Abstract
Background
The transcription factor homeobox C9 (HOXC9) plays a crucial role in developmental regulatory systems, where it determines the specific positional identities of cells along the anteroposterior axis. The expression of HOXC9 has been found to be dysregulated in some cancers such as lung cancer, breast cancer, and neuroblastoma. Here, we report for the first time that HOXC9 is a novel autophagy regulator and reveal its oncogenic role in cell survival and its usefulness as a prognostic marker in glioblastoma patients.
Methods
Kaplan-Meier analysis was performed to evaluate the possible prognostic value of HOXC9 in glioblastoma. Growth curve assays, subcutaneous, and orthotopic implantations were used to analyze cell viability and tumor formation, respectively. Luciferase and chromatin immunoprecipitation assays were employed to explore the mechanisms involved in the association between HOXC9 and its downstream effector, death-associated protein kinase 1 (DAPK1).
Results
High expression of HOXC9 was found to be an indicator of a poor prognosis in glioblastoma. HOXC9 knockdown resulted in a significant reduction of cell viability, migration, invasion, and tumorigenicity and a marked increase in autophagy. During the autophagy process, HOXC9 inhibited DAPK1 transcription by directly binding to its promoter. The downregulation of HOXC9 releases its transcriptional inhibition of DAPK1, resulting in the activation of the DAPK1-Beclin1 pathway, which induces autophagy in glioblastoma cells.
Conclusions
Collectively, our data indicate that HOXC9 is an oncogene in glioblastoma. We have revealed its role in the control of autophagy, and we suggest that HOXC9 is a novel and promising therapeutic target.
Keywords: autophagy, DAPK1, glioblastoma, HOXC9, tumorigenesis
Glioblastoma, which is classified as a grade IV glioma, is the most common and deadly type of brain tumor.1,2 It exhibits a high rate of cellular proliferation and highly aggressive and infiltrative metastasis.3,4 Despite advances in surgical and pharmacological therapies, glioblastoma remains incurable and has a low survival rate. Therefore, identifying alternative therapeutic approaches is critical but cannot be achieved without understanding the molecular mechanisms underlying the initiation and progression of glioblastoma.
HOX genes belong to the homeobox family, which encodes evolutionarily conserved transcription factors that provide positional information during differentiation and morphogenesis along the anteroposterior axis.5–7 They also control other cellular processes (eg, cell proliferation, cell migration, the determination of cell morphology, and apoptosis) in cancer cells.8 The dysregulation of HOX genes has been reported to cause abnormal development and malignancy (eg, HOXA9 dysregulation in leukemia, HOXB13 dysregulation in ovarian cancer, and HOXB5 dysregulation in breast cancer).9–11 In some cases, HOX genes act as tumor suppressors, including HOXC6 in serous ovarian cancer and HOXC9 in neuroblastoma.12–14
As a member of the HOX family, homeobox C9 (HOXC9) has been reported to induce cell differentiation and reduce self-renewal capabilities in neuroblastoma.14 However, it has also been reported that HOXC9 is highly expressed in CD133(+) astrocytomas and that it promotes cell proliferation.15 The abnormal expression of HOXC9 has also been observed in non–small cell lung cancer, breast cancer, and canine mammary tumors,7,16,17 but the role that HOXC9 plays in these cancers has not been identified. The role of HOXC9 in tumorigenesis and tumor progression is therefore complicated and needs to be further clarified.
Recently, Yacine Graba et al found that HOX genes are key factors that regulate autophagy during metamorphosis in Drosophila larvae.18,19 Autophagy, an evolutionarily conserved catabolic process, involves the transport of damaged, degenerated, senescent, or nonessential cellular proteins and organelles to lysosomes for digestion and recycling, which is beneficial for homeostatic maintenance under normal physiological conditions. Under stress, this highly regulated, multistep process prevents poisonous or carcinogenic proteins and organelles from accumulating and thus inhibits cell cancerization. However, autophagy shows 2 sides in cancer cells.20,21 Normal autophagy provides nutrition, which promotes cancer cell survival under stressed conditions. However, excessive and sustained induction of autophagy may limit cancer cell survival.22,23 Hence, determining the molecular mechanism underlying the regulation of autophagy is vital for cancer research and exploiting new anticancer therapies.
In this study, we demonstrated that HOXC9 plays an oncogenic role in glioblastoma. Knockdown of HOXC9 significantly reduced cell proliferation and induced autophagy, which indicates that HOXC9 may be a potential therapeutic target for glioblastoma treatment.
Materials and Methods
Reagents and Antibodies
The HOXC9 antibody (sc-81100) and polybrene (sc-134220) were purchased from Santa Cruz Biotechnology. The death-associated protein kinase 1 (DAPK1) (D1319) and pSer (308)-DAPK1 (D4941) antibodies, 3-[4, 5-dimethylthiazol-2-yl]-2, 5-diphenyltetrazolium bromide (MTT) (M 5655), 5′-bromo-2-deoxyuridine (BrdU), and dimethyl sulfoxide (DMSO) (D5879) were obtained from Sigma-Aldrich. The HOXC9 antibody (ARP35813_T100), which was used for immunohistochemistry, was acquired from Aviva Systems Biology. The tubulin antibody, 4′, 6-diamidino-2-phenylindole (DAPI) and 3, 3′-diaminobenzidine (DAB) were purchased from Beyotime. The autophagy antibody sampler kit (#4445) was purchased from Cell Signaling Technologies. HRP goat anti-mouse and goat anti-rabbit antibodies were purchased from KPL. Alexa Fluor 488 goat anti-rabbit IgG (H+L), lipofectamine 2000, and puromycin (A1113803) were obtained from Life Technologies. The BrdU antibody (ab6326) was purchased from Abcam.
Cell Culture
All glioblastoma cell lines were purchased from ATCC and cultured in Dulbecco's modified Eagle's medium (DMEM) except for U-251 MG, which was cultured in DMEM/F-12. All cultures were supplemented with 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin (P/S). 293FT cells were cultured in 293FT growth medium consisting of DMEM with 10% FBS, 1% P/S, 0.5 mg/mL G418, 4 mM L-glutamine, 0.1 mM nonessential amino acids (MEM), and 1 mM sodium pyruvate. The 293FT transfection medium, 293FT growth medium without P/S, and G418 were used. All cell lines were cultured at 37°C in a humidified atmosphere of 5% CO2. The growth media, FBS, antibiotics, and supplements were acquired from Thermo Fisher.
Transfection and Infection
Lentiviral constructs expressing HOXC9 short hairpin (sh) RNA (RHS4533-NM_006897) were purchased from Open Biosystems. The negative control pLKO.1-shGFP plasmid was purchased from Addgene. shDAPK1 and shBeclin1 sequences (Supplementary material, Table S1) were inserted into the pLKO.1 vector. The GFP-LC3B plasmid was a gift from Prof. Ning Gao at the Third Military Medical University, China. Lentiviruses were generated by co-transfecting the 293FT cell line with the packaging plasmids pLP1, pLP2, and pLP/VSVG (Invitrogen) and the corresponding shRNA plasmids. Lipofectamine 2000 was used for all transfections according to the manufacturer's instructions. Virus-containing supernatants were harvested and used for cell infections with a final concentration of 4 μg/mL of polybrene. At 24 hours after the final round of infection, the cells were cultured in the presence of 2 mg/mL puromycin for 3 days, and the drug-resistant cells were selected and pooled for subsequent experiments.
Patient Data Analysis
Patient and gene expression data were downloaded from the R2: genomics analysis and visualization platform (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi). Kaplan-Meier survival analysis was conducted based on the high versus low HOXC9 expression cutoff determined using the R2 algorithm. P values (log-rank test) were obtained from the website. Primary tumor specimens were from Third Military Medical University, China, with written approval obtained from patients before experimentation.
Western Blot Analysis
Cells and tissues were lysed, and protein was extracted. Western blot analyses were then performed, as previously described.24
Immunohistochemistry Assay
Immunohistochemistry assays were performed on paraffin-embedded sections. Tissue sections were incubated with HOXC9 antibodies (1:50) at 4°C overnight, followed by incubation with HRP-conjugated secondary antibodies (CST) at room temperature for 3 hours. Staining was visualized using DAB, and tissues were counterstained with hematoxylin.
Cell Viability and Proliferation Assays
Cell growth curves were obtained using the MTT assay. Cells were seeded at a density of 8 × 102/per well in 200 μL of medium in 96-well plates. At the indicated time points after seeding, 20 μL of MTT solution (5 mg/mL in phosphate-buffered saline [PBS] from Sigma) were added to each well, and cells were incubated at 37°C for another 3 hours. At the end of the incubation period, the medium was replaced with 150 μL of DMSO to dissolve the formazan crystals. After agitation on a shaker for 10 minutes, absorbance was measured at 560 nm using a Modulus Microplate Multimode Reader (Promega). BrdU staining was performed, as previously described.25
In Vitro and in Vivo Tumorigenic Assays
For the in vitro soft agar assays, growth medium with 0.6% agarose was added to 6-well plates to form the lower layer. Growth medium with 0.3% agarose and 1 × 103 cells was added to form the upper layer. Colonies were photographed and recorded after 2–3 weeks of growth.
Subcutaneous xenograft and orthotopic implantations were used for the in vivo study. Six NOD/SCID female mice (5 weeks old) were subcutaneously injected into the lateral backside with 2–4 × 106 cells. Cells in which HOXC9 was stably silenced were implanted into the right side of each mouse, and control cells were implanted into the left side of each mouse. Calipers were used to measure the tumor volume, which was calculated using the formula Volume = (π/6) × length × width2. After 4 weeks of growth, the mice were euthanized, and tumors were removed and weighed. For orthotopic implantations, 1 × 105 cells were intracranially injected into the brains of NOD/SCID mice (2 mm lateral and 1 mm anterior to the bregma, 3.5 mm deep) and placed into a stereotaxic frame. Signs of disease progression were followed until the last mouse in the control group had died, at which time the shHOXC9 mice were all euthanatized. At the endpoint, the original tumors were photographed, and brains were collected, fixed in neutral buffered formalin, and embedded in paraffin. Hematoxylin and eosin (H&E) staining and immunohistochemical analysis were performed for histopathological evaluations of the tissues.
All mice were raised and monitored under specific pathogen-free (SPF) conditions. Animal welfare and experimental procedures were carried out in accordance with the Guide for the Care and Use of Laboratory Animals (Ministry of Science and Technology of China, 2006) and approved by the animal ethics committee of Southwest University.
Flow Cytometry Analysis
Cells were plated at a density of 3 × 105 cells into 60 mm plates. After lentivirus transfection, the cells were cultivated for 3 days and then harvested, washed 3 times with ice-cold PBS, and centrifuged at 1000 × g for 5 minutes. Cells were then resuspended in 1 × binding buffer containing fluorescein isothiocyanate (FITC)-labeled Annexin V and propidium iodide. After incubation at room temperature for 25 minutes, the data were analyzed using a BD Accuri C6 flow cytometer according to the manufacturer's directions.
Immunofluorescence Staining Assay
For immunofluorescence staining, 2 × 104 cells were seeded on coverslips in 24-well plates and cultured for 24 hours. The cells were then washed with PBS, fixed in 4% paraformaldehyde for 15 minutes at room temperature, and permeabilized using 0.1% Triton X-100 for 10 minutes. Coverslips were blocked in 10% goat serum for 1 hour, and the cells were then incubated with primary antibodies against microtubule-associated protein light chain 3 beta (LC3B) at a dilution of 1:500, followed by incubation with Alexa Fluor 488 goat anti-rabbit IgG (H+L), which was used as the secondary antibody. Finally, 300 nM DAPI in PBS was used for nuclear staining.
Luciferase Reporter Assay
Different DAPK1 promoter fragments were amplified using PCR and confirmed by gene sequencing (BGI, Shenzhen). They were then ligated into the pGL3-basic vector, which was purchased from Promega. The empty pGL3-basic vector served as a negative control. A total of 1.5–3 × 105 cells/well were placed in 24-well plates for cell transfection. A total of 1 μg pGL3 plasmid and 100 ng pRL-TK internal control vector (Promega) were co-transfected into cells in serum-free Opti-MEM Reduced-Serum Medium (Life Technologies). After 4–6 hours, culture medium was added to each well to make the final volume 1 mL. After a further incubation for 48 hours, a luciferase reporter assay was performed according to the manufacturer's instructions (Promega). Luciferase activity was normalized to pRL-TK activity. Each experiment was performed in triplicate.
Chromatin Immunoprecipitation
A chromatin immunoprecipitation (ChIP) assay was performed using a ChIP assay kit (Millipore) according to the manufacturer's instructions. Briefly, LN-229 cells were cross-linked and lysed, and DNA was sheared into 200–800-bp fragments using sonication. Precleared chromatin was immunoprecipitated with a HOXC9 antibody (Santa Cruz), and DNA was isolated after reverse cross-linking for quantitative real-time PCR (qRT-PCR). The relevant primer sequences are presented in Table S3 in the Supplemental Materials.
Results
High HOXC9 Expression Is an Indicator of a Poor Prognosis in Patients with Glioblastoma
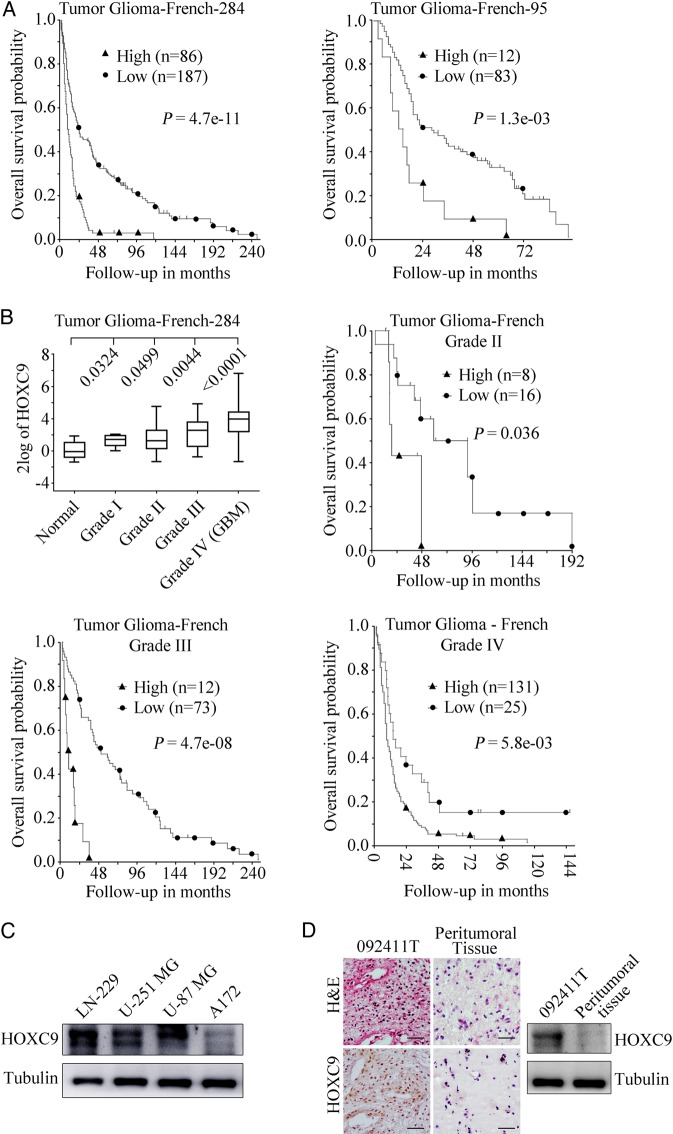
HOXC9 gene expression dysregulation has been reported in many types of cancer14–17,26; however, it has not yet been well defined in glioblastoma. To investigate whether HOXC9 is a prognostic marker for glioblastoma, we conducted a microarray-based search using 2 databases (Tumor Glioma-French-284 and Tumor Glioma-French-95) that are available online using the R2: genomics analysis and visualization platform. A Kaplan-Meier analysis of progression-free survival using these 2 databases showed that high HOXC9 expression was strongly associated with a poor outcome, whereas low HOXC9 expression was correlated with good overall survival (Fig. 1A). Moreover, increased HOXC9 expression was significantly correlated with advanced tumor stages and was a meaningful prognostic indicator for the different stages of glioma that were analyzed (grades II-IV) (Fig. 1B). A Kaplan-Meier graph could not be drawn for grade I glioma due to the small sample size.
Fig. 1.
High HOXC9 expression is a poor prognostic indicator in patients with glioblastoma. (A) Results of the Kaplan-Meier analysis of progression-free survival and the log rank test P values are indicated for the Tumor Glioma-French-284 dataset (left) and the Tumor Glioma French-95 dataset (right). (B) Top left, box plot of HOXC9 expression levels in the peritumoral tissues of tumors (Normal) and grade I-IV gliomas. The data were analyzed using 2-tailed Student t tests with P values indicated. Top right and bottom, Kaplan-Meier analysis of the Tumor Glioma-French-284 dataset according to tumor grade with the log-rank test P values indicated. (C) Western blot analysis was used to detect HOXC9 levels in different glioblastoma cell lines. Tubulin was used as the loading control. (D) Hematoxylin and eosin staining and immunohistochemistry assays (left) and Western blot analysis (right) of the representative tumor specimen 092411T and its peritumoral tissue. Scale bar = 20 μm.
To further confirm our results, we examined the expression of HOXC9 in several glioblastoma cell lines and primary tumor specimens and their corresponding peritumoral tissues. HOXC9 was commonly expressed in all 4 cell lines. The A172 cell line, which is a relatively benign cell line with no tumor-formation ability, had a relatively low level of HOXC9 expression (Fig. 1C). Moreover, the expression levels of HOXC9 in primary tumor specimens were much higher than those in their corresponding peritumoral tissues, in which HOXC9 was barely expressed. Data from a representative primary tumor specimen, 092411T, and its peritumoral tissue are shown in Fig. 1D (right). H&E staining and immunohistochemistry (IHC) assays, which were performed on the representative primary specimen and associated peritumoral tissue, confirmed the results of the Western blot analysis (Fig. 1D, left). Our data suggest that a higher expression of HOXC9 predicts a poor prognosis for patients with glioblastoma, indicating that HOXC9 may play an oncogenic role in tumor development.
HOXC9 Is Essential for Sustaining Glioblastoma Cell Proliferation, Migration, and Invasion
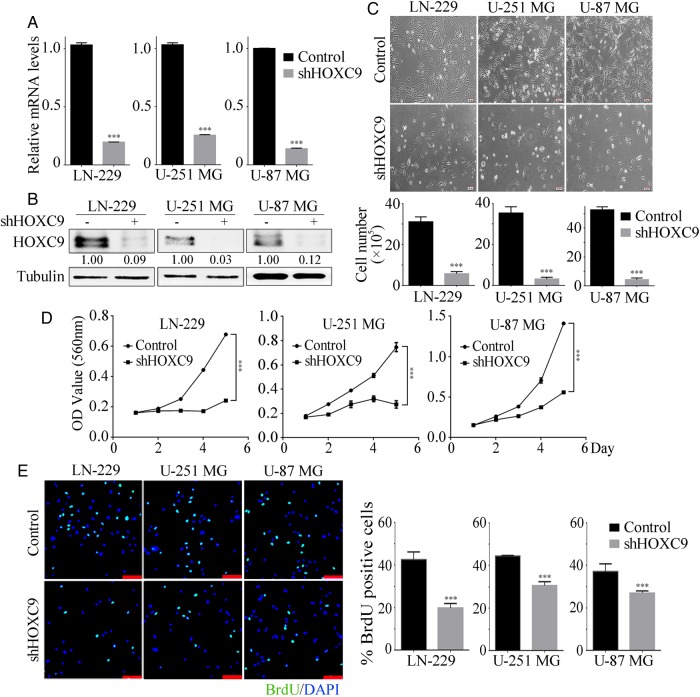
Next, HOXC9 was knocked down in 3 glioblastoma cell lines (LN-229, U-251 MG, and U-87 MG) using shRNA. For these experiments, 3 lentiviral plasmids that expressed shRNA sequences against HOXC9 were used, and all of them successfully knocked down HOXC9 expression. shHOXC9#10 exhibited the most significant reduction efficiency (Supplementary material, Fig. S1) and highly downregulated HOXC9 levels in all 3 cell lines (Fig. 2A and B). Therefore, the subsequent experiments were all performed using shHOXC9#10, which is referred to as shHOXC9. shGFP was used as a negative control. Using microscopy, we found that shHOXC9 cells showed a clear decrease in cell numbers and severe morphological changes compared with the corresponding control cells (Fig. 2C), suggesting an inhibitory effect on cell growth. Accordingly, we analyzed cell growth and proliferation in these cells using MTT and BrdU assays, and the results showed that silencing HOXC9 led to sharp declines in growth curves and the proportion of BrdU-positive cells (Fig. 2D and E). These results indicate that HOXC9 accelerates glioblastoma cell growth and proliferation.
Fig. 2.
HOXC9 is essential for sustaining glioblastoma cell proliferation and survival. After HOXC9 knockdown by shRNA in 3 glioblastoma cell lines, HOXC9 expression was detected at the mRNA level using qRT-PCR (A) and at the protein level using Western blot analysis (B). shGFP was used as the control. (C) Morphological examinations and cell number quantifications after HOXC9 knockdown. (D) Cell growth was monitored using MTT assays after the downregulation of HOXC9. (E) BrdU assays were performed after HOXC9 knockdown. Representative images show immunofluorescence and the quantification of BrdU-positive cells (scale bars, 20 μm). Data were analyzed using 2-tailed Student t tests. Error bars, SEM, n = 3, *** P < .001.
To determine whether cell migration and invasion were affected by HOXC9 silencing, wound healing and transwell assays were performed. The results of the wound healing assays showed that gap widths were much wider in the shHOXC9 group than in the control group after 24 hours (Supplementary material, Fig. S2A). Consistent with these results, shHOXC9 cells showed a much weaker ability to migrate across the membrane of transwell chambers, either with or without Matrigel, demonstrating that the loss of HOXC9 led to a significant reduction in migration and invasion (Supplementary material, Fig. S2B and C). These results suggest that HOXC9 is essential to sustaining cell migration and invasion.
HOXC9 Is Required for Self-renewal and Tumorigenesis of Glioblastoma Cells
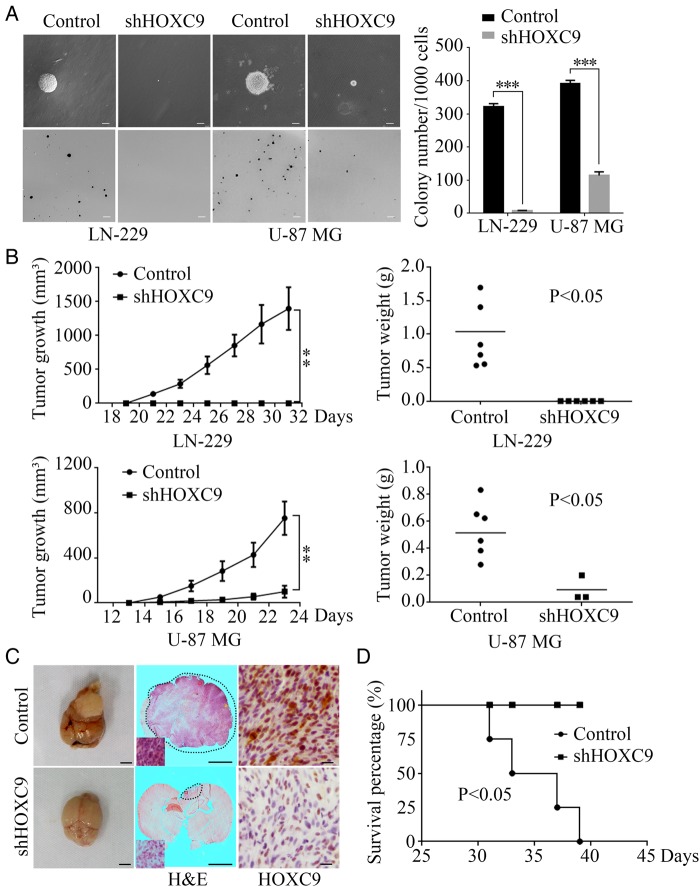
Next, the roles of HOXC9 in the self-renewal and tumorigenesis of glioblastoma cells were evaluated. In vitro colony formation was first examined using soft agar assays. Two glioblastoma cell lines, LN-229 and U-87 MG, were investigated; the results showed that the HOXC9-silenced group formed substantially fewer and smaller colonies than the control group (Fig. 3A), suggesting that HOXC9 functions to promote clonogenic and self-renewal abilities. Next, a subcutaneous xenograft mouse model was used to examine the effect of HOXC9 knockdown on tumor formation in NOD/SCID mice. As shown, the volumes and weights of shHOXC9 U87-MG tumors were much smaller than those in the corresponding control group. The shHOXC9 LN-229 group lost its tumorigenic activity during the same time course (Fig. 3B and C).
Fig. 3.
HOXC9 is required for glioblastoma cell self-renewal and tumorigenesis of glioblastoma cells. (A) Soft agar assays were performed after HOXC9 knockdown in LN-229 and U-87 MG cell lines. The quantification of colony numbers is also presented (error bars, SEM, n = 3; scale bars, upper = 50 μm, lower = 200 μm). (B) Subcutaneous xenograft tumor growth was monitored after HOXC9 knockdown in LN-229 and U-87 MG cells. Tumor volumes were measured using a caliper every 2 days (error bars, SEM, n = 6). Tumor weights are presented in scatterplots with horizontal lines indicating the mean. (C) Orthotopic implantation was performed after HOXC9 knockdown in U-87 MG cells. shGFP was used as the control. Representative images of the original tumor formation (left), hematoxylin and eosin(H&E) staining (middle), and immunohistochemistry analysis of HOXC9 expression (right) are presented. Scale bars: left and middle, 3 mm; right, 20 μm. (D) Survival rates were analyzed after orthotopic implantation of shHOXC9 U-87 MG cells. n = 4, P < .05 by the log-rank test for significance. Statistical analysis was performed using 2-tailed Student t tests, ** P < .01, *** P < .001.
Furthermore, to provide more relevant in vivo data, orthotopic implantations were performed. The data showed that the shHOXC9 group had smaller tumors by volume (Fig. 3C, left and middle) and longer survival times than the shGFP control group (Fig. 3D), which confirms the results of the subcutaneous xenograft experiments. Consistent with these results, HOXC9 expression (detected by IHC) in the orthotopic xenografts was lower in the shHOXC9 group than in the control group (Fig. 3C, right).
These results indicate an oncogenic function of HOXC9 in glioblastoma development.
Downregulation of HOXC9 Induces Autophagy but not Apoptosis
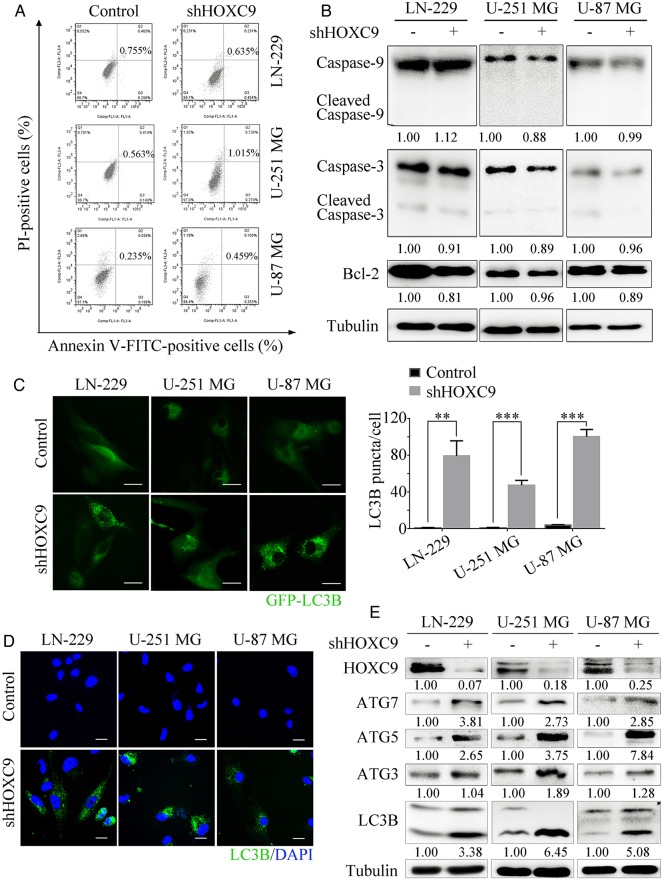
It has been reported that HOXC9 is associated with the intrinsic pathway of apoptosis.13 We therefore aimed to determine whether the decrease in cell numbers observed after HOXC9 silencing was caused by apoptosis. Flow cytometry analysis showed that no apoptosis was detected (Fig. 4A). Furthermore, apoptosis-associated proteins were examined by Western blot analysis. As shown in Fig. 4B, neither caspase-3 nor caspase-9 was activated, and the antiapoptosis protein Bcl-2 exhibited no significant decrease.
Fig. 4.
Downregulation of HOXC9 induces autophagy but not apoptosis. (A) The rate of apoptosis was analyzed using flow cytometry assays after HOXC9 knockdown. (B) Caspases and Bcl-2 were detected by Western blot analysis after the downregulation of HOXC9. (C) Cells transfected with the GFP-LC3B plasmid after HOXC9 knockdown were examined using fluorescence microscopy (scale bars, 10 μm). The quantification of LC3B-positive puncta is presented as a histogram (error bars, SEM, n = 3). (D) Immunofluorescence staining with a LC3B antibody was performed to confirm the induction of autophagy after HOXC9 downregulation. Representative LC3B-positive cells are shown (scale bars, 10 μm). (E) The level of autophagy was evaluated by autophagy-associated proteins and LC3B expression, as determined using Western blot analysis after HOXC9 knockdown. For Western blot analyses, tubulin was used as the loading control. ** P < .01, *** P < .001.
HOX genes have been reported to be key factors in the regulation of autophagy in Drosophila.18,19 These studies inspired us to investigate the effect of HOXC9 on autophagy status in cancer cells. LC3B is a specific marker of autophagy initiation. Upon the induction of autophagy, LC3B-I is converted to its lipidated LC3B-II form, which is then incorporated into the autophagosomal membrane and results in the cellular redistribution of LC3B from a diffuse pattern to a punctate pattern. Conversion of the cytoplasmic form of LC3B (LC3B-I, 16 kDa) to its pre-autophagosomal and autophagosomal membrane-bound form, LC3B (LC3B-II, 14 kDa) is closely correlated with the steady-state levels of autophagosomes.27,28 According to these, we transiently transfected glioblastoma cells with GFP-LC3B plasmid and examined the cells under a fluorescence microscope. shHOXC9 cells showed a punctate pattern of GFP-LC3B fluorescence that represented the recruitment of LC3B-II to autophagosomes and the formation of autophagic vacuoles. This result differed from the diffuse LC3B-associated green fluorescence observed in the control cells (Fig. 4C). To further confirm our results, immunofluorescence and Western blot analysis were performed to detect the conversion of LC3B-I to LC3B-II and the progression of autophagy. As predicted, after HOXC9 was downregulated, the formation of LC3B-positive puncta increased markedly (Fig. 4D). These results are in agreement with the biochemical results as demonstrated by the increased conversion of LC3B and enhancement of autophagy-associated proteins (Fig. 4E). Moreover, the induction of autophagy was further supported by Western blot analysis of xenograft tumor tissues (Supplementary material, Fig. S3B). Together, our results provide evidence that HOXC9 silencing induces autophagy but not apoptosis in glioblastoma cells.
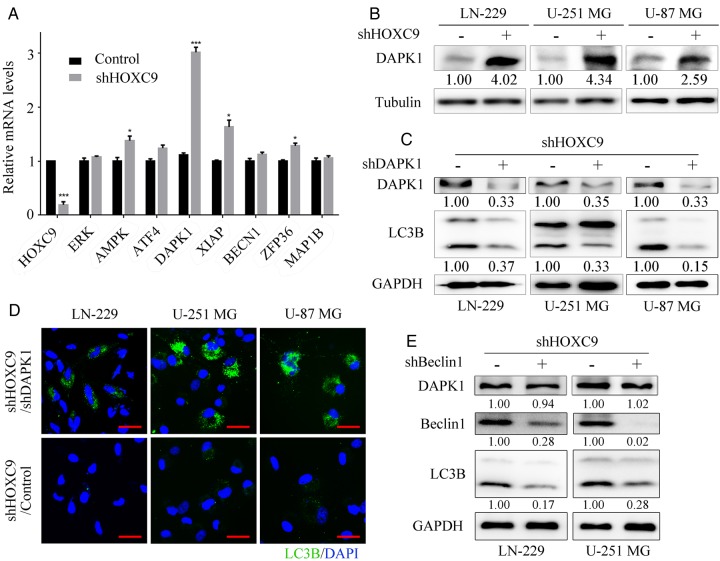
DAPK1 Is a Critical Downstream Effector of HOXC9
To further explore the mechanism by which HOXC9 controls autophagy, we focused on the expression of downstream genes involved in regulating autophagy, in view of the transcriptional role of HOXC9. The expression levels of a panel of 8 genes were analyzed using qRT-PCR. As demonstrated in Fig. 5A and B, DAPK1 showed the most significant increase in its mRNA level and an obvious increase in its protein level after HOXC9 was silenced. DAPK1 is generally known as a calcium/calmodulin-dependent serine/threonine kinase that is involved in multiple cellular signaling pathways including autophagy regulation.29 It is activated by the dephosphorylation of Ser308, which is required for autophagy induction.30,31 We therefore detected the amount of DAPK1 that was phosphorylated at Ser308 using Western blot analysis and found that the phosphorylation of DAPK1 was not altered. However, the total amount of DAPK1 protein was significantly increased after HOXC9 downregulation, indicating that DAPK1 was activated (Supplementary material, Fig. S3A). Similar results were also obtained using xenograft tumor tissues (Supplementary material, Fig. S3B). To gain further insight into the role of DAPK1 in HOXC9-silencing–induced autophagy, we knocked down DAPK1 after HOXC9 was silenced to examine whether this would reduce autophagy. The results showed that the formation of LC3B-positive puncta and the conversion of LC3B-I to LC3B-II were clearly reduced after the knockdown of DAPK1 (Fig. 5C and D). These results suggest that DAPK1 plays a vital role in mediating the autophagy induced by the downregulation of HOXC9.
Fig. 5.
DAPK1 is a critical downstream effector of HOXC9. (A) Genes involved in the regulation of autophagy were analyzed using qRT-PCR after knockdown of HOXC9 in the LN-229 cell line (error bars, SEM, n = 3). (B) DAPK1 protein expression levels were verified using Western blot analysis after knockdown of HOXC9. (C) DAPK1 and LC3B protein levels were detected by Western blot analysis after knockdown of DAPK1 by shRNA following HOXC9 downregulation. (D) Autophagy status was examined using immunofluorescence staining with LC3B antibodies after knockdown of DAPK1 (scale bars, 10 μm). (E) Beclin1 and LC3B protein levels were detected using Western blot analysis after knockdown of Beclin1 by shRNA following HOXC9 downregulation. For Western blot analyses, tubulin or GAPDH was used as the loading control. * P < .05, ** P < .01, *** P < .001.
It has been reported that Beclin1 is an indispensable downstream effector of DAPK1 during the regulation of autophagy.20,32 We knocked down Beclin1 following HOXC9 silencing to assess whether autophagy would consequently be reduced. The level of autophagy, as indicated by the conversion of LC3B-I to LC3B-II, was markedly decreased in the shBeclin1 group compared with the shGFP group (Fig. 5E). Our results collectively demonstrated that HOXC9 suppresses autophagy through its effect on the DAPK1-Beclin1 pathway.
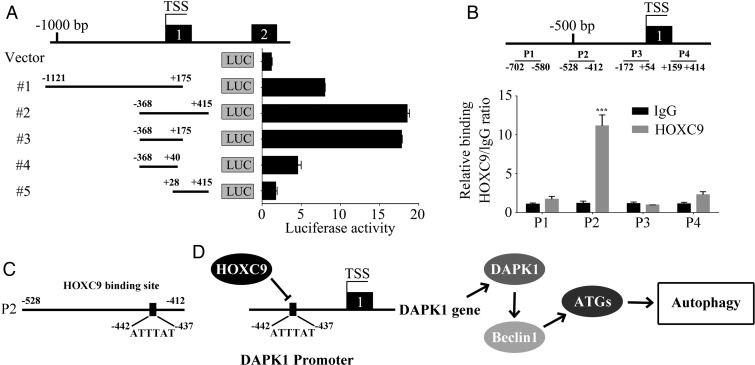
HOXC9 Binds to DAPK1 Promoter and Inhibits its Transcription
Next, a dual-luciferase reporter assay was performed to explore the mechanism by which HOXC9 modulates DAPK1 expression in glioblastoma cells. A total of 5 fragments from different DAPK1 promoter regions were designed and inserted into the pGL3 basic vector. These constructs were then co-transfected with the pRL-TK plasmid. The empty pGL3 basic vector was used as the control. As shown in Fig. 6A, DAPK1 promoter activity was significantly increased by the deletion of its upstream region from −1121 bp to −368 bp before the transcription start site, indicating that this region is associated with the inhibition of DAPK1 transcription. To determine whether the suppression of DAPK1 promoter activity was caused by HOXC9 binding, a ChIP assay was used to map the HOXC9-binding locus on the DAPK1 promoter. As shown in Fig. 6B, HOXC9-binding sites were enriched in the promoter-proximal region p2 (−528 bp to −412 bp). It was previously reported that HOXC9, a transcription factor, can recognize and bind to the DNA sequence T/ATTTAT.33 We identified an ATTTAT sequence in the DAPK1 promoter region from −442 bp to −437 bp (Fig. 6C). This finding confirmed our previous results showing that HOXC9 inhibits DAPK1 transcription by directly binding to its promoter.
Fig. 6.
HOXC9 binds to DAPK1 promoter and inhibits its transcription. (A) Different DAPK1 promoter regions (#1–#5) were ligated into the pGL3 plasmid and co-transfected with pRL-TK. Luciferase activity was examined 48 hours after transfection. The pGL3-basic vector, without the insert, was used as the negative control. (B) A total of 4 sets of primers were designed within the human DAPK1 promoter, and a chromatin immunoprecipitation ChIP assay was performed using HOXC9 antibodies. IgG was used as the negative control. Error bars, SEM; n = 3. (C) A HOXC9 conserved recognition sequence was identified in the DAPK1 promoter in a region within −528 bp to 412 bp. D, Schematic diagram of the role of HOXC9 in autophagy regulation. *** P < .001.
Discussion
HOXC9 participates in regulating and controlling many cellular processes including the cell cycle, differentiation, and apoptosis.13,14,33,34 There has recently been an increase in the evidence indicating that HOXC9 dysregulation is closely related to tumor initiation and development.14–16 However, the function of HOXC9 in glioblastoma remains unknown, and the molecular mechanisms contributing to its effects remain undetermined. In this study, we demonstrated that a high level of HOXC9 predicts a poor prognosis in glioblastoma patients. Knockdown of HOXC9 significantly reduced cell proliferation, migration, invasion, self-renewal, and tumorigenesis in glioblastoma cells, indicating that HOXC9 functions as an oncogene in glioblastoma cells.
Autophagy plays crucial roles in various biological responses and has been shown to have 2 functions in cancer whereby it either promotes or limits cancer cell survival under different conditions. Appropriate and excessive autophagy can lead to contrasting consequences.22,23 It has been reported that HOX genes are crucial to the regulation of autophagy during the process of Drosophila metamorphosis,18,19 but whether HOX genes perform the same functions in mammals remains unknown. Our study presents, for the first time, evidence showing that HOXC9 is an autophagy regulator in cancer cells. Moreover, different form that the silencing of all HOX genes is required to initiate autophagy in Drosophila,19 we found that silencing of HOXC9 alone was able to trigger autophagy in glioblastoma cells.
The molecular mechanisms whereby HOX genes regulate autophagy have been previously unknown. Our study provides original evidence showing that HOXC9 reduces autophagy by transcriptionally inhibiting DAPK1. The downregulation of HOXC9 significantly increases and activates DAPK1, which triggers autophagy, and knocking down DAPK1 after HOXC9 silencing markedly reduces autophagy. DAPK1 is reported to mediate cell death,35 including autophagic cell death.36 Kimchi et al verified that there is a positive correlation between DAPK1 and autophagy.37 DAPK1 phosphorylates Beclin1, causing it to dissociate from Bcl-XL and result in the induction of autophagy.32 Our data show that the downregulation of Beclin1 significantly reduced DAPK1-induced autophagy. For the first time, we have identified the link between HOXC9 and DAPK1 in the autophagy pathway (Fig. 6D).
In summary, we present evidence showing that HOXC9 functions as an oncogene and an inhibitor of autophagy in glioblastoma. The downregulation or inhibition of HOXC9 may be a potential therapeutic strategy for glioblastoma treatment.
Supplementary Material
Funding
This work was supported by the National Basic Research Program of China (No. 2012CB114603), the National Natural Science Foundation of China (No. 31501100, 81502574, 81201551), and the Research Fund for the Doctoral Program of Higher Education of China (20130182110003).
Conflict of interest statement. The authors disclose no potential conflicts of interest.
Supplementary Material
References
- 1.Benjamin R, Capparella J, Brown A. Classification of glioblastoma multiforme in adults by molecular genetics. Cancer J. 2003;9(2):82–90. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason W, van den Bent M et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 3.Weller M, Cloughesy T, Perry JR, Wick W. Standards of care for treatment of recurrent glioblastoma–are we there yet? Neuro Oncol. 2013;15(1):4–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie Q, Mittal S, Berens ME. Targeting adaptive glioblastoma: an overview of proliferation and invasion. Neuro Oncol. 2014;16(12):1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGinnis W, Krumiauft R. Homeobox genes and axial patterning. Cell. 1992;68(2):283–302. [DOI] [PubMed] [Google Scholar]
- 6.Mann RS, Lelli KM, Joshi R. Hox specificity: unique roles for cofactors and collaborators. Curr Top Dev Biol. 2009;88:63–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merabet S, Hudry B, Saadaoui M, Graba Y. Classification of sequence signatures: a guide to Hox protein function. Bioessays. 2009;31(5):500–511. [DOI] [PubMed] [Google Scholar]
- 8.Daniels TR, Neacato II, Rodriguez JA, Pandha HS, Morgan R, Penichet ML. Disruption of HOX activity leads to cell death that can be enhanced by the interference of iron uptake in malignant B cells. Leukemia. 2010;24(9):1555–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10(5):361–371. [DOI] [PubMed] [Google Scholar]
- 10.Argiropoulos B, Humphries R. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26(47):6766–6776. [DOI] [PubMed] [Google Scholar]
- 11.Lee J, Hur H, Yun H et al. HOXB5 Promotes the Proliferation and Invasion of Breast Cancer Cells. Int J Biol Sci. 2015;11(6):701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tait D, Bahrani-Mostafavi Z, Vestal C, Richardson C, Mostafavi M. Downregulation of HOXC6 in Serous Ovarian Cancer. Cancer Invest. 2015;33(7):303–311. [DOI] [PubMed] [Google Scholar]
- 13.Kocak H, Ackermann S, Hero B et al. Hox-C9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. Cell Death Dis. 2013;4: e586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mao L, Ding J, Zha Y et al. HOXC9 links cell-cycle exit and neuronal differentiation and is a prognostic marker in neuroblastoma. Cancer Res. 2011;71(12):4314–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okamoto O, Oba-Shinjo S, Lopes L, Nagahashi Marie S. Expression of HOXC9 and E2F2 are up-regulated in CD133(+) cells isolated from human astrocytomas and associate with transformation of human astrocytes. Biochim Biophys Acta. 2007;1769(7–8):437–442. [DOI] [PubMed] [Google Scholar]
- 16.Hur H, Lee J, Yun H, Park B, Kim M. Analysis of HOX gene expression patterns in human breast cancer. Mol Biotechnol. 2014;56(1):64–71. [DOI] [PubMed] [Google Scholar]
- 17.Deinnocentes P, Perry A, Graff E, Lutful Kabir F, Curtis Bird R. Characterization of HOX gene expression in canine mammary tumour cell lines from spontaneous tumours. Vet Comp Oncol. 2013;13(3):322–336. [DOI] [PubMed] [Google Scholar]
- 18.Campello S, Cecconi F. Ho(a)xing autophagy to regulate development. Dev Cell. 2014;28(1):3–4. [DOI] [PubMed] [Google Scholar]
- 19.Banreti A, Hudry B, Sass M, Saurin AJ, Graba Y. Hox proteins mediate developmental and environmental control of autophagy. Dev Cell. 2014;28(1):56–69. [DOI] [PubMed] [Google Scholar]
- 20.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Xu HL, Liu YX, An N, Zhao S, Bao JK. Autophagy modulation as a target for anticancer drug discovery. Acta Pharmacol Sin. 2013;34(5):612–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12(6):401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brech A, Ahlquist T, Lothe R, Stenmark H. Autophagy in tumour suppression and promotion. Mol Oncol. 2009;3(4):366–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang M, Liu Y, Zou J et al. Transcriptional co-activator TAZ sustains proliferation and tumorigenicity of neuroblastoma by targeting CTGF and PDGF-β. Oncotarget. 2015;6(11):9517–9530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang R, Wu Y, Wang M et al. HDAC9 promotes glioblastoma growth via TAZ-mediated EGFR pathway activation. Oncotarget. 2015;6(10):7644–7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin Q, Geng J, Ma K et al. RASSF1A, APC, ESR1, ABCB1 and HOXC9, but not p16INK4A, DAPK1, PTEN and MT1G genes were frequently methylated in the stage I non-small cell lung cancer in China. J Cancer Res Clin Oncol. 2009;135(12):1675–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klionsky D, Abdalla F, Abeliovich H et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bialik S, Kimchi A. Lethal weapons: DAP-kinase, autophagy and cell death: DAP-kinase regulates autophagy. Curr Opin Cell Biol. 2010;22(2):199–205. [DOI] [PubMed] [Google Scholar]
- 30.Levin-Salomon V, Bialik S, Kimchi A. DAP-kinase and autophagy. Apoptosis. 2014;19(2):346–356. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Hu M, Shu H, S L.. Death-associated protein kinase 1 is an IRF3/7-interacting protein that is involved in the cellular antiviral immune response. Cell Mol Immunol. 2014;11(3):245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zalckvar E, Berissi H, Mizrachy L et al. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10(3):285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Choi J, Ding J et al. HOXC9 directly regulates distinct sets of genes to coordinate diverse cellular processes during neuronal differentiation. BMC Genomics. 2013;14:830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Yang L, Choi J et al. Genome-wide analysis of HOXC9-induced neuronal differentiation of neuroblastoma cells. Genom Data. 2014;2:50–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen O, Feinstein E, Kimchi A. DAP-kinase is a Ca2+/calmodulin-dependent, cytoskeletal-associated protein kinase, with cell death-inducing functions that depend on its catalytic activity. EMBO J. 1997;16(5):998–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bialik S, Kimchi A. The death-associated protein kinases: structure, function, and beyond. Annu Rev Biochem. 2006;75:189–210. [DOI] [PubMed] [Google Scholar]
- 37.Inbal B, Bialik S, Sabanay I, Shani G, Kimchi A. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J Cell Biol. 2002;157(3):455–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.