Abstract
Invariant Natural Killer T (iNKT) cells represent a population of innate T lymphocytes which act as ‘first-responders’ to infection. While they have long been considered a versatile cell, capable of secretion of multiple cytokines upon activation, recent evidence now indicates that distinct lineages of iNKT cells with unique transcriptional and cytokine profiles exist in different peripheral tissue and as such represent ‘fine-tuning’ of these cells, which act as mediators between the innate and adaptive immune systems. Here we discuss the molecules regulating the differentiation of iNKT cell lineages, the transcription factors associated with their development, and the role of E protein transcription factors and their negative regulators the Id proteins, as these cells develop from immature progenitor cells to terminally differentiated cells in peripheral tissue.
Keywords: Lineage differentiation, T cell, Innate immunity
1. Introduction
Although they develop in the thymus, invariant Natural Killer T (iNKT) cells are commonly considered innate-like T lymphocytes and indeed their cytokine response upon activation can occur in a matter of hours. Type I NKT cells possess a semi-invariant T cell receptor (TCR) in which the variable region Vα14 joins with Jα14 in the mouse, making a Vα14Jα18 TCRα chain (Vα24Jα18 in humans) which in turn pairs with a limited number of TCRβ chains including Vβ2, Vβ7 or Vβ8.2 (Vβ11 in humans) (reviewed in Ref. [1]). Thus, these cells are known as invariant or iNKT cells and will be the topic of this review. Unlike conventional T cells, which recognize peptides in the context of the MHC complex, the invariant TCR enables iNKT cells to recognize glycolipids presented in the context of CD1d, an MHC class I-like complex. Commonly used in the laboratory is the synthetic glycolipid known as α-galactosylceramide (α-GalCer), which can elicit a strong immune response and cause rapid cytokine production from iNKT cells. Recent evidence, however, suggested that α-GalCer can also induce a distinct lineage of iNKT cells known as NKT10 cells (discussed below) [2]. iNKT cells can also be activated indirectly through cytokines produced from recently activated antigen presenting cells (APCs) [3–5]. As such, despite their highly conserved TCR, iNKT cells can either directly or indirectly influence the immune response and play roles in response to infectious disease, cancer and autoimmunity (reviewed in Ref. [6–9]).
For many years, iNKT cell developmental biology focused on sequential step-wise developmental stages with iNKT cells, upon rearrangement of their invariant T cell receptor (TCR) at the double positive (DP) stage, reported to transition through stage 0 (CD24+), stage 1 (CD44− NK1.1−), stage 2 (CD44+ NK1.1−) and finally completing maturation at stage 3 (CD44+ NK1.1+) [1]. However, recent studies have shown that iNKT cell development is more complex and in fact, results in a number of distinct lineages each with a unique transcriptional and cytokine profile. It has now become clear that iNKT cells can be subdivided into NKT1, NKT2, NKT17 and NKT10 subsets; lineages that naturally develop in the thymus and home to specific sites in peripheral tissue [2,10–15]. Here we will review some of the key iNKT cell lineages now identified and the transcriptional regulators that likely regulate their differentiation and maintenance.
2. NKT1 and NKT2 cells
NKT1 cells are in many ways equivalent to Th1 cells in that they express high levels of TBET and produce IFN-γ upon activation (Fig. 1). Importantly though, these cells can also produce IL-4 albeit at lower levels than during their development in the thymus. NKT1 cells are considered to make up the majority of stage 3 CD44+ NK1.1+ iNKT cells in C57BL/6 mice and perhaps not surprisingly, TBET deficiency resulted in a reduction in this cell population [13,16]. NKT1 cells express IL-15Rα (CD122) and loss of IL-15 also resulted in loss of these cells [17]. Interestingly, loss of TBET or functional IL-15 also led to an increase in NKT2 and NKT17 cell frequency and number (discussed below) [13,15].
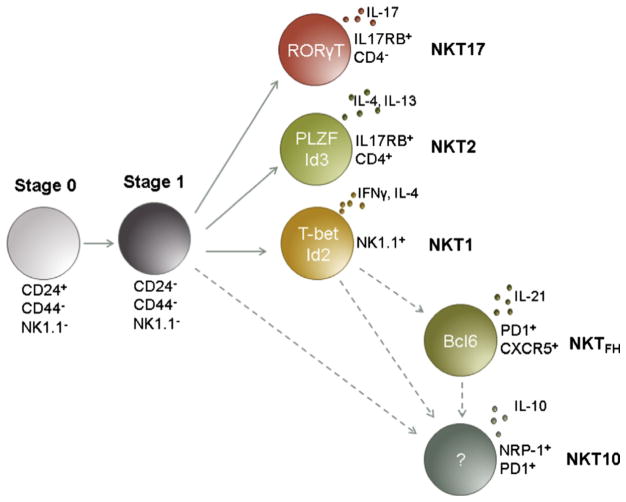
Fig. 1.
There are at least 5 distinct known lineages of iNKT cells that arise from iNKT precursor cells (stage 0/1). Each terminally differentiated lineage is defined by expression of a unique selection of cytokines and transcription factors. NKT1 cells express high levels of TBET and release IFNγ and some IL-4 upon activation. NKT2 cells continually express PLZF and predominantly release IL-4 and IL-13 upon activation. NKT17 cells express RORγt, and release IL-17 upon activation. NKTFH express the transcription factor Bcl6 and develop after activation although it is not clear whether these cells develop from NKT1 precursors. Also unknown are the transcription factors regulating NKT10 cells, although, these cells can be defined by IL-10 production as well as expression of NRP-1 and PD1. The DNA binding proteins ID2 and ID3 have been shown to be major factors in the differentiation of NKT1 and NKT2 cells, respectively. Evidence points to a role for E proteins in NKT17 and NKT10 lineage differentiation, although further investigation will likely clarify how these cells are regulated.
A number of recent data have indicated an important role for the E protein transcription factors and their negative regulators, the Id proteins, in the regulation of iNKT cell lineage differentiation. E proteins bind DNA as homo- or heterodimers at specific E box sites (CANNTG) where they can initiate or repress transcription of their target gene(s), although their function is more often associated with positive regulation [18]. They are negatively regulated by the Inhibitor of DNA (Id) proteins, which do not contain a basic DNA binding domain but have a Helix-Loop-Helix (HLH) domain, allowing the Id proteins to form heterodimers with the E proteins and so remove them from their E box DNA binding sites [18].
In terms of iNKT cell biology, Id2 was initially described as essential for the survival of iNKT cells specifically in peripheral hepatic tissue [19]. More recent data indicate Id2 expression is highly upregulated in the NKT1 sublineage [20]. Using Id2-YFP reporter mice, Id2 expression was shown to be low in stage 1 and 2 iNKT cells before being upregulated in stage 3 cells, which correspond to the NKT1 sublineage [13,20]. Interestingly, by gating on stage 2 NK1.1− cells, a subset of TBET+ cells could be identified, leading to speculation that these cells represent immature NKT1 cells that had yet to upregulate Id2 expression [20]. Loss of Id2 expression in turn led to a corresponding loss of NKT1 cells [20]. Given that TBET is the dominant transcription factor associated with NKT1 cells and that loss of Id2 expression would presumably give rise to uncontrolled E protein DNA binding activity, it is tempting to speculate that E proteins might negatively regulate TBET expression. Certainly, E box sites are present in the Tbx21 (TBET) promoter (Stradner, personal communication). A separate study in which E protein activity was maintained throughout iNKT cell development (using ET2, a fusion protein that competes with Id proteins to bind E proteins and results in their sustained activity at their DNA binding sites) also showed a decrease in TBET expression in total iNKT cells and a corresponding decrease in IFN-γ production upon activation [21]. However, when subdivided into their different lineages, a decrease in TBET expression in the NKT1 population was not observed within the ET2 model but rather a loss in the number of NKT1 cells [21]. Here it was suggested that rather than negatively regulating TBET expression, E proteins might act to regulate a checkpoint at which different iNKT cell lineages develop [21]. Further work in this area will no doubt elucidate these possibilities.
Along with the identification of NKT1 cells, NKT2 cells (analogous with Th2 cells), were identified and confirmed to represent a final differentiation stage within the heterogenous stage 2 CD44+ NK1.1− iNKT cell population [13,15,22]. Interestingly, NKT2 cells were initially identified based on cell surface expression of CD4 and interleukin 17 receptor B (IL-17RB), the receptor for IL-25 [22]. Importantly, the authors identified these cells as IL-4 and IL-13 producers: hallmarks of NKT2 cells. This work was confirmed and further elaborated on by Watarai and colleagues, who showed that IL-17RB was expressed by both NKT2 and NKT17 cells (discussed below) and that CD4 expression could identify NKT2 cells, while CD4− IL-17RB+ cells identified NKT17 cells [15]. Loss of IL-17RB led to a loss of NKT2 and NKT17 cells with a corresponding increase in stage 3 NKT1 cells, indicating that many so-called stage 2 cells have terminally differentiated [15].
As in the case of NKT1 cells, a unique transcriptional profile is also associated with NKT2 cells. Although it had previously been suggested, Lee and colleagues were the first to show that NKT2 cells could be identified by continuous maintenance of PLZF expression [10,11,13]. Using BALB/c mice, which have an abundance of NKT2 IL-4-producing cells, it was shown through intrathymic transfer that NKT2 cells represent terminally differentiated PLZF+ TBET− cells that could not give rise to NKT1 cells. These data help reconcile previous work investigating the role of the transcription factor GATA3 on the development of iNKT cells [23]. With loss of GATA3 expression, the authors observed loss of NK1.1− iNKT cells with a corresponding increase in mature NK1.1+ cells strongly suggesting that like Th2 cells, NKT2 cells rely on GATA3 expression for terminal differentiation.
Id protein transcriptional regulators also play a role in differentiation of NKT2 cells. Using Id3-GFP reporter mice, it was shown that PLZF+ NKT2 cells express high levels of Id3 [20] (Fig. 1). Surprisingly, loss of Id3 actually led to an increase in NKT2 cells and loss of NKT1 cells, most likely due to unrestrained E protein activity in the absence of Id3. Both E2A and HEB (E protein transcription factors) were shown to bind E box sites at the PLZF promoter, further supporting this possibility [20]. In a separate model in which Id3 conditional floxed mice were crossed to a CD4-Cre deleting line, a similar increase in the frequency and number of NKT2 cells was observed along with a corresponding loss in TBET+ NKT1 cells [24]. In the ET2 model, in which E protein activity is unimpeded, an increase in NKT2 and NKT17 cell differentiation was observed [21].
So what are the factors likely to regulate Id2 and Id3 during iNKT cell lineage differentiation? Previous data indicate E proteins themselves can bind to the Id3 promoter [25]. This implies a negative feedback loop in which E proteins expressed early in iNKT cell upregulate Id3. This feedback loop would still allow some E protein activity to maintain Id3 and PLZF expression, as observed in NKT2 cells. What is less clear are the factors that might regulate E protein expression, although microarray data from the Goldrath group indicated that cytokine signaling could potentially regulate their expression [26]. As NKT2 cells express IL-17RB, the receptor for IL-25, it is possible that IL-25 signaling might also regulate E protein expression.
The factors regulating Id2 expression have been explored in more detail in the CD8+ effector T cell immune response and evidence exists that the transcription factors STAT4 and STAT5 can both influence Id2 expression in these cells [27]. Given that NKT1 cells are CD122+ and dependent on IL-15 for their survival, there is the intriguing possibility that IL-15 signaling could regulate Id2 expression. In support of this are data showing the transcription factor Egr2 is necessary for CD122 expression and that Id2 expression is down regulated in the absence of Egr2 [28]. The Goldrath group also previously demonstrated that Id2 expression was upregulated through IL-12 signaling and STAT4 activity [27]. NKT1 cells have higher Stat4 mRNA levels in comparison to NKT2 cells, which may indicate that Id2 expression is regulated by multiple cytokines [15]. Thus, the current data strongly suggest Id2 expression is regulated by cell extrinsic factors, perhaps dictated by the peripheral organ in which the NKT lineage resides. While future research will no doubt further elaborate how Id2 and Id3 expression is regulated, current data indicate Id3 expression is cell-intrinsically regulated by E proteins while Id2 expression is regulated extrinsically, most likely by cytokines.
3. NKT17 cells
This subset of iNKT cells, now typically termed NKT17 cells, was initially identified in 2007 and is considered a distinct lineage of iNKT cells developing in the thymus [14]. These cells are NK1.1− and as their name suggests, produce IL-17A (as opposed to IFN-γ or IL-4) upon stimulation with α-GalCer or during infection [14,29]. NKT17 cells depend on the transcription factor ROR-γt for development in the thymus and differentiate as iNKT cells move through stage 2 during development [30,31]. As well as ROR-γt expression, NKT17 cells can be identified by low expression of NK1.1 and CD4 and high expression of CCR6 and IL-17RB [15]. Mature NKT17 cells predominantly reside in peripheral tissue, in particular in the peripheral lymph nodes, the skin and in the lung [12,14]. In terms of disease, NKT17 cells have been associated with airway neutrophilia and play a role in both ozone- and viral-induced airway hyperreactivity (AHR) [14,15,32]. They have also been shown to play a detrimental role in collagen-induced arthritis [33]. In response to infection with Streptococcus pneumoniae, which has a cognate ligand for iNKT cells or to Staphylococcus aureus, which has no known ligand for iNKT cells, NKT17 cells respond and produce IL-17A, indicating the importance of these cells in the primary immune response [34,35].
Aside from ROR-γt, what are the extrinsic and intrinsic factors required for the differentiation of NKT17 cells? During development in the thymus, it has been reported that TGF-β signaling is required for survival of NKT17 cells [36]. Here it was shown that NKT17 cells have higher surface expression of TBFβRII as well as phosphorylation of SMAD2/3, proteins known to be downstream of the TGF-β signaling pathway. Loss of SMAD4 during development led to a significant reduction in NKT17 cells due to increased cell death as these cells developed in the thymus [36]. Moreover, NKT17 cells express the surface protein Neuropilin-1 (Nrp-1) [37], a receptor for TGF-β [38]. Thus, it is likely that TGF-β signaling, either through its own receptor or via interaction with Nrp-1, is required for differentiation of NKT17 cells in the thymus.
The transcription factor ZBTB7B (also called Th-POK and cKrox) was recently shown to negatively regulate differentiation of the NKT17 population. With loss of ZBTB7B expression, while overall numbers of iNKT cells remained unaffected, the majority of iNKT cells express ROR-γt and produce IL-17A, essentially differentiating into NKT17 cell [39,40]. Given that these cells bear all the hallmarks of NKT17 cells including expression of ROR-γt, Nrp-1, CCR6 and Ahr, these data suggest NKT cell differentiation is exquisitely sensitive to ZBTB7B and that low levels of this transcription factor are required to permit NKT17 cell development [39,40].
Recent data delineating the role of the transcriptional regulators Id2 and Id3 in NKT cell lineage differentiation showed that both Id2 and Id3 expression was low/intermediate in NKT17 cells [20] (Fig. 1). Given that Id2 and Id3 negatively regulate the E protein transcription factors, it is possible that E proteins may promote development of the NKT17 sublineage. Indeed, in a model of sustained E protein activity, an increase in the frequency of NKT17 cells (as well as NKT2 cells) was observed [21]. However, loss of Id3 alone did not lead to a significant increase in NKT17 cells [20]. Thus it is more likely that NKT17 cell differentiation is coordinated by a complex interplay of transcription factors including ROR-γt, ZBTB7B and E proteins.
4. NKTFH cells
Recent data have provided evidence that NKT cell interaction with B cells during the course of infection induces differentiation of a unique subset of NKT cells now known as NKTFH (follicular helper) cells [41,42]. It was previously known that NKT cells could directly and indirectly interact with B cells during the course of infection, inducing a robust antibody response as a result [43–45]. These recent data however, extend the initial findings to show that during the course of their interaction with B cells, NKT cells can upregulate the transcriptional regulator Bcl-6, the definitive transcription factor associated with TFH cells. Both initial studies coupled α-GalCer to a B cell specific antigen: hen egg lysozyme in the case of Chang et al. and 4-hydroxy-3-nitrophenylacetate (NP) or NP linked to keyhole limpet hemocyanin in the case of King et al. In response to these antigens, NKT cells became activated and upregulated PD-1 and CXCR5, typical hallmarks of TFH cells [41,42]. Both groups went on to show the activated NKT cells upregulated the transcriptional repressor Bcl-6 and that IL-21 production by the NKTFH cells was necessary for efficient germinal center formation, B cell proliferation and antibody secretion by responding B cells [41,42]. Surprisingly, while NKTFH cells elicited a robust primary B cell response, they were not effective at initiating long term B cell memory, giving rise to the idea that NKTFH help is ‘fast but does not last’ [41,42,46].
This idea that NKTFH cells induce a B cell response that does not induce B cell memory formation was recently challenged, although the antigens tested varied from those used in the original reports [47]. In this study, nanoparticles expressing synthetic lipid as well as polysaccharide antigens from the pathogen S. pneumoniae could induce not only a robust primary response from both NKT cells and B cells but could also induce long-lasting B cell memory response to subsequent infection [47]. Furthermore, it was shown that NKT cells first interacted with dendritic cells (DCs), which induced activation of the NKT cells before interacting with B cells to induce affinity maturation, isotype switching and finally robust B cell memory [47]. Thus it appears that this subset of NKT cells, the NKTFH cells, represent a sublineage of cells that differentiate in response to infection and represent not only a first line of protection from disease but also a way to potentially influence vaccine design and outcome.
5. NKT10 regulatory cells
Once activated with a strong stimulus through their TCR, iNKT cells were shown to undergo what was initially termed iNKT cell ‘anergy’, a differentiation step resulting in unresponsiveness, lack of proliferation and an inability to produce IFN-γ upon restimulation [48]. In particular, alpha-galactosylceramide (α-GalCer), delivers a strong TCR stimulus resulting in iNKT cell ‘anergy’ [48,49]. Use of α-GalCer is currently being investigated in a number of clinical trials, however given the induction of iNKT cell unresponsiveness, the efficacy of such a strategy is called into question [50,51]. Similarly, iNKT cell unresponsiveness has been described in the context of microbial infection. Here, upon infection of mice with Mycobacterium bovis, the iNKT cell response became blunted to restimulation during the course of the primary infection [52]. It was postulated that while iNKT cells participate in the initial response to infection, their contraction and unresponsiveness as the infection proceeds, enables the adaptive immune response to ‘take over’ and eventually clear the infection [52].
Recently, the ‘anergic’ phenotype itself has been called into question [2]. Sag et al. showed that iNKT cells previously stimulated with α-GalCer divide more quickly than unstimulated iNKT cells. Furthermore, these cells remained cytotoxic and could respond to restimulation. Perhaps most interesting was the discovery that so-called ‘anergic’ iNKT cells had properties indicative of regulatory T cells including increased expression of CTLA4, Nrp-1 and folate receptor 4 (FR4) as well as constitutive IL-10 expression, prompting the authors to rename these cells NKT10 cells [2] (Fig. 1). Surprisingly, NKT10 cells could be identified in the adipose tissue of unstimulated mice as well as in human peripheral blood. Moreover, NKT10 cells were detrimental in anti-tumor response to B16 melanoma and helped control disease in Experimental Autoimmune Encephalomyelitis (EAE), a mouse model of multiple sclerosis [2].
The identification of this new subset of iNKT cells raises certain questions. It is not yet clear if this subset develops in the thymus and expands upon stimulation, or if this subset differentiates from existing subsets of NKT cells such as NKT1, NKT2 and NKT17 cells. Similarly, the relationship between NKT10 and NKTFH cells is unclear. Certainly, iNKT cells upregulate Bcl-6 expression on day 6 post-stimulation with α-GalCer but at later time points it was not immediately obvious if the NKTFH cells population converted to NKT10 cells or if the NKT10 cells represent proliferation of endogenous NKT10 cells [2]. Moreover, the molecular mechanisms regulating NKT10 cell development and differentiation are not yet known. Recent data from studies of effector CD8+ T cells, indicate the E protein E2A regulates IL-10 expression perhaps in collaboration with IRF4 [53]. Our unpublished data indicate Id2 expression is downregulated with strong TCR stimulus (Stradner, personnel communication). It is possible that E proteins not only regulate the early stages of iNKT cell development but also regulate differentiation into the NKT10 lineage. Future work in this area will no doubt clarify the role of E protein transcription factors in the regulation of NKT10 cell differentiation.
Identification of the NKT10 subset does however provide some answers to the anti-inflammatory role attributed to iNKT cells in various disease settings. In an allogenic skin transplantation model, repeated activation of iNKT cells using α-GalCer resulted in reduced transplant rejection [54]. Most noteworthy was that the authors identified high IL-10 mRNA expression associated with the post-activated host iNKT cells, perhaps providing early insight into the NKT10 sublineage [54]. Identification of endogenous NKT10 cells also helps explain the regulatory role attributed to iNKT cells found in the subcutaneous adipose tissue [55–58]. Here activation of iNKT cells isolated from the subcutaneous adipose tissue of healthy mice, led to increased IL-10 and IL-4 production which in turn promoted suppressive M2 macrophage expansion [55–58]. Thus, naturally occurring NKT10 cells may play a tolerogenic role in the maintenance of healthy adipose tissue.
6. Foxp3+ iNKT cells
Although NKT10 cells do not express Foxp3, two groups previously showed that iNKT cells could be induced to express Foxp3 in certain settings [2,59,60]. Under EAE-inducing conditions with addition of α-GalCer stimulation, iNKT cells expressing Foxp3 could be detected in draining lymph nodes from the CNS [59]. Moreover, both human and mouse iNKT cells stimulated in vitro with TGF-β can be induced to express Foxp3 [59,60]. Similarly, using iNKT cell human clones isolated from PBMCs, Foxp3 mRNA and protein expression could be detected at varying levels [61]. In mice, induced Foxp3+ iNKT cells maintained expression of the iNKT cell-associated transcription factor PLZF, but also expressed markers associated with regulatory T (Treg) cells including CTLA4, CD25 and GITR [59]. In contrast, human iNKT cells cultured with TGF-β upregulated CD25 but not CTLA4 or GITR, indicating an unconventional Treg phenotype [60]. Unlike conventional Tregs, which home to the peripheral LNs, Foxp3+ iNKT cells preferentially homed to the liver [59]. Interestingly, Foxp3+ iNKT cells from humans could only suppress proliferation of conventional T cells when generated in vitro in the presence of both TGF-β and rapamycin [60]. Future work will likely illuminate if these suppressive iNKT cells can be identified in healthy individuals and what their role might be in regulation of tolerance and autoimmune disease.
7. Conclusions
iNKT cells represent a diverse population of lymphocytes, capable of responding directly to cognate glycolipids or indirectly to cytokine stimulation, resulting in release of a unique cohort of cytokines depending on the lineage activated and the site of activation. iNKT cells can elicit a strong response to the synthetic glycolipid αGalCer, which has been tested in a variety of clinical trials, from non-small cell lung cancer treatment to hepatitis B [62,63]. New studies have shown that treatment with αGalCer can result in differentiation of an immunomodulatory lineage called NKT10 cells and so it is critical to determine the role iNKT cells play in response to treatment with this molecule for the success of these and future clinical trials.
NKT1 and NKT2 cells, upon release of IFNγ and IL-4, respectively, have demonstrated their function as key players in the first line of defense in disease. Meanwhile, NKT17 cells, located primarily in peripheral lymph nodes and skin, represent a tissue specific component to the iNKT response. As suggested above, the regulatory role of iNKT cells is becoming clearer with the discovery of NKT10 cells that release IL-10 in response to re-stimulation with αGalCer. Further, interactions between NKTFH cells and B cells, represent an increase in understanding of the diversity and versatility of this important cell type. Still unclear at present are the origins of NKT10 and NKTFH cells: they could independently originate from NKT1 cells (as these cells essentially disappear after stimulation through the TCR). Another possibility is that NKTFH cells differentiate from NKT1 cells before transitioning to NKT10 cells. A third intriguing possibility is that both populations arise from an undefined precursor pool. No doubt future work will endeavor to elucidate the lineage differentiation of these subsets.
Id2, Id3 and E protein transcription factors are iNKT cell regulators that, in the future, could be manipulated to alter the makeup of NKT lineages, further fine tuning the immune response of this population of cells to help in the ongoing fight against a variety of pathogens and human diseases.
Acknowledgments
This work was supported by the University of Pittsburgh Immunology Department and R00AI097286 to L.M.D. We thank members of the D’Cruz group for critical review of the manuscript.
References
- 1.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 2.Sag D, et al. IL-10-producing NKT10 cells are a distinct regulatory invariant NKT cell subset. J Clin Invest. 2014;124(9):3725–40. doi: 10.1172/JCI72308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brigl M, et al. Mechanism of CD1d-restricted natural killer T cell activation during microbial infection. Nat Immunol. 2003;4(12):1230–7. doi: 10.1038/ni1002. [DOI] [PubMed] [Google Scholar]
- 4.Fox L, Hegde S, Gumperz JE. Natural killer T cells: innate lymphocytes positioned as a bridge between acute and chronic inflammation? Microbes Infect. 2010;12(14–15):1125–33. doi: 10.1016/j.micinf.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mattner J, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434(7032):525–9. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 6.Kronenberg M, Kinjo Y. Innate-like recognition of microbes by invariant natural killer T cells. Curr Opin Immunol. 2009;21(4):391–6. doi: 10.1016/j.coi.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Motohashi S, Nakayama T. Invariant natural killer T cell-based immunotherapy for cancer. Immunotherapy. 2009;1(1):73–82. doi: 10.2217/1750743X.1.1.73. [DOI] [PubMed] [Google Scholar]
- 8.Van Kaer L, Parekh VV, Wu L. Invariant NK T cells: potential for immunotherapeutic targeting with glycolipid antigens. Immunotherapy. 2011;3(1):59–75. doi: 10.2217/imt.10.85. [DOI] [PubMed] [Google Scholar]
- 9.Wu L, Van Kaer L. Natural killer T cells and autoimmune disease. Curr Mol Med. 2009;9(1):4–14. doi: 10.2174/156652409787314534. [DOI] [PubMed] [Google Scholar]
- 10.Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol. 2013;13(2):101–17. doi: 10.1038/nri3369. [DOI] [PubMed] [Google Scholar]
- 11.Constantinides MG, Bendelac A. Transcriptional regulation of the NKT cell lineage. Curr Opin Immunol. 2013;25(2):161–7. doi: 10.1016/j.coi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doisne JM, et al. Skin and peripheral lymph node invariant NKT cells are mainly retinoic acid receptor-related orphan receptor (γ)t+ and respond preferentially under inflammatory conditions. J Immunol. 2009;183(3):2142–9. doi: 10.4049/jimmunol.0901059. [DOI] [PubMed] [Google Scholar]
- 13.Lee YJ, et al. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. 2013;14(11):1146–54. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michel ML, et al. Identification of an IL-17-producing NK1. 1(neg) iNKT cell population involved in airway neutrophilia. J Exp Med. 2007;204(5):995–1001. doi: 10.1084/jem.20061551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watarai H, et al. Development and function of invariant natural killer T cells producing T(h)2- and T(h)17-cytokines. PLoS Biol. 2012;10(2):e1001255. doi: 10.1371/journal.pbio.1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Townsend MJ, et al. T-bet regulates the terminal maturation and homeostasis of NK and Vα14i NKT cells. Immunity. 2004;20(4):477–94. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 17.Matsuda JL, et al. Homeostasis of Vα14i NKT cells. Nat Immunol. 2002;3(10):966–74. doi: 10.1038/ni837. [DOI] [PubMed] [Google Scholar]
- 18.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol. 2005;6(11):1079–86. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 19.Monticelli LA, et al. Transcriptional regulator Id2 controls survival of hepatic NKT cells. Proc Natl Acad Sci USA. 2009;106(46):19461–6. doi: 10.1073/pnas.0908249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D’Cruz LM, et al. E and Id proteins influence invariant NKT cell sublineage differentiation and proliferation. J Immunol. 2014;192(5):2227–36. doi: 10.4049/jimmunol.1302904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu T, et al. Increased level of E protein activity during invariant NKT development promotes differentiation of invariant NKT2 and invariant NKT17 subsets. J Immunol. 2013;191(10):5065–73. doi: 10.4049/jimmunol.1301546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terashima A, et al. A novel subset of mouse NKT cells bearing the IL-17 receptor B responds to IL-25 and contributes to airway hyperreactivity. J Exp Med. 2008;205(12):2727–33. doi: 10.1084/jem.20080698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim PJ, et al. GATA-3 regulates the development and function of invariant NKT cells. J Immunol. 2006;177(10):6650–9. doi: 10.4049/jimmunol.177.10.6650. [DOI] [PubMed] [Google Scholar]
- 24.Verykokakis M, et al. Essential functions for ID proteins at multiple checkpoints in invariant NKT cell development. J Immunol. 2013;191(12):5973–83. doi: 10.4049/jimmunol.1301521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitz R, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490(7418):116–20. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D’Cruz LM, et al. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat Immunol. 2010;11(3):240–9. doi: 10.1038/ni.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang CY, et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol. 2011;12(12):1221–9. doi: 10.1038/ni.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seiler MP, et al. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat Immunol. 2012;13(3):264–71. doi: 10.1038/ni.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coquet JM, et al. Diverse cytokine production by NKT cell subsets and identification of an IL-17-producing CD4-NK1. 1- NKT cell population. Proc Natl Acad Sci USA. 2008;105(32):11287–92. doi: 10.1073/pnas.0801631105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michel ML, et al. Critical role of ROR-γt in a new thymic pathway leading to IL-17-producing invariant NKT cell differentiation. Proc Natl Acad Sci USA. 2008;105(50):19845–50. doi: 10.1073/pnas.0806472105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rachitskaya AV, et al. Cutting edge: NKT cells constitutively express IL-23 receptor and RORγt and rapidly produce IL-17 upon receptor ligation in an IL-6-independent fashion. J Immunol. 2008;180(8):5167–71. doi: 10.4049/jimmunol.180.8.5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pichavant M, et al. Ozone exposure in a mouse model induces airway hyperreactivity that requires the presence of natural killer T cells and IL-17. J Exp Med. 2008;205(2):385–93. doi: 10.1084/jem.20071507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshiga Y, et al. Invariant NKT cells produce IL-17 through IL-23-dependent and -independent pathways with potential modulation of Th17 response in collagen-induced arthritis. Int J Mol Med. 2008;22(3):369–74. [PubMed] [Google Scholar]
- 34.Doisne JM, et al. Cutting edge: crucial role of IL-1 and IL-23 in the innate IL-17 response of peripheral lymph node NK1. 1- invariant NKT cells to bacteria. J Immunol. 2011;186(2):662–6. doi: 10.4049/jimmunol.1002725. [DOI] [PubMed] [Google Scholar]
- 35.Kinjo Y, et al. Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria. Nat Immunol. 2011;12(10):966–74. doi: 10.1038/ni.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Havenar-Daughton C, et al. Development and function of murine RORγt+ iNKT cells are under TGF-β signaling control. Blood. 2012;119(15):3486–94. doi: 10.1182/blood-2012-01-401604. [DOI] [PubMed] [Google Scholar]
- 37.Milpied P, et al. IL-17-producing invariant NKT cells in lymphoid organs are recent thymic emigrants identified by neuropilin-1 expression. Blood. 2011;118(11):2993–3002. doi: 10.1182/blood-2011-01-329268. [DOI] [PubMed] [Google Scholar]
- 38.Glinka Y, Prud’homme GJ. Neuropilin-1 is a receptor for transforming growth factor β-1, activates its latent form, and promotes regulatory T cell activity. J Leukoc Biol. 2008;84(1):302–10. doi: 10.1189/jlb.0208090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Enders A, et al. ZBTB7B (Th-POK) regulates the development of IL-17-producing CD1d-restricted mouse NKT cells. J Immunol. 2012;189(11):5240–9. doi: 10.4049/jimmunol.1201486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engel I, et al. The transcription factor Th-POK negatively regulates Th17 differentiation in Vα14i NKT cells. Blood. 2012;120(23):4524–32. doi: 10.1182/blood-2012-01-406280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang PP, et al. Identification of Bcl-6-dependent follicular helper NKT cells that provide cognate help for B cell responses. Nat Immunol. 2012;13(1):35–43. doi: 10.1038/ni.2166. [DOI] [PubMed] [Google Scholar]
- 42.King IL, et al. Invariant natural killer T cells direct B cell responses to cognate lipid antigen in an IL-21-dependent manner. Nat Immunol. 2012;13(1):44–50. doi: 10.1038/ni.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barral P, et al. B cell receptor-mediated uptake of CD1d-restricted antigen augments antibody responses by recruiting invariant NKT cell help in vivo. Proc Natl Acad Sci USA. 2008;105(24):8345–50. doi: 10.1073/pnas.0802968105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leadbetter EA, et al. NK T cells provide lipid antigen-specific cognate help for B cells. Proc Natl Acad Sci USA. 2008;105(24):8339–44. doi: 10.1073/pnas.0801375105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tonti E, et al. NKT-cell help to B lymphocytes can occur independently of cognate interaction. Blood. 2009;113(2):370–6. doi: 10.1182/blood-2008-06-166249. [DOI] [PubMed] [Google Scholar]
- 46.Lehuen A, Fazilleau N. Innate iNKT cell help to B cells: fast but does not last. Nat Immunol. 2012;13(1):11–3. doi: 10.1038/ni.2186. [DOI] [PubMed] [Google Scholar]
- 47.Bai L, et al. Natural killer T (NKT)-B-cell interactions promote prolonged antibody responses and long-term memory to pneumococcal capsular polysaccharides. Proc Natl Acad Sci USA. 2013;110(40):16097–102. doi: 10.1073/pnas.1303218110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parekh VV, et al. Glycolipid antigen induces long-term natural killer T cell anergy in mice. J Clin Invest. 2005;115(9):2572–83. doi: 10.1172/JCI24762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sullivan BA, Kronenberg M. Activation or anergy: NKT cells are stunned by α-galactosylceramide. J Clin Invest. 2005;115(9):2328–9. doi: 10.1172/JCI26297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cerundolo V, et al. Harnessing invariant NKT cells in vaccination strategies. Nat Rev Immunol. 2009;9(1):28–38. doi: 10.1038/nri2451. [DOI] [PubMed] [Google Scholar]
- 51.Salio M, et al. Biology of CD1- and MR1-restricted T cells. Annu Rev Immunol. 2014;32:323–66. doi: 10.1146/annurev-immunol-032713-120243. [DOI] [PubMed] [Google Scholar]
- 52.Chiba A, et al. Rapid NKT cell responses are self-terminating during the course of microbial infection. J Immunol. 2008;181(4):2292–302. doi: 10.4049/jimmunol.181.4.2292. [DOI] [PubMed] [Google Scholar]
- 53.Masson F, et al. Id2 represses E2A-mediated activation of IL-10 expression in T cells. Blood. 2014;123(22):3420–8. doi: 10.1182/blood-2014-03-561456. [DOI] [PubMed] [Google Scholar]
- 54.Oh K, et al. Direct regulatory role of NKT cells in allogeneic graft survival is dependent on the quantitative strength of antigenicity. J Immunol. 2005;174(4):2030–6. doi: 10.4049/jimmunol.174.4.2030. [DOI] [PubMed] [Google Scholar]
- 55.Ji Y, et al. Short term high fat diet challenge promotes alternative macrophage polarization in adipose tissue via natural killer T cells and interleukin-4. J Biol Chem. 2012;287(29):24378–86. doi: 10.1074/jbc.M112.371807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ji Y, et al. Activation of natural killer T cells promotes M2 Macrophage polarization in adipose tissue and improves systemic glucose tolerance via interleukin-4 (IL-4)/STAT6 protein signaling axis in obesity. J Biol Chem. 2012;287(17):13561–71. doi: 10.1074/jbc.M112.350066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lynch L, et al. Adipose tissue invariant NKT cells protect against diet-induced obesity and metabolic disorder through regulatory cytokine production. Immunity. 2012;37(3):574–87. doi: 10.1016/j.immuni.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huh JY, et al. A novel function of adipocytes in lipid antigen presentation to iNKT cells. Mol Cell Biol. 2013;33(2):328–39. doi: 10.1128/MCB.00552-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Monteiro M, et al. Identification of regulatory Foxp3+ invariant NKT cells induced by TGF-β. J Immunol. 2010;185(4):2157–63. doi: 10.4049/jimmunol.1000359. [DOI] [PubMed] [Google Scholar]
- 60.Moreira-Teixeira L, et al. Rapamycin combined with TGF-β converts human invariant NKT cells into suppressive Foxp3+ regulatory cells. J Immunol. 2012;188(2):624–31. doi: 10.4049/jimmunol.1102281. [DOI] [PubMed] [Google Scholar]
- 61.Engelmann P, et al. Characterization of human invariant natural killer T cells expressing FoxP3. Int Immunol. 2011;23(8):473–84. doi: 10.1093/intimm/dxr040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Motohashi S, et al. A phase I-II study of α-galactosylceramide-pulsed IL-2/GM-CSF- cultured peripheral blood mononuclear cells in patients with advanced and recurrent non-small cell lung cancer. J Immunol. 2009;182(4):2492–501. doi: 10.4049/jimmunol.0800126. [DOI] [PubMed] [Google Scholar]
- 63.Woltman AM, et al. Alpha-galactosylceramide in chronic hepatitis B infection: results from a randomized placebo-controlled Phase I/II trial. Antivir Ther. 2009;14(6):809–18. doi: 10.3851/IMP1295. [DOI] [PubMed] [Google Scholar]