation of 0.125 μg/ml ciprofloxacin or 0.025 μg/ml mitomycin C or temperature was decreased for 5°C or the cell suspension was exposed to aerobic conditions. Differences in fraction of competent cells at t1 or t2 due to change in incubation conditions are shown in the inserted diagram (at least three experiments for each condition; error bars, standard deviation). For aerobic stress conditions data are depicted from time point 1h after t0.
In order to establish the parameters triggering competence development, we changed conditions at time point t0 and monitored alterations of the amount of the competent fraction at time points t1 and t2. Exchange of supernatant by fresh medium at t0 before the switch into the competent state reduced the fraction of competent cells at time point t2 (when the cells reached an OD of ~0.7, Fig 4, lower panel). Exchange of medium with fresh medium containing DNA-damaging compounds 0.125 μg/ml of ciprofloxacin or 0.025 μg/ml of mitomycin C, concentrations, which slowed down growth, did not lead to enhanced competence development. Instead, these conditions were similar to exchange with fresh medium without supplements (Fig 4, lower panel).
Interestingly, addition of 0.5 mM adenine at t0 inhibited the switch into competence state (competent fraction decrease of -36.3 ± 11.2% at t2), while the addition of 0.5 mM glutamine had no effect (Fig 4, lower part). Note that lower concentrations of adenine (0.05 mM), which were sufficient for purine complementation in minimal medium, had no effect on competence development (S1 Appendix, Fig E). This indicated that the effect of adenine on competence development was distinct from that of purine deficiency. A stimulating effect on competence development was observed by lowering incubation temperature by 5°C to non-optimal growth conditions (competence fraction increase of +37.2 ± 16.6% at t1 and +14.3 ± 17.3% at t2 relative to control cells). Compared to the control, the above mentioned conditions did not alter pH during incubation relative to the control. Exchange of medium by fresh BB-FBS accounted on average for -0.03 ± 0.03 pH units (maximum -0.07) at t2 relative to the control, while -0.01 ± 0.03 pH units deviation (maximum -0.05) were observed for addition of adenine and -0.07 ± 0.03 pH units (maximum -0.15) upon lowering the temperature for 5°C. These small pH deviations have unlikely caused the observed effects on competence development, in particular, because lowering the temperature by 5°C resulted in an increase rather than a decrease of the amount of competent cells at t2.
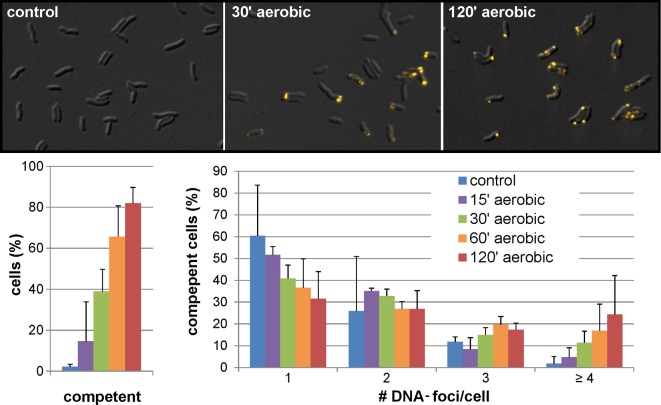
The most pronounced effect on competence development was detected upon exposure to aerobic conditions (competent fraction increase of +67 ± 14% within the much shorter time period of 1 h compared to ~3–4 hours at time point t1 (Fig 4, lower panel, black bar)). Under the latter condition, we did observe a significant rise in pH, probably due to loss of CO2 from the medium upon switch from microaerobic to aerobic conditions (initial culture pH at t0 of 6.7 ± 0.06 and a final pH of 7.2 ± 0.06 after 1 h of aerobic incubation, see also below). In order to get an idea of the kinetics of competence development under aerobic conditions, samples were analyzed after 15, 30, 60 and 120 min (Fig 5), starting with a competent fraction of 2.2 ± 1.7%. H. pylori cannot grow under normal atmospheric conditions but survival was observed at least 2 h without loss of CFU (log CFU/ml at t0 was 8.2 ± 0.1 and at t2h 8.0 ± 0.7; n = 5). After 30 min of exposure to oxidative stress, already 38.9 ± 8.1% of the cells were capable of import of external DNA. After 60 min this fraction increased to 65.6 ± 14.8% and reached a maximum of 81.9 ± 7.9% after 120 min. We observed that upon 30 min of oxidative stress most of the competent cells exhibited more than one DNA focus, with over 20% of the competent cells harboring more than four visually distinct locations of active DNA uptake after 2 h of oxidative stress. This confirmed enormous DNA uptake capacity. Selected samples were measured for transformation rate, using the point mutation marker rpsL(A128G). We confirmed integration of the marker into the chromosome by recombination in agreement with the data in Fig 1.
Fig 5. Kinetics of competence development under aerobic conditions.
H. pylori N6 was grown microaerobically in BB-FBS overnight at t0 before competence development (OD600~0.2). Cells were exposed to aerobic conditions at 37°C for 120 min. At the indicated time points the competence fraction of the cells was monitored. Control cells were kept under microaerobic atmosphere. Upper panel, DIC/Cy3 images of bacteria at indicated timepoints, with DNA stained in yellow; lower panel (left), fraction of competent cells; lower panel (right), number of distinct DNA foci per competent cell. Data stem from at least three experiments; error bars, standard deviation.
In order to dissect the role of pH and oxidative stress in triggering competence development, non-competent cells were exposed to aerobic conditions in BB-FBS for 1 h establishing various initial pH. As observed before, 64.3 ± 13.1% of the cells developed competence in non-titrated BB-FBS, with an initial culture pH at t0 of 6.7 ± 0.06 and a final pH of 7.2 ± 0.06 after 1 h of aerobic incubation (Fig 6). When the initial pH was titrated to 6.2 ± 0.09 before exposure to aerobic conditions and reached a final value of 6.6 ± 0.06 after 1 h, competence development was substantially inhibited (only 6.4 ± 7.5% competent cells). These results suggested that slight acidic pH prevented competence development. Furthermore, when we exchanged the medium with fresh BB-FBS titrated to an initial pH of 7.5 and incubated microaerobically for 1 h (corresponding to pH 7.2 ± 0.04 after equilibration of CO2), a significantly lower fraction of 33.3 ± 3.5% switched into the competence state compared to control cells under aerobic atmosphere within 1 hour (Fig 6). Note that the initial pH under microaerobic conditions was higher (pH 7.2 after CO2 equilibration) than in case of the aerobic condition (pH 6.7 rising to 7.2 during 1 h). From these data we conclude that competence development was not only pH-dependent but also triggered by the level of oxidative stress. Consistently, when oxidative stress was further reduced by using an atmosphere of 1% O2, 10% CO2 and 89% N2 and the same pH medium as for the microaerobic condition, the fraction of competent bacteria was further reduced (23.3 ± 2.5%, Fig 6).
Fig 6. Neutral pH opens the opportunity for competence development triggered by oxidative stress.
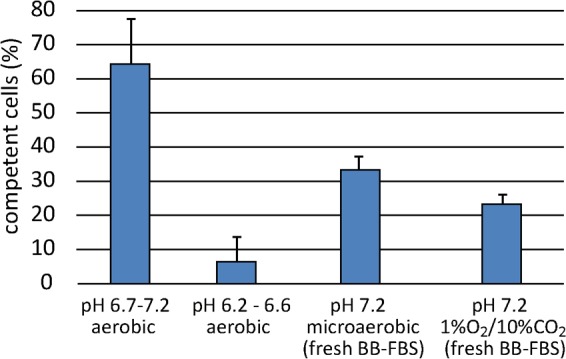
H. pylori N6 was grown microaerobically in BB-FBS overnight until growth phase before competence development (OD600~0.2). Cells were exposed to different pH and atmospheric conditions for 1 hour. Data stem from at least three experiments; error bars, standard deviation.
Likewise, using TSB-FBS as a different growth medium, which had a slightly higher pH value (pH 6.8 ± 0.04 without H. pylori preincubation) than BB-FBS (pH 6.5 ± 0.04 without H. pylori preincubation), competence development under microaerobic conditions occurred earlier and was also inhibited by 0.5 mM adenine. At OD600 of 0.18 ± 0.05 (corresponding to t0) already 30.3% ± 15.1% of the cells were competent in TSB-FBS compared to 11.4% ± 8.3% in the presence of adenine (OD600 of 0.20 ± 0.07). At t1 (OD600 of 0.37 ± 0.05) the majority of bacteria (62.6% ± 6.2%) had switched into the competent state, whereas a reduced fraction of 37% ± 5.7% displayed competence when adenine was present (OD600 of 0.35 ± 0.03). Growth rates were comparable in both media.
A final experiment was conducted confirming that pH was a prerequisite of competence development and fine-tuned by the degree of oxidative stress. In this experiment, we titrated BB-FBS to distinct initial pH values (pH 6.6, 6.8, 6.9 and 7.0 after equilibration of CO2 under microaerobic conditions) and exchanged the medium of a non-competent cell suspension at t0 with these fresh media for further microaerobic incubation (S1 Appendix, Fig F). Note that the overall competence development under microaerobic conditions is slower than under aerobic conditions, confirming the results depicted in Fig 6. As observed for the control cells, the relative pH increased for about 0.2 units ± 0.05 during microaerobic growth over 6 hours. We observed that the increase of competent cells rose with increasing initial pH values (S1 Appendix, Fig F). For each pH condition, addition of 0.5 mM adenine inhibited competence development as observed before. The absolute effect of adenine inhibition was less with rising pH, putatively indicating that sensitivity towards oxidative stress might be increased with rising pH.
Taking together, the combined results are consistent with a competence window opening at a pH above 6.5. Subsequently, the combination of pH and oxidative stress appeared to define the kinetics of competence development in H. pylori.
Growth of an oxygen-sensitive H. pylori mutant (sodB) is dependent on adenine
We further wanted to confirm that oxidative stress is indeed part of the trigger for competence development. Therefore, we intended to pinpoint the effect of adenine as radical quencher and constructed an oxygen-sensitive mutant of H. pylori. The oxidative stress enzyme superoxide dismutase (sodB) detoxifies superoxide radicals [46]. A mutant lacking SodB is confronted with higher oxidative stress because of oxygen radicals produced during microaerobic respiration. Likewise, sodB mutants of H. pylori were oxygen-sensitive and defective in mice colonization [10]. We constructed a mutant lacking SodB in H. pylori N6. Resuscitation of transformants at common microaerobic atmosphere (5% O2, 10% CO2, 85% N2) failed. However, transformants were obtained on Columbia blood agar at lower oxygen tension (1% O2, 10% CO2, 89% N2). Also subculturing sodB mutants on Columbia blood agar under 5% oxygen were unsuccessful, suggesting that indeed the mutant exhibited an oxygen-sensitive phenotype.
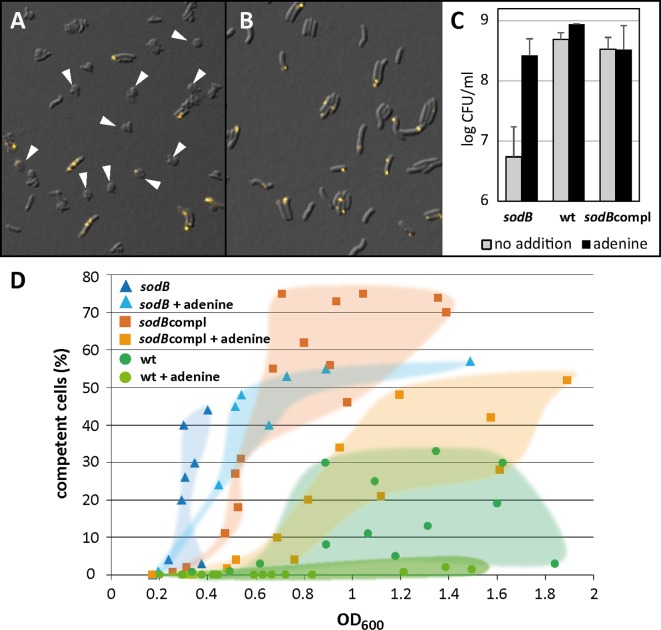
In liquid culture (BB-FBS), where cells are dispersed and oxygen stress might be increased the mutant ceased to grow after some doublings and transformed into non-cultivable coccoid forms even at reduced oxygen level of 1% (Fig 7A). When we added 0.5 mM of adenine to BB-FBS, growth was partially complemented; the sodB mutant maintained its rod-shaped morphology and was re-cultivable (Fig 7B). After 18–24 h in BB-FBS at 1% oxygen, CFU were log 6.7 ± 0.5 in the absence of adenine and partially restored to 8.4 ± 0.3 in the presence of adenine (Fig 7C), which is consistent with the microscopic observation of coccoid formation in the absence of adenine. Complementation of the sodB deletion was performed by integration of sodB into the fliP locus (sodBcompl). The fliP locus was previously used for insertion of a gentamycin cassette for the creation of a non-motile mutant and proven not to interfere with competence [18]. CFU of the wild type and the sodBcompl in the absence or presence of adenine was similar ranging between log 8.5 and 8.9 (Fig 7C). Competence development occurred at lower OD values in the sodB mutant than in the wild type under similar conditions (Fig 7D). The sodBcompl mutant showed an intermediate competence phenotype between the deletion mutant and the wild type, indicating that the deletion of sodB at its native locus was only partially complemented by insertion of sodB in the fliP locus. As shown before, addition of adenine resulted in a decrease of the competent fraction of cells. Note that consistently, the wild type also displayed lower competence development at 1% oxygen (Fig 7) as compared to 5% oxygen (Fig 4). The data support the following conclusions: (i) competence development during growth seemed to be triggered by oxidative stress and, (ii), adenine appeared to have an oxygen-protective role, probably as radical quencher.
Fig 7. Phenotype of an oxygen-sensitive mutant sodB lacking superoxide dismutase.
H. pylori were grown in BB-FBS at reduced oxygen atmosphere (1% O2, 10% CO2, 89% N2) in the absence and presence of 0.5 mM adenine. A and B, overlay images of DIC/Cy3 of sodB after 10 min of DNA uptake with DNA stained in yellow; arrowheads indicate coccoid formation of the sodB mutant in the absence of adenine (A), while cells kept their rod-shaped morphology in the presence of adenine (B). C, log CFU/ml after 18–24 h of growth of sodB, the wild type and the sodBcompl. D, competence development occurred with lower OD600 values in the sodB mutant compared to the wild type; the sodBcompl showed an intermediate phenotype. Addition of adenine reduced competence development. Data in C and D stem from at least three independent experiments; datapoints of the respective strain/condition were highlighted in D for better visualization.
Taking together, the data indicate that the opportunity for competence development was established at pH values above 6.5. Within this window, the combination of pH and oxidative stress triggered competence development. This might support the hypothesis that intrinsic (caused by respiration) as well as extrinsic oxidative stress (caused by oxygen tension and/or host inflammatory response) contribute to the impressive genetic diversity of the pathogen, colonizing close to the neutral gastric mucosa.
Discussion
Using single cell analysis, we demonstrated that H. pylori cells import huge amounts of non-homologous DNA within short periods of time, the median amount ranging between 124 kb and 351 kb in two different strains tested (Fig 2). Individual cells were shown to import the equivalent of a complete genome (~1.6 Mb). This DNA amount far exceeded the previously quantified imported DNA in other Gram-negative bacteria, which was limited to 40 kb by the binding capacity of the periplasmic protein ComE (ComEA homologue) [21]. A homologue of comEA is missing in the H. pylori genome [23] and our data indicate an unknown alternative periplasmic protein with much higher binding capacity or higher abundance. However, compared to the natural packaging of λ DNA in the icosahedron capsid of the bacteriophage, DNA density in H. pylori periplasm was around 100-fold less. Yet the capsid of the bacteriophage λ spatially limits DNA packaging, resulting in high internal pressure from DNA bending, essential for DNA ejection [47]. The velocity of a DNA uptake complex in H. pylori was 1.26 kb/s [18], corresponding to a maximum import activity of 75.6 kb/min. We measured a maximum amount of 758 kb DNA per focus in J99 after 10 min of import. Although DNA uptake is processive in H. pylori, it is limited by the size of single λ DNA molecules and probably interrupted before binding and start of uptake of a new molecule. Therefore, it seems likely that a fluorescent DNA focus might also be the result of DNA import activity of multiple DNA uptake complexes in close proximity. This notion is corroborated by our previous observation of multiple concomitant uptake events into a single cell [18].
Impact of oxidative stress on competence development
We observed that competence development occurred in liquid medium with increasing optical density. Exchange of the medium by fresh medium before the switch to the competent state inhibited competence development. A regulation by quorum sensing, however is not likely, as a similar effect was observed by adding adenine, conditions during which a putative autoinducer is not eliminated (Fig 4). In order to understand the effect of adenine (or fresh medium) on inhibition of competence development, we characterized a mutant deficient in superoxide dismutase, an important enzyme for oxidative stress defense [10]. The mutant was only able to grow at reduced oxygen levels. In BB-FBS, we observed that addition of adenine was crucial for prolonged growth and survival of the mutant, which turned into coccoids in its absence (Fig 7). Since superoxide dismutase is crucial for superoxide radical inactivation [46], this suggested that the effect of adenine on competence development in the wild type probably results from its radical quenching capacity [48]. Also, fresh medium might restore quenching capacity, which is depleted during growth. Note that the pH in the presence of adenine or after exchange of the medium by fresh medium at t0 was comparable to that of the control. Moreover, the sodB mutant displayed enhanced competence under similar conditions compared to the wild type (Fig 7), which supports the hypothesis that oxidative stress triggered competence development. The sodBcompl mutant showed an intermediate competence phenotype between wild type and deletion mutant, indicating that the deletion of sodB at the native locus could only partially be complemented. Likewise, the most pronounced competence development was observed under aerobic conditions. Competence developed quite rapidly; within only 30 min of exposure, nearly 40% of the cells had switched into competent state and after 2 hours this fraction had increased to over 80%, with 20% of the competent cells expressing more than four distinct DNA uptake locations (Fig 5). Concomitantly, a pH elevation of around 0.5 units by loss of dissolved CO2 from the medium upon shift from microaerobic to aerobic atmosphere was observed, with implications discussed below.
We also tested other putative quencher molecules, like vitamin C and vitamin E. However, initial experiments showed that at those concentrations, at which competence development was inhibited, growth was drastically reduced, too. Therefore, these experiments were not followed up and data were not included in this manuscript, since substantial growth inhibition is expected to indifferently reduce protein biosynthesis. It might hint at the fact that quenchers can also impair the function of the respiratory chain. Instead, we showed by three independent experiments, (i) addition of adenine, (ii) variation of oxygen level and (iii) competence development in an oxygen-sensitive mutant that oxidative stress might play a crucial role in natural transformation.
Impact of pH on competence development
Transcriptome analysis previously indicated transcriptional repression of two essential genes for natural transformation (comB8 and comH) at pH 5.4 relative to neutral growth conditions [49]. A recent study also hinted towards an effect of pH on natural transformation [38]. The authors conducted the experiments on agar plates, supplemented with a pH indicator. They could observe a 1- to 2-log increase in transformation rate, when the visually estimated pH was increased by about 0.2 units or when low CO2 concentrations were used. In our study we detected a comparable rise in pH by 0.2 units during growth in broth (Fig 4). Alteration of the culture conditions before switch into competent state and direct monitoring of single cell DNA uptake activity enabled us to gain more insight into the interplay of pH and oxidative stress and their effect on competence development.
The prerequisite of detection of fluorescent DNA taken up over the outer membrane is at least one fully active ComB-dependent DNA uptake complex per cell. ComB dependence was demonstrated by lack of DNA uptake activity in mutants harboring a deletion of one of the essential comB genes, either comB4 or comB6 [18]. The short-term DNA uptake assay (10 min of incubation in the presence of DNA) was performed at pH 6.8–7.5, a pH range at which we detected maximal DNA uptake activity of a competent cell suspension (Fig 3). Upon variation of pH during this short-time incubation, the majority of competent cells showed DNA uptake at pH above 6.5, while slight acidic pH did not lead to detectable DNA uptake. In addition, we were able to reactivate DNA uptake after short-time incubation at slight acidic pH values, indirectly demonstrating that the DNA uptake complexes were transiently inactive at pH below 6.5 (Fig 3). This led us to the conclusion that pH regulation took place at the level of DNA uptake activity (mediated by ComB transporter complexes).
However, pH regulation also occurred at the level of competence development. Starting with a practically non-competent cell suspension that did not show DNA uptake at DNA uptake assay conditions of pH 6.8–7.5 we monitored competence development under different incubation conditions over time (Figs 4–6). We conclude that the prerequisite for detection of DNA uptake activity over the outer membrane (at least one fully active DNA uptake complex per cell) was developed in response to pH alteration and oxidative stress in these experiments. Monitoring transcription of a subset of comB genes does not completely answer the question of whether the bacterium is fully equipped for DNA uptake. In contrast, visualization of DNA uptake activity during the experiment most directly shows competence of the bacteria, which is based on successful expression of all necessary genes (including comB genes), assembly of the proteins and eventual activation of the ComB transport system.
Does the tested pH range itself impact on chemical modification of DNA and, thereby, influence DNA uptake? Extreme acidic conditions might lead to some depurination, i. e. the release of purine bases from nucleic acids even at physiological salt concentrations and temperature [50]. The pH range used (pH 5-7.5) only covered slight acidic conditions. The cytoplasmic pH in H. pylori was approximately 5 at an external pH of 1 in the presence of urea [51], conditions under which survival of the bacterium was fully supported [52] and, thus, cytoplasmic DNA unaffected in vivo. At pH 5 DNA is expected to be more protonated compared to neutral conditions. Considering physiochemical properties, transport of a negatively charged macromolecule against the transmembrane potential (with negatively charged cytoplasm) might be facilitated at lower pH, at which net charge of DNA is reduced by protonation. However, our data show that under these pH values DNA uptake did not occur. Therefore, we conclude that biologically relevant pH regulation occurred, which might play a crucial role for the lifestyle of the gastric pathogen.
Is DNA damage a trigger for competence development?
We showed competence development in BB-FBS under normal microaerobic growth conditions without external stress (Fig 4). From the overall data, we are tempted to conclude that (i) a slight increase in pH by approximately 0.2 units (above pH 6.5) in combination with (ii) decreasing quenching capacity of the medium led to competence development. It was suggested that the presence of antibiotics triggered natural transformation in H. pylori [37], as shown for Streptococcus and Legionella [27, 53]. In the study of Dorer and collegues the presence of ciprofloxacin resulted in a four-fold increase in transformation rate. This increase is rather marginal, considering dynamics of log 5 demonstrated in this study or around log 2 in the study of Moore et al. [38]. We tested ciprofloxacin and also mitomycin C under our atmosphere-, pH-, temperature- and growth phase controlled conditions. Fluoroquinoles are thought to generate double strand breaks in DNA [54, 55]. Mitomycin C crosslinks to DNA and causes DNA damage [56, 57]. We chose sublethal concentrations that slow down growth. Additionally, we tested higher concentrations and confirmed lack of competence induction as shown previously [37]. However, sublethal intermediate concentrations of either ciprofloxacin or mitomycin C also did not show any impact on competence development under our conditions (Fig 4). In contrast, our results demonstrated that small variations in atmosphere, external pH and temperature largely influence competence development. This might be an important reason for the relatively high variation in published transformation rates in different laboratories using various media and gas-generation systems. Furthermore, alterations of these parameters during processing of the sample within the experimental setup, including a drop of temperature have to be taken into account in order to prevent biased results.
Identification of a pH-defined competence window in H. pylori
In B. subtilis the term “competence window” was used to define a time period, during which the likelihood for switching into competence state is increased by rising the basal level of the master competence regulator [58, 59]. Our data suggest that in H. pylori a “competence window” is opened and closed by external pH. In case the pH reached near neutral values (> pH 6.5), competence development was triggered by the combination of pH increase and oxidative stress. The kinetics of competence development was modifiable by oxygen tension (Fig 6) or by the addition of the radical quencher adenine (Fig 4) under comparable pH conditions. Drop in temperature to non-optimal growth conditions also led to increase in competence development, confirming that competence development was not purely regulated by external pH. The temperature effect might be explained by an imbalance of metabolic processes, putatively also leading to sub-optimal (oxidative?) stress defense.
From our data we conclude that DNA uptake in H. pylori is dispensable and eventually even deleterious under acidic conditions. In contrast, the process appears to be limited to neutral pH values. From transcriptome data it was suggested that H. pylori colonizes an acidic niche [60]. However, that study focused on early colonization (5–10 days after infection) of H. pylori in gerbils and not only surface epithelial cells with attached bacteria but also gastric mucus was collected as pool samples. Since the majority of H. pylori is thought to reside in the mucus layer, where slight acidic pH values are likely to occur, detection of acid induced genes may be explainable. However, these data do not necessarily reflect pH in direct contact with the host. Measurements using pH microelectrodes showed that H. pylori is confronted with a pH gradient lining from the near neutral gastric mucosa to the extreme acidic pH in the stomach lumen [61, 62]. Therefore, our data suggest that DNA uptake is most active in direct contact with the host, a site with near neutral pH. In early stages of H. pylori gastritis, the bacterium is preferentially localized at the antrum; with continued inflammation, gastrin producing cells are lost in the antrum, probably leading to decrease in acid secretion by the parietal cells and spread of bacteria and inflammation to the corpus [63]. Likewise, the pH value of the gastric mucosa in patients with ulcers was higher than for a control group [64]. At the sites of chronic severe inflammation, increased oxidative stress due to ROS production by the host might be expected [65].
We observed that, in contrast to B. subtilis, the majority of the H. pylori population acquired the competent state and growth of the bacterium was not interrupted. In B. subtilis natural transformation probably enables the bacterium to survive and adapt to changing unfavorable conditions. In this respect, transiently interrupting growth might serve for increased survival and enhanced persistence. Our data suggest that natural transformation in H. pylori is established under the most favorable conditions within the hostile stomach, namely at the multiplication site close to the near neutral mucosa. Triggering chronic gastritis, ROS-production by the host cell inflammatory system will be a daily reality for the gastric pathogen.
Role/s of natural transformation in H. pylori
What is the purpose of this high energy hungry process? Natural transformation was suggested to play a role in nutrient supply in other bacteria [45]. However, when we exposed highly competent H. pylori with external DNA in a minimal medium without purine source, growth was not supported (S1 Appendix, Fig C). Intriguingly, the bacterium was capable of import of DNA into the cytoplasm under these conditions (S1 Appendix, Fig D) but it was not able to recycle nucleotides for replication. Interestingly, H. pylori did not discriminate between own and foreign DNA, as demonstrated by the import of λ DNA ([18] and Fig 2). In principle, there are two explanations: First, there is no need to select for its own DNA in an ecological niche where DNA of siblings prevails [66]. Abundant human DNA will probably exert a limited risk for deleterious, non-homologous transformation events. By demonstrating that active DNA complexes were completely shut down at a pH ≤ 5.5, it is likely that DNA uptake is prevented in the acidic lumen, where the presence of DNA of foodborne bacteria is expected. In this scenario, a high level of DNA uptake from siblings can serve for DNA repair under oxygen stress conditions and/or for exchange of genetic material. The immune system exerts selection pressure on H. pylori, for which an environmental niche or other host reservoirs besides humans are not known. DNA repair systems were shown to be crucial for optimal colonization at the gastric mucosa and it was concluded that pathogen DNA is a target for host-generated oxidative stress [67]. Extensive horizontal gene transfer probably provides high adaptive capacity to create H. pylori diversity for enhanced fitness, in particular, for novel host infection.
Second and highly recommended to be considered, DNA uptake might serve a purpose beyond DNA repair and genetic diversification. We could imagine that the import of huge amounts of non-homologous DNA might establish a dilution effect. In this scenario, imported DNA might create a reservoir of oxidizable nucleotides, protecting own cytoplasmic chromosomal DNA against oxidative stress, in particular during host immune response. In this regard, genetic diversity would be a side product of oxidative stress defence. However, while genetic diversification was shown to be extraordinary in H. pylori, leading to a panmictic population structure, the latter putative hypothesis will require further analysis.
The in vivo relevance of the comB system for colonization was clearly demonstrated for comB4 in the gerbil model [68]. However, since a H. pylori mutant, defective in the comB8-B10 operon was only attenuated but not excluded from the gerbil stomach, the conclusion was that natural transformation was not essential for initial (3-weeks) colonization. Active chronic gastritis was detected at 3 to 8 weeks after infection in Mongolian gerbils [69–71], whereas ulcers were observed after approximately 6 months [70, 72] and gastric cancer after 18 months of infection [72]. In another study, a comB10 mutant colonized mice during one week like the wild type, but colonization was somewhat decreased after 8 weeks in a competition experiment [73]. Interestingly, a dprA mutant, intact for DNA uptake but impaired in recombination of the DNA into the chromosome, did not show a defect in colonization after 8 weeks but displayed reduction after 12 weeks. A future systematic investigation of essential functions of natural transformation (outer membrane transport, inner membrane transport and recombination) in long-term colonization experiments—most appropriate in the gerbil model with human-like inflammatory response—will provide more insight into the multiple roles of natural transformation.
Conclusions
In conclusion, we showed for the first time that during microaerobic growth neutral pH values defined a competence window in which DNA uptake capacity was triggered by a combination of pH and oxidative stress. In consequence, this led to the import of staggering amounts of DNA into the majority of cells within the population. Concomitantly, we detected high transformation rates under these conditions. Hence, our data deliver explanations for the observed extraordinary genetic diversity of this microaerobic pathogen, persisting lifelong at a near neutral multiplication site under conditions of chronic inflammation.
Materials and Methods
Strains and growth conditions
H. pylori strains N6 [74] and J99 [75] were grown either on Columbia blood agar base (Oxoid) supplemented with 5% defibrinated sheep blood (Oxoid, Heidelberg, Germany) or in liquid culture (shaking at 140 rpm) using tryptic soy broth (TSB, Becton Dickinson, USA) or brucella broth (BB, Becton Dickinson) with 5% fetal calf serum (PAN-Biotech GmbH, Aidenbach, Germany), in the following referred to as TSB-FBS or BB-FBS, respectively. Antibiotics were used at the following final concentrations per milliliter: 12.5 μg vancomycin, 0.31 μg polymyxin B, 6.25 μg trimethoprim, and 2.5 μg amphotericin B. If applicable, kanamycin or chloramphenicol was added at 20 μg/mL or 8 μg/ml, respectively. Plates were incubated at 37°C in a microaerobic incubator (Binder, Tuttlingen, Germany) at 1% or 5% O2, 10% CO2 and 85% N2. Atmosphere for liquid cultures was established in jars by evacuating air and refilling the volume with defined gas mixture (1% or 5% O2, 10% CO2, rest N2).
The minimal medium was composed according to [76] but lacked the sole purine source adenine, which was added at concentrations of 5 or 50 μg/ml (corresponding to 0.037 or 0.37 mM, respectively). Instead of addition of single amino acids, 100 x MEM non-essential amino acids solution (Life Technologies) was used as a 20-fold stock solution and 50 x MEM amino acids solution (Life Technologies) as a 10-fold stock solution, matching the amino acid concentrations of the original minimal medium except for 66 mg/l asparagine, 210 mg/l histidine and 180 mg/l tyrosine. Glutamine was added separately according to the original protocol. The pH of the medium was adjusted to 7.4. For supplementation of the medium with 10 μg/ml DNA, either DNA of salmon sperm (Sigma Aldrich) or purified chromosomal H. pylori N6 DNA was used.
H. pylori mutants used in this study
Oligonucleotides used in this study are depicted in S1 Appendix, Tab A. Genomic DNA of a streptomycin resistant mutant of H. pylori strain 26695, bearing an A128G mutation in rpsL was used as template for the amplification of a 1,022-bp rpsL(A128G) PCR fragment using oligos H3 and H6. N6 comB4 and N6 comB6 were constructed by transformation of a PCR fragment amplified on genomic DNA from the respective J99 mutants published previously [18] and selected on 20 μg/ml kanamycin.
Approximately 500-bp flanking regions of sodB (HP0389; ORF definition of 26695) were amplified using oligos H167/H168 (5′ region including start codon of sodB) and H169/H170 (3′ region including 39 bp of sodB) and genomic DNA of N6 as template [77]. The nonpolar kanamycin resistance cassette was amplified from pUC18ΔK2 [78] using oligos H13 and H14. Since the oligo H168 contains 22 bp, which are reverse complementary to H13, and H169 contains 21 bp, which are reverse complementary to H14, the nonpolar kanamycin cassette was inserted between the flanking regions by fusion PCR. This led to a fusion PCR fragment, suitable for nonpolar inactivation of sodB in N6 by direct transformation and selection on 20 μg/mL kanamycin. For complementation of the sodB mutant, a 1.2 kb fragment comprising sodB including the annotated transcriptional start site [79] and approximately 500 bp 5’ region was amplified using H167 and H177 and genomic DNA of N6 as template. The gentamicin cassette was amplified using H11 and H12 from pUC1813apra [80]. Approximately 600-bp flanking regions of the insertion locus fliP were amplified using oligos H7/H178 (5’ region) and H9/H10 (3’ region). Since the oligo H178 contains a 20 bp reverse complementary region to H167, the oligo H177 a 21 bp reverse complementary region to H11 and the oligo H9 a 22 bp reverse complementary region to H12, all four fragments were fused by PCR using oligos H7 and H10, resulting in an approximately 3.2 kb fragment. Correct allelic exchange of all mutants was checked by PCR and sequencing.
Fluorescence labeling of DNA and bacterial uptake
λ DNA (dam- and dcm-; Thermo Fisher Scientific, USA) was covalently fluorescently labeled applying Mirus Label IT Cy3 (MoBiTec GmbH, Goettingen, Germany) according to the manufacturer’s protocol using a 1:1 (volume: weight) ratio of Label IT reagent to nucleic acid.
λ DNA was non-covalently labeled with the fluorescent dye YOYO-1 (Invitrogen, USA) at a bp:dye ratio of 1:50 in phosphate-buffered saline a pH 7.2. YOYO-1 specifically stains dsDNA by intercalation at bp:dye ratios of >8 and is virtually non-fluorescent in aqueous solutions [81]. One microgram per ml of labeled DNA was added to a H. pylori cell suspension, which had been centrifuged and resuspended in TSB-FBS (100 μl OD600 ∼0.2–0.8). After incubation for 10 min at 37°C under microaerobic atmosphere, the suspension was centrifuged, resuspended in 25–50 μl TSB-FBS and incubated for 5 min at 37°C in the presence of 10 units of DNaseI (Roche, Basel, Switzerland).
Quantification of imported DNA
For quantification of fluorescence intensity from single λ DNA molecules, a drop of freshly labelled Cy3 λ DNA diluted in TSB was spread out on a 1.5% low melting agarose pad by air flow and immediately sealed with a cover slip. In parallel, H. pylori cells were incubated for 10 min in the presence of the same lot of Cy3 λ DNA. Fluorescence microscopy was performed using a Zeiss Axio Observer Z1 microscope with a plan apochromatic 63x/1.4 objective and differential interference contrast (DIC). Cy3 was visualised using a metal halide light source (HXP120C) and a filter set with excitation at 550 nm (bandwidth 25 nm), emission at 605 nm (bandwidth 70 nm) and a dichroic beamsplitter at 570 nm. YOYO-1 that intercalated into dsDNA was detected with a filter set providing excitation at 470 ± 20 nm and emission at 525 ± 25 nm (dichroic beamsplitter at 495 nm). Images were acquired with the 12-bit monochromatic AxioCam MRm camera, using exposure times between 60 ms and 2.5 s. Intensity values of Cy3 were checked for linearity between different exposure times and bleaching was found negligible (S1 Appendix, Fig A). Regions of interest (ROIs) were set manually and analyzed by ZEN software (blue edition) version 2.0.0.0. In order to determine the fluorescence of one single λ DNA molecule, fluorescence intensity was determined from spread out DNA molecules with a minimal length of ½ of maximal size of λ DNA (8.6 μm, [43]) and defined as one molecule. Fluorescence intensity in single DNA foci and in whole cells were determined, compared to fluorescence intensity of λ DNA molecules and expressed as imported kb. The cut-off value for the analysis of DNA foci was set to a signal to background ratio of ≥ 2.
Determination of transformation rate
For the determination of transformation rate, bacterial cells grown in liquid culture were centrifuged and resuspended in 100 μl TSB-FBS to OD600 ∼1. Subsequently, 1 μg/ml of a 1,022-bp rpsL(A128G) PCR fragment was added and incubated for 1 h at 37°C under microaerobic atmosphere. Cells were centrifuged, resuspended in 50 μl TSB-FBS in the presence of 10 U of DNaseI (Roche, Basel, Switzerland) and directly spotted on Columbia blood agar. After incubation under microaerobic conditions at 37°C for 20 ± 4 h, bacteria were harvested in TSB-FBS. Total colony forming units as well as the number of streptomycin resistant transformants were determined according to the serial dilution method using Columbia blood agar with and without 20 μg/mL streptomycin and a selection time of 4–5 days. As alternative resistance marker, the chloramphenicol cassette of pILL2157 [40] was used within the context of this replicative plasmid or amplified using oligos H108 and H109 and fused to approximately 500 bp 5’ region (oligos H110 and H111) and 3’ region (oligos H112 and H113) of nucT in order to create a 1886 bp PCR fusion fragment (H110/H113) for homologous recombination into the nucT locus.
Acceleration/deceleration of foci development
Cells were inoculated at OD600 ~0.003 and grown over night under microaerobic conditions in BB-FBS to time point t0, at which the culture had reached approximately OD600 between 0.15 and 0.25. At t0 either one of the different conditions was established; (i) one of the effectors (see below) was added or (ii), the culture was temperature downshifted by 5°C or (iii), the culture was exposed to aerobic conditions at 37°C or (iv), the medium was exchanged for fresh medium with or without supplementation of either one of the DNA-damaging agents 0.125 μg/ml ciprofloxacin or 0.025 μg/ml mitomycin C. It was guaranteed that the procedure for changing the condition was completed within 10 min. For exchanging the medium, cells were centrifuged at 9000 x g for 2 min at room temperature and immediately resuspended in the same volume of prewarmed (37°C) fresh medium with or without DNA-damaging agents. At t1 (3–4 h after t0) and t2 (6–8 h after t0) a test aliquot was taken from the cultures and checked for the number of cells harboring DNA foci as mentioned above. For aerobic exposure, samples were taken after 15, 30, 60 and 120 min, respectively. If indicated, pH was measured using a temperature-controlled pH electrode with an accuracy of 0.01 pH units (pH/ATC, Denver Instrument, New York, USA). In order not to disturb competence development, parallel culture aliquots served for pH measurements.
Supporting Information
Table A: Oligonucleotides used in this study. Figure A: Bleaching curve of Cy3 labelled λ DNA. Figure B: Distribution of estimated area per λ molecule in periplasmic DNA foci of H. pylori. Figure C: Growth of competent H. pylori N6 in minimal medium without purine source cannot be restored by addition of external DNA. Figure D: Competent H. pylori did not grow in minimal medium in the absence of a purine source but were still capable of DNA uptake. Figure E: Competence development is not influenced by lower concentrations of adenine. Figure F: pH is a prerequisite of competence development and fine-tuned by the degree of oxidative stress.
(PDF)
Acknowledgments
We thank Thomas Wassmer for critical reading of the manuscript and for valuable discussions. pILL2157 was a gift of Ivo G. Boneca.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
NJK was financed by the Deutsche Forschungsgemeinschaft (DFG) project STI 201/1-3. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bhattacharyya A, Chattopadhyay R, Mitra S, Crowe SE. Oxidative stress: an essential factor in the pathogenesis of gastrointestinal mucosal diseases. Physiol Rev. 2014;94(2):329–54. 10.1152/physrev.00040.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hanada K, Uchida T, Tsukamoto Y, Watada M, Yamaguchi N, Yamamoto K, et al. Helicobacter pylori infection introduces DNA double-strand breaks in host cells. Infect Immun. 2014;82(10):4182–9. 10.1128/IAI.02368-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Raza Y, Khan A, Farooqui A, Mubarak M, Facista A, Akhtar SS, et al. Oxidative DNA damage as a potential early biomarker of Helicobacter pylori associated carcinogenesis. Pathol Oncol Res. 2014;20(4):839–46. 10.1007/s12253-014-9762-1 [DOI] [PubMed] [Google Scholar]
- 4. Brown LM. Helicobacter pylori: epidemiology and routes of transmission. Epidemiol Rev. 2000;22(2):283–97. [DOI] [PubMed] [Google Scholar]
- 5. Ernst PB, Gold BD. The disease spectrum of Helicobacter pylori: the immunopathogenesis of gastroduodenal ulcer and gastric cancer. Annu Rev Microbiol. 2000;54:615–40. 10.1146/annurev.micro.54.1.615 [DOI] [PubMed] [Google Scholar]
- 6. Bury-Mone S, Skouloubris S, Labigne A, De Reuse H. The Helicobacter pylori UreI protein: role in adaptation to acidity and identification of residues essential for its activity and for acid activation. Mol Microbiol. 2001;42(4):1021–34. [DOI] [PubMed] [Google Scholar]
- 7. Weeks DL, Eskandari S, Scott DR, Sachs G. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science. 2000;287(5452):482–5. [DOI] [PubMed] [Google Scholar]
- 8. Harris AG, Wilson JE, Danon SJ, Dixon MF, Donegan K, Hazell SL. Catalase (KatA) and KatA-associated protein (KapA) are essential to persistent colonization in the Helicobacter pylori SS1 mouse model. Microbiology. 2003;149(Pt 3):665–72. [DOI] [PubMed] [Google Scholar]
- 9. Olczak AA, Seyler RW Jr., Olson JW, Maier RJ. Association of Helicobacter pylori antioxidant activities with host colonization proficiency. Infect Immun. 2003;71(1):580–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seyler RW Jr., Olson JW, Maier RJ. Superoxide dismutase-deficient mutants of Helicobacter pylori are hypersensitive to oxidative stress and defective in host colonization. Infect Immun. 2001;69(6):4034–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kennemann L, Didelot X, Aebischer T, Kuhn S, Drescher B, Droege M, et al. Helicobacter pylori genome evolution during human infection. Proc Natl Acad Sci U S A. 2011;108(12):5033–8. 10.1073/pnas.1018444108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hofreuter D, Odenbreit S, Haas R. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol Microbiol. 2001;41(2):379–91. [DOI] [PubMed] [Google Scholar]
- 13. Backert S, Kwok T, Konig W. Conjugative plasmid DNA transfer in Helicobacter pylori mediated by chromosomally encoded relaxase and TraG-like proteins. Microbiology. 2005;151(Pt 11):3493–503. [DOI] [PubMed] [Google Scholar]
- 14. Fischer W, Windhager L, Rohrer S, Zeiller M, Karnholz A, Hoffmann R, et al. Strain-specific genes of Helicobacter pylori: genome evolution driven by a novel type IV secretion system and genomic island transfer. Nucleic Acids Res. 2010;38(18):6089–101. 10.1093/nar/gkq378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuipers EJ, Israel DA, Kusters JG, Blaser MJ. Evidence for a conjugation-like mechanism of DNA transfer in Helicobacter pylori . J Bacteriol. 1998;180(11):2901–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fernandez-Gonzalez E, Backert S. DNA transfer in the gastric pathogen Helicobacter pylori . J Gastroenterol. 2014;49(4):594–604. 10.1007/s00535-014-0938-y [DOI] [PubMed] [Google Scholar]
- 17. Karnholz A, Hoefler C, Odenbreit S, Fischer W, Hofreuter D, Haas R. Functional and topological characterization of novel components of the comB DNA transformation competence system in Helicobacter pylori . J Bacteriol. 2006;188(3):882–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stingl K, Müller S, Scheidgen-Kleyboldt G, Clausen M, Maier B. Composite system mediates two-step DNA uptake into Helicobacter pylori . Proc Natl Acad Sci U S A. 2010;107(3):1184–9. 10.1073/pnas.0909955107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Inamine GS, Dubnau D. ComEA, a Bacillus subtilis integral membrane protein required for genetic transformation, is needed for both DNA binding and transport. J Bacteriol. 1995;177(11):3045–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yeh YC, Lin TL, Chang KC, Wang JT. Characterization of a ComE3 homologue essential for DNA transformation in Helicobacter pylori . Infect Immun. 2003;71(9):5427–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gangel H, Hepp C, Müller S, Oldewurtel ER, Aas FE, Koomey M, et al. Concerted spatio-temporal dynamics of imported DNA and ComE DNA uptake protein during gonococcal transformation. PLoS Pathog. 2014;10(4):e1004043 10.1371/journal.ppat.1004043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seitz P, Pezeshgi Modarres H, Borgeaud S, Bulushev RD, Steinbock LJ, Radenovic A, et al. ComEA is essential for the transfer of external DNA into the periplasm in naturally transformable Vibrio cholerae cells. PLoS Genet. 2014;10(1):e1004066 10.1371/journal.pgen.1004066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori . Nature. 1997;388(6642):539–47. 10.1038/41483 [DOI] [PubMed] [Google Scholar]
- 24. Havarstein LS, Coomaraswamy G, Morrison DA. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae . Proc Natl Acad Sci U S A. 1995;92(24):11140–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Magnuson R, Solomon J, Grossman AD. Biochemical and genetic characterization of a competence pheromone from B. subtilis . Cell. 1994;77(2):207–16. [DOI] [PubMed] [Google Scholar]
- 26. Suckow G, Seitz P, Blokesch M. Quorum sensing contributes to natural transformation of Vibrio cholerae in a species-specific manner. J Bacteriol. 2011;193(18):4914–24. 10.1128/JB.05396-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae . Science. 2006;313(5783):89–92. 10.1126/science.1127912 [DOI] [PubMed] [Google Scholar]
- 28. Lo Scrudato M, Blokesch M. A transcriptional regulator linking quorum sensing and chitin induction to render Vibrio cholerae naturally transformable. Nucleic Acids Res. 2013;41(6):3644–58. 10.1093/nar/gkt041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Goodman SD, Scocca JJ. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae . Proc Natl Acad Sci U S A. 1988;85(18):6982–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Berry JL, Cehovin A, McDowell MA, Lea SM, Pelicic V. Functional analysis of the interdependence between DNA uptake sequence and its cognate ComP receptor during natural transformation in Neisseria species. PLoS Genet. 2013;9(12):e1004014 10.1371/journal.pgen.1004014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smith HO, Tomb JF, Dougherty BA, Fleischmann RD, Venter JC. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science. 1995;269(5223):538–40. [DOI] [PubMed] [Google Scholar]
- 32. Mell JC, Hall IM, Redfield RJ. Defining the DNA uptake specificity of naturally competent Haemophilus influenzae cells. Nucleic Acids Res. 2012;40(17):8536–49. 10.1093/nar/gks640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Redfield RJ, Cameron AD, Qian Q, Hinds J, Ali TR, Kroll JS, et al. A novel CRP-dependent regulon controls expression of competence genes in Haemophilus influenzae . J Mol Biol. 2005;347(4):735–47. 10.1016/j.jmb.2005.01.012 [DOI] [PubMed] [Google Scholar]
- 34. Israel DA, Lou AS, Blaser MJ. Characteristics of Helicobacter pylori natural transformation. FEMS Microbiol Lett. 2000;186(2):275–80. [DOI] [PubMed] [Google Scholar]
- 35. Saunders NJ, Peden JF, Moxon ER. Absence in Helicobacter pylori of an uptake sequence for enhancing uptake of homospecific DNA during transformation. Microbiology. 1999;145 (Pt 12):3523–8. 10.1099/00221287-145-12-3523 [DOI] [PubMed] [Google Scholar]
- 36. Baltrus DA, Guillemin K. Multiple phases of competence occur during the Helicobacter pylori growth cycle. FEMS Microbiol Lett. 2006;255(1):148–55. 10.1111/j.1574-6968.2005.00066.x [DOI] [PubMed] [Google Scholar]
- 37. Dorer MS, Fero J, Salama NR. DNA damage triggers genetic exchange in Helicobacter pylori . PLoS Pathog. 2010;6(7):e1001026 10.1371/journal.ppat.1001026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moore ME, Lam A, Bhatnagar S, Solnick JV. Environmental determinants of transformation efficiency in Helicobacter pylori . J Bacteriol. 2014;196(2):337–44. 10.1128/JB.00633-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Levine SM, Lin EA, Emara W, Kang J, DiBenedetto M, Ando T, et al. Plastic cells and populations: DNA substrate characteristics in Helicobacter pylori transformation define a flexible but conservative system for genomic variation. FASEB J. 2007;21(13):3458–67. 10.1096/fj.07-8501com [DOI] [PubMed] [Google Scholar]
- 40. Boneca IG, Ecobichon C, Chaput C, Mathieu A, Guadagnini S, Prevost MC, et al. Development of inducible systems to engineer conditional mutants of essential genes of Helicobacter pylori . Appl Environ Microbiol. 2008;74(7):2095–102. 10.1128/AEM.01348-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang XS, Blaser MJ. Natural transformation of an engineered Helicobacter pylori strain deficient in type II restriction endonucleases. J Bacteriol. 2012;194(13):3407–16. 10.1128/JB.00113-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Falush D, Kraft C, Taylor NS, Correa P, Fox JG, Achtman M, et al. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age. Proc Natl Acad Sci U S A. 2001;98(26):15056–61. 10.1073/pnas.251396098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. MacHattie LA, Thomas CA Jr. DNA from Bacteriophage Lambda: Molecular Length and Conformation. Science. 1964;144(3622):1142–4. [DOI] [PubMed] [Google Scholar]
- 44. Dokland T, Murialdo H. Structural transitions during maturation of bacteriophage lambda capsids. J Mol Biol. 1993;233(4):682–94. 10.1006/jmbi.1993.1545 [DOI] [PubMed] [Google Scholar]
- 45. Sinha S, Mell J, Redfield R. The availability of purine nucleotides regulates natural competence by controlling translation of the competence activator Sxy. Mol Microbiol. 2013;88(6):1106–19. 10.1111/mmi.12245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Klug D, Rabani J, Fridovich I. A direct demonstration of the catalytic action of superoxide dismutase through the use of pulse radiolysis. J Biol Chem. 1972;247(15):4839–42. [PubMed] [Google Scholar]
- 47. Evilevitch A, Lavelle L, Knobler CM, Raspaud E, Gelbart WM. Osmotic pressure inhibition of DNA ejection from phage. Proc Natl Acad Sci U S A. 2003;100(16):9292–5. 10.1073/pnas.1233721100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cohn CA, Fisher SC, Brownawell BJ, Schoonen MA. Adenine oxidation by pyrite-generated hydroxyl radicals. Geochem Trans. 2010;11(1):2 10.1186/1467-4866-11-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bury-Mone S, Thiberge JM, Contreras M, Maitournam A, Labigne A, De Reuse H. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori . Mol Microbiol. 2004;53(2):623–38. 10.1111/j.1365-2958.2004.04137.x [DOI] [PubMed] [Google Scholar]
- 50. An R, Jia Y, Wan B, Zhang Y, Dong P, Li J, et al. Non-enzymatic depurination of nucleic acids: factors and mechanisms. PLoS One. 2014;9(12):e115950 10.1371/journal.pone.0115950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stingl K, Uhlemann EM, Schmid R, Altendorf K, Bakker EP. Energetics of Helicobacter pylori and its implications for the mechanism of urease-dependent acid tolerance at pH 1. J Bacteriol. 2002;184(11):3053–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stingl K, Uhlemann Em EM, Deckers-Hebestreit G, Schmid R, Bakker EP, Altendorf K. Prolonged survival and cytoplasmic pH homeostasis of Helicobacter pylori at pH 1. Infect Immun. 2001;69(2):1178–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Charpentier X, Kay E, Schneider D, Shuman HA. Antibiotics and UV radiation induce competence for natural transformation in Legionella pneumophila . J Bacteriol. 2011;193(5):1114–21. 10.1128/JB.01146-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Drlica K, Malik M, Kerns RJ, Zhao X. Quinolone-mediated bacterial death. Antimicrob Agents Chemother. 2008;52(2):385–92. 10.1128/AAC.01617-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kampranis SC, Maxwell A. The DNA gyrase-quinolone complex. ATP hydrolysis and the mechanism of DNA cleavage. J Biol Chem. 1998;273(35):22615–26. [DOI] [PubMed] [Google Scholar]
- 56. Tomasz M, Palom Y. The mitomycin bioreductive antitumor agents: cross-linking and alkylation of DNA as the molecular basis of their activity. Pharmacol Ther. 1997;76(1–3):73–87. [DOI] [PubMed] [Google Scholar]
- 57. Wang G, Maier RJ. Critical role of RecN in recombinational DNA repair and survival of Helicobacter pylori . Infect Immun. 2008;76(1):153–60. 10.1128/IAI.00791-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Leisner M, Stingl K, Radler JO, Maier B. Basal expression rate of comK sets a 'switching-window' into the K-state of Bacillus subtilis . Mol Microbiol. 2007;63(6):1806–16. 10.1111/j.1365-2958.2007.05628.x [DOI] [PubMed] [Google Scholar]
- 59. Maamar H, Raj A, Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis . Science. 2007;317(5837):526–9. 10.1126/science.1140818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Scott DR, Marcus EA, Wen Y, Oh J, Sachs G. Gene expression in vivo shows that Helicobacter pylori colonizes an acidic niche on the gastric surface. Proc Natl Acad Sci U S A. 2007;104(17):7235–40. 10.1073/pnas.0702300104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Quigley EM, Turnberg LA. pH of the microclimate lining human gastric and duodenal mucosa in vivo. Studies in control subjects and in duodenal ulcer patients. Gastroenterology. 1987;92(6):1876–84. [DOI] [PubMed] [Google Scholar]
- 62. Schade C, Flemstrom G, Holm L. Hydrogen ion concentration in the mucus layer on top of acid-stimulated and -inhibited rat gastric mucosa. Gastroenterology. 1994;107(1):180–8. [DOI] [PubMed] [Google Scholar]
- 63. El-Zimaity HMT. Recent advances in the histopathology of gastritis. Curr Diag Pathol. 2007;13:340–8. [Google Scholar]
- 64. Rawlings JW, Danesh BJ, Lucas ML, Morgan RJ, Main AN, Russell RI. Gastroduodenal mucosal surface and luminal pH in gastric ulcer. Dig Dis Sci. 1991;36(11):1543–9. [DOI] [PubMed] [Google Scholar]
- 65. Ohshima H, Tatemichi M, Sawa T. Chemical basis of inflammation-induced carcinogenesis. Archives of Biochemistry and Biophysics. 2003;417(1):3–11. 10.1016/S0003-9861(03)00283-2 [DOI] [PubMed] [Google Scholar]
- 66. Zhang C, Cleveland K, Schnoll-Sussman F, McClure B, Bigg M, Thakkar P, et al. Identification of low abundance microbiome in clinical samples using whole genome sequencing. Genome Biol. 2015;16:265 10.1186/s13059-015-0821-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. O'Rourke EJ, Chevalier C, Pinto AV, Thiberge JM, Ielpi L, Labigne A, et al. Pathogen DNA as target for host-generated oxidative stress: role for repair of bacterial DNA damage in Helicobacter pylori colonization. Proc Natl Acad Sci U S A. 2003;100(5):2789–94. 10.1073/pnas.0337641100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kavermann H, Burns BP, Angermuller K, Odenbreit S, Fischer W, Melchers K, et al. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J Exp Med. 2003;197(7):813–22. 10.1084/jem.20021531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nakagawa S, Osaki T, Fujioka Y, Yamaguchi H, Kamiya S. Long-term infection of Mongolian gerbils with Helicobacter pylori: microbiological, histopathological, and serological analyses. Clin Diagn Lab Immunol. 2005;12(2):347–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Watanabe T, Tada M, Nagai H, Sasaki S, Nakao M. Helicobacter pylori infection induces gastric cancer in mongolian gerbils. Gastroenterology. 1998;115(3):642–8. [DOI] [PubMed] [Google Scholar]
- 71. Yoshizawa N, Takenaka Y, Yamaguchi H, Tetsuya T, Tanaka H, Tatematsu M, et al. Emergence of spasmolytic polypeptide-expressing metaplasia in Mongolian gerbils infected with Helicobacter pylori . Lab Invest. 2007;87(12):1265–76. 10.1038/labinvest.3700682 [DOI] [PubMed] [Google Scholar]
- 72. Honda S, Fujioka T, Tokieda M, Satoh R, Nishizono A, Nasu M. Development of Helicobacter pylori-induced gastric carcinoma in Mongolian gerbils. Cancer Res. 1998;58(19):4255–9. [PubMed] [Google Scholar]
- 73. Dorer MS, Cohen IE, Sessler TH, Fero J, Salama NR. Natural competence promotes Helicobacter pylori chronic infection. Infect Immun. 2013;81(1):209–15. 10.1128/IAI.01042-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ferrero RL, Cussac V, Courcoux P, Labigne A. Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. J Bacteriol. 1992;174(13):4212–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori . Nature. 1999;397(6715):176–80. 10.1038/16495 [DOI] [PubMed] [Google Scholar]
- 76. Reynolds DJ, Penn CW. Characteristics of Helicobacter pylori growth in a defined medium and determination of its amino acid requirements. Microbiology. 1994;140 (Pt 10):2649–56. [DOI] [PubMed] [Google Scholar]
- 77. Behrens W, Bonig T, Suerbaum S, Josenhans C. Genome sequence of Helicobacter pylori hpEurope strain N6. J Bacteriol. 2012;194(14):3725–6. 10.1128/JB.00386-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Skouloubris S, Thiberge JM, Labigne A, De Reuse H. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infect Immun. 1998;66(9):4517–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, et al. The primary transcriptome of the major human pathogen Helicobacter pylori . Nature. 2010;464(7286):250–5. 10.1038/nature08756 [DOI] [PubMed] [Google Scholar]
- 80. Bury-Mone S, Skouloubris S, Dauga C, Thiberge JM, Dailidiene D, Berg DE, et al. Presence of active aliphatic amidases in Helicobacter species able to colonize the stomach. Infect Immun. 2003;71(10):5613–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Larsson A, Carlsson C, Jonsson M. Characterization of the binding of YO to [poly(dA-dT)]2 and [poly(dG-dC)]2, and of the fluorescent properties of YO and YOYO complexed with the polynucleotides and double-stranded DNA. Biopolymers. 1995;36(2):153–67. 10.1002/bip.360360205 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table A: Oligonucleotides used in this study. Figure A: Bleaching curve of Cy3 labelled λ DNA. Figure B: Distribution of estimated area per λ molecule in periplasmic DNA foci of H. pylori. Figure C: Growth of competent H. pylori N6 in minimal medium without purine source cannot be restored by addition of external DNA. Figure D: Competent H. pylori did not grow in minimal medium in the absence of a purine source but were still capable of DNA uptake. Figure E: Competence development is not influenced by lower concentrations of adenine. Figure F: pH is a prerequisite of competence development and fine-tuned by the degree of oxidative stress.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.