Abstract
Local vascular immune response is primarily initiated via Toll-like receptors (TLRs) and triggering receptor expressed on myeloid cells-1 (TREM-1). We previously showed that certain TLR and TREM-1 gene polymorphisms are associated with coronary artery disease (CAD). Therefore, we hypothesized that these gene polymorphisms are associated with atherosclerosis severity. This study included 292 consecutive patients with CAD who were admitted to the Research Institute for Complex Issues of Cardiovascular Diseases (Kemerovo, Russian Federation) during 2011–2012. Sample genotyping was performed in 96-well format using the TaqMan SNP genotyping assay. We found that C/C genotype of the rs3804099 polymorphism within TLR2 gene and T/T genotype of the rs4711668 polymorphism within TREM-1 gene were significantly associated with severe coronary atherosclerosis while C allele of the rs5743551 polymorphism within TLR1 gene, A/G genotype of the rs4986790 polymorphism and C/T genotype of the rs4986791 polymorphism within TLR4 gene, and C allele of the rs3775073 polymorphism within TLR6 gene were significantly associated with severe noncoronary atherosclerosis. However, A/A genotype of the rs5743810 polymorphism within TLR6 gene was significantly associated with mild noncoronary atherosclerosis. We conclude that certain TLR and TREM-1 gene polymorphisms are significantly associated with atherosclerosis severity in a Russian population.
Keywords: Atherosclerosis, Coronary artery disease, Toll-like receptors, Triggering receptor expressed on myeloid cells-1, Gene polymorphisms
Highlights
-
•
Rs3804099 polymorphism of TLR2 gene is associated with severe coronary atherosclerosis.
-
•
Rs4711668 polymorphism of TREM-1 gene is associated with severe coronary atherosclerosis.
-
•
Rs5743551 polymorphism of TLR1 gene is associated with severe noncoronary atherosclerosis.
-
•
Rs4986790 and rs4986791 polymorphisms of TLR4 gene are associated with severe noncoronary atherosclerosis.
-
•
Rs3775073 polymorphism of TLR6 gene is associated with severe noncoronary atherosclerosis.
1. Introduction
Atherosclerosis, manifesting itself as acute coronary syndrome, stroke, and peripheral artery disease (Bentzon et al., 2014), is a chronic progressive inflammatory disease characterized by the accumulation of lipid and fibrous elements in arterial walls, which is driven by innate and adaptive immune response (Shah et al., 2014). The underlying mechanism of the chronic inflammatory process in atherosclerosis is still unknown in a significant extent (Ammirati et al., 2015). However, it is known that local vascular immune response is primarily initiated through the pattern recognition receptors, particularly Toll-like receptors (TLRs) (Pelham and Agrawal, 2014), and via triggering receptor expressed on myeloid cells-1 (TREM-1) (Eguchi and Manabe, 2014). Moreover, it was recently demonstrated that TREM-1 has complex signal integration with certain TLRs; in particular, it was observed that TREM-1 is able to enhance TLR-induced inflammatory response (Eguchi and Manabe, 2014).
Widespread distribution of genotyping technologies resulted in the emergence of studies examining the association of gene polymorphisms with various diseases (Yuzhalin and Kutikhin, 2012). Gene polymorphisms can result in various effects according to their location in the genome (Bakhtiar et al., 2014). For instance, gene polymorphisms within noncoding regions may influence transcription initiation or mRNA splicing (Bakhtiar et al., 2014). Nonsynonymous (i.e. those causing amino acid change) gene polymorphisms are able to alter protein expression, stability, and folding, or affect post-translational modifications (Bakhtiar et al., 2014).
Previously, we demonstrated that certain TLR and TREM-1 gene polymorphisms are associated with coronary artery disease (CAD) in a Russian population (Golovkin et al., 2014). In this study we asked whether these polymorphisms are associated with atherosclerosis severity in patients with CAD.
2. Material and methods
2.1. Study population
The criteria of inclusion into the study were Russian ethnicity, inhabitance in Kemerovo Region during at least two generations, angiographically proved coronary artery stenosis, and written informed consent to participate in the study after a full explanation of its aims and design. The criteria of exclusion included past medical history of malignant tumors, concomitant autoimmune disorders, chronic infectious diseases, and mental disorders. The study was approved by the local ethical committee.
A total of 946 consecutive patients who were admitted to the Research Institute for Complex Issues of Cardiovascular Diseases (Kemerovo, Russian Federation) during 2011–2012 and underwent coronary artery bypass graft (CABG) surgery due to CAD were involved in the study. 244 patients were excluded from the study in accordance with the above-mentioned criteria. Clinical data sufficient for the statistical analysis were obtained for 292 out of remaining 702 patients (239 males, 53 females) between 40 and 70 years of age (mean age 57.75 years, 95% confidence interval (CI) for the mean 57.04–58.45 years, standard deviation 6.14 years).
The diagnosis of CAD was based on the Russian Society of Cardiology (RSC) National Guidelines on Stable Angina and was further revisited according to 2014 American College of Cardiology/American Heart Association/American Association for Thoracic Surgery/Preventive Cardiovascular Nurses Association/Society for Cardiovascular Angiography and Interventions/Society of Thoracic Surgeons guideline for the diagnosis and management of patients with stable ischemic heart disease (Fihn et al., 2014). Coronary angiography was performed using GE Healthcare Innova 3100 Cardiac Angiography System (General Electric Healthcare, USA). Luminal stenosis ≥ 50% was defined as hemodynamically significant coronary stenosis. For the further assessment of coronary stenosis severity, we used widely accepted SYNTAX Score (Sianos et al., 2005). Median SYNTAX score was 19.50 (95%CI for the median 19.00–21.50, interquartile range 14.00–26.50). Color duplex screening of the extracranial arteries (ECA) and lower extremity arteries (LEA) was performed at the 5th–7th day of hospitalization in all patients using the cardiovascular ultrasound system Vivid 7 Dimension (General Electric Healthcare, USA) with a 5.7 MHz linear array transducer (for ECA), a 2.5-3 MHz curved array transducer, and a 5 MHz linear array transducer (for LEA). The extent of arterial stenosis was assessed in B regimen and by dopplerography (visualizing the local hemodynamics in the stenosis zone). Common and internal carotid arteries, vertebral, and subclavial arteries were visualized from both sides during the ECA screening; common and deep femoral arteries, popliteal, anterior and posterior tibial arteries were visualized from both sides during the LEA screening. The intima-media thickness (IMT) of the common carotid artery was measured in automatic mode (the value up to 1 mm was considered normal). Polyvascular disease (PVD) was defined as IMT increase ≥ 1 mm or ECA and/or LEA stenosis. The clinicopathological features of the patients are represented in Table 1. Fig. 1 demonstrates the study pipeline.
Table 1.
Clinicopathological features of the patients who underwent CABG surgery.
| Feature | Value, N (%) |
|---|---|
| Male gender | 239 (81.85%) |
| Age > 55 years | 185 (63.36%) |
| SYNTAX score > 22 | 112 (38.36%) |
| Number of coronary arteries affected by atherosclerosis > 1 | 223 (76.37%) |
| Polyvascular disease | 253 (86.64%) |
| Hemodynamically significant (≥ 50%) stenosis of extracranial or/and lower extremity arteries | 83 (28.42%) |
| New York Heart Association functional class III–IV symptoms | 139 (47.60%) |
| Past medical history of myocardial infarction | 224 (76.71%) |
| Past medical history of stroke | 31 (10.62%) |
| Arterial hypertension | 262 (89.73%) |
| Carbohydrate metabolism disorders | 87 (29.79%) |
| Dyslipidemia | 228 (78.08%) |
| Smoking | 196 (67.12%) |
| Overweight and obesity (body mass index > 25) | 220 (75.34%) |
Fig. 1.
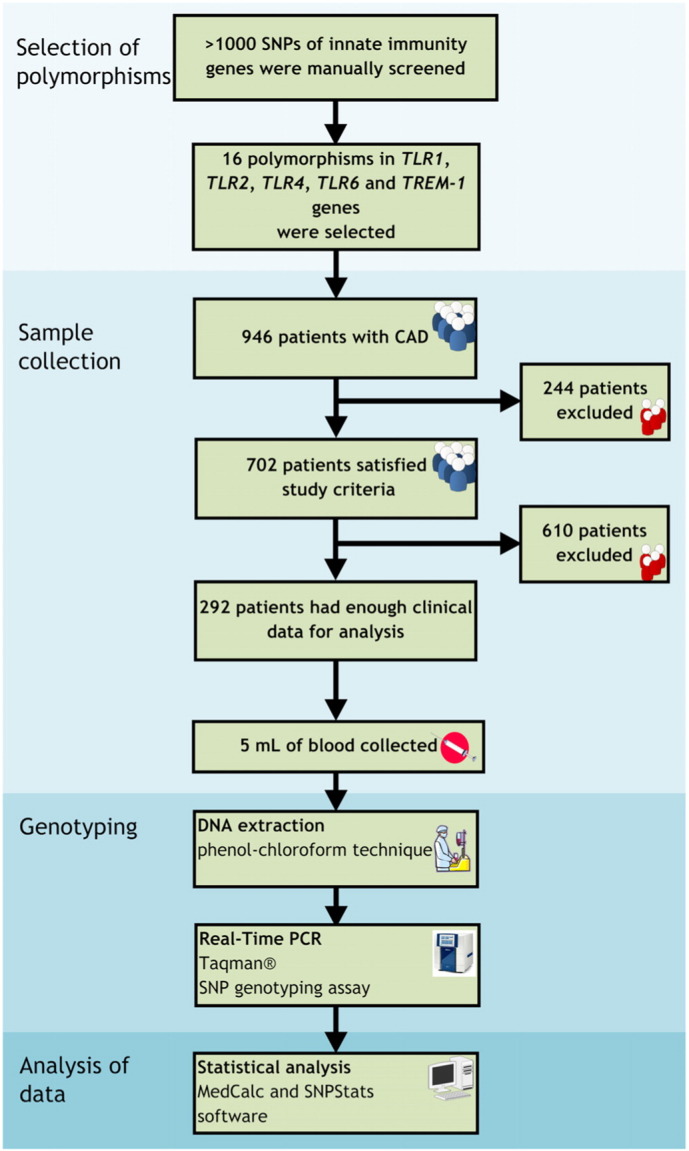
Study pipeline.
2.2. Gene polymorphism selection and genotyping
Three selection criteria for the gene polymorphisms were: (1) high prevalence in a population (minor allele frequency ≥ 5% for Russian population according to HapMap), (2) suggested or proven functional consequence on a molecular level, and (3) few or no studies investigating the role of the gene polymorphism with respect to atherosclerosis severity. The National Center for Biotechnology Information dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP), SNPinfo (http://snpinfo.niehs.nih.gov/snpinfo/snpfunc.htm) (Xu and Taylor, 2009), and SNPnexus (http://www.snp-nexus.org/) (Dayem Ullah et al., 2012) databases were used for the selection of the gene polymorphisms for the study. A total of 16 polymorphisms in 5 genes were investigated: TLR1 (rs5743551 and rs5743611), TLR2 (rs3804099 and rs5743708), TLR4 (rs4986790 and rs4986791), TLR6 (rs3775073 and rs5743810), and TREM-1 (rs1817537, rs3804277, rs6910730, rs7768162, rs2234246, rs4711668, rs9471535, and rs2234237). The data on investigated gene polymorphisms are represented in Table 2.
Table 2.
Some features of the gene polymorphisms investigated in the study.
| Polymorphism | Nucleotide substitution | Chromosomal position | Amino acid substitution | Forward 5′-3′ and reverse 3′-5′ polymerase chain reaction primers |
|---|---|---|---|---|
| TLR1 gene | ||||
| rs5743551 | T > C | 38807654 | 5′-upstream | F: agtgggcagggcagtaagggaagct R: ctcagcactctgaattcctgttttt |
| rs5743611 | C > G | 38800214 | Arg80Thr | F: aacactgatatcaagatactggatt R: tattatgagaaattatcaaaatcct |
| TLR2 gene | ||||
| rs3804099 | T > C | 154624656 | Asn199Asn | F: caaaaagtttgaagtcaattcagaa R: gtaagtcatctgatccttcatatga |
| rs5743708 | G > A | 154626317 | Arg753Gln | F: aagccattccccagcgcttctgcaagctgc R: gaagataatgaacaccaagacctacctgga |
| TLR4 gene | ||||
| rs4986790 | A > G | 120475302 | Asp299Gly | F: gattagcatacttagactactacctcgatg R: attattgacttatttaattgtttgacaaat |
| rs4986791 | C > T | 120475602 | Thr399Ile | F: gttgctgttctcaaagtgattttgggacaa R: agcctaaagtatttagatctgagcttcaat |
| TLR6 gene | ||||
| rs3775073 | T > C | 38829832 | Lys421Lys | F: cactatactctcaacccaagtgcagttttc R: tatgtctaccagattccaaagaattccagc |
| rs5743810 | A > G | 38830350 | Ser249Pro | F: ttgagggtaaaattcagtaaggttg R: acctctggtgagttctgataaaaat |
| TREM-1 gene | ||||
| rs1817537 | C > G | 41244567 | Intronic | F: acacagggacagacagatggcaatggaaca R:aaggccagatgcagagccagtgctatgcag |
| rs3804277 | C > T | 41245172 | Intronic | F: ccagcatctctctcacccctcacatggtgg R: cactcagcatcctcagcatctgccccgatt |
| rs6910730 | A > G | 41246633 | 3′-downstream | F: catggagcaacaccaaggtctaggggcaag R: aatctaggatggattcgtgctgacttccca |
| rs7768162 | A > G | 41255511 | 5′-upstream | F: aaagattcctactgctaaataaacaaaaaa R: taacttggtttcttcaaaggaattgaaata |
| rs2234246 | C > T | 41243740 | 3′-UTR | F: ggaaggtgagacgctgactttagaaatagc R: ggtgattacagatttaattcatgttattaa |
| rs4711668 | T > C | 41246473 | 3′-downstream | F: gctagtgtggattccactttccagactgga R: ttggctgaaaggatagttcatattagatga |
| rs9471535 | T > C | 41255490 | 5′-upstream | F: aaaatttttaaatttaaataaaaagattcc R: ctgctaaataaacaaaaaaataacttggtt |
| rs2234237 | T > A | 41250466 | Thr25Ser | F: gcccctctttcagttcatacttttcctcag R: aatttagttgcagctcggagttctataagc |
All study participants provided 5 mL of peripheral venous blood which was collected into a tube containing ethylenediaminetetraacetic acid. Then, 0.5 mL of blood was immediately transferred into a fresh tube following addition of 1 mL of saline–sodium citrate buffer (Promega). Further, the tube was vortexed before being spun at 12,000 rpm for 2 min. The pellet was digested in a buffer containing 10% sodium dodecyl sulfate (Sigma) and 100 μg/mL Proteinase K (Helicon) for 3 h at 50 °C. One volume of phenol:chloroform:isoamyl alcohol (25:24:1) was added to the tube before being vortexed for 20 s and spun at 12,000 rpm for 15 min. A viscous interphase layer was transferred into a fresh tube, and 70% ethanol was added to precipitate genomic DNA from the sample. The sample was spun down at 12,000 rpm for 5 min. DNA pellet was incubated overnight in deionized water at room temperature and was further stored at − 70 °C until use.
Sample genotyping was performed in 96-well format using the TaqMan SNP genotyping assay on the ViiA™ 7 Real-Time PCR System (Life Technologies, USA) in accordance with the manufacturer's instructions (https://tools.lifetechnologies.com/content/sfs/manuals/cms_095288.pdf). The amplification was performed in the final volume of 10 μL of reaction mixture, containing 100 ng of DNA, 1.25 μL of each primer, 2.5 mM of MgCl2, 1 mM of dNTPs, and 1 U of Taq polymerase (Life Technologies). The following PCR cycle conditions were used: hold stage 50 °C for 120 s and 95 °C for 10 min; PCR stage 95 °C for 15 s and 60 °C for 1 min repeated in 40 cycles. Table 2 shows the data on sequence-specific primers for all gene polymorphisms investigated in the study. Laboratory staff engaged in genotyping was blinded to patient status, and 10% of the samples were genotyped in duplicates for quality control purposes.
2.3. Statistical analysis
The sampling distribution was assessed by D'Agostino–Pearson test for normal distribution (MedCalc, MedCalc Software). Mean, standard deviation, median, interquartile range (25th and 75th percentiles), and 95% confidence intervals (CIs) for both mean and median were calculated to describe the quantitative data whereas proportions were used to describe the qualitative data (MedCalc, MedCalc Software, Belgium). The statistical analysis of gene polymorphisms was performed utilizing the SNPStats, a web tool for the analysis of genetic association studies (Solé et al., 2006). Hardy–Weinberg equilibrium (HWE) was examined with a chi-square goodness-of-fit test with one degree of freedom to compare the observed and expected genotype frequencies. To estimate the risk conferred by a certain allele or genotype, odds ratios (ORs) with 95% CIs for ORs were calculated. Calculation of ORs was conducted in accordance with the five common models of inheritance (codominant, dominant, recessive, overdominant, and log-additive). The choice of the most probable model was carried out in accordance with the Akaike information criterion (AIC); the model with the least AIC value was defined as the most probable. Linkage disequilibrium was calculated for all possible combination of pairs of gene polymorphisms located on different chromosomes. Adjustment for multiple comparisons was performed using the false discovery rate (http://users.ox.ac.uk/~npike/fdr/) to determine the final threshold of statistical significance. P-values, or q-values if FDR was applied (q-values are the name given to the adjusted p-values found using an optimized FDR approach), ≤ 0.05 were regarded as statistically significant.
3. Results
First, we assessed an association of TLR and TREM-1 gene polymorphisms with severity of coronary atherosclerosis. We found that the C/C genotype of the rs3804099 polymorphism within the TLR2 gene was significantly associated with SYNTAX score > 22 (OR = 2.03, 95%CI = 1.09–3.77, P = 0.025, according to the recessive model, Table 3) while the T/T genotype of the rs4711668 polymorphism within the TREM-1 gene was significantly associated with 2 or 3 affected coronary arteries (OR = 2.09, 95%CI = 1.02–4.27, P = 0.034, according to the recessive model, Table 4).
Table 3.
Association of polymorphisms within the genes encoding Toll-like receptors (TLRs) and Triggering receptor expressed on myeloid cells-1 (TREM-1) with coronary atherosclerosis regarding SYNTAX score (after the adjustments by age and gender).
| Polymorphism | Inheritance model | Genotype | SYNTAX score 0–22 | SYNTAX score > 22 | OR (95% CI) | P value | HWE |
|---|---|---|---|---|---|---|---|
| TLR1 gene | |||||||
| rs5743551 | Codominant | T/T | 114 (63.3%) | 76 (69.1%) | 1.00 | 0.62 | 0.85 |
| C/T | 60 (33.3%) | 31 (28.2%) | 0.78 (0.46–1.32) | ||||
| C/C | 6 (3.3%) | 3 (2.7%) | 0.75 (0.18–3.10) | ||||
| Dominant | T/T | 114 (63.3%) | 76 (69.1%) | 1.00 | 0.33 | ||
| C/T-C/C | 66 (36.7%) | 34 (30.9%) | 0.78 (0.47–1.29) | ||||
| Recessive | T/T-C/T | 174 (96.7%) | 107 (97.3%) | 1.00 | 0.77 | ||
| C/C | 6 (3.3%) | 3 (2.7%) | 0.81 (0.20–3.32) | ||||
| Overdominant | T/T-C/C | 120 (66.7%) | 79 (71.8%) | 1.00 | 0.37 | ||
| C/T | 60 (33.3%) | 31 (28.2%) | 0.79 (0.47–1.33) | ||||
| Log-additive | – | – | – | 0.81 (0.52–1.26) | 0.34 | ||
| rs5743611 | Codominant | C/C | 107 (59.4%) | 65 (58.6%) | 1.00 | 0.99 | 0.87 |
| C/G | 63 (35%) | 40 (36%) | 1.03 (0.62–1.71) | ||||
| G/G | 10 (5.6%) | 6 (5.4%) | 0.97 (0.33–2.79) | ||||
| Dominant | C/C | 107 (59.4%) | 65 (58.6%) | 1.00 | 0.92 | ||
| C/G-G/G | 73 (40.6%) | 46 (41.4%) | 1.02 (0.63–1.66) | ||||
| Recessive | C/C-C/G | 170 (94.4%) | 105 (94.6%) | 1.00 | 0.93 | ||
| G/G | 10 (5.6%) | 6 (5.4%) | 0.95 (0.34–2.71) | ||||
| Overdominant | C/C-G/G | 117 (65%) | 71 (64%) | 1.00 | 0.89 | ||
| C/G | 63 (35%) | 40 (36%) | 1.04 (0.63–1.70) | ||||
| Log-additive | – | – | – | 1.01 (0.68–1.50) | 0.96 | ||
| TLR2 gene | |||||||
| rs5743708 | — | G/G | 164 (91.1%) | 104 (93.7%) | 1.00 | 0.43 | 0.99 |
| A/G | 16 (8.9%) | 7 (6.3%) | 0.69 (0.28–1.74) | ||||
| rs3804099 | Codominant | T/T | 82 (45.6%) | 41 (36.9%) | 1.00 | 0.066 | 0.32 |
| C/T | 74 (41.1%) | 44 (39.6%) | 1.19 (0.70–2.03) | ||||
| C/C | 24 (13.3%) | 26 (23.4%) | 2.21 (1.13–4.35) | ||||
| Dominant | T/T | 82 (45.6%) | 41 (36.9%) | 1.00 | 0.14 | ||
| C/T-C/C | 98 (54.4%) | 70 (63.1%) | 1.44 (0.88–2.34) | ||||
| Recessive | T/T-C/T | 156 (86.7%) | 85 (76.6%) | 1.00 | 0.025 | ||
| C/C | 24 (13.3%) | 26 (23.4%) | 2.03 (1.09–3.77) | ||||
| Overdominant | T/T-C/C | 106 (58.9%) | 67 (60.4%) | 1.00 | 0.79 | ||
| C/T | 74 (41.1%) | 44 (39.6%) | 0.94 (0.58–1.52) | ||||
| Log-additive | – | – | – | 1.43 (1.03–1.99) | 0.031 | ||
| TLR4 gene | |||||||
| rs4986790 | Codominant | A/A | 155 (86.1%) | 90 (81.1%) | 1.00 | 0.46 | 0.99 |
| A/G | 24 (13.3%) | 20 (18%) | 1.49 (0.77–2.87) | ||||
| G/G | 1 (0.6%) | 1 (0.9%) | 1.78 (0.10–30.27) | ||||
| Dominant | A/A | 155 (86.1%) | 90 (81.1%) | 1.00 | 0.22 | ||
| A/G-G/G | 25 (13.9%) | 21 (18.9%) | 1.50 (0.79–2.85) | ||||
| Recessive | A/A-A/G | 179 (99.4%) | 110 (99.1%) | 1.00 | 0.71 | ||
| G/G | 1 (0.6%) | 1 (0.9%) | 1.71 (0.10–29.03) | ||||
| Overdominant | A/A-G/G | 156 (86.7%) | 91 (82%) | 1.00 | 0.24 | ||
| A/G | 24 (13.3%) | 20 (18%) | 1.48 (0.77–2.86) | ||||
| Log-additive | – | – | – | 1.46 (0.80–2.67) | 0.22 | ||
| rs4986791 | Codominant | C/C | 154 (85.6%) | 89 (80.2%) | 1.00 | 0.43 | 0.99 |
| C/T | 25 (13.9%) | 21 (18.9%) | 1.51 (0.79–2.87) | ||||
| T/T | 1 (0.6%) | 1 (0.9%) | 1.78 (0.10–30.30) | ||||
| Dominant | C/C | 154 (85.6%) | 89 (80.2%) | 1.00 | 0.2 | ||
| C/T-T/T | 26 (14.4%) | 22 (19.8%) | 1.52 (0.81–2.86) | ||||
| Recessive | C/C-C/T | 179 (99.4%) | 110 (99.1%) | 1.00 | 0.71 | ||
| T/T | 1 (0.6%) | 1 (0.9%) | 1.71 (0.10–29.03) | ||||
| Overdominant | C/C-T/T | 155 (86.1%) | 90 (81.1%) | 1.00 | 0.22 | ||
| C/T | 25 (13.9%) | 21 (18.9%) | 1.50 (0.79–2.86) | ||||
| Log-additive | – | – | – | 1.48 (0.82–2.67) | 0.2 | ||
| TLR6 gene | |||||||
| rs3775073 | Codominant | T/T | 58 (32.2%) | 33 (29.7%) | 1.00 | 0.83 | 0.91 |
| T/C | 88 (48.9%) | 54 (48.6%) | 1.06 (0.62–1.84)– | ||||
| C/C | 34 (18.9%) | 24 (21.6%) | 1.24 (0.63–2.43) | ||||
| Dominant | T/T | 58 (32.2%) | 33 (29.7%) | 1.00 | 0.69 | ||
| T/C-C/C | 122 (67.8%) | 78 (70.3%) | 1.11 (0.66–1.86) | ||||
| Recessive | T/T-T/C | 146 (81.1%) | 87 (78.4%) | 1.00 | 0.56 | ||
| C/C | 34 (18.9%) | 24 (21.6%) | 1.19 (0.66–2.14) | ||||
| Overdominant | T/T-C/C | 92 (51.1%) | 57 (51.4%) | 1.00 | 0.93 | ||
| T/C | 88 (48.9%) | 54 (48.6%) | 0.98 (0.61–1.57) | ||||
| Log-additive | – | – | – | 1.11 (0.79–1.55) | 0.55 | ||
| rs5743810 | Codominant | G/G | 78 (43.3%) | 45 (40.5%) | 1.00 | 0.8 | 0.52 |
| A/G | 79 (43.9%) | 49 (44.1%) | 1.06 (0.64–1.78) | ||||
| A/A | 23 (12.8%) | 17 (15.3%) | 1.29 (0.62–2.66) | ||||
| Dominant | G/G | 78 (43.3%) | 45 (40.5%) | 1.00 | 0.66 | ||
| A/G-A/A | 102 (56.7%) | 66 (59.5%) | 1.11 (0.69–1.80) | ||||
| Recessive | G/G-A/G | 157 (87.2%) | 94 (84.7%) | 1.00 | 0.53 | ||
| A/A | 23 (12.8%) | 17 (15.3%) | 1.25 (0.63–2.46) | ||||
| Overdominant | G/G-A/A | 101 (56.1%) | 62 (55.9%) | 1.00 | 0.99 | ||
| A/G | 79 (43.9%) | 49 (44.1%) | 1.00 (0.63–1.61) | ||||
| Log-additive | – | – | – | 1.12 (0.79–1.57) | 0.53 | ||
| TREM-1 gene | |||||||
| rs2234246 | Codominant | C/C | 61 (33.9%) | 49 (44.1%) | 1.00 | 0.18 | 0.14 |
| C/T | 86 (47.8%) | 42 (37.8%) | 0.61 (0.36–1.03) | ||||
| T/T | 33 (18.3%) | 20 (18%) | 0.75 (0.38–1.47) | ||||
| Dominant | C/C | 61 (33.9%) | 49 (44.1%) | 1.00 | 0.082 | ||
| C/T-T/T | 119 (66.1%) | 62 (55.9%) | 0.65 (0.40–1.06) | ||||
| Recessive | C/C-C/T | 147 (81.7%) | 91 (82%) | 1.00 | 0.93 | ||
| T/T | 33 (18.3%) | 20 (18%) | 0.97 (0.53–1.80) | ||||
| Overdominant | C/C-T/T | 94 (52.2%) | 69 (62.2%) | 1.00 | 0.1 | ||
| C/T | 86 (47.8%) | 42 (37.8%) | 0.67 (0.41–1.08) | ||||
| Log-additive | – | – | – | 0.81 (0.58–1.13) | 0.22 | ||
| rs4711668 | Codominant | C/C | 60 (33.5%) | 39 (35.1%) | 1.00 | 0.14 | 0.23 |
| T/C | 80 (44.7%) | 38 (34.2%) | 0.75 (0.43–1.31) | ||||
| T/T | 39 (21.8%) | 34 (30.6%) | 1.37 (0.74–2.53) | ||||
| Dominant | C/C | 60 (33.5%) | 39 (35.1%) | 1.00 | 0.85 | ||
| T/C-T/T | 119 (66.5%) | 72 (64.9%) | 0.95 (0.58–1.57) | ||||
| Recessive | C/C-T/C | 140 (78.2%) | 77 (69.4%) | 1.00 | 0.09 | ||
| T/T | 39 (21.8%) | 34 (30.6%) | 1.60 (0.93–2.74) | ||||
| Overdominant | C/C-T/T | 99 (55.3%) | 73 (65.8%) | 1.00 | 0.088 | ||
| T/C | 80 (44.7%) | 38 (34.2%) | 0.65 (0.40–1.07) | ||||
| Log-additive | – | – | – | 1.14 (0.84–1.56) | 0.39 | ||
| rs3804277 | Codominant | C/C | 66 (36.7%) | 49 (44.1%) | 1.00 | 0.36 | 0.11 |
| C/T | 83 (46.1%) | 42 (37.8%) | 0.68 (0.40–1.15) | ||||
| T/T | 31 (17.2%) | 20 (18%) | 0.86 (0.44–1.69) | ||||
| Dominant | C/C | 66 (36.7%) | 49 (44.1%) | 1.00 | 0.2 | ||
| C/T-T/T | 114 (63.3%) | 62 (55.9%) | 0.73 (0.45–1.18) | ||||
| Recessive | C/C-C/T | 149 (82.8%) | 91 (82%) | 1.00 | 0.89 | ||
| T/T | 31 (17.2%) | 20 (18%) | 1.05 (0.56–1.95) | ||||
| Overdominant | C/C-T/T | 97 (53.9%) | 69 (62.2%) | 1.00 | 0.17 | ||
| C/T | 83 (46.1%) | 42 (37.8%) | 0.71 (0.44–1.16) | ||||
| Log-additive | – | – | – | 0.88 (0.63–1.22) | 0.43 | ||
| rs2234237 | Codominant | T/T | 138 (76.7%) | 86 (77.5%) | 1.00 | 0.9 | 0.19 |
| A/T | 37 (20.6%) | 21 (18.9%) | 0.91 (0.50–1.66) | ||||
| A/A | 5 (2.8%) | 4 (3.6%) | 1.23 (0.32–4.74) | ||||
| Dominant | T/T | 138 (76.7%) | 86 (77.5%) | 1.00 | 0.85 | ||
| A/T-A/A | 42 (23.3%) | 25 (22.5%) | 0.95 (0.54–1.67) | ||||
| Recessive | T/T-A/T | 175 (97.2%) | 107 (96.4%) | 1.00 | 0.74 | ||
| A/A | 5 (2.8%) | 4 (3.6%) | 1.25 (0.33–4.81) | ||||
| Overdominant | T/T-A/A | 143 (79.4%) | 90 (81.1%) | 1.00 | 0.73 | ||
| A/T | 37 (20.6%) | 21 (18.9%) | 0.90 (0.50–1.64) | ||||
| Log-additive | – | – | – | 0.99 (0.62–1.58) | 0.97 | ||
| rs6910730 | Codominant | A/A | 140 (77.8%) | 87 (78.4%) | 1.00 | 0.75 | 0.31 |
| A/G | 36 (20%) | 20 (18%) | 0.89 (0.48–1.63) | ||||
| G/G | 4 (2.2%) | 4 (3.6%) | 1.55 (0.38–6.40) | ||||
| Dominant | A/A | 140 (77.8%) | 87 (78.4%) | 1.00 | 0.87 | ||
| A/G-G/G | 40 (22.2%) | 24 (21.6%) | 0.95 (0.54–1.69) | ||||
| Recessive | A/A-A/G | 176 (97.8%) | 107 (96.4%) | 1.00 | 0.52 | ||
| G/G | 4 (2.2%) | 4 (3.6%) | 1.59 (0.39–6.53) | ||||
| Overdominant | A/A-G/G | 144 (80%) | 91 (82%) | 1.00 | 0.66 | ||
| A/G | 36 (20%) | 20 (18%) | 0.87 (0.47–1.60) | ||||
| Log-additive | – | – | – | 1.02 (0.63–1.65) | 0.94 | ||
| rs1817537 | Codominant | C/C | 66 (36.7%) | 49 (44.1%) | 1.00 | 0.36 | 0.64 |
| C/G | 83 (46.1%) | 42 (37.8%) | 0.68 (0.40–1.15) | ||||
| G/G | 31 (17.2%) | 20 (18%) | 0.86 (0.44–1.69) | ||||
| Dominant | C/C | 66 (36.7%) | 49 (44.1%) | 1.00 | 0.2 | ||
| C/G-G/G | 114 (63.3%) | 62 (55.9%) | 0.73 (0.45–1.18) | ||||
| Recessive | C/C-C/G | 149 (82.8%) | 91 (82%) | 1.00 | 0.89 | ||
| G/G | 31 (17.2%) | 20 (18%) | 1.05 (0.56–1.95) | ||||
| Overdominant | C/C-G/G | 97 (53.9%) | 69 (62.2%) | 1.00 | 0.17 | ||
| C/G | 83 (46.1%) | 42 (37.8%) | 0.71 (0.44–1.16) | ||||
| Log-additive | – | – | – | 0.88 (0.63–1.22) | 0.43 | ||
| rs9471535 | Codominant | T/T | 137 (76.5%) | 84 (76.4%) | 1.00 | 0.94 | 0.19 |
| C/T | 37 (20.7%) | 22 (20%) | 0.97 (0.53–1.75) | ||||
| C/C | 5 (2.8%) | 4 (3.6%) | 1.24 (0.32–4.77) | ||||
| Dominant | T/T | 137 (76.5%) | 84 (76.4%) | 1.00 | 0.99 | ||
| C/T-C/C | 42 (23.5%) | 26 (23.6%) | 1.00 (0.57–1.75) | ||||
| Recessive | T/T-C/T | 174 (97.2%) | 106 (96.4%) | 1.00 | 0.75 | ||
| C/C | 5 (2.8%) | 4 (3.6%) | 1.25 (0.33–4.78) | ||||
| Overdominant | T/T-C/C | 142 (79.3%) | 88 (80%) | 1.00 | 0.89 | ||
| C/T | 37 (20.7%) | 22 (20%) | 0.96 (0.53–1.73) | ||||
| Log-additive | – | – | – | 1.03 (0.64–1.64) | 0.91 | ||
| rs7768162 | Codominant | G/G | 63 (35.2%) | 41 (37.3%) | 1.00 | 0.21 | 0.63 |
| A/G | 90 (50.3%) | 45 (40.9%) | 0.79 (0.46–1.35) | ||||
| A/A | 26 (14.5%) | 24 (21.8%) | 1.42 (0.72–2.81) | ||||
| Dominant | G/G | 63 (35.2%) | 41 (37.3%) | 1.00 | 0.8 | ||
| A/G-A/A | 116 (64.8%) | 69 (62.7%) | 0.94 (0.57–1.54) | ||||
| Recessive | G/G-A/G | 153 (85.5%) | 86 (78.2%) | 1.00 | 0.13 | ||
| A/A | 26 (14.5%) | 24 (21.8%) | 1.62 (0.88–3.01) | ||||
| Overdominant | G/G-A/A | 89 (49.7%) | 65 (59.1%) | 1.00 | 0.15 | ||
| A/G | 90 (50.3%) | 45 (40.9%) | 0.70 (0.43–1.14) | ||||
| Log-additive | – | – | – | 1.12 (0.80–1.57) | 0.51 | ||
Table 4.
Association of polymorphisms within the genes encoding Toll-like receptors (TLRs) and Triggering receptor expressed on myeloid cells-1 (TREM-1) with coronary atherosclerosis regarding the number of affected coronary arteries (after the adjustments by age and gender).
| Polymorphism | Inheritance model | Genotype | One affected coronary artery | Two or three affected coronary arteries | OR (95% CI) | P value | HWE |
|---|---|---|---|---|---|---|---|
| TLR1 gene | |||||||
| rs5743551 | Codominant | T/T | 40 (58%) | 150 (67.9%) | 1.00 | 0.16 | 0.85 |
| C/T | 28 (40.6%) | 63 (28.5%) | 0.61 (0.35–1.08) | ||||
| C/C | 1 (1.4%) | 8 (3.6%) | 2.13 (0.26–17.76) | ||||
| Dominant | T/T | 40 (58%) | 150 (67.9%) | 1.00 | 0.16 | ||
| C/T-C/C | 29 (42%) | 71 (32.1%) | 0.66 (0.38–1.17) | ||||
| Recessive | T/T-C/T | 68 (98.5%) | 213 (96.4%) | 1.00 | 0.33 | ||
| C/C | 1 (1.4%) | 8 (3.6%) | 2.54 (0.31–20.90) | ||||
| Overdominant | T/T-C/C | 41 (59.4%) | 158 (71.5%) | 1.00 | 0.077 | ||
| C/T | 28 (40.6%) | 63 (28.5%) | 0.60 (0.34–1.05) | ||||
| Log-additive | – | – | – | 0.79 (0.48–1.28) | 0.34 | ||
| rs5743611 | Codominant | C/C | 43 (62.3%) | 129 (58.1%) | 1.00 | 0.86 | 0.87 |
| C/G | 23 (33.3%) | 80 (36%) | 1.11 (0.62–1.99) | ||||
| G/G | 3 (4.3%) | 13 (5.9%) | 1.36 (0.37–5.07) | ||||
| Dominant | C/C | 43 (62.3%) | 129 (58.1%) | 1.00 | 0.65 | ||
| C/G-G/G | 26 (37.7%) | 93 (41.9%) | 1.14 (0.65–2.00) | ||||
| Recessive | C/C-C/G | 66 (95.7%) | 209 (94.1%) | 1.00 | 0.68 | ||
| G/G | 3 (4.3%) | 13 (5.9%) | 1.31 (0.36–4.80) | ||||
| Overdominant | C/C-G/G | 46 (66.7%) | 142 (64%) | 1.00 | 0.78 | ||
| C/G | 23 (33.3%) | 80 (36%) | 1.08 (0.61–1.93) | ||||
| Log-additive | – | – | – | 1.13 (0.71–1.81) | 0.59 | ||
| TLR2 gene | |||||||
| rs5743708 | – | G/G | 62 (89.9%) | 206 (92.8%) | 1.00 | 0.46 | 0.99 |
| A/G | 7 (10.1%) | 16 (7.2%) | 0.69 (0.27–1.79) | ||||
| rs3804099 | Codominant | T/T | 27 (39.1%) | 96 (43.2%) | 1.00 | 0.46 | 0.99 |
| C/T | 32 (46.4%) | 86 (38.7%) | 0.73 (0.40–1.33) | ||||
| C/C | 10 (14.5%) | 40 (18%) | 1.11 (0.49–2.54) | ||||
| Dominant | T/T | 27 (39.1%) | 96 (43.2%) | 1.00 | 0.49 | ||
| C/T-C/C | 42 (60.9%) | 126 (56.8%) | 0.82 (0.47–1.44) | ||||
| Recessive | T/T-C/T | 59 (85.5%) | 182 (82%) | 1.00 | 0.48 | ||
| C/C | 10 (14.5%) | 40 (18%) | 1.31 (0.61–2.80) | ||||
| Overdominant | T/T-C/C | 37 (53.6%) | 136 (61.3%) | 1.00 | 0.22 | ||
| C/T | 32 (46.4%) | 86 (38.7%) | 0.71 (0.41–1.23) | ||||
| Log-additive | – | – | – | 0.98 (0.67–1.42) | 0.91 | ||
| TLR4 gene | |||||||
| rs4986790 | Codominant | A/A | 62 (89.9%) | 183 (82.4%) | 1.00 | 0.14 | 0.99 |
| A/G | 7 (10.1%) | 37 (16.7%) | 2.07 (0.87–4.98) | ||||
| G/G | 0 (0%) | 2 (0.9%) | NA (0.00–NA) | ||||
| Dominant | A/A | 62 (89.9%) | 183 (82.4%) | 1.00 | 0.064 | ||
| A/G-G/G | 7 (10.1%) | 39 (17.6%) | 2.17 (0.91–5.17) | ||||
| Recessive | A/A-A/G | 69 (100%) | 220 (99.1%) | 1.00 | 0.34 | ||
| G/G | 0 (0%) | 2 (0.9%) | NA (0.00–NA) | ||||
| Overdominant | A/A-G/G | 62 (89.9%) | 185 (83.3%) | 1.00 | 0.086 | ||
| A/G | 7 (10.1%) | 37 (16.7%) | 2.06 (0.86–4.95) | ||||
| Log-additive | – | – | – | 2.17 (0.93–5.08) | 0.055 | ||
| rs4986791 | Codominant | C/C | 61 (88.4%) | 182 (82%) | 1.00 | 0.21 | 0.99 |
| C/T | 8 (11.6%) | 38 (17.1%) | 1.84 (0.80–4.22) | ||||
| T/T | 0 (0%) | 2 (0.9%) | NA (0.00–NA) | ||||
| Dominant | C/C | 61 (88.4%) | 182 (82%) | 1.00 | 0.11 | ||
| C/T-T/T | 8 (11.6%) | 40 (18%) | 1.92 (0.84–4.38) | ||||
| Recessive | C/C-C/T | 69 (100%) | 220 (99.1%) | 1.00 | 0.34 | ||
| T/T | 0 (0%) | 2 (0.9%) | NA (0.00–NA) | ||||
| Overdominant | C/C-T/T | 61 (88.4%) | 184 (82.9%) | 1.00 | 0.14 | ||
| C/T | 8 (11.6%) | 38 (17.1%) | 1.83 (0.79–4.20) | ||||
| Log-additive | – | – | – | 1.93 (0.86–4.33) | 0.091 | ||
| TLR6 gene | |||||||
| rs3775073 | Codominant | T/T | 24 (34.8%) | 67 (30.2%) | 1.00 | 0.8 | 0.91 |
| T/C | 31 (44.9%) | 111 (50%) | 1.24 (0.66–2.30) | ||||
| C/C | 14 (20.3%) | 44 (19.8%) | 1.13 (0.52–2.43) | ||||
| Dominant | T/T | 24 (34.8%) | 67 (30.2%) | 1.00 | 0.54 | ||
| T/C-C/C | 45 (65.2%) | 155 (69.8%) | 1.20 (0.67–2.14) | ||||
| Recessive | T/T-T/C | 55 (79.7%) | 178 (80.2%) | 1.00 | 0.98 | ||
| C/C | 14 (20.3%) | 44 (19.8%) | 0.99 (0.50–1.96) | ||||
| Overdominant | T/T-C/C | 38 (55.1%) | 111 (50%) | 1.00 | 0.55 | ||
| T/C | 31 (44.9%) | 111 (50%) | 1.18 (0.68–2.04) | ||||
| Log-additive | – | – | – | 1.08 (0.73–1.59) | 0.69 | ||
| rs5743810 | Codominant | G/G | 31 (44.9%) | 92 (41.4%) | 1.00 | 0.52 | 0.52 |
| A/G | 31 (44.9%) | 97 (43.7%) | 1.01 (0.56–1.80) | ||||
| A/A | 7 (10.1%) | 33 (14.9%) | 1.64 (0.65–4.12) | ||||
| Dominant | G/G | 31 (44.9%) | 92 (41.4%) | 1.00 | 0.68 | ||
| A/G-A/A | 38 (55.1%) | 130 (58.6%) | 1.12 (0.65–1.95) | ||||
| Recessive | G/G-A/G | 62 (89.9%) | 189 (85.1%) | 1.00 | 0.25 | ||
| A/A | 7 (10.1%) | 33 (14.9%) | 1.63 (0.68–3.92) | ||||
| Overdominant | G/G-A/A | 38 (55.1%) | 125 (56.3%) | 1.00 | 0.72 | ||
| A/G | 31 (44.9%) | 97 (43.7%) | 0.90 (0.52–1.57) | ||||
| Log-additive | – | – | – | 1.19 (0.80–1.77) | 0.39 | ||
| TREM-1 gene | |||||||
| rs2234246 | Codominant | C/C | 20 (29%) | 90 (40.5%) | 1.00 | 0.23 | 0.14 |
| C/T | 36 (52.2%) | 92 (41.4%) | 0.58 (0.31–1.09) | ||||
| T/T | 13 (18.8%) | 40 (18%) | 0.68 (0.31–1.52) | ||||
| Dominant | C/C | 20 (29%) | 90 (40.5%) | 1.00 | 0.096 | ||
| C/T-T/T | 49 (71%) | 132 (59.5%) | 0.61 (0.34–1.10) | ||||
| Recessive | C/C-C/T | 56 (81.2%) | 182 (82%) | 1.00 | 0.85 | ||
| T/T | 13 (18.8%) | 40 (18%) | 0.94 (0.46–1.89) | ||||
| Overdominant | C/C-T/T | 33 (47.8%) | 130 (58.6%) | 1.00 | 0.15 | ||
| C/T | 36 (52.2%) | 92 (41.4%) | 0.67 (0.38–1.15) | ||||
| Log-additive | – | – | – | 0.79 (0.54–1.16) | 0.23 | ||
| rs4711668 | Codominant | C/C | 24 (34.8%) | 75 (33.9%) | 1.00 | 0.093 | 0.99 |
| T/C | 34 (49.3%) | 84 (38%) | 0.86 (0.46–1.60) | ||||
| T/T | 11 (15.9%) | 62 (28.1%) | 1.92 (0.86–4.26) | ||||
| Dominant | C/C | 24 (34.8%) | 75 (33.9%) | 1.00 | 0.7 | ||
| T/C-T/T | 45 (65.2%) | 146 (66.1%) | 1.12 (0.63–2.00) | ||||
| Recessive | C/C-T/C | 58 (84.1%) | 159 (72%) | 1.00 | 0.034 | ||
| T/T | 11 (15.9%) | 62 (28.1%) | 2.09 (1.02–4.27) | ||||
| Overdominant | C/C-T/T | 35 (50.7%) | 137 (62%) | 1.00 | 0.15 | ||
| T/C | 34 (49.3%) | 84 (38%) | 0.67 (0.38–1.16) | ||||
| Log-additive | – | – | – | 1.30 (0.90–1.88) | 0.16 | ||
| rs3804277 | Codominant | C/C | 24 (34.8%) | 91 (41%) | 1.00 | 0.63 | 0.11 |
| C/T | 33 (47.8%) | 92 (41.4%) | 0.74 (0.41–1.36) | ||||
| T/T | 12 (17.4%) | 39 (17.6%) | 0.85 (0.38–1.88) | ||||
| Dominant | C/C | 24 (34.8%) | 91 (41%) | 1.00 | 0.37 | ||
| C/T-T/T | 45 (65.2%) | 131 (59%) | 0.77 (0.44–1.36) | ||||
| Recessive | C/C-C/T | 57 (82.6%) | 183 (82.4%) | 1.00 | 0.99 | ||
| T/T | 12 (17.4%) | 39 (17.6%) | 1.00 (0.48–2.05) | ||||
| Overdominant | C/C-T/T | 36 (52.2%) | 130 (58.6%) | 1.00 | 0.38 | ||
| C/T | 33 (47.8%) | 92 (41.4%) | 0.78 (0.45–1.36) | ||||
| Log-additive | – | – | – | 0.89 (0.61–1.30) | 0.54 | ||
| rs2234237 | Codominant | T/T | 48 (69.6%) | 176 (79.3%) | 1.00 | 0.22 | 0.41 |
| A/T | 18 (26.1%) | 40 (18%) | 0.61 (0.32–1.16) | ||||
| A/A | 3 (4.3%) | 6 (2.7%) | 0.44 (0.10–1.87) | ||||
| Dominant | T/T | 48 (69.6%) | 176 (79.3%) | 1.00 | 0.09 | ||
| A/T-A/A | 21 (30.4%) | 46 (20.7%) | 0.58 (0.31–1.08) | ||||
| Recessive | T/T-A/T | 66 (95.7%) | 216 (97.3%) | 1.00 | 0.35 | ||
| A/A | 3 (4.3%) | 6 (2.7%) | 0.49 (0.12–2.07) | ||||
| Overdominant | T/T-A/A | 51 (73.9%) | 182 (82%) | 1.00 | 0.17 | ||
| A/T | 18 (26.1%) | 40 (18%) | 0.63 (0.33–1.20) | ||||
| Log-additive | – | – | – | 0.63 (0.38–1.05) | 0.083 | ||
| rs6910730 | Codominant | A/A | 49 (71%) | 178 (80.2%) | 1.00 | 0.23 | 0.67 |
| A/G | 18 (26.1%) | 38 (17.1%) | 0.56 (0.29–1.08) | ||||
| G/G | 2 (2.9%) | 6 (2.7%) | 0.68 (0.13–3.54) | ||||
| Dominant | A/A | 49 (71%) | 178 (80.2%) | 1.00 | 0.087 | ||
| A/G-G/G | 20 (29%) | 44 (19.8%) | 0.57 (0.31–1.07) | ||||
| Recessive | A/A-A/G | 67 (97.1%) | 216 (97.3%) | 1.00 | 0.76 | ||
| G/G | 2 (2.9%) | 6 (2.7%) | 0.77 (0.15–3.99) | ||||
| Overdominant | A/A-G/G | 51 (73.9%) | 184 (82.9%) | 1.00 | 0.096 | ||
| A/G | 18 (26.1%) | 38 (17.1%) | 0.57 (0.30–1.09) | ||||
| Log-additive | – | – | – | 0.66 (0.39–1.11) | 0.12 | ||
| rs1817537 | Codominant | C/C | 24 (34.8%) | 91 (41%) | 1.00 | 0.63 | 0.11 |
| C/G | 33 (47.8%) | 92 (41.4%) | 0.74 (0.41–1.36) | ||||
| G/G | 12 (17.4%) | 39 (17.6%) | 0.85 (0.38–1.88) | ||||
| Dominant | C/C | 24 (34.8%) | 91 (41%) | 1.00 | 0.37 | ||
| C/G-G/G | 45 (65.2%) | 131 (59%) | 0.77 (0.44–1.36) | ||||
| Recessive | C/C-C/G | 57 (82.6%) | 183 (82.4%) | 1.00 | 0.99 | ||
| G/G | 12 (17.4%) | 39 (17.6%) | 1.00 (0.48–2.05) | ||||
| Overdominant | C/C-G/G | 36 (52.2%) | 130 (58.6%) | 1.00 | 0.38 | ||
| C/G | 33 (47.8%) | 92 (41.4%) | 0.78 (0.45–1.36) | ||||
| Log-additive | – | – | – | 0.89 (0.61–1.30) | 0.54 | ||
| rs9471535 | Codominant | T/T | 48 (69.6%) | 173 (78.6%) | 1.00 | 0.26 | 0.41 |
| C/T | 18 (26.1%) | 41 (18.6%) | 0.63 (0.33–1.21) | ||||
| C/C | 3 (4.3%) | 6 (2.7%) | 0.45 (0.11–1.90) | ||||
| Dominant | T/T | 48 (69.6%) | 173 (78.6%) | 1.00 | 0.11 | ||
| C/T-C/C | 21 (30.4%) | 47 (21.4%) | 0.60 (0.33–1.12) | ||||
| Recessive | T/T-C/T | 66 (95.7%) | 214 (97.3%) | 1.00 | 0.36 | ||
| C/C | 3 (4.3%) | 6 (2.7%) | 0.50 (0.12–2.09) | ||||
| Overdominant | T/T-C/C | 51 (73.9%) | 179 (81.4%) | 1.00 | 0.2 | ||
| C/T | 18 (26.1%) | 41 (18.6%) | 0.65 (0.34–1.24) | ||||
| Log-additive | – | – | – | 0.65 (0.39–1.08) | 0.1 | ||
| rs7768162 | Codominant | G/G | 25 (36.2%) | 79 (35.9%) | 1.00 | 0.21 | 0.63 |
| A/G | 37 (53.6%) | 98 (44.5%) | 0.93 (0.51–1.69) | ||||
| A/A | 7 (10.1%) | 43 (19.6%) | 1.97 (0.78–4.96) | ||||
| Dominant | G/G | 25 (36.2%) | 79 (35.9%) | 1.00 | 0.74 | ||
| A/G-A/A | 44 (63.8%) | 141 (64.1%) | 1.10 (0.62–1.95) | ||||
| Recessive | G/G-A/G | 62 (89.9%) | 177 (80.5%) | 1.00 | 0.079 | ||
| A/A | 7 (10.1%) | 43 (19.6%) | 2.06 (0.88–4.85) | ||||
| Overdominant | G/G-A/A | 32 (46.4%) | 122 (55.5%) | 1.00 | 0.34 | ||
| A/G | 37 (53.6%) | 98 (44.5%) | 0.76 (0.44–1.33) | ||||
| Log-additive | – | – | – | 1.26 (0.84–1.88) | 0.26 | ||
Second, we investigated whether TLR and TREM-1 gene polymorphisms are associated with severity of noncoronary (ECA and/or LEA) atherosclerosis. We found that the C allele of the rs5743551 polymorphism within the TLR1 gene, the G allele of the rs4986790 polymorphism within the TLR4 gene, and the C allele of the rs3775073 polymorphism within the TLR6 gene were significantly associated with increased PVD risk (OR = 2.32, 95%CI = 1.06–5.07, P = 0.022; OR = 4.36, 95%CI = 1.01–18.80, P = 0.016, both according to the log-additive model; OR = 2.09, 95%CI = 1.05–4.17, P = 0.039, according to the dominant model, respectively) whereas the A/A genotype of the rs5743810 polymorphism within the TLR6 gene was significantly associated with decreased PVD risk (OR = 0.34, 95%CI = 0.15–0.76, P = 0.013, according to the recessive model, Table 5).
Table 5.
Association of polymorphisms within the genes encoding Toll-like receptors (TLRs) and Triggering receptor expressed on myeloid cells-1 (TREM-1) with polyvascular disease (after the adjustments by age and gender; PVD was defined as intima-media thickness increase ≥ 1 mm or any stenosis of extracranial or/and lower extremity arteries).
| Polymorphism | Inheritance model | Genotype | No PVD | PVD | OR (95% CI) | P value | HWE |
|---|---|---|---|---|---|---|---|
| TLR1 gene | |||||||
| rs5743551 | Codominant | T/T | 31 (79.5%) | 159 (63.4%) | 1.00 | 0.05 | 0.85 |
| C/T | 8 (20.5%) | 83 (33.1%) | 2.09 (0.91–4.78) | ||||
| C/C | 0 (0%) | 9 (3.6%) | NA (0.00–NA) | ||||
| Dominant | T/T | 31 (79.5%) | 159 (63.4%) | 1.00 | 0.035 | ||
| C/T-C/C | 8 (20.5%) | 92 (36.6%) | 2.31 (1.01–5.26) | ||||
| Recessive | T/T-C/T | 39 (100%) | 242 (96.4%) | 1.00 | 0.11 | ||
| C/C | 0 (0%) | 9 (3.6%) | NA (0.00–NA) | ||||
| Overdominant | T/T-C/C | 31 (79.5%) | 168 (66.9%) | 1.00 | 0.089 | ||
| C/T | 8 (20.5%) | 83 (33.1%) | 1.98 (0.87–4.52) | ||||
| Log-additive | – | – | – | 2.32 (1.06–5.07) | 0.022 | ||
| rs5743611 | Codominant | C/C | 22 (56.4%) | 150 (59.5%) | 1.00 | 0.15 | 0.87 |
| C/G | 12 (30.8%) | 91 (36.1%) | 1.07 (0.50–2.27) | ||||
| G/G | 5 (12.8%) | 11 (4.4%) | 0.31 (0.10–0.98) | ||||
| Dominant | C/C | 22 (56.4%) | 150 (59.5%) | 1.00 | 0.63 | ||
| C/G-G/G | 17 (43.6%) | 102 (40.5%) | 0.85 (0.43–1.68) | ||||
| Recessive | C/C-C/G | 34 (87.2%) | 241 (95.6%) | 1.00 | 0.051 | ||
| G/G | 5 (12.8%) | 11 (4.4%) | 0.30 (0.10–0.93) | ||||
| Overdominant | C/C-G/G | 27 (69.2%) | 161 (63.9%) | 1.00 | 0.58 | ||
| C/G | 12 (30.8%) | 91 (36.1%) | 1.23 (0.59–2.56) | ||||
| Log-additive | – | – | – | 0.71 (0.42–1.23) | 0.23 | ||
| TLR2 gene | |||||||
| rs5743708 | – | G/G | 36 (92.3%) | 232 (92.1%) | 1.00 | 0.94 | 0.99 |
| A/G | 3 (7.7%) | 20 (7.9%) | 1.05 (0.30–3.74) | ||||
| rs3804099 | Codominant | T/T | 18 (46.1%) | 105 (41.7%) | 1.00 | 0.84 | 0.17 |
| C/T | 14 (35.9%) | 104 (41.3%) | 1.23 (0.58–2.62) | ||||
| C/C | 7 (17.9%) | 43 (17.1%) | 1.01 (0.39–2.61) | ||||
| Dominant | T/T | 18 (46.1%) | 105 (41.7%) | 1.00 | 0.67 | ||
| C/T-C/C | 21 (53.9%) | 147 (58.3%) | 1.16 (0.59–2.29) | ||||
| Recessive | T/T-C/T | 32 (82%) | 209 (82.9%) | 1.00 | 0.84 | ||
| C/C | 7 (17.9%) | 43 (17.1%) | 0.91 (0.37–2.22) | ||||
| Overdominant | T/T-C/C | 25 (64.1%) | 148 (58.7%) | 1.00 | 0.56 | ||
| C/T | 14 (35.9%) | 104 (41.3%) | 1.23 (0.61–2.49) | ||||
| Log-additive | – | – | – | 1.05 (0.65–1.67) | 0.85 | ||
| TLR4 gene | |||||||
| rs4986790 | Codominant | A/A | 37 (94.9%) | 208 (82.5%) | 1.00 | 0.054 | 0.99 |
| A/G | 2 (5.1%) | 42 (16.7%) | 4.30 (0.99–18.77) | ||||
| G/G | 0 (0%) | 2 (0.8%) | NA (0.00–NA) | ||||
| Dominant | A/A | 37 (94.9%) | 208 (82.5%) | 1.00 | 0.016 | ||
| A/G-G/G | 2 (5.1%) | 44 (17.5%) | 4.42 (1.02–19.20) | ||||
| Recessive | A/A-A/G | 39 (100%) | 250 (99.2%) | 1.00 | 0.55 | ||
| G/G | 0 (0%) | 2 (0.8%) | NA (0.00–NA) | ||||
| Overdominant | A/A-G/G | 37 (94.9%) | 210 (83.3%) | 1.00 | 0.02 | ||
| A/G | 2 (5.1%) | 42 (16.7%) | 4.29 (0.98–18.72) | ||||
| Log-additive | – | – | – | 4.36 (1.01–18.80) | 0.016 | ||
| rs4986791 | Codominant | C/C | 36 (92.3%) | 207 (82.1%) | 1.00 | 0.14 | 0.99 |
| C/T | 3 (7.7%) | 43 (17.1%) | 2.86 (0.83–9.84) | ||||
| T/T | 0 (0%) | 2 (0.8%) | NA (0.00–NA) | ||||
| Dominant | C/C | 36 (92.3%) | 207 (82.1%) | 1.00 | 0.053 | ||
| C/T-T/T | 3 (7.7%) | 45 (17.9%) | 2.94 (0.86–10.07) | ||||
| Recessive | C/C-C/T | 39 (100%) | 250 (99.2%) | 1.00 | 0.55 | ||
| T/T | 0 (0%) | 2 (0.8%) | NA (0.00–NA) | ||||
| Overdominant | C/C-T/T | 36 (92.3%) | 209 (82.9%) | 1.00 | 0.062 | ||
| C/T | 3 (7.7%) | 43 (17.1%) | 2.85 (0.83–9.82) | ||||
| Log-additive | – | – | – | 2.91 (0.86–9.89) | 0.05 | ||
| TLR6 gene | |||||||
| rs3775073 | Codominant | T/T | 18 (46.1%) | 73 (29%) | 1.00 | 0.081 | 0.91 |
| T/C | 13 (33.3%) | 129 (51.2%) | 2.41 (1.11–5.22) | ||||
| C/C | 8 (20.5%) | 50 (19.8%) | 1.57 (0.63–3.91) | ||||
| Dominant | T/T | 18 (46.1%) | 73 (29%) | 1.00 | 0.039 | ||
| T/C-C/C | 21 (53.9%) | 179 (71%) | 2.09 (1.05–4.17) | ||||
| Recessive | T/T-T/C | 31 (79.5%) | 202 (80.2%) | 1.00 | 0.98 | ||
| C/C | 8 (20.5%) | 50 (19.8%) | 0.99 (0.43–2.29) | ||||
| Overdominant | T/T-C/C | 26 (66.7%) | 123 (48.8%) | 1.00 | 0.044 | ||
| T/C | 13 (33.3%) | 129 (51.2%) | 2.05 (1.00–4.18) | ||||
| Log-additive | – | – | – | 1.41 (0.86–2.30) | 0.17 | ||
| rs5743810 | Codominant | G/G | 13 (33.3%) | 110 (43.6%) | 1.00 | 0.042 | 0.52 |
| A/G | 15 (38.5%) | 113 (44.8%) | 0.86 (0.39–1.89) | ||||
| A/A | 11 (28.2%) | 29 (11.5%) | 0.32 (0.13–0.78) | ||||
| Dominant | G/G | 13 (33.3%) | 110 (43.6%) | 1.00 | 0.19 | ||
| A/G-A/A | 26 (66.7%) | 142 (56.4%) | 0.63 (0.31–1.28) | ||||
| Recessive | G/G-A/G | 28 (71.8%) | 223 (88.5%) | 1.00 | 0.013 | ||
| A/A | 11 (28.2%) | 29 (11.5%) | 0.34 (0.15–0.76) | ||||
| Overdominant | G/G-A/A | 24 (61.5%) | 139 (55.2%) | 1.00 | 0.53 | ||
| A/G | 15 (38.5%) | 113 (44.8%) | 1.25 (0.62–2.50) | ||||
| Log-additive | – | – | – | 0.58 (0.36–0.94) | 0.026 | ||
| TREM-1 gene | |||||||
| rs2234246 | Codominant | C/C | 13 (33.3%) | 97 (38.5%) | 1.00 | 0.68 | 0.14 |
| C/T | 20 (51.3%) | 108 (42.9%) | 0.76 (0.36–1.61) | ||||
| T/T | 6 (15.4%) | 47 (18.6%) | 1.08 (0.38–3.04) | ||||
| Dominant | C/C | 13 (33.3%) | 97 (38.5%) | 1.00 | 0.61 | ||
| C/T-T/T | 26 (66.7%) | 155 (61.5%) | 0.83 (0.41–1.71) | ||||
| Recessive | C/C-C/T | 33 (84.6%) | 205 (81.3%) | 1.00 | 0.61 | ||
| T/T | 6 (15.4%) | 47 (18.6%) | 1.27 (0.50–3.21) | ||||
| Overdominant | C/C-T/T | 19 (48.7%) | 144 (57.1%) | 1.00 | 0.38 | ||
| C/T | 20 (51.3%) | 108 (42.9%) | 0.74 (0.37–1.46) | ||||
| Log-additive | – | – | – | 0.98 (0.61–1.58) | 0.95 | ||
| rs4711668 | Codominant | C/C | 15 (38.5%) | 84 (33.5%) | 1.00 | 0.68 | 0.11 |
| T/C | 14 (35.9%) | 104 (41.4%) | 1.42 (0.64–3.14) | ||||
| T/T | 10 (25.6%) | 63 (25.1%) | 1.16 (0.49–2.78) | ||||
| Dominant | C/C | 15 (38.5%) | 84 (33.5%) | 1.00 | 0.45 | ||
| T/C-T/T | 24 (61.5%) | 167 (66.5%) | 1.31 (0.65–2.66) | ||||
| Recessive | C/C-T/C | 29 (74.4%) | 188 (74.9%) | 1.00 | 0.93 | ||
| T/T | 10 (25.6%) | 63 (25.1%) | 0.97 (0.44–2.11) | ||||
| Overdominant | C/C-T/T | 25 (64.1%) | 147 (58.6%) | 1.00 | 0.42 | ||
| T/C | 14 (35.9%) | 104 (41.4%) | 1.33 (0.66–2.71) | ||||
| Log-additive | – | – | – | 1.10 (0.70–1.73) | 0.67 | ||
| rs3804277 | Codominant | C/C | 16 (41%) | 99 (39.3%) | 1.00 | 0.92 | 0.11 |
| C/T | 17 (43.6%) | 108 (42.9%) | 1.05 (0.50–2.20) | ||||
| T/T | 6 (15.4%) | 45 (17.9%) | 1.23 (0.45–3.38) | ||||
| Dominant | C/C | 16 (41%) | 99 (39.3%) | 1.00 | 0.79 | ||
| C/T-T/T | 23 (59%) | 153 (60.7%) | 1.10 (0.55–2.19) | ||||
| Recessive | C/C-C/T | 33 (84.6%) | 207 (82.1%) | 1.00 | 0.7 | ||
| T/T | 6 (15.4%) | 45 (17.9%) | 1.20 (0.47–3.05) | ||||
| Overdominant | C/C-T/T | 22 (56.4%) | 144 (57.1%) | 1.00 | 0.97 | ||
| C/T | 17 (43.6%) | 108 (42.9%) | 0.99 (0.50–1.96) | ||||
| Log-additive | – | – | – | 1.10 (0.68–1.76) | 0.7 | ||
| rs2234237 | Codominant | T/T | 27 (69.2%) | 197 (78.2%) | 1.00 | 0.17 | 0.12 |
| A/T | 9 (23.1%) | 49 (19.4%) | 0.76 (0.34–1.74) | ||||
| A/A | 3 (7.7%) | 6 (2.4%) | 0.22 (0.05–0.98) | ||||
| Dominant | T/T | 27 (69.2%) | 197 (78.2%) | 1.00 | 0.23 | ||
| A/T-A/A | 12 (30.8%) | 55 (21.8%) | 0.63 (0.30–1.32) | ||||
| Recessive | T/T-A/T | 36 (92.3%) | 246 (97.6%) | 1.00 | 0.076 | ||
| A/A | 3 (7.7%) | 6 (2.4%) | 0.24 (0.05–1.02) | ||||
| Overdominant | T/T-A/A | 30 (76.9%) | 203 (80.6%) | 1.00 | 0.65 | ||
| A/T | 9 (23.1%) | 49 (19.4%) | 0.83 (0.37–1.87) | ||||
| Log-additive | – | – | – | 0.59 (0.33–1.08) | 0.1 | ||
| rs6910730 | Codominant | A/A | 28 (71.8%) | 199 (79%) | 1.00 | 0.14 | 0.078 |
| A/G | 8 (20.5%) | 48 (19.1%) | 0.84 (0.36–1.96) | ||||
| G/G | 3 (7.7%) | 5 (2%) | 0.19 (0.04–0.86) | ||||
| Dominant | A/A | 28 (71.8%) | 199 (79%) | 1.00 | 0.3 | ||
| A/G-G/G | 11 (28.2%) | 53 (21%) | 0.66 (0.31–1.42) | ||||
| Recessive | A/A-A/G | 36 (92.3%) | 247 (98%) | 1.00 | 0.052 | ||
| G/G | 3 (7.7%) | 5 (2%) | 0.20 (0.04–0.88) | ||||
| Overdominant | A/A-G/G | 31 (79.5%) | 204 (81%) | 1.00 | 0.83 | ||
| A/G | 8 (20.5%) | 48 (19.1%) | 0.91 (0.39–2.12) | ||||
| Log-additive | – | – | – | 0.60 (0.32–1.11) | 0.11 | ||
| rs1817537 | Codominant | C/C | 16 (41%) | 99 (39.3%) | 1.00 | 0.92 | 0.11 |
| C/G | 17 (43.6%) | 108 (42.9%) | 1.05 (0.50–2.20) | ||||
| G/G | 6 (15.4%) | 45 (17.9%) | 1.23 (0.45–3.38) | ||||
| Dominant | C/C | 16 (41%) | 99 (39.3%) | 1.00 | 0.79 | ||
| C/G-G/G | 23 (59%) | 153 (60.7%) | 1.10 (0.55–2.19) | ||||
| Recessive | C/C-C/G | 33 (84.6%) | 207 (82.1%) | 1.00 | 0.7 | ||
| G/G | 6 (15.4%) | 45 (17.9%) | 1.20 (0.47–3.05) | ||||
| Overdominant | C/C-G/G | 22 (56.4%) | 144 (57.1%) | 1.00 | 0.97 | ||
| C/G | 17 (43.6%) | 108 (42.9%) | 0.99 (0.50–1.96) | ||||
| Log-additive | – | – | – | 1.10 (0.68–1.76) | 0.7 | ||
| rs9471535 | Codominant | T/T | 26 (68.4%) | 195 (77.7%) | 1.00 | 0.15 | 0.13 |
| C/T | 9 (23.7%) | 50 (19.9%) | 0.75 (0.33–1.73) | ||||
| C/C | 3 (7.9%) | 6 (2.4%) | 0.21 (0.05–0.93) | ||||
| Dominant | T/T | 26 (68.4%) | 195 (77.7%) | 1.00 | 0.22 | ||
| C/T-C/C | 12 (31.6%) | 56 (22.3%) | 0.62 (0.29–1.31) | ||||
| Recessive | T/T-C/T | 35 (92.1%) | 245 (97.6%) | 1.00 | 0.068 | ||
| C/C | 3 (7.9%) | 6 (2.4%) | 0.23 (0.05–0.98) | ||||
| Overdominant | T/T-C/C | 29 (76.3%) | 201 (80.1%) | 1.00 | 0.64 | ||
| C/T | 9 (23.7%) | 50 (19.9%) | 0.82 (0.36–1.86) | ||||
| Log-additive | – | – | – | 0.58 (0.32–1.06) | 0.09 | ||
| rs7768162 | Codominant | G/G | 14 (36.8%) | 90 (35.9%) | 1.00 | 0.95 | 0.63 |
| A/G | 18 (47.4%) | 117 (46.6%) | 1.11 (0.52–2.38) | ||||
| A/A | 6 (15.8%) | 44 (17.5%) | 1.14 (0.41–3.18) | ||||
| Dominant | G/G | 14 (36.8%) | 90 (35.9%) | 1.00 | 0.76 | ||
| A/G-A/A | 24 (63.2%) | 161 (64.1%) | 1.12 (0.55–2.29) | ||||
| Recessive | G/G-A/G | 32 (84.2%) | 207 (82.5%) | 1.00 | 0.88 | ||
| A/A | 6 (15.8%) | 44 (17.5%) | 1.07 (0.42–2.74) | ||||
| Overdominant | G/G-A/A | 20 (52.6%) | 134 (53.4%) | 1.00 | 0.86 | ||
| A/G | 18 (47.4%) | 117 (46.6%) | 1.07 (0.53–2.14) | ||||
| Log-additive | – | – | – | 1.08 (0.65–1.77) | 0.77 | ||
Moreover, the A/G genotype of the rs4986790 polymorphism and the C/T genotype of the rs4986791 polymorphism within the TLR4 gene were associated with significantly higher risk of ≥ 50% stenosis of ECA and/or LEA (OR = 1.70, 95%CI = 0.85–3.41, P = 0.015 and OR = 1.56, 95%CI = 0.78–3.09, P = 0.02, both according to the codominant model, Table 6).
Table 6.
Association of polymorphisms within the genes encoding Toll-like receptors (TLRs) and Triggering receptor expressed on myeloid cells-1 (TREM-1) with hemodynamically significant (≥ 50%) stenosis of extracranial or/and lower extremity arteries (after the adjustments by age and gender).
| Polymorphism | Inheritance model | Genotype | No stenosis or < 50% stenosis | ≥ 50% stenosis | OR (95% CI) | P value | HWE |
|---|---|---|---|---|---|---|---|
| TLR1 gene | |||||||
| rs5743551 | Codominant | T/T | 136 (65.4%) | 54 (65.8%) | 1.00 | 0.43 | 0.85 |
| C/T | 64 (30.8%) | 27 (32.9%) | 1.08 (0.62–1.89) | ||||
| C/C | 8 (3.8%) | 1 (1.2%) | 0.32 (0.04–2.61) | ||||
| Dominant | T/T | 136 (65.4%) | 54 (65.8%) | 1.00 | 0.99 | ||
| C/T-C/C | 72 (34.6%) | 28 (34.1%) | 1.00 (0.58–1.72) | ||||
| Recessive | T/T-C/T | 200 (96.2%) | 81 (98.8%) | 1.00 | 0.21 | ||
| C/C | 8 (3.8%) | 1 (1.2%) | 0.31 (0.04–2.53) | ||||
| Overdominant | T/T-C/C | 144 (69.2%) | 55 (67.1%) | 1.00 | 0.68 | ||
| C/T | 64 (30.8%) | 27 (32.9%) | 1.13 (0.65–1.96) | ||||
| Log-additive | – | – | – | 0.91 (0.56–1.47) | 0.7 | ||
| rs5743611 | Codominant | C/C | 126 (60.6%) | 46 (55.4%) | 1.00 | 0.64 | 0.87 |
| C/G | 70 (33.6%) | 33 (39.8%) | 1.26 (0.73–2.16) | ||||
| G/G | 12 (5.8%) | 4 (4.8%) | 0.85 (0.26–2.81) | ||||
| Dominant | C/C | 126 (60.6%) | 46 (55.4%) | 1.00 | 0.49 | ||
| C/G-G/G | 82 (39.4%) | 37 (44.6%) | 1.20 (0.71–2.02) | ||||
| Recessive | C/C-C/G | 196 (94.2%) | 79 (95.2%) | 1.00 | 0.67 | ||
| G/G | 12 (5.8%) | 4 (4.8%) | 0.78 (0.24–2.51) | ||||
| Overdominant | C/C-G/G | 138 (66.3%) | 50 (60.2%) | 1.00 | 0.37 | ||
| C/G | 70 (33.6%) | 33 (39.8%) | 1.28 (0.75–2.17) | ||||
| Log-additive | – | – | – | 1.09 (0.71–1.67) | 0.69 | ||
| TLR2 gene | |||||||
| rs5743708 | – | G/G | 192 (92.3%) | 76 (91.6%) | 1.00 | 0.82 | 0.99 |
| A/G | 16 (7.7%) | 7 (8.4%) | 1.12 (0.44–2.85) | ||||
| rs3804099 | Codominant | T/T | 88 (42.3%) | 35 (42.2%) | 1.00 | 0.92 | 0.075 |
| C/T | 85 (40.9%) | 33 (39.8%) | 0.99 (0.56–1.74) | ||||
| C/C | 35 (16.8%) | 15 (18.1%) | 1.15 (0.55–2.38) | ||||
| Dominant | T/T | 88 (42.3%) | 35 (42.2%) | 1.00 | 0.91 | ||
| C/T-C/C | 120 (57.7%) | 48 (57.8%) | 1.03 (0.61–1.74) | ||||
| Recessive | T/T-C/T | 173 (83.2%) | 68 (81.9%) | 1.00 | 0.68 | ||
| C/C | 35 (16.8%) | 15 (18.1%) | 1.15 (0.59–2.27) | ||||
| Overdominant | T/T-C/C | 123 (59.1%) | 50 (60.2%) | 1.00 | 0.84 | ||
| C/T | 85 (40.9%) | 33 (39.8%) | 0.95 (0.56–1.60) | ||||
| Log-additive | – | – | – | 1.05 (0.74–1.50) | 0.77 | ||
| TLR4 gene | |||||||
| rs4986790 | Codominant | A/A | 180 (86.5%) | 65 (78.3%) | 1.00 | 0.015 | 0.99 |
| A/G | 28 (13.5%) | 16 (19.3%) | 1.70 (0.85–3.41) | ||||
| G/G | 0 (0%) | 2 (2.4%) | NA (0.00–NA) | ||||
| Dominant | A/A | 180 (86.5%) | 65 (78.3%) | 1.00 | 0.053 | ||
| A/G-G/G | 28 (13.5%) | 18 (21.7%) | 1.96 (1.00–3.84) | ||||
| Recessive | A/A-A/G | 208 (100%) | 81 (97.6%) | 1.00 | 0.012 | ||
| G/G | 0 (0%) | 2 (2.4%) | NA (0.00–NA) | ||||
| Overdominant | A/A-G/G | 180 (86.5%) | 67 (80.7%) | 1.00 | 0.16 | ||
| A/G | 28 (13.5%) | 16 (19.3%) | 1.66 (0.83–3.32) | ||||
| Log-additive | – | – | – | 2.12 (1.13–3.96) | 0.02 | ||
| rs4986791 | Codominant | C/C | 178 (85.6%) | 65 (78.3%) | 1.00 | 0.02 | 0.99 |
| C/T | 30 (14.4%) | 16 (19.3%) | 1.56 (0.78–3.09) | ||||
| T/T | 0 (0%) | 2 (2.4%) | NA (0.00–NA) | ||||
| Dominant | C/C | 178 (85.6%) | 65 (78.3%) | 1.00 | 0.088 | ||
| C/T-T/T | 30 (14.4%) | 18 (21.7%) | 1.80 (0.92–3.49) | ||||
| Recessive | C/C-C/T | 208 (100%) | 81 (97.6%) | 1.00 | 0.012 | ||
| T/T | 0 (0%) | 2 (2.4%) | NA (0.00–NA) | ||||
| Overdominant | C/C-T/T | 178 (85.6%) | 67 (80.7%) | 1.00 | 0.23 | ||
| C/T | 30 (14.4%) | 16 (19.3%) | 1.52 (0.77–3.02) | ||||
| Log-additive | – | – | – | 1.96 (1.06–3.63) | 0.036 | ||
| TLR6 gene | |||||||
| rs3775073 | Codominant | T/T | 68 (32.7%) | 23 (27.7%) | 1.00 | 0.65 | 0.91 |
| T/C | 101 (48.6%) | 41 (49.4%) | 1.15 (0.63–2.11) | ||||
| C/C | 39 (18.8%) | 19 (22.9%) | 1.41 (0.68–2.94) | ||||
| Dominant | T/T | 68 (32.7%) | 23 (27.7%) | 1.00 | 0.48 | ||
| T/C-C/C | 140 (67.3%) | 60 (72.3%) | 1.23 (0.70–2.16) | ||||
| Recessive | T/T-T/C | 169 (81.2%) | 64 (77.1%) | 1.00 | 0.42 | ||
| C/C | 39 (18.8%) | 19 (22.9%) | 1.29 (0.69–2.42) | ||||
| Overdominant | T/T-C/C | 107 (51.4%) | 42 (50.6%) | 1.00 | 0.99 | ||
| T/C | 101 (48.6%) | 41 (49.4%) | 1.00 (0.60–1.68) | ||||
| Log-additive | – | – | – | 1.19 (0.82–1.71) | 0.36 | ||
| rs5743810 | Codominant | G/G | 86 (41.4%) | 37 (44.6%) | 1.00 | 0.8 | 0.52 |
| A/G | 92 (44.2%) | 36 (43.4%) | 0.89 (0.51–1.54) | ||||
| A/A | 30 (14.4%) | 10 (12.1%) | 0.77 (0.34–1.76) | ||||
| Dominant | G/G | 86 (41.4%) | 37 (44.6%) | 1.00 | 0.56 | ||
| A/G-A/A | 122 (58.6%) | 46 (55.4%) | 0.86 (0.51–1.44) | ||||
| Recessive | G/G-A/G | 178 (85.6%) | 73 (88%) | 1.00 | 0.62 | ||
| A/A | 30 (14.4%) | 10 (12.1%) | 0.82 (0.38–1.78) | ||||
| Overdominant | G/G-A/A | 116 (55.8%) | 47 (56.6%) | 1.00 | 0.82 | ||
| A/G | 92 (44.2%) | 36 (43.4%) | 0.94 (0.56–1.58) | ||||
| Log-additive | – | – | – | 0.88 (0.61–1.28) | 0.51 | ||
| TREM-1 gene | |||||||
| rs2234246 | Codominant | C/C | 77 (37%) | 33 (39.8%) | 1.00 | 0.72 | 0.14 |
| C/T | 91 (43.8%) | 37 (44.6%) | 0.95 (0.54–1.68) | ||||
| T/T | 40 (19.2%) | 13 (15.7%) | 0.74 (0.35–1.57) | ||||
| Dominant | C/C | 77 (37%) | 33 (39.8%) | 1.00 | 0.66 | ||
| C/T-T/T | 131 (63%) | 50 (60.2%) | 0.89 (0.52–1.50) | ||||
| Recessive | C/C-C/T | 168 (80.8%) | 70 (84.3%) | 1.00 | 0.42 | ||
| T/T | 40 (19.2%) | 13 (15.7%) | 0.76 (0.38–1.51) | ||||
| Overdominant | C/C-T/T | 117 (56.2%) | 46 (55.4%) | 1.00 | 0.86 | ||
| C/T | 91 (43.8%) | 37 (44.6%) | 1.05 (0.63–1.76) | ||||
| Log-additive | – | – | – | 0.88 (0.61–1.25) | 0.47 | ||
| rs4711668 | Codominant | C/C | 72 (34.6%) | 27 (32.9%) | 1.00 | 0.83 | 0.0021 |
| T/C | 83 (39.9%) | 35 (42.7%) | 1.20 (0.66–2.19) | ||||
| T/T | 53 (25.5%) | 20 (24.4%) | 1.07 (0.54–2.12) | ||||
| Dominant | C/C | 72 (34.6%) | 27 (32.9%) | 1.00 | 0.62 | ||
| T/C-T/T | 136 (65.4%) | 55 (67.1%) | 1.15 (0.66–1.99) | ||||
| Recessive | C/C-T/C | 155 (74.5%) | 62 (75.6%) | 1.00 | 0.91 | ||
| T/T | 53 (25.5%) | 20 (24.4%) | 0.97 (0.53–1.75) | ||||
| Overdominant | C/C-T/T | 125 (60.1%) | 47 (57.3%) | 1.00 | 0.56 | ||
| T/C | 83 (39.9%) | 35 (42.7%) | 1.17 (0.69–1.97) | ||||
| Log-additive | – | – | – | 1.04 (0.74–1.46) | 0.81 | ||
| rs3804277 | Codominant | C/C | 82 (39.4%) | 33 (39.8%) | 1.00 | 0.8 | 0.11 |
| C/T | 88 (42.3%) | 37 (44.6%) | 1.04 (0.60–1.83) | ||||
| T/T | 38 (18.3%) | 13 (15.7%) | 0.82 (0.38–1.74) | ||||
| Dominant | C/C | 82 (39.4%) | 33 (39.8%) | 1.00 | 0.92 | ||
| C/T-T/T | 126 (60.6%) | 50 (60.2%) | 0.97 (0.58–1.65) | ||||
| Recessive | C/C-C/T | 170 (81.7%) | 70 (84.3%) | 1.00 | 0.52 | ||
| T/T | 38 (18.3%) | 13 (15.7%) | 0.80 (0.40–1.60) | ||||
| Overdominant | C/C-T/T | 120 (57.7%) | 46 (55.4%) | 1.00 | 0.69 | ||
| C/T | 88 (42.3%) | 37 (44.6%) | 1.11 (0.66–1.86) | ||||
| Log-additive | – | – | – | 0.93 (0.65–1.33) | 0.69 | ||
| rs2234237 | Codominant | T/T | 161 (77.4%) | 63 (75.9%) | 1.00 | 0.79 | 0.064 |
| A/T | 40 (19.2%) | 18 (21.7%) | 1.13 (0.60–2.14) | ||||
| A/A | 7 (3.4%) | 2 (2.4%) | 0.66 (0.13–3.29) | ||||
| Dominant | T/T | 161 (77.4%) | 63 (75.9%) | 1.00 | 0.85 | ||
| A/T-A/A | 47 (22.6%) | 20 (24.1%) | 1.06 (0.58–1.94) | ||||
| Recessive | T/T-A/T | 201 (96.6%) | 81 (97.6%) | 1.00 | 0.57 | ||
| A/A | 7 (3.4%) | 2 (2.4%) | 0.64 (0.13–3.19) | ||||
| Overdominant | T/T-A/A | 168 (80.8%) | 65 (78.3%) | 1.00 | 0.66 | ||
| A/T | 40 (19.2%) | 18 (21.7%) | 1.15 (0.61–2.17) | ||||
| Log-additive | – | – | – | 0.99 (0.60–1.64) | 0.97 | ||
| rs6910730 | Codominant | A/A | 163 (78.4%) | 64 (77.1%) | 1.00 | 0.93 | 0.059 |
| A/G | 39 (18.8%) | 17 (20.5%) | 1.07 (0.56–2.04) | ||||
| G/G | 6 (2.9%) | 2 (2.4%) | 0.78 (0.15–4.02) | ||||
| Dominant | A/A | 163 (78.4%) | 64 (77.1%) | 1.00 | 0.92 | ||
| A/G-G/G | 45 (21.6%) | 19 (22.9%) | 1.03 (0.56–1.91) | ||||
| Recessive | A/A-A/G | 202 (97.1%) | 81 (97.6%) | 1.00 | 0.75 | ||
| G/G | 6 (2.9%) | 2 (2.4%) | 0.77 (0.15–3.95) | ||||
| Overdominant | A/A-G/G | 169 (81.2%) | 66 (79.5%) | 1.00 | 0.81 | ||
| A/G | 39 (18.8%) | 17 (20.5%) | 1.08 (0.57–2.06) | ||||
| Log-additive | – | – | – | 0.99 (0.59–1.67) | 0.98 | ||
| rs1817537 | Codominant | C/C | 82 (39.4%) | 33 (39.8%) | 1.00 | 0.8 | 0.11 |
| C/G | 88 (42.3%) | 37 (44.6%) | 1.04 (0.60–1.83) | ||||
| G/G | 38 (18.3%) | 13 (15.7%) | 0.82 (0.38–1.74) | ||||
| Dominant | C/C | 82 (39.4%) | 33 (39.8%) | 1.00 | 0.92 | ||
| C/G-G/G | 126 (60.6%) | 50 (60.2%) | 0.97 (0.58–1.65) | ||||
| Recessive | C/C-C/G | 170 (81.7%) | 70 (84.3%) | 1.00 | 0.52 | ||
| G/G | 38 (18.3%) | 13 (15.7%) | 0.80 (0.40–1.60) | ||||
| Overdominant | C/C-G/G | 120 (57.7%) | 46 (55.4%) | 1.00 | 0.69 | ||
| C/G | 88 (42.3%) | 37 (44.6%) | 1.11 (0.66–1.86) | ||||
| Log-additive | – | – | – | 0.93 (0.65–1.33) | 0.69 | ||
| rs9471535 | Codominant | T/T | 159 (76.8%) | 62 (75.6%) | 1.00 | 0.81 | 0.068 |
| C/T | 41 (19.8%) | 18 (21.9%) | 1.10 (0.59–2.08) | ||||
| C/C | 7 (3.4%) | 2 (2.4%) | 0.65 (0.13–3.26) | ||||
| Dominant | T/T | 159 (76.8%) | 62 (75.6%) | 1.00 | 0.92 | ||
| C/T-C/C | 48 (23.2%) | 20 (24.4%) | 1.03 (0.56–1.89) | ||||
| Recessive | T/T-C/T | 200 (96.6%) | 80 (97.6%) | 1.00 | 0.57 | ||
| C/C | 7 (3.4%) | 2 (2.4%) | 0.64 (0.13–3.17) | ||||
| Overdominant | T/T-C/C | 166 (80.2%) | 64 (78%) | 1.00 | 0.72 | ||
| C/T | 41 (19.8%) | 18 (21.9%) | 1.12 (0.60–2.11) | ||||
| Log-additive | – | – | – | 0.97 (0.59–1.62) | 0.92 | ||
| rs7768162 | Codominant | G/G | 75 (36.2%) | 29 (35.4%) | 1.00 | 0.75 | 0.63 |
| A/G | 95 (45.9%) | 40 (48.8%) | 1.18 (0.76–2.11) | ||||
| A/A | 37 (17.9%) | 13 (15.8%) | 0.92 (0.43–1.99) | ||||
| Dominant | G/G | 75 (36.2%) | 29 (35.4%) | 1.00 | 0.71 | ||
| A/G-A/A | 132 (63.8%) | 53 (64.6%) | 1.11 (0.64–1.90) | ||||
| Recessive | G/G-A/G | 170 (82.1%) | 69 (84.2%) | 1.00 | 0.62 | ||
| A/A | 37 (17.9%) | 13 (15.8%) | 0.84 (0.42–1.69) | ||||
| Overdominant | G/G-A/A | 112 (54.1%) | 42 (51.2%) | 1.00 | 0.47 | ||
| A/G | 95 (45.9%) | 40 (48.8%) | 1.22 (0.72–2.05) | ||||
| Log-additive | – | – | – | 1.00 (0.69–1.44) | 0.99 | ||
There were no other statistically significant associations of TLR and TREM-1 gene polymorphisms with severity of atherosclerosis. All these results are represented here after the adjustments by age and gender. No statistically significant differences were identified among the age and gender subgroups and concerning the haplotype frequencies (data not shown).
4. Discussion
There has been little research to examine whether polymorphisms within TLR genes are associated with severity of atherosclerosis. Several groups demonstrated that the G allele of the rs4986790 polymorphism within the TLR4 gene (the 299Gly allele) was not associated with CAD severity (Boekholdt et al., 2003, Yang et al., 2003, Mandal et al., 2006, Manolakis et al., 2011, Guven et al., 2015); however, a protective effect of this allele was also reported (Kiechl et al., 2002, Hernesniemi et al., 2006, Berg et al., 2009). Regarding noncoronary atherosclerosis, an association of the 299Gly allele with a higher number of vascular territories affected by clinically relevant atherosclerosis was demonstrated (Vainas et al., 2006); however, no association with aortic atherosclerosis was found (Hommels et al., 2007). Although early studies showed an association of the 299Gly allele with decreased intima-media thickness (Kiechl et al., 2002), this was not confirmed in further investigations (Netea et al., 2004, Norata et al., 2005, Labrum et al., 2007, Hernesniemi et al., 2008). A relatively recent meta-analysis demonstrated an overall lack of association between the rs4986790 (Asp299Gly) polymorphism and atherosclerosis (Zhang et al., 2012). In this study, we did not find a statistically significant association of the 299Gly allele with coronary atherosclerosis severity; nevertheless, we demonstrated for the first time that it is significantly associated with higher risk of PVD and ≥ 50% stenosis of ECA and/or LEA.
Previously, a protective role of the A allele of the rs5743810 polymorphism within the TLR6 gene (the Ser249 allele) with relation to the risk of restenosis after percutaneous transluminal coronary angioplasty was shown (Hamann et al., 2013). This matches with our findings, where the A/A genotype of this polymorphism was significantly associated with decreased PVD risk.
We for the first time detected that the C/C genotype of the rs3804099 polymorphism within the TLR2 gene is significantly associated with higher coronary atherosclerosis severity. In addition, three markers within TLR1, TLR4, and TLR6 genes (the C allele of the rs5743551 polymorphism, the C/T genotype of the rs4986791 polymorphism, and the C allele of the rs3775073 polymorphism, respectively) were for the first time found being associated with increased severity of noncoronary atherosclerosis.
Regarding TREM-1 gene polymorphisms, their impact on the susceptibility to diseases has been poorly investigated up to now. To the best of our knowledge, we carried out a first investigation devoted to the role of TREM-1 gene polymorphisms in atherosclerosis severity. It was previously shown that the rs9471535 and rs2234237 polymorphisms are associated with elevated risk of intestinal Behcet's disease (Jung et al., 2011). Also, the rs2234237 polymorphism was reported to be associated with sepsis prognosis according to three inheritance models (Su et al., 2012) and with risk of ventilator-associated pneumonia in burn-injured patients (Rivera-Chávez et al., 2013) but the conflicting results were also published (Chen et al., 2008). No association of eight TREM-1 gene polymorphisms with infective endocarditis was revealed in another study from our group (Golovkin et al., 2015).
However, we previously found that the A/A genotype of the rs2234237 polymorphism, the G/G genotype of the rs6910730 polymorphism, the C/C genotype of the rs9471535 polymorphism, and the T/T genotype of the rs4711668 polymorphism within the TREM-1 gene were significantly associated with elevated CAD risk (Golovkin et al., 2014). Conversely, the G allele of the rs1817537 polymorphism, the T allele of the rs2234246 polymorphism, and the T allele of the rs3804277 polymorphism within the TREM-1 gene were significantly associated with similarly decreased risk of CAD (Golovkin et al., 2014). Out of them, only T/T genotype of the rs4711668 polymorphism was significantly associated with higher severity of coronary atherosclerosis.
The main advantage of our research is a large number of polymorphisms taken into account. We investigated eight polymorphisms within TLR genes and eight polymorphisms within the TREM-1 gene while other authors studied not more than four TLR gene polymorphisms. Moreover, this is the first study regarding the association of TLR and TREM-1 gene polymorphisms with atherosclerosis severity carried out on Russian population. However, we were unable to exclude certain shortcomings. This is a single-center study; in addition, we could also neglect gene polymorphisms which are in causal association with atherosclerosis severity.
Reasons for the discrepancies between the results of our investigation and other relevant genetic association studies may include confounding host or environmental factors in different populations modifying the penetrance of the variant allele, differences in the sample size and in various clinicopathological characteristics between the study samples, and also disparities in diagnostics, stratification, genotyping methods, and chance. In addition, certain studies in which negative results were obtained could have never been published (so-called file drawer effect) that may create a significant bias and distort a picture that we can observe at the moment. However, only widely established methods of diagnostics such as coronary angiography or ultrasonography were used in all considered studies, so this factor does not seem to impact on the disparities between them significantly. Moreover, genotyping methods are also unlikely to be the cause of differences between distinct studies.
Finally, we conclude that certain TLR and TREM-1 gene polymorphisms are significantly associated with atherosclerosis severity in a Russian population. Further studies are needed to confirm these gene polymorphisms as predictive and pathogenic markers of atherosclerosis.
Funding Sources
There was no financial assistance with the project.
Disclosure
On behalf of all the authors, the corresponding author states that there is no conflict of interest.
References
- Ammirati E., Moroni F., Magnoni M., Camici P.G. The role of T and B cells in human atherosclerosis and atherothrombosis. Clin. Exp. Immunol. 2015;179:173–187. doi: 10.1111/cei.12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhtiar S.M., Ali A., Baig S.M., Barh D., Miyoshi A., Azevedo V. Identifying human disease genes: advances in molecular genetics and computational approaches. Genet. Mol. Res. 2014;13:5073–5087. doi: 10.4238/2014.July.4.23. [DOI] [PubMed] [Google Scholar]
- Bentzon J.F., Otsuka F., Virmani R., Falk E. Mechanisms of plaque formation and rupture. Circ. Res. 2014;114:1852–1866. doi: 10.1161/CIRCRESAHA.114.302721. [DOI] [PubMed] [Google Scholar]
- Berg K.K., Madsen H.O., Garred P., Wiseth R., Gunnes S., Videm V. The additive contribution from inflammatory genetic markers on the severity of cardiovascular disease. Scand. J. Immunol. 2009;69:36–42. doi: 10.1111/j.1365-3083.2008.02187.x. [DOI] [PubMed] [Google Scholar]
- Boekholdt S.M., Agema W.R., Peters R.J., Zwinderman A.H., van der Wall E.E., Reitsma P.H. Variants of toll-like receptor 4 modify the efficacy of statin therapy and the risk of cardiovascular events. Circulation. 2003;107:2416–2421. doi: 10.1161/01.CIR.0000068311.40161.28. [DOI] [PubMed] [Google Scholar]
- Chen Q., Zhou H., Wu S., Wang H., Lv C., Cheng B. Lack of association between TREM-1 gene polymorphisms and severe sepsis in a Chinese Han population. Hum. Immunol. 2008;69:220–226. doi: 10.1016/j.humimm.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Dayem Ullah A.Z., Lemoine N.R., Chelala C. SNPnexus: a web server for functional annotation of novel and publicly known genetic variants (2012 update) Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gks364. (Web Server issue):W65-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi K., Manabe I. Toll-like receptor, lipotoxicity and chronic inflammation: the pathological link between obesity and cardiometabolic disease. J. Atheroscler. Thromb. 2014;21:629–639. doi: 10.5551/jat.22533. [DOI] [PubMed] [Google Scholar]
- Fihn S.D., Blankenship J.C., Alexander K.P., Bittl J.A., Byrne J.G., Fletcher B.J. ACC/AHA/AATS/PCNA/SCAI/STS focused update of the guideline for the diagnosis and management of patients with stable ischemic heart disease: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines, and the American Association for Thoracic Surgery, Preventive Cardiovascular Nurses Association, Society for Cardiovascular Angiography and Interventions, and Society of Thoracic Surgeons. Circulation. 2014;130:1749–1767. doi: 10.1161/CIR.0000000000000095. (2014) [DOI] [PubMed] [Google Scholar]
- Golovkin A.S., Ponasenko A.V., Khutornaya M.V., Kutikhin A.G., Salakhov R.R., Yuzhalin A.E. Association of TLR and TREM-1 gene polymorphisms with risk of coronary artery disease in a Russian population. Gene. 2014;550:101–109. doi: 10.1016/j.gene.2014.08.022. [DOI] [PubMed] [Google Scholar]
- Golovkin A.S., Ponasenko A.V., Yuzhalin A.E., Salakhov R.R., Khutornaya M.V., Kutikhin A.G. An association between single nucleotide polymorphisms within TLR and TREM-1 genes and infective endocarditis. Cytokine. 2015;71:16–21. doi: 10.1016/j.cyto.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Guven M., Ismailoglu Z., Batar B., Unal S., Onaran I., Karadag B. The effect of genetic polymorphisms of TLR2 and TLR4 in Turkish patients with coronary artery disease. Gene. 2015;558:99–102. doi: 10.1016/j.gene.2014.12.047. [DOI] [PubMed] [Google Scholar]
- Hamann L., Koch A., Sur S., Hoefer N., Glaeser C., Schulz S. Association of a common TLR-6 polymorphism with coronary artery disease — implications for healthy ageing? Immun. Ageing. 2013;10:43. doi: 10.1186/1742-4933-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernesniemi J., Lehtimäki T., Rontu R., Islam M.S., Eklund C., Mikkelsson J. Toll-like receptor 4 polymorphism is associated with coronary stenosis but not with the occurrence of acute or old myocardial infarctions. Scand. J. Clin. Lab. Invest. 2006;66:667–675. doi: 10.1080/00365510600933011. [DOI] [PubMed] [Google Scholar]
- Hernesniemi J.A., Raitakari O.T., Kähönen M., Juonala M., Hutri-Kähönen N., Marniemi J. Toll-like receptor 4 gene (Asp299Gly) polymorphism associates with carotid artery elasticity. The cardiovascular risk in young Finns study. Atherosclerosis. 2008;198:152–159. doi: 10.1016/j.atherosclerosis.2007.09.024. [DOI] [PubMed] [Google Scholar]
- Hommels M.J., Kroon A.A., Netea M.G., de Leeuw P.W., Bruggeman C.A., Leiner T. The Asp299Gly Toll-like receptor 4 polymorphism in advanced aortic atherosclerosis. Neth J. Med. 2007;65:203–207. [PubMed] [Google Scholar]
- Jung E.S., Kim S.W., Moon C.M., Shin D.J., Son N.H., Kim E.S. Relationships between genetic polymorphisms of triggering receptor expressed on myeloid cells-1 and inflammatory bowel diseases in the Korean population. Life Sci. 2011;89:289–294. doi: 10.1016/j.lfs.2011.06.018. [DOI] [PubMed] [Google Scholar]
- Kiechl S., Lorenz E., Reindl M., Wiedermann C.J., Oberhollenzer F., Bonora E. Toll-like receptor 4 polymorphisms and atherogenesis. N. Engl. J. Med. 2002;347:185–192. doi: 10.1056/NEJMoa012673. [DOI] [PubMed] [Google Scholar]
- Labrum R., Bevan S., Sitzer M., Lorenz M., Markus H.S. Toll receptor polymorphisms and carotid artery intima-media thickness. Stroke. 2007;38:1179–1184. doi: 10.1161/01.STR.0000260184.85257.2b. [DOI] [PubMed] [Google Scholar]
- Mandal K., Afzal A.R., Brecker S.J., Poloniecki J., Xu Q., Jahangiri M. Association of serum soluble heat shock protein 60 with toll-like receptor 4 polymorphism and severity of coronary artery disease. Heart. 2006;92:683–685. doi: 10.1136/hrt.2004.059170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolakis A.C., Kapsoritakis A.N., Tiaka E.K., Sidiropoulos A., Gerovassili A., Satra M. TLR4 gene polymorphisms: evidence for protection against type 2 diabetes but not for diabetes-associated ischaemic heart disease. Eur. J. Endocrinol. 2011;165:261–267. doi: 10.1530/EJE-11-0280. [DOI] [PubMed] [Google Scholar]
- Netea M.G., Hijmans A., van Wissen S., Smilde T.J., Trip M.D., Kullberg B.J. Toll-like receptor-4 Asp299Gly polymorphism does not influence progression of atherosclerosis in patients with familial hypercholesterolaemia. Eur. J. Clin. Investig. 2004;34:94–99. doi: 10.1111/j.1365-2362.2004.01303.x. [DOI] [PubMed] [Google Scholar]
- Norata G.D., Garlaschelli K., Ongari M., Raselli S., Grigore L., Benvenuto F. Effect of the Toll-like receptor 4 (TLR-4) variants on intima-media thickness and monocyte-derived macrophage response to LPS. J. Intern. Med. 2005;258:21–27. doi: 10.1111/j.1365-2796.2005.01509.x. [DOI] [PubMed] [Google Scholar]
- Pelham C.J., Agrawal D.K. Emerging roles for triggering receptor expressed on myeloid cells receptor family signaling in inflammatory diseases. Expert. Rev. Clin. Immunol. 2014;10:243–256. doi: 10.1586/1744666X.2014.866519. [DOI] [PubMed] [Google Scholar]
- Rivera-Chávez F.A., Huebinger R.M., Burris A., Liu M.M., Minei J.P., Hunt J.L. A TREM-1 polymorphism A/T within the Exon 2 is associated with pneumonia in burn-injured patients. ISRN Inflamm. 2013;2013:431739. doi: 10.1155/2013/431739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P.K., Chyu K.Y., Dimayuga P.C., Nilsson J. Vaccine for atherosclerosis. J. Am. Coll. Cardiol. 2014;64:2779–2791. doi: 10.1016/j.jacc.2014.10.018. [DOI] [PubMed] [Google Scholar]
- Sianos G., Morel M.A., Kappetein A.P., Morice M.C., Colombo A., Dawkins K. The SYNTAX Score: an angiographic tool grading the complexity of coronary artery disease. EuroIntervention. 2005;1:219–227. [PubMed] [Google Scholar]
- Solé X., Guinó E., Valls J., Iniesta R., Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- Su L., Liu C., Li C., Jiang Z., Xiao K., Zhang X. Dynamic changes in serum soluble triggering receptor expressed on myeloid cells-1 (sTREM-1) and its gene polymorphisms are associated with sepsis prognosis. Inflammation. 2012;35:1833–1843. doi: 10.1007/s10753-012-9504-z. [DOI] [PubMed] [Google Scholar]
- Vainas T., Stassen F.R., Bruggeman C.A., Welten R.J., van den Akker L.H., Kitslaar P.J. Synergistic effect of Toll-like receptor 4 and CD14 polymorphisms on the total atherosclerosis burden in patients with peripheral arterial disease. J. Vasc. Surg. 2006;44:326–332. doi: 10.1016/j.jvs.2006.04.035. [DOI] [PubMed] [Google Scholar]
- Xu Z., Taylor J.A. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37 doi: 10.1093/nar/gkp290. (Web Server issue):W600-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang I.A., Holloway J.W., Ye S., Southampton Atherosclerosis Study (SAS) Group TLR4 Asp299Gly polymorphism is not associated with coronary artery stenosis. Atherosclerosis. 2003;170:187–190. doi: 10.1016/s0021-9150(03)00286-7. [DOI] [PubMed] [Google Scholar]
- Yuzhalin A.E., Kutikhin A.G. Integrative systems of genomic risk markers for cancer and other diseases: future of predictive medicine. Cancer Manag. Res. 2012;4:131–135. doi: 10.2147/CMAR.S30855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Zhang L., Zhou B., Wang Y., Song Y., Rao L. Lack of association between TLR4 Asp299Gly polymorphism and atherosclerosis: evidence from meta-analysis. Thromb. Res. 2012;130:e203–e208. doi: 10.1016/j.thromres.2012.07.008. [DOI] [PubMed] [Google Scholar]


