Abstract
This unit contains several protocols to determine the energy utilization of T cells in real-time using a Seahorse Extracellular Flux Analyzer (www.seahorsebio.com). The advantages to using this machine over traditional metabolic assays include the simultaneous measurement of glycolysis and mitochondrial respiration, in real-time, on relatively small numbers of cells, without any radioactivity. The Basic Protocol describes a standard mitochondrial stress test on the XFe96, which yields information about oxidative phosphorylation and glycolysis, two energy-generating pathways. The alternate protocols provide examples of adaptations to the Basic Protocol, including adjustments for the use of the XFe24. A protocol for real-time bioenergetic responses to T cell activation allows for the analysis of immediate metabolic changes after T cell receptor stimulation. Specific substrate utilization can be determined by the use of differential assay media, or the injection of drugs that specifically affect certain metabolic processes. Accurate cell numbers, purity, and viability are critical to obtain reliable results.
Keywords: T cells, bioenergetics, glycolysis, oxidative phosphorylation
Introduction
The advantages to using a Seahorse Extracellular Flux Analyzer (EFA) to measure metabolic phenotypes of cells, as opposed to other more traditional metabolism assays, are considerable. For example, by using fiber-optic probes containing two sensors the Seahorse EFA allows the simultaneous measurement of the extracellular acidification rate (ECAR; an indicator of glycolysis) and the oxygen consumption rate (OCR; an indicator of oxidative phosphorylation (OXPHOS) from relatively low numbers of cells, in real-time, without any radioactivity (for detailed instruction videos see www.seahorsebio.com). Assays that directly measure substrate usage, such as traditional glycolysis or beta-oxidation assays, require radioactively labeled substrates, and much larger numbers of cells than that needed by the Seahorse EFA. While measuring the flux of heavy labeled substrates through metabolic pathways is highly informative, directly shows engagement of a pathway, and does not require radioactivity, this technique requires specialized instrumentation (mass spectrometry) and training to analyze the data, which can come at a great expense. Furthermore, this type of metabolic flux analysis often requires large numbers of cells (1×106–107 T cells) that are cultured with heavy labeled substrates for extended periods of time. In contrast, the Seahorse EFA assay allows magnitudes fewer cells to be measured (as low as 1×105 for T cells). The need for less starting material makes this technique amenable for measuring metabolism in rare subsets of T cells, as well as T cells directly ex vivo, without any extended culture period. In addition to these aspects, as a Seahorse EFA assay is running, compounds can be injected through ports, exposing cells to various substrates, inhibitors, drugs, or stimuli, to evaluate immediate effects on cellular bioenergetics. Finally, the Seahorse machine is easy to use after minimal training, and as will become evident in the following protocols, is well within the capability of any immunologist to use.
**It is worth noting that using labeled substrates, and measuring how that substrate is metabolized, is considered the ‘gold’ standard in terms of proving the engagement of a particular pathway (e.g. tracing glucose carbon through the glycolysis pathway shows engagement of glycolysis). However, the flexibility and ability to perform real-time measurements afforded by the Seahorse technology can often allow one to deduce pathway engagement, and depending on the particular needs of the researcher and the hypothesis to be tested, this can be ample evidence of particular metabolic activity in cells.
With the launch of the extracellular flux analyzer, Seahorse Bioscience described the ‘mitochondrial stress test’; a detailed protocol for this assay and background information about the Seahorse EFA can be found on their website (www.seahorsebio.com). The protocols described in this unit have simply been adapted from the Seahorse protocols for use with lymphocytes. Importantly, all protocols describe the use of the newest machines (XFe), but can also be performed on the older models (XF machines), using the same materials as described (XFe FluxPak). Basic Protocol 1 describes a standard measurement of T cell bioenergetics on the XFe96 EFA, establishing ECAR and OCR in basal conditions, and in response to drugs that interrogate mitochondrial function. The Alternate Protocols describe the standard mitochondrial stress test on the XFe24 EFA (Alternate Protocol 1), the assessment of real-time bioenergetics in response to T cell activation (Alternate Protocol 2), and examples of how to test substrate utilization in T cells (Alternate Protocols 3 and 4). The XFe96 requires fewer cells per well than the XFe24, so when working with low cell numbers the XFe96 provides an advantage.
CAUTION: When working with animal or human derived material all biosafety practices must be followed.
Basic Protocol 1: ‘Mitochondrial Stress Test’ on the XFe96 EFA
In this protocol the bioenergetics of T cell populations are measured in the basal state, and in response to drugs that affect mitochondrial function, i.e. oligomycin, FCCP, and rotenone + antimycin A. This protocol uses only 3 of the 4 injection ports, if an additional injection is used, 26 μl can be injected in Port D.
Materials
XFe96 extracellular flux analyzer (Seahorse bioscience)
37°C non-CO2 incubator
XFe96 FluxPak (Seahorse bioscience: including sensor cartridges and cell culture microplates)
Calibrant (included in the XFe96 FluxPak)
XF media pH 7.4 (non-buffered RPMI 1640 containing 25 mM glucose, 2 mM L-glutamine and 1mM sodium pyruvate)
Defined murine T cell subset suspension (cultured or freshly isolated) in XF media
Poly-D lysine (PDL; Sigma, use at 50 μg/ml)
Mitochondria interrogating drugs (1 mM oligomycin, 1.5 mM Fluoro-carbonyl cynade phenylhydrazon (FCCP), 100 μM rotenone, and 1 mM antimycin A; Sigma)
Additional reagents and equipment for counting cells
NOTE: A sensor cartridge is packaged on top of a microplate, and delivered with loading guides for easy drug loading.
NOTE: It is recommended to use at least 4 replicate wells/group for accuracy of the run. However, acquiring repeated results from independent experiments is critical to validate conclusions (also see Background information).
NOTE: If cultured cells are assayed, it is important to realize that cell concentration, nutrient availability, and the surface area of the media for gas exchange (also dependent on the depth of the media) can affect the metabolic phenotype of cells. Keeping these parameters consistent is critical for comparisons between experiments.
NOTE: Standard XF media lacks serum, so cells should not be left in this media for long periods of time. Depending on the question to be answered, serum can be added to the XF media. It is important to note that in that case drug concentrations, in particular FCCP, may need to be adjusted (see Background information).
Carefully lift up and put aside (upside down) the sensor cartridge.
Add 200 μl of calibrant into each well of the microplate and put the sensor cartridge (with lid) back up in a correct orientation to hydrate all probes for 4–24 hours at 37°C in a non-CO2 incubator.
Coat the XF cell culture microplate plate with 20 μl poly-D-lysine for 1–2 hours.
- Aspirate off the poly-D-lysine, wash the wells with H2O, and let them air-dry.Coating of the plate can be done the day before. Keep the plate at 4°C overnight and make sure it warms up to room temperature before plating the cells.
- Harvest the cells, wash in XF media, and then resuspend in XF media.Estimate the volume of XF media so that the cell concentration will be at least 5×106 cells/ml.
- Count the cells and adjust the concentration to 5×106 cells/ml with XF media.If the cell concentration is lower than 5×106 cells/ml, a lower concentration of cells can be used, but do not use a concentration lower than 2×106 cells/ml.
Plate 40 μl of cells into the bottom of the wells (= 200,000 cells/well). Be sure to use at least 4 wells (typically wells A1, A12, H1, and H12) for background correction; put only XF media in these wells (no cells).
Centrifuge 5 min at 400 × g to allow the cells to collect into a monolayer at the bottom of the plate.
- Slowly add 140 μl of XF media to each well making sure not to disrupt the cell monolayer.Placing the pipet tip against the side at the higher end in the well helps not to disrupt the monolayer.
Observe cells under a microscope to determine that a monolayer of cells is present in all groups, and that no cells were dislodged.
Incubate cell plate at 37°C in a non-CO2 incubator for 30–60 minutes.
Prepare 10× stocks of drugs in XF media (2.8 ml for one plate). Final concentrations after injections will be: 1 μM oligomycin, 1.5 μM FCCP, 100 nM rotenone, and 1 μM antimycin A
-
Without removing the microplate underneath, pipet the drugs directly into the drug delivery ports on top of the sensor cartridge as follows:
Port A) 20 μl oligomycin
Port B) 22 μl FCCP
Port C) 24 μl rotenone + antimycin AUsing a multichannel pipette to load the drugs will reduce the process time. Ensure that the order of the ports (see user's manual) is correct. Incubate cartridge at 37°C in a non-CO2 incubator while setting up the program.
-
Standard seahorse run:
Calibrate
Equilibrate
Base line readings (Loop 3 times):
Mix – 3 min Wait – 2 min Measure – 3 min End loop
Inject port A (Loop 3 times):
Mix – 3 min Wait – 2 min Measure – 3 min End loop
Inject Port B (Loop 3 times):
Mix – 3 min Wait – 2 min Measure – 3 min End loop
Inject Port C (Loop 3 times):
Mix – 3 min Wait – 2 min Measure – 3 min End loop
End programAfter the calibration step the user will be asked to replace the calibrant plate for the cell plate. Make sure that the cell plate is loaded in the correct orientation.
Alternate Protocol 1: ‘Mitochondrial Stress Test’ on the Xfe24 EFA
This protocol is designed for the same readouts as Basic Protocol 1, now using the Seahorse XFe24 EFA. The choice between using this protocol (XFe24) and Basic protocol 1 (XFe96) depends on the availability of the respective machines, the number of required wells, and the number of available cells. If the 4th port is used, inject 95 μl in Port D.
Additional or Alternate Materials (also see Basic Protocol 1)
XFe24 extracellular flux analyzer (Seahorse bioscience)
XFe24 FluxPak (Seahorse bioscience)
Perform steps 1–4 as described in Basic Protocol 1.
- Harvest the cells, wash in XF media, and then resuspend in XF media.Estimate the volume of XF media so that the cell concentration will be around 1×107 cells/ml.
- Count the cells and adjust the concentration to 5–8×106 cells/ml with XF media.If the cell concentration is lower than 5×106 cells/ml, centrifuge the cells again, resuspend in a lower volume and re-count.
- Plate 100 μl of cells into the bottom of the wells (= 5–8×105 cells/well).It is important not to plate the cells in more than 100 μl. If more volume is used not all cells will collect into the bottom of the wells, thereby reducing accuracy of the measurements.
Centrifuge 5 min at 400 × g to allow for cells to collect into a monolayer at the bottom of the plate.
- Slowly add 530 μl of XF media to each well making sure not to disrupt the cell monolayer.Placing the pipet tip against the side at the higher end in the well helps not to disrupt the monolayer.
Observe cells under a microscope to determine that a monolayer of cells is present in all groups and that no cells were dislodged.
Incubate cell plate at 37°C in a non-CO2 incubator for 30–60 minutes.
Prepare 10× stocks of drugs in XF media (2.8 ml for one plate). Final concentrations after injections will be: 1 μM oligomycin, 1.5 uM FCCP, 100 nM rotenone, and 1 μM antimycin A
-
Pipet the drugs into the drug delivery ports on top of the sensor cartridge as follows:
Port A) 70 μl oligomycin
Port B) 75 μl FCCP
Port C) 85 μl rotenone + antimycin AUsing a multichannel pipette to load the drugs will reduce the process time (use every other channel). Ensure that the order of the ports (see user's manual) is correct. Perform steps 13 and 14 as described in Basic Protocol 1.
Alternate Protocol 2: Real-Time Bioenergetic Measurements in Response to Activation
In this protocol real-time bioenergetic changes are measured in T cells in response to activation (using PMA/ionomycin or anti-CD3/CD28 coated beads). Subsequently, a mitochondrial stress test is performed in the same run. This protocol is designed for the Seahorse XFe96, but can also be performed on the XFe24 when volumes and cell numbers are adapted accordingly (see Alternate Protocol 1).
Additional Materials (also see Basic Protocol 1)
PMA and ionomycin (Sigma)
anti-CD3/CD28 coated beads (Gibco)
Perform steps 1–11 as described in Basic Protocol 1.
Prepare 10× stocks of drugs or beads in XF media (2.8 ml for one plate). Final concentrations after injections will be: 5–50 ng/ml PMA, 500 ng/ml ionomycin, 1 μM oligomycin, 1.5 μM FCCP, 100 nM rotenone, and 1 μM antimycin A. For bead injection use 2 beads/cell.
-
Plate the drugs into the cartridge plate as follows:
Port A) 20 μl PMA+ionomycin or anti-CD3/CD28 beads or XF media
Port B) 22 μl oligomycin
Port C) 24 μl FCCP
Port D) 26 μl rotenone + antimycin AUsing a multichannel pipette to load the drugs will reduce the process time. Ensure that the order of the ports (see user's manual) is correct.
6. Incubate cartridge at 37°C in a non-CO2 incubator while setting up the program.
-
7. Program:
Calibrate
Equilibrate
Base line readings (Loop 3 times):
Mix – 3 min Wait – 2 min Measure – 3 min End loop
Inject port A (Loop 6 – 15 times):
Mix – 3 min Wait – 2 min Measure – 3 min End loop
Inject Port B (Loop 3 times):
Mix – 3 min Wait – 2 min Measure – 3 min End loop
Inject Port C (Loop 3 times):
Mix – 3 min Wait – 2 min Measure – 3 min End loop
Inject Port D (Loop 3 times):
Mix – 3 min Wait – 2 min Measure – 3 min End loop
End programAfter the calibration step the user will be asked to replace the calibrant plate for the cell plate. Make sure that the cell plate is loaded in the correct orientation.
Alternate Protocol 3: Substrate Utilization; Galactose
Cells will rely on mitochondrial metabolism, rather than glycolysis, when grown in galactose. This protocol is designed for the Seahorse X96e EFA, but can also be performed on the XFe24 when volumes and cell numbers are adapted accordingly (see Alternate Protocol 1).
Additional Materials (also see Basic Protocol 1)
Glucose XF media pH 7.4 (non-buffered RPMI 1640 containing 25 mM glucose, 2 mM L-glutamine and 1mM sodium pyruvate)
Galactose XF media pH 7.4 (non-buffered RPMI 1640 containing 25 mM galactose, 2 mM L-glutamine and 1mM sodium pyruvate)
NOTE: Activated T cells cultured in glucose will engage aerobic glycolysis and OXPHOS, however, T cells cultured in galactose will not engage aerobic glycolysis, and instead will primarily use mitochondrial respiration. If cells are cultured in galactose-containing media before being assayed on the Seahorse EFA, it is important to use glucose-free media and to dialyze the serum to ensure a complete glucose-free medium. All cell groups, whether cultured in glucose or galactose, must be cultured in media that contains the same dialyzed serum.
Perform steps 1–4 as described in Basic Protocol 1.
Harvest the cells and split cells into two groups: glucose and galactose.
Wash cells with either Glucose or Galactose XF media and resuspend in corresponding XF media.
Perform steps 5–7 as described in Basic Protocol 1.
Slowly add 140 μl of corresponding XF media to each well making sure not to disrupt the cell monolayer.
Observe cells under a microscope to determine that a monolayer of cells is present in all groups and that no cells were dislodged.
Incubate cell plate at 37°C in a non-CO2 incubator for 30–60 minutes.
Prepare 10× stocks of drugs in corresponding XF media (2.8 ml for one plate). Final concentrations after injections will be: 1 μM oligomycin, 1.5 μM FCCP, 100 nM rotenone, and 1 μM antimycin A
Perform steps 12–14 as described in Basic Protocol 1.
Alternate Protocol 4: Substrate Utilization; Long-Chain Fatty Acids
In this protocol the relative contribution of mitochondrial long-chain fatty acid oxidation to T cell respiration is determined. The injection of etomoxir in port A will reveal to what extent fatty acid oxidation contributes to basal oxygen consumption. Alternatively, the injection of etomoxir in port C will show the contribution of fatty acid oxidation to spare respiratory capacity (SRC, also see Background information). This protocol is designed for the Seahorse X96 EFA, but can also be performed on the XFe24 when volumes and cell numbers are adapted accordingly (see Alternate Protocol 1).
Additional Materials (also see Basic Protocol 1)
20 mM Etomoxir
Perform steps 1–11 as described in Basic Protocol 1.
Prepare 10× stocks of drugs in XF media (2.8 ml for one plate). Final concentrations after injections will be: 200 μM etomoxir, 1 μM oligomycin, 1.5 μM FCCP, 100 nM rotenone, and 1 μM antimycin A
-
Pipet the drugs into the drug delivery ports on top of the sensor cartridge as follows:
To determine if fatty acid oxidation contributes to basal respiration;
Port A) 20 μl etomoxir or XF media
Port B) 22 μl oligomycin
Port C) 24 μl FCCP
Port D) 26 μl rotenone + antimycin A
To determine if fatty acid oxidation contributes to SRC;
Port A) 20 μl oligomycin
Port B) 22 μl FCCP
Port C) 24 μl etomoxir or XF media
Port D) 26 μl rotenone + antimycin AUsing a multichannel pipette to load the drugs will reduce the process time. Ensure that the order of the ports (see user's manual) is correct. Incubate cartridge at 37°C in a non-CO2 incubator while setting up the program.
-
Program:
Calibrate
Equilibrate
Base line readings (Loop 3 times)
Mix – 3 min Wait – 2 min Measure – 3 min End loop
Inject port A (Loop 3 times):
Mix – 3 min Wait – 2 min Measure – 3 min End loop
Inject Port B (Loop 3 times):
Mix – 3 min Wait – 2 min Measure – 3 min End loop
Inject Port C (Loop 3 times):
Mix – 3 min Wait – 2 min Measure – 3 min End loop
Inject Port D (Loop 3 times):
Mix – 3 min Wait – 2 min Measure – 3 min End loop
End programAfter the calibration step the user will be asked to replace the calibrant plate for the cell plate. Make sure that the cell plate is loaded in the correct orientation.
Reagents and Solutions
XF media (Basic Protocol 1)
Non-buffered RPMI 1640 (Sigma) supplemented with:
25 mM glucose
2 mM L-glutamine
1 mM sodium pyruvate
pH 7.4, pass through sterile filter
Store at 4°C as recommended by the manufacturer
Galactose XF media
Non-buffered RPMI 1640 (Sigma) supplemented with:
25 mM galactose
2 mM L-glutamine
1 mM sodium pyruvate
pH 7.4, pass through sterile filter
Store at 4°C as recommended by the manufacturer
Poly-D-lysine (PDL)
Reconstitute lyophilized PDL (Sigma) in sterile tissue culture grade H2O, making a working concentration of 50 μg/ml. Store current aliquot at 4°C, and additional aliquots at −20°C.
Oligomycin, 1000×
Dissolve oligomycin (Sigma) in fresh molecular biology grade DMSO at 1 mM. Make 30 μl aliquots and store at −20°C.
Fluoro-carbonyl cynade phenylhydrazon (FCCP), 1000×
Dissolve FCCP (Sigma) in fresh molecular biology grade DMSO at 1.5 mM. Make 30 ul aliquots and store at −20°C.
Rotenone, 1000×
Dissolve rotenone (Sigma) in fresh molecular biology grade DMSO at 100 μM. Make 30 ul aliquots and store at −20°C.
Antimycin A, 1000×
Dissolve antimycin A (Sigma) in fresh molecular biology grade DMSO at 1 mM. Make 30 μl aliquots and store at −20°C.
Etomoxir, 100×
Reconstitute lyophilized etomoxir (Sigma) in sterile tissue culture grade H2O at 20 mM. Make 120 μl aliquots and store at −20°C.
Phorbol 12-myristate 13-acetate (PMA)
Dissolve PMA (Sigma) in fresh molecular biology grade DMSO at 5 mg/ml. Make 20 μl aliquots and store at −20°C.
Ionomycin
Dissolve ionomycin (Sigma) in fresh molecular biology grade DMSO at 1 mg/ml. Make 20 μl aliquots and store at −20°C.
Commentary
Background information
The metabolic phenotype of T cells varies with their activation status; activated T cells highly engage aerobic glycolysis, as well as OXPHOS, while resting T cells mainly use OXPHOS to meet their energy demands (Krauss et al., 2001; Pearce et al., 2013). OXPHOS takes place in mitochondria where NADH, which is generated in the tricarboxylic acid (TCA) cycle, donates electrons to Complex I of the electron transport chain (ETC) (Saraste, 1999). The electron flow from Complex I to Complex IV (where oxygen is used as the final electron acceptor) builds a proton gradient in the intermembrane space (between the inner and outer membranes of the mitochondria). This gradient enables protons to flux back into the matrix through Complex V (ATP synthase), resulting in ATP synthesis. Aerobic glycolysis is the process where pyruvate, derived from glucose breakdown, is fermented to lactate in the cytoplasm, even when sufficient oxygen is present to utilize OXPHOS, often referred to as the Warburg effect (Frauwirth et al., 2002; Gerriets and Rathmell, 2012; Pearce et al., 2013; Vander Heiden et al., 2009; Warburg, 1956).
Several techniques are available to measure cellular metabolism. The process of glycolysis results in the excretion of protons outside the cell. As such, often the simplest indicator that cells are using glycolysis is a color change of culture media that contains phenol red. Quantitative methods to rate glycolysis include the measurement of glucose or lactate concentrations in culture media (Everts et al., 2014), assessment of glucose uptake by means of a fluorescent glucose analog 2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino]-2-deoxy-D-glucose (2-NBDG) (Everts et al., 2014), intracellular mass spectrometry (Dupuy et al., 2013), or glucose tracing using heavy-labeled glucose and the rate of conversion into lactate (Faubert et al., 2014). Measuring OXPHOS in T cells can be achieved by determining oxygen consumption using an oxygen electrode (Hynes et al., 2006), or an oxygen consumption rate assay kit (Cayman chemical). To address specific substrate utilization for OXPHOS, heavy-labeled substrates can be used, such as galactose, glutamine, pyruvate, or palmitate (Chang et al., 2013; Pearce et al., 2009; Wang et al., 2011). The advantages of the protocols described in this unit over the other techniques described above, include the simultaneous real-time measurements of glycolysis and OXPHOS rates, the ease of screening numerous samples in a 24 or 96 well format, the requirement of relatively smaller cell numbers, and no need for specialized equipment to analyze heavy-labeled products. Importantly, the ability to inject drugs during the measurement allows for the evaluation of immediate effects.
The mitochondrial stress test as described in Basic Protocol 1 uses four drugs that interrogate mitochondrial function (Figure 1). Oligomycin inhibits Complex V (ATP synthase) of the ETC, and injection of this compound shows how much of the OCR is due to ATP synthesis. Subsequent injection of the uncoupler FCCP, a protonophore, allows protons to move into the mitochondrial matrix independent of the ATP synthase. In order to maintain the membrane potential, protons are moved back into the intermembrane space by increasing the flow of electrons across the ETC to the maximum speed. Thus, injection of FCCP reveals the maximal respiratory capacity (maximal OCR) of the cells. The ratio of maximal OCR as compared to basal OCR is known as spare respiratory capacity (SRC). SRC is the extra mitochondrial capacity available in a cell to produce energy under conditions of increased work or stress and is thought to be important for long-term cellular survival and function (Choi et al., 2009; Ferrick et al., 2008; Nicholls, 2009; Nicholls et al., 2010; van der Windt et al., 2012; Yadava and Nicholls, 2007). Rotenone and antimycin A block Complex I and III, respectively, thereby completely inhibiting the ETC, and as such any remaining oxygen consumption after these drugs is non-mitochondrial. The difference between OCR after oligomycin and OCR after rotenone and antimycin A indicates mitochondrial proton-leak.
Figure 1. Bioenergetic Profile: a schematic overview of the effects of mitochondrial inhibitors on the electron transport chain.
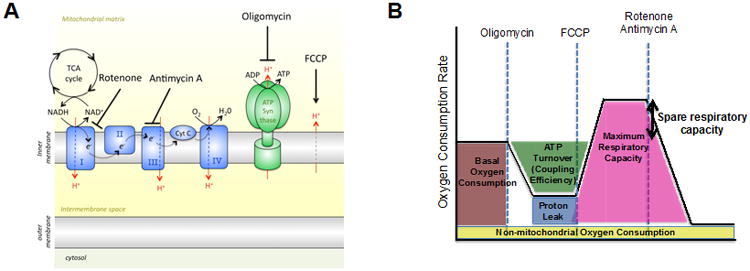
(A) Oligomycin is an ATP synthase inhibitor, FCCP is a protonophore that uncouples ATP synthesis from oxygen consumption, rotenone is a Complex I inhibitor, and antimycin A is a Complex III inhibitor. (B) A schematic of the mitochondrial stress test using the extracellular flux analyzer: O2 consumption rate (OCR) is measured prior to the addition of drugs (basal OCR) and then following the addition of the indicated drugs. Reduction in OCR after oligomycin indicates the amount of O2 consumed for mitochondrial ATP generation. FCCP allows H+ back into the matrix independent of the ATP synthase; cells attempt to maintain the chemiosmotic gradient after FCCP by moving H+ back out to the intermembrane space, which requires the use of the ETC and the consumption of O2 as the final electron acceptor. After FCCP the maximum capacity of the mitochondria to use OXPHOS is revealed. Spare respiratory capacity (SRC) is the difference between maximal OCR and basal OCR and as such is an indicator of how close to its bioenergetic limit the cell is functioning (Nicholls et al., 2010). Rotenone and Antimycin A together render a complete shutdown of the ETC, and thus mitochondrial oxygen consumption. Figure was originally published in Immunity (van der Windt et al., 2012).
To determine the extent of the utilization of a particular substrate, drugs that interfere with specific metabolic reactions can be injected, e.g. etomoxir (see Alternate Protocol 4), a specific inhibitor of long-chain fatty acid oxidation, or 2-deoxy-D-glucose (2-DG), a glycolysis inhibitor. Alternatively, specific compounds can be added or excluded from the standard XF media to address questions about substrate utilization. While most traditional substrate utilization assays are typically performed in serum-free conditions to maximize the utilization of an added substrate, some cell types, and depending on the question asked by the experiment, may require serum in the XF media. Of note, if 10% fetal calf serum is added to the XF media, a higher concentration of FCCP is required to determine maximum OCR (approximately a 5× higher concentration).
The choice between Basic Protocol 1 and Alternate Protocol 1 will be determined by the availability of the XFe96 and XFe24 EFAs, the number of cells available per sample (200,000 for XFe96 versus 500,000–800,000 for XFe24), and the number of sample groups to compare. Alternate Protocols 2–4 are simple adaptations to the Basic Protocol and can be run on either the XFe96 or XFe24 EFAs, and should be chosen when the basic mitochondrial stress test will not suffice to address the hypothesis to be tested. The methods described in this unit are not limited to murine T cells, and can also be applied to human T cells, and other (immune) cell types, such as macrophages, dendritic cells, B cells, and tumor cells. It is important to note that adaptations to the protocols may be required when human T cells and other cell types are studied. For instance, macrophages and dendritic cells often do not require coating of the cell plate with PDL, as in certain conditions these cells will adhere to the cell plate during culture (Everts et al., 2014; Everts et al., 2012). In addition, the number of cells plated per well will vary with different cell types, and should be optimized by the investigator. A guideline is to aim for a basal OCR signal between 100-300 pMoles/min and a basal ECAR reading above 3 mpH/min, and not to exceed a monolayer of cells. Repeatability of the results in multiple runs will be important to determine whether readings are valid and above background, and will also reduce the possibility that differences between groups resulted from plating different numbers of cells per group, or differences in culturing of the cells. The concentrations of oligomycin and FCCP may also need to be optimized for different cell types. For oligomycin a first test-range of 0.5–2 μM is recommended, for FCCP 0.1–2 μM (unless serum is used, see above). The optimal concentration of oligomycin or FCCP is the lowest concentration that maximally decreases or increases OCR, respectively.
**Cells that have been shipped overnight on wet ice (not cold packs as the cells can freeze!) in complete media can be assayed in the Seahorse EFA. It is always important to recount cells before plating.
Critical parameters and troubleshooting
All cell preparations should be done as quickly as possible, and all cells must be kept on ice while working with one cell population at a time. Accurate cell counting is critical for adequate data comparison between groups. If T cells are isolated using a cell sorter, they should be re-counted after sorting, as during sorting, cells may die, thus sorter counts are not sufficiently accurate. In addition, a fixed number of cells should be seeded in each well, and it is recommended to have a minimum of 4 wells per group. If one would like to compare data between different plates/runs, a common control, such as naïve T cells, or a group of cells treated in the same condition as previous runs could possibly be included with each plate to normalize the data between the different runs.
Before seeding the cells, ensure that the cell plate is at room temperature. After plating the cells and the addition of XF media, wells should be checked under a microscope to confirm an intact monolayer of cells in all wells. In case only part of the plate is used, it is critical to fill all injection ports (of the ports that will be injected, for Basic Protocol 1 that is ports A–C). This will avoid inconsistent injection of drugs into wells due to unbalanced pressure. It is recommended to fill all unused wells in the cell plate with XF media as these can function as additional background wells. In order to eliminate CO2 perturbations and to reduce potential measurement variation during the run, it is important to incubate the cell plate in a 37°C non-CO2 incubator for at least 30 minutes but not more than 60 minutes before putting it in the EFA. Table 1 lists some potential problems and suggestions.
Table 1. Troubleshooting Guide for Measuring Bioenergetics with an Extracellular Flux Analyzer.
| Problem | Comments and Suggestions |
|---|---|
| Poor basal signal | |
| - Low cell number | Increase the number of cells per well. |
| - Low cell viability | Keep cells on ice for all steps before plating and/or reduce processing time. |
| High variation between replicates | |
| - Uneven cell number plated | Make sure the correct volume of cells is added in each well of the cell plate, and that the volume of cells is maximal 100 μl. |
| - A certain probe does not detect signals properly | Make sure all probes are hydrated for 4 – 24 hours with sufficient volume of Calibrant XF, and use cartridges before the expiration date. |
| - Cell monolayer was disturbed during XF media addition | Make sure the cell plate is PDL-coated and the plate has been spun down after adding the cells. Using too many cells, as well as having |
| substantial numbers of dead cells in the well, results in multiple cell layers, and will increase disturbance of the cells after injection. | |
| High variation between repeated measurements within a loop | |
| - Decline in basal measurements | Make sure to keep the cell plate in a 37°C non-CO2 incubator for 30–60 minutes before putting the cells in the machine. |
| - Decline in measurements after drug injections | This is sometimes, but not exclusively, seen after FCCP injection. The drug concentration may not be optimal. Titration of the drug might eliminate this problem, however, other biological factors, independent of drug dose, could also contribute to a drop. |
| - Cell monolayer was disturbed after injection | Make sure the cell plate is PDL-coated and the plate is spun down after plating the cells. Using too many cells, as well as having substantial numbers of dead cells in the well, results in multiple cell layers, and will increase disturbance of the cells after injection. |
| Minimal or unexpected changes in OCR after drug injection | |
| - No drug is injected | Make sure all drugs are added directly into the correct ports in the right orientation without contaminating other ports. |
| - Inconsistent injection of drugs between wells | Fill all injection ports that will be injected with drugs of interest. Make sure all injection ports that will be injected are filled with the proper volume to ensure balanced injection. |
| - Concentration of drug is too low | Titration of drug concentration may be required. |
| - Incorrect orientation of cartridge or cell plate during plating or running | The drugs will be injected in a wrong order if the orientation of the cartridge is incorrect. Ensure correct orientation (see user's manual of the EFA) of the cell plate and the cartridge, both during plating/drug loading and during the run. |
Anticipated results
Measurement of T cell bioenergetics using the Seahorse EFA will vary depending on the type and doses of drugs, measuring time, cell numbers and the pre-treatment of cells. Figure 2 demonstrates typical OCR curves of naïve, effector and memory CD8+ T cells from wild-type mice infected with Listeria monocytogenes using drugs described in Basic Protocol 1. OCR should decrease after the addition of oligomycin, and increase after FCCP injection. The extent of the increase in OCR after FCCP depends on the subset of T cells (van der Windt et al., 2012). SRC can be calculated as the ratio of maximum OCR after FCCP injection/basal OCR. Figure 3 shows an immediate increase in ECAR in T cells upon PMA/ionomycin injection, indicating the engagement of glycolysis after T cell stimulation (van der Windt et al., 2013).
Figure 2.
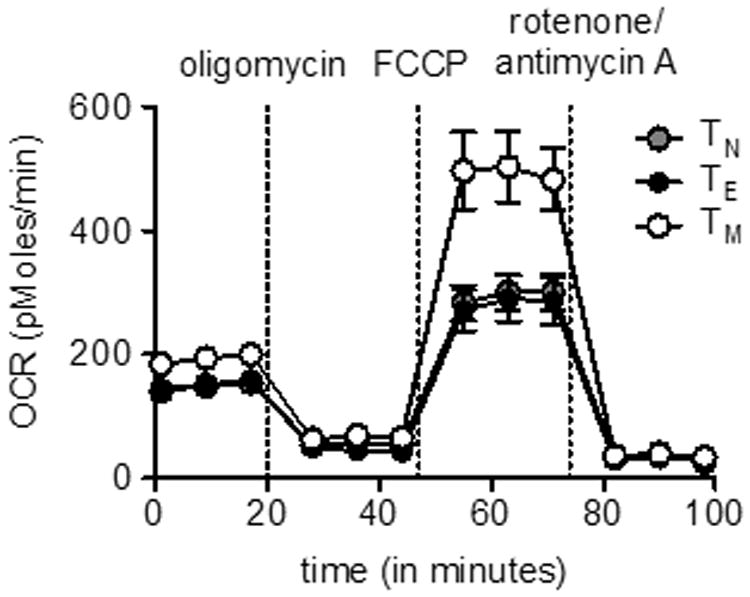
CD8+ T cells were isolated from spleens and lymph nodes harvested from naïve (for naïve T cells: TN) and Listeria monocytogenes infected mice (effector cells : TE, and memory cells: TM). O2 consumption rates (OCR) of CD8+ T cells were measured in real-time under basal conditions and in response to indicated mitochondrial inhibitors. Notice that TM cells have considerable spare respiratory capacity (SRC). Figure was originally published in Immunity (van der Windt et al., 2012).
Figure 3.
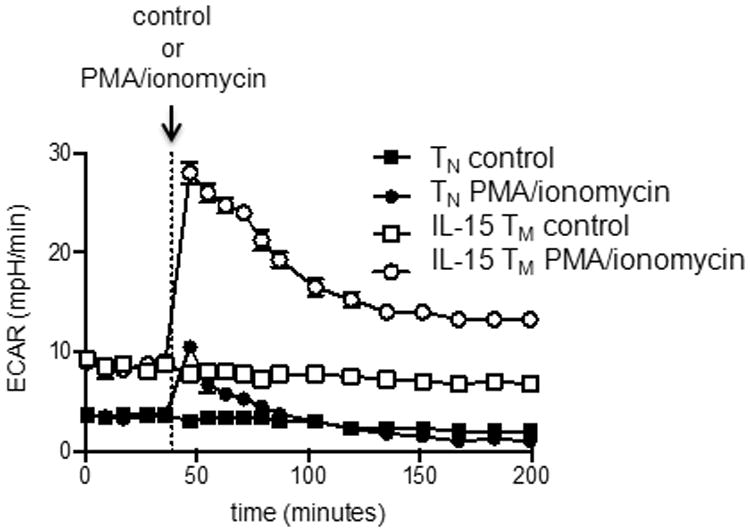
OT-I cells were activated with OVA-peptide and IL-2 for 3 days and subsequently cultured in the presence of IL-15 for 3 days to generate IL-15 memory (TM) cells. Extracellular acidification rates of naive (TN) and IL-15 TM cells, in resting conditions, and after stimulation with PMA/ionomycin or control (injected after 5 measurements). Figure was originally published in PNAS (van der Windt et al., 2013).
Time considerations
The preparation of the cells for bioenergetic measurement depends strongly on the number of groups. Typically this process takes 30–120 min, including the harvesting and counting of cells, the adjustment to the desired cell concentrations, the plating of cells and the addition of XF media. Plating the drugs into the ports of the cartridge usually requires 20 minutes, but this also depends on the number and combinations of drugs used in the experiment. As such, the time considerations are rough guidelines and it is recommended that an investigator becomes familiar with the technique before assessing many groups on a plate. In addition, the waiting time after mixing and before measurement is crucial for the XFe24 EFA, but it is optional for the XFe96 model. Finally, the Seahorse EFA running time for a standard mitochondrial stress test (as described in Basic Protocol 1) takes 160 minutes, including the time for the cartridge calibration and equilibration steps.
Acknowledgments
This work was supported by a Veni grant from the Netherlands Organisation for Scientific Research (G.J.W.W.), and grants from the National Institutes of Health (E.L.P.).
Literature Cited
- Chang CH, Curtis JD, Maggi LB, Jr, Faubert B, Villarino AV, O'Sullivan D, Huang SC, van der Windt GJ, Blagih J, Qiu J, Weber JD, Pearce EJ, Jones RG, Pearce EL. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SW, Gerencser AA, Nicholls DG. Bioenergetic analysis of isolated cerebrocortical nerve terminals on a microgram scale: spare respiratory capacity and stochastic mitochondrial failure. J Neurochem. 2009;109:1179–1191. doi: 10.1111/j.1471-4159.2009.06055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy F, Griss T, Blagih J, Bridon G, Avizonis D, Ling C, Dong Z, Siwak DR, Annis MG, Mills GB, Muller WJ, Siegel PM, Jones RG. LKB1 is a central regulator of tumor initiation and pro-growth metabolism in ErbB2-mediated breast cancer. Cancer Metab. 2013;1:18. doi: 10.1186/2049-3002-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts B, Amiel E, Huang SC, Smith AM, Chang CH, Lam WY, Redmann V, Freitas TC, Blagih J, van der Windt GJ, Artyomov MN, Jones RG, Pearce EL, Pearce EJ. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKvarepsilon supports the anabolic demands of dendritic cell activation. Nat Immunol. 2014;15:323–332. doi: 10.1038/ni.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts B, Amiel E, van der Windt GJ, Freitas TC, Chott R, Yarasheski KE, Pearce EL, Pearce EJ. Commitment to glycolysis sustains survival of NO-producing inflammatory dendritic cells. Blood. 2012;120:1422–1431. doi: 10.1182/blood-2012-03-419747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubert B, Vincent EE, Griss T, Samborska B, Izreig S, Svensson RU, Mamer OA, Avizonis D, Shackelford DB, Shaw RJ, Jones RG. Loss of the tumor suppressor LKB1 promotes metabolic reprogramming of cancer cells via HIF-1alpha. Proc Natl Acad Sci U S A. 2014;111:2554–2559. doi: 10.1073/pnas.1312570111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrick DA, Neilson A, Beeson C. Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov Today. 2008;13:268–274. doi: 10.1016/j.drudis.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, Elstrom RL, June CH, Thompson CB. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16:769–777. doi: 10.1016/s1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- Gerriets VA, Rathmell JC. Metabolic pathways in T cell fate and function. Trends in immunology. 2012;33:168–173. doi: 10.1016/j.it.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes J, Marroquin LD, Ogurtsov VI, Christiansen KN, Stevens GJ, Papkovsky DB, Will Y. Investigation of drug-induced mitochondrial toxicity using fluorescence-based oxygen-sensitive probes. Toxicological sciences : an official journal of the Society of Toxicology. 2006;92:186–200. doi: 10.1093/toxsci/kfj208. [DOI] [PubMed] [Google Scholar]
- Krauss S, Brand MD, Buttgereit F. Signaling takes a breath--new quantitative perspectives on bioenergetics and signal transduction. Immunity. 2001;15:497–502. doi: 10.1016/s1074-7613(01)00205-9. [DOI] [PubMed] [Google Scholar]
- Nicholls DG. Spare respiratory capacity, oxidative stress and excitotoxicity. Biochem Soc Trans. 2009;37:1385–1388. doi: 10.1042/BST0371385. [DOI] [PubMed] [Google Scholar]
- Nicholls DG, Darley-Usmar VM, Wu M, Jensen PB, Rogers GW, Ferrick DA. Bioenergetic profile experiment using C2C12 myoblast cells. J Vis Exp. 2010 doi: 10.3791/2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science. 2013;342:1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, Choi Y. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste M. Oxidative phosphorylation at the fin de siecle. Science. 1999;283:1488–1493. doi: 10.1126/science.283.5407.1488. [DOI] [PubMed] [Google Scholar]
- van der Windt GJ, Everts B, Chang CH, Curtis JD, Freitas TC, Amiel E, Pearce EJ, Pearce EL. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Windt GJ, O'Sullivan D, Everts B, Huang SC, Buck MD, Curtis JD, Chang CH, Smith AM, Ai T, Faubert B, Jones RG, Pearce EJ, Pearce EL. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1221740110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, Green DR. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Yadava N, Nicholls DG. Spare respiratory capacity rather than oxidative stress regulates glutamate excitotoxicity after partial respiratory inhibition of mitochondrial complex I with rotenone. J Neurosci. 2007;27:7310–7317. doi: 10.1523/JNEUROSCI.0212-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]


