Abstract
Dramatic genome dynamics, such as chromosome instability, contribute to the remarkable genomic heterogeneity among the blastomeres comprising a single embryo during human preimplantation development. This heterogeneity, when compatible with life, manifests as constitutional mosaicism, chimerism, and mixoploidy in live-born individuals. Chimerism and mixoploidy are defined by the presence of cell lineages with different parental genomes or different ploidy states in a single individual, respectively. Our knowledge of their mechanistic origin results from indirect observations, often when the cell lineages have been subject to rigorous selective pressure during development. Here, we applied haplarithmisis to infer the haplotypes and the copy number of parental genomes in 116 single blastomeres comprising entire preimplantation bovine embryos (n = 23) following in vitro fertilization. We not only demonstrate that chromosome instability is conserved between bovine and human cleavage embryos, but we also discovered that zygotes can spontaneously segregate entire parental genomes into different cell lineages during the first post-zygotic cleavage division. Parental genome segregation was not exclusively triggered by abnormal fertilizations leading to triploid zygotes, but also normally fertilized zygotes can spontaneously segregate entire parental genomes into different cell lineages during cleavage of the zygote. We coin the term “heterogoneic division” to indicate the events leading to noncanonical zygotic cytokinesis, segregating the parental genomes into distinct cell lineages. Persistence of those cell lines during development is a likely cause of chimerism and mixoploidy in mammals.
A variety of post-zygotic events—including mitotic non-disjunction, anaphase lagging, breakage–fusion–bridge cycles, centric fission, and the fusion of chromosome-containing cellular fragments with blastomeres—can contribute to the generation of mosaic genomic architectures in early embryogenesis (Vanneste et al. 2009; Voet et al. 2011a,b; Chavez et al. 2012; Zamani Esteki et al. 2015). This chromosomal instability may lead to mosaicism, which persists during embryonic development causing chromosomal mosaicism in the placenta, the fetus, or the live-born. Human conceptuses are often burdened with developmental anomalies (Campbell et al. 2014).
A peculiar form of mosaicism comprises the presence of cells with different parental genomic constitutions within a single individual, called chimeric or mixoploid individuals. According to the accepted theoretical model, chimeric individuals are the developmental outcome of the fusion of two different zygotes into a single embryo, and hence are also known as tetragametic chimeras (Yu et al. 2002; Yunis et al. 2007). In addition to tetragametic chimeras, also parthenogenetic/gynogenetic and androgenetic chimeras exist. Parthenogenesis refers to asexual reproduction, whereby offspring results from an unfertilized oocyte undergoing mitotic divisions, and gynogenesis indicates the development of an embryo with only maternal DNA due to activation of the egg by a sperm that does not unite with the egg's nucleus. Although parthenotes/gynogenotes are not viable in humans, several cases of mosaic individuals, exhibiting a mixture of aberrant diploid cells with only maternal DNA and normal diploid biparental cells do exist (Strain et al. 1995; Giltay et al. 1998; Yamazawa et al. 2010; Xia et al. 2014). Those individuals are usually ascertained by cytogenetic and molecular marker analysis, as they for instance may be afflicted with disorders of sex development caused by the presence of 46,XX and 46,XY cell lineages. Similar to these parthenogenetic/gynogenetic chimeras, androgenetic chimeras are individuals comprised of cells with only paternally derived genomes and cells that are normal diploid. Clinical manifestations of human androgenetic chimerism include placental mesenchymal dysplasia (PMD) (Kaiser-Rogers et al. 2006), complete hydatidiform moles (CHM) (Ford et al. 1986; Weaver et al. 2000), and partial hydatidiform moles (Surti et al. 2005; Kaiser-Rogers et al. 2006).
A variety of theoretical models have been invoked to explain the origin of parthenogenetic/gynogenetic and androgenetic chimeras (Strain et al. 1995; Giltay et al. 1998; Robinson et al. 2007), which however remain largely unvalidated because they rely on genome analyses of live-born individuals. As a consequence, the cells containing the signature genomes that may illuminate the mechanism(s) of chimerism may be eradicated early during prenatal development by selection.
Mixoploidy refers to individuals harboring cell lines with different ploidy states. Most described mixoploids contain a mixture of triploid and diploid cells (van de Laar et al. 2002; Boonen et al. 2011). Similar to chimeras, the origin of mixoploids is not well known, including speculation on fusion of a diploid with a triploid zygote (van de Laar et al. 2002), fusion of a polar body with a diploid blastomere (Quigley et al. 2005), or fertilization errors, e.g., dispermic fertilization, leading to a triploid conceptus that subsequently loses an entire haplotype of a parental genome in part of the cells (Uchida and Freeman 1985; Zaragoza et al. 2000). In addition, the frequency of mixoploidy following fertilization may be underestimated as most analyses are performed on postnatal tissue that has been subjected to developmental selection likely eliminating haploid or polyploid cell lineages early in development. Equally important in this context is the fact that conventional (single-cell) DNA copy-number analysis methods used for aneuploidy screening of in vitro fertilized (IVF) cleavage-stage embryos are unable to detect violations to ploidy states.
We have recently developed a novel methodology—termed haplarithmisis—enabling concurrent haplotyping and copy-number typing of single cells (Zamani Esteki et al. 2015). In this study, we applied haplarithmisis to investigate the genome-wide haplotype architecture of all single blastomeres comprising bovine cleavage-stage embryos following in vitro fertilization. We find that chromosome instability is conserved between human and bovine preimplantation embryos and, unexpectedly, disclose spontaneous segregations of entire parental genomes into distinct cell lineages, a phenomenon that we term “heterogoneic cell divisions” (Greek, meaning different parental origin). These heterogoneic cell divisions, occurring in the first cleavage division of the mammalian zygote, are the likely cradle of chimerism and mixoploidy.
Results
Single-cell haplarithmisis on bovine cleavage-stage embryos
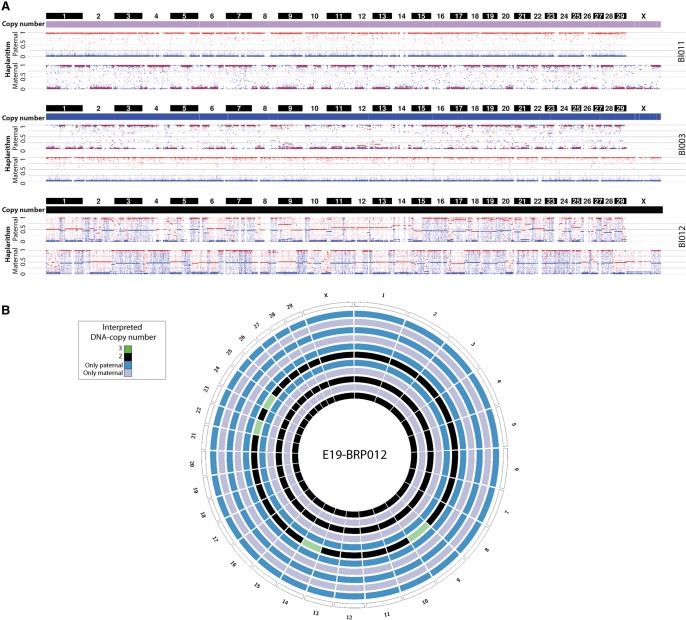
We genotyped about 777,000 uniformly spanned SNPs across the genomes of 160 single blastomeres derived from 25 in vitro–derived bovine cleavage-stage embryos, of which 72% (n = 116) were analyzed by haplarithmisis following quality control (Supplemental Table S1; Supplemental Fig. S1). For crosses BRP004 to BRP009, we applied haplarithmisis using the parents of the bull as a seed for paternal genotype phasing (option 1) (Methods). For BRP010 and BRP011, lacking grandparental DNA samples, expanded blastocysts (approximately 100–125 cells) were used as a seed for parental genotype phasing (option 2) (Methods); for BRP012, following an initial analysis, a single euploid blastomere from each of the two embryos was chosen as a seed (option 2) (Methods). The latter analysis option was also implemented in selected embryos from BRP004, BRP005, and BRP006 to obtain, apart from paternal, also maternal haplarithm profiles, illuminating the mechanistic origin of aberrations affecting the maternal lineage.
Chromosome instability is comparable in bovine and human cleavage-stage embryos following IVF
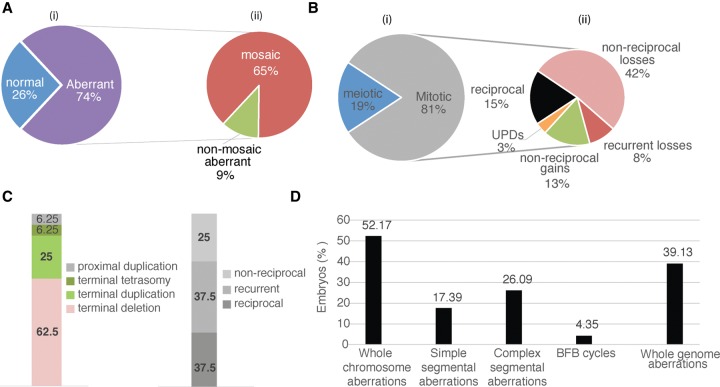
We first assessed whether chromosome instability is comparable between bovine and human IVF cleavage-stage embryos (Iwasaki and Nakahara 1990; Viuff et al. 2000; King 2008; Vanneste et al. 2009). To determine the genomic constitution of embryos, we included only those embryos of which more than half of the blastomeres passed our QC filters (Methods). Twenty-three of 25 embryos met this criterion. In our cohort, six embryos (26%) were normal diploid in all analyzed cells, and 17 (74%) were abnormal containing at least one blastomere with a segmental or a full chromosome aberration. Of the abnormal embryos, 15 (88%) were mosaic, including four mosaic normal/aberrant and 11 mosaic aberrant/aberrant embryos (Fig. 1; Supplemental Fig. S1B).
Figure 1.
Incidence and nature of full chromosome and segmental aberrations. (A) General characteristics of the embryos involved in this study, including (i) proportion of normal euploid and abnormal embryos, and (ii) of the abnormal embryos proportion of mosaic and nonmosaic embryos. (B) The proportion of different whole-chromosome imbalances, including (i) mechanistic origin of whole-chromosome abnormalities, and (ii) frequency and nature of the mitotic whole-chromosome anomalies. (C) The proportion of different natures of segmental imbalances. (D) The proportion of embryos with different genomic anomalies.
A staggering 39% of embryos (9/23) contain blastomeres with full genome anomalies, including the presence of either a single parental haplotype leading to genome-wide loss of heterozygosity in a blastomere, or polyploid blastomeres (e.g., two maternal genomes: one paternal genome) (Fig. 1D; Supplemental Fig. S1B). Based on the detected copies of the parental haplotypes in a blastomere, we could ascertain both the type and the origin of a blastomere's ploidy—e.g., diandric or digynic triploidy. Similarly, those blastomeres containing only paternal or maternal genomes could be detected and were termed androgenetic and gynogenetic, respectively.
Among the embryos with blastomeres affected by genome-wide anomalies, E20_BRP005 is a parthenote or a gynogenote, containing only one maternal haplotype genome wide in all analyzed blastomeres (Supplemental Fig. S1A). Interestingly, two of the eight blastomeres of E10_BRP011 (blastomeres Bl005 and Bl008) demonstrated a similar signature of scattered fragments of the paternal genome and no maternal inherited DNA, demonstrating that remnants of sperm (pro)nuclei can replicate and divide into two daughter cells (Supplemental Figs. S1, S3).
Incidence and nature of whole-chromosome imbalances
To calculate the incidence of whole-chromosome and segmental anomalies, we analyzed a total of 75 blastomeres, excluding those single blastomeres (n = 44) with a uniform genome-wide ploidy anomaly (cells with genome-wide ploidy anomalies having signatures of whole-chromosome or segmental aberrations were included) (Supplemental Fig. S1A; Supplemental Table S2).
We detected 92 whole-chromosome imbalances, including losses (monosomies and nullisomies), gains (trisomies, tetrasomies, and amplifications), and uniparental disomies (UPDs), at a frequency of 66%, 30%, and 3% of all numerical haplotype anomalies of entire chromosomes, respectively.
The majority of these whole-chromosome imbalances are mitotic in origin (81%) (Fig. 1B; Supplemental Fig. S2). Whole-chromosome losses, which had a reciprocal duplication of the same parental chromosome in a sister blastomere represented 15% of the mitotic numerical chromosome anomalies. The majority of the mitotic chromosome losses are, however, neither reciprocal nor recurrent (42%) in sister blastomeres. The latter is likely due to merotelic attachments of microtubules to kinetochores leading to chromosome anaphase lagging and exclusion of one chromatid from one of both sister blastomeres. Whole-chromosome gains entail trisomies, tetrasomies, and amplifications (more than four copies), which represent 26%, 3%, and 1% of all mitotic numerical chromosome anomalies, respectively. Nonreciprocal gains account for 13%—most likely, these have a reciprocal loss in a sister blastomere that failed to be included in the analysis.
An exception to this assumption involves a tetrasomy of Chr 4 in one blastomere of a four-cell-stage embryo E03_BRP010 of which all blastomeres were analyzed (Supplemental Fig. S2). The maternal haplarithm profile for this autosome indicates the presence of a maternal haplotype, which is different from the one detected in the remaining three blastomeres comprising the embryo (Supplemental Fig. S2). This indicates that the anomaly may have arisen due to a maternal meiotic error followed by a post-fertilization mitotic error restoring the euploid number of chromosomes.
Meiotic losses, characterized by monosomies, were detected in two embryos (8.7%; embryos E03_BRP010 and E08_BRP011), whereas meiotic trisomy was detected only in embryo E18_BRP004 for three chromosomes (Chr 8, Chr 20, and Chr 24) in all analyzed blastomeres. The haplarithms for the trisomies demonstrate a signature typical for a maternal meiosis II non-disjunction error. In addition, haplarithmisis revealed two maternal UPDs for embryo E12_BRP010 on Chr 7 (Bl006 and Bl007), and a paternal UPD for Chr 7 was also detected in embryo E19_BRP005 (Bl006) (Supplemental Table S2; Supplemental Figs. S1A, S2).
In conclusion, haplarithmisis discloses the diversity of whole-chromosome anomalies, of which some would be missed by conventional single-cell DNA copy number profiling methods.
Incidence of segmental chromosome aberrations
We observed 30 segmental imbalances (Supplemental Table S2; Supplemental Fig. S1A). Terminal deletions, duplications, UPDs, or amplifications in isolation were termed simple segmental aberrations, whereas terminal imbalances coexisting with other segmental aberration(s) for the remaining part of the same chromosome were labeled complex.
Simple segmental copy number imbalances represent 53% of the detected segmental abnormalities. Of the simple segmental anomalies, 62.5% are terminal deletions, 25% terminal duplications, 6.25% terminal tetrasomies, and 6.25% involve a proximal duplication (Fig. 1C). Of the simple segmental aberrations, 37.5% are reciprocal—i.e., a terminal deletion in one blastomere has a reciprocal gain with the same breakpoints on the same parental allele in a sister blastomere. Recurrent segmental imbalances (37.5%) entail only terminal losses.
Complex aberrations represent 47% of all segmental aberrations. Signatures characteristic for deletion–inversion–duplication chromosomes were detected in three embryos (13%), e.g., Chr 20 of E08-BRP011 (Fig. 2), which may spark breakage–fusion–bridge (BFB) cycles (e.g., Chr 1 in Embryo E13_BRP006) (Supplemental Fig. S1B).
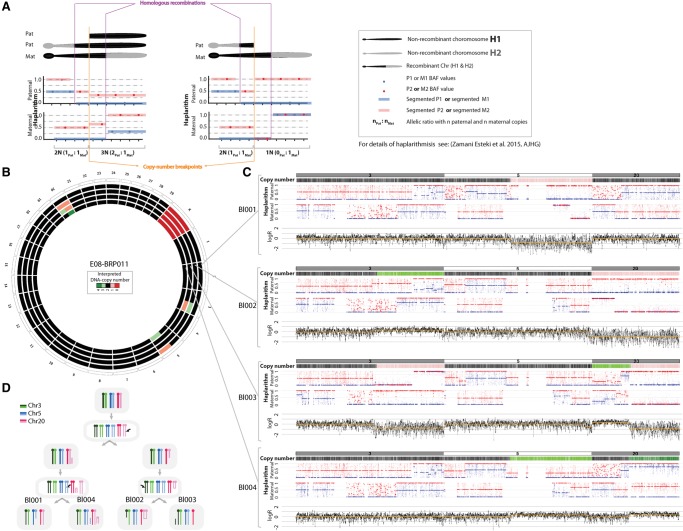
Figure 2.
Haplarithmisis unveils segmental imbalances across the genome. (A) Expected haplarithm patterns for segmental duplication (left) and deletion (right) of paternal in origin. For a detailed overview of haplarithmisis, see Zamani Esteki et al. (2015). (B) A Circos plot representing interpreted copy number imbalances for each cell. (C) siCHILD-determined profiles for Chr 3, Chr 5, and Chr 20. In each panel from top to bottom, respectively, we depict the interpreted copy-number profile, the paternal and maternal haplarithms, and the normalized logR-values. (D) A model illustrating how the detected segmental aberrations may arise.
In summary, chromosome instability is prominent in bovine cleavage-stage embryo following IVF, and a similar frequency and nature of whole-chromosome and segmental DNA anomalies have been observed in human cleavage-stage embryos following IVF (Vanneste et al. 2009; Johnson et al. 2010; Voet et al. 2011b; Mertzanidou et al. 2013; Zamani Esteki et al. 2015). This suggests that the bovine embryo is a valuable model for studying the mechanisms underlying genome dynamics during early embryonic life.
Parental genomes segregate into androgenetic and gynogenetic cell lineages with different ploidy levels in abnormally fertilized embryos
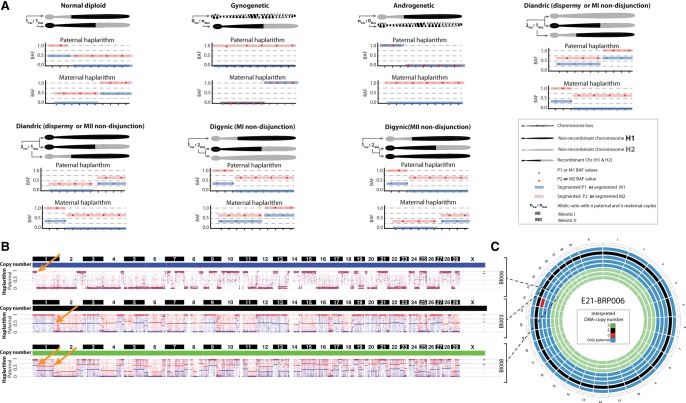
Haplarithmisis disclosed aberrant inheritance patterns of parental haplotypes to which conventional genome analysis methods are blind (Fig. 3A). In five embryos (22%; n = 23), the paternal haplarithm profiles revealed dispermic fertilization as two different paternal haplotypes with distinct homologous recombination sites were detected in the same cell (Fig. 3A–C). For instance, in embryo E21_BRP006, blastomere Bl008 carries two different paternal haplotypes genome wide, whereas blastomeres Bl003 and Bl006 carry either of these two paternal haplotypes (Fig. 3B). Within the same cells, haplarithmisis revealed that Bl008 is triploid, Bl003 is diploid, and Bl006 is an androgenetic cell carrying exclusively paternal DNA.
Figure 3.
Haplarithmisis reveals abnormal fertilization (dispermy) and segregation of parental genomes in distinct cell lineages. (A) Schematic overview of haplarithm pattern for different genomic constitutions. The segmented M1, M2, P1, and P2 single-cell BAF values, as well as the distances between “M1 and M2” and “P1 and P2” values in maternal and paternal haplarithms, respectively, and the positioning of breakpoints, i.e., homologous recombinations (HR), represent inheritance of different parental homologous chromosomes in a single blastomere (for details, see Zamani Esteki et al. 2015). Note that for simplicity's sake, we show only one chromosome, and each pattern should be seen in all (or the majority) of chromosomes to be considered as a genome-wide event. (B) Genome-wide paternal haplarithms of three blastomeres derived from the same embryo (E21_BRP006) uncover different HR-sites in two cells (top and middle) and a combination thereof in the other cell (bottom). The orange arrows point to HR sites on Chr 1. Note that here, option 1 of siCHILD (see Methods) was applied as the paternal grandparents were used for phasing the paternal genome. (C) Circos plot illustrating the genome-wide interpreted copy-number profiles of all the single blastomeres derived from embryo E21_BRP006.
Surprisingly, none of the dispermic embryos were uniformly triploid. Instead, they were mixoploid, demonstrating cells with different numerical ploidy states and different parental genome contributions across the blastomeres of the same embryo (Supplemental Figs. S1B, S2, S4). In all dispermic embryos, we observed that one of the two paternal genomes always segregated in a distinct cell lineage—a process we term heterogoneic divisions—giving rise to androgenetic blastomeres. Of the five dispermic embryos, four had triploid cell lineages. In E17_BRP011, which does not exhibit triploid cells, a biparental cell and an androgenetic cell lineage presented with two different paternal genomes (Supplemental Figs. S2, S4C). However, since four of seven blastomeres could not be analyzed, the possibility for a triploid lineage could not be excluded.
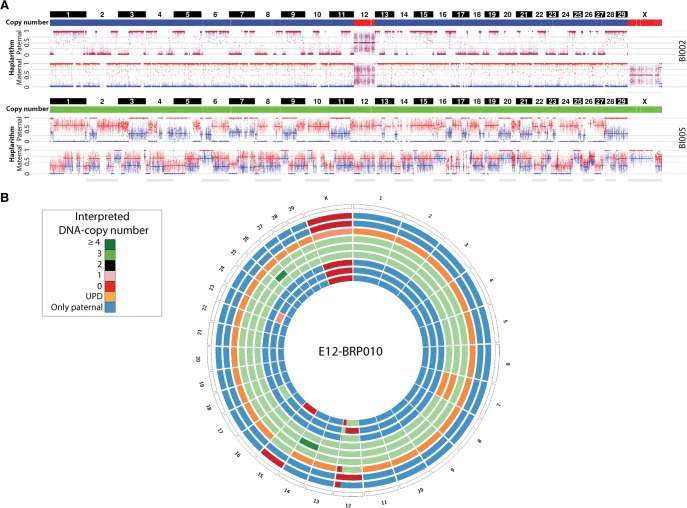
Triploid blastomeres containing both paternal genomes—i.e., triploid diandric cell lineages—were detected in three of those four dispermic embryos (embryos E21_BRP006, E16_BRP011, E04_BRP010), whereas E12_BRP010 demonstrates haplarithms for digynic triploid cells (e.g., blastomere Bl005), showing an additional set of maternal alleles genome wide, possibly due to nonextrusion of the second polar body (PB2) or reabsorption of the latter into the zygote or a blastomere (Fig. 4). Interestingly, in the same embryo, which is both dispermic and digynic, the androgenetic cells had extra whole-chromosome abnormalities affecting Chr 12, Chr 15, and Chr X, and segmental anomalies on Chr 12 (Fig. 4; Supplemental Figs. S2, S4B). Since these anomalies are an exclusive feature of the androgenetic cell line, this corroborates a model in which those blastomeres arose from a common progenitor.
Figure 4.
Segregation of the parental genomes in distinct cell lineages following dispermic fertilization. (A) Genome-wide paternal and maternal haplarithms reveal dispermic fertilization and maternal MII triploidy (consult also the digynic haplarithm profile in Fig. 3A). Blastomere 2 (top) is androgenetic. This paternal genome is absent from the triploid blastomere (bottom, Bl005) as different paternal genome-wide HR-sites were detected in this cell. Note that here, option 2 of siCHILD (see Methods) was applied as an expanded blastocyst was used for phasing the parental genomes. (B) Circos plot illustrating the genome-wide interpreted copy-number profiles of all the single blastomeres derived from embryo E12_BRP010.
Biparental blastomeres—i.e., normal diploid blastomeres—were detected in two of the four embryos (embryos E21_BRP006 and E16_BRP011); and in one dispermic embryo, also a gynogenetic blastomere was observed (E04_BRP010) (Supplemental Fig. S4D). An extraordinary example of heterogoneic segregation dynamics can be seen in embryo E16_BRP011. This 10-cell embryo comprises gross mixoploidy, as haplarithmisis unveiled pure androgenetic cells containing one paternal genome (three cells), two tetraploid cells (3Paternal:1Maternal allelic ratio), one compound androgenetic cell (containing two different paternal genomes), and two biparental blastomeres (Supplemental Figs. S1B, S2).
All in all, dispermic embryos commonly segregate parental genomes resulting in uniparental and different ploidy level cell lineages. Interestingly, in every dispermic embryo, one paternal genome was consistently allocated in a distinct cell lineage, suggesting a common mechanism.
Spontaneous segregation of the parental genomes can occur in the absence of fertilization errors
Haplarithmisis revealed two embryos, E19_BRP005 and E19_BRP012, whose zygotes also underwent a heterogoneic division. These embryos comprise androgenetic, gynogenetic, and biparental blastomeres, but without any signs of dispermy or digyny (Supplemental Figs. S1B, S2). In E19_BRP005, we detected one gynogenetic, three androgenetic, and two biparental blastomeres. Additional copy-number anomalies, affecting specific segments or entire chromosomes, were also detected (Supplemental Figs. S1B, S2, S5). The allele of maternal Chr 9 that was lost in the gynogenetic blastomere was detected in the androgenetic blastomeres, demonstrating a diploid biparental profile for the chromosome. Interestingly, additional segmental copy-number aberrations affect exclusively the paternal Chr 3, Chr 25, and Chr X, and are reciprocal between the androgenetic and the biparental blastomeres. A simple explanation would be the segregation of the parental genomes into three blastomere lineages following a trichotomous division of the zygote (Supplemental Fig. S5). Similarly, the second normally fertilized embryo, E19_BRP012, is comprised of an androgenetic (four cells), a gynogenetic (four cells), and a biparental (three cells) cell lineage (Fig. 5; Supplemental Fig. S2). Copy-number abnormalities were detected only in Bl006, where haplarithmisis confirms four mitotic maternal gains for Chr 9, Chr 14, Chr 22, and Chr 24 (Supplemental Fig. S2). We speculate that the reciprocal losses may trace to the cell that could not be analyzed (Bl008).
Figure 5.
Parental genomes are segregated in distinct cell lineages following normal fertilization. (A) Genome-wide paternal and maternal haplarithms of three blastomeres derived from the same monospermic embryo (E19_BRP012) demonstrate that the same parental genomes (same HR profiles across each parental genome) segregated into three distinct lineages (top: gynogenetic; middle: androgenetic; bottom: biparental) (see also Fig. 3A). Note that here, option 2 of siCHILD was applied (see Methods). (B) Circos plot illustrating the genome-wide interpreted copy-number profiles of all the single blastomeres derived from embryo E19_BRP012.
Discussion
The mechanisms underlying the origins of chimerism and mixoploidy in natural conceptions remain highly speculative. Using haplarithmisis, a method reconstituting both haplotypes and copy numbers in single cells, we demonstrate that the parental genomes within a single zygote can segregate independently, leading to mixoploidy and/or chimerism in early cleavage-stage embryos. We term this process a heterogoneic cell division.
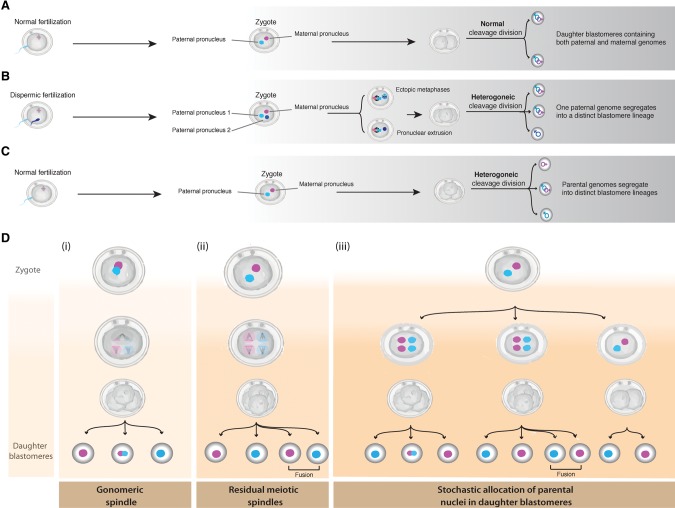
A triploid zygote undergoing a heterogoneic cytokinesis segregates the parental genomes into mixoploid blastomere lineages, which can in turn instigate chimerism. Although mixoploidy has been observed in human IVF preimplantation embryos following dispermic fertilization (Kola et al. 1987; Plachot and Crozet 1992; Staessen and Van Steirteghem 1997), the allocation of specific parental genomes into distinct blastomeres has never been demonstrated. Indeed, our data demonstrate that in the dispermic bovine embryos, a paternal genome is always segregated into an androgenetic blastomere lineage (Fig. 6; Supplemental Fig. S4).
Figure 6.
Fertilization processes and variants of heterogoneic divisions. (A–C) Sequence of events and their outcomes following the first post-zygotic division. (A) Normal fertilization followed by post-zygote mitosis, resulting in two diploid biparental daughter blastomeres. (B) Dispermic fertilization followed by ectopic metaphases or the extrusion of the paternal pronucleus, leading to the segregation of one paternal genome in an androgenetic cell line. (C) Normal fertilization followed by the segregation of the maternal and the paternal genomes into distinct cell lineages (see D). (D) The different hypothetical models, which might cause the first zygotic cleavage into heterogoneic cell lineages in E19_BRP012: (i) One pole of the gonomeric spindle loses its integrity and becomes tripolar; subsequently the zygote divides directly in three cells; (ii) the maternally and paternally derived spindles operate separately in the zygote (Van Blerkom et al. 2004) and segregate the paternal and the maternal genomes into four haploid blastomeres; and (iii) following failure of cytokinesis, the zygote contains four nuclei, which are allocated stochastically into daughter blastomeres following a noncanonical first zygotic division (direct cleavage into three or four cells), or the maternal and the paternal pronuclei are allocated in two different daughter cells following a noncanonical zygotic division.
In bovine zygotes, the first mitotic spindle is organized by the paternally inherited centrosome (Navara et al. 1994; Sathananthan et al. 1997) similar to humans (Sathananthan et al. 1996). Therefore, a heterogoneic division in the case of dispermy could entail the independent operation of two astral spindles leading to two cell lines, including an androgenetic and a biparental (Fig. 6B). This hypothesis is indirectly supported by the observation of two distinct astral spindles in dispermic bovine zygotes (Navara et al. 1994); one organizing a diploid metaphase, and a second ectopic spindle organizing an additional haploid metaphase. Ectopic metaphases were also described in human triploid zygotes (Macas et al. 1988; Pieters et al. 1992; Rosenbusch et al. 1997), including either a diploid metaphase and an ectopic set of haploid chromosomes or three separate haploid metaphases. These violations from the expected congression of chromosomes on a single metaphase plate indicate failure of karyogamy (close apposition of the parental pronuclei), providing a mechanistic basis for the separation of a haploid genome during the first post-zygotic division, resulting in diploid/triploid (2n/3n) and haploid/diploid (1n/2n) mosaics. Since only 25% of human triploid dispermic zygotes resulted in uniformly triploid blastomeres, while 2n lineages and 1n/2n or 2n/3n derivatives were often detected, Golubovsky (2003) proposed that a genome diploidization pathway underlies mixoploidy. Golubovsky (2003) further associated the developmental variants of this pathway with chimeric and mixoploid phenotypes in humans. By comparing our data to Golubovsky's observations, we argue that the perceived diploidization pathway in triploid dispermic embryos involves a heterogoneic division mediated either by the operation of two distinct astral spindles or by the allocation of one paternal pronucleus in a distinct lineage through noncanonical cytokinesis (Fig. 6B). The second mechanism is supported by the detection of a pronuclear extrusion, originally described as an expelled third cell-like structure suggested to contain a single haploid chromosome complement during the first cleavage of human triploid zygotes (Kola et al. 1987; Pieters et al. 1992). Subsequently, it was proposed that the extrusion would occasionally fuse with a blastomere, leading to 3n/2n mixoploidy, or degenerate, leading to uniform diploidization. Although in human triploids, the pronuclear extrusion could provide a biological basis for mixoploidy, it did not provide insights on the mode of parental genome segregation and the parental origin of mixoploidy. Our findings provide deeper insight, showing that all five dispermic zygotes generate a pure androgenetic cell line, which is consistent with the pronuclear extrusion model (Fig. 6B; Supplemental Fig. S4).
Most intriguingly, we found that the separation of the parental genomes into distinct lineages is not exclusive to abnormally fertilized embryos but it was also observed twice in monospermic embryos, which contained three cell lines, including an androgenetic, a gynogenetic, and a normal diploid biparental line. The nature and parental origin of the chromosomal imbalances detected in the blastomeres of E19_BRP005 (Supplemental Fig. S5) allowed us to reconstitute the chromosome segregational history of this embryo from the fertilized egg. We envision that the zygote was partitioned into three blastomeres in a single cleavage, instigating three distinct lineages through the operation of a tripolar gonomeric spindle, i.e., distinct bundles of microtubules associated with the maternal and the paternal genomes with distinct topology in metaphase (Fig. 6D; Supplemental Fig. S5). Up to now, there is no mechanistic insight in the formation of tripolar spindles in normally fertilized zygotes. However, several pathways underlying the loss of centrosome integrity leading to spindle tripolarity have been described in cancer cells (Maiato and Logarinho 2014). Gonomeric spindles have been described in crustaceans and insects (Lin and Wolfner 1991; Kawamura 2001; Tram et al. 2006). In mammals, the spatial segregation of the parental genomes has been reported in murine preimplantation embryos (Mayer et al. 2000).
The aberrant gonomeric spindle model could also underlie the emergence of distinct blastomere lineages in the second embryo E19_BRP012. However, for this embryo, alternative mechanisms could explain the heterogoneic segregation during the first zygote division (Fig. 6D). This normally fertilized embryo, contrary to E19_BRP005, lacks whole or segmental chromosome anomalies, which would facilitate the identification of sister blastomeres and the reconstruction of its cleavage history up to the zygote stage. Indeed, we hypothesize that the segregation of the parental genomes into distinct lineages in monospermic zygotes could be a consequence of a putative concurrent function of a sperm centrosome-mediated as well as a residual meiotic spindle organizing the segregation of the paternal and maternal chromosomes in the zygote, respectively. Previously, Van Blerkom et al. (2004) observed coexisting distinct, ectopic, astral, and barrel-shaped anastral spindles around chromatin structures in the so-called “silent fertilizations” (Van Blerkom et al. 2004).
In the same scenario for E19_BRP012, if cytokinesis were not disrupted in the zygote, where both the maternally and paternally derived spindles operate, we would expect a subsequent segregation of the maternal and the paternal chromatin into four haploid blastomeres (two gynogenetic and two androgenetic) (Fig. 6D). Alternatively, in the case of cytokinesis failure, a multikaryon zygote would form (Fig. 6D). The resulting multikaryon zygote could subsequently undergo a noncanonical cytoplasmic division, whereby the parental pronuclei would be stochastically allocated in distinct cell lines. The stochastic allocation of the paternal and the maternal nuclei to different blastomeres through noncanonical cytokinesis could also occur in a zygote where the parental pronuclei replicate but fail to closely appose (Fig. 6D).
Our proposed models are consistent with the live-imaging studies, which have uncovered atypical pronuclear evolution and aberrant natures of zygotic cytokinesis (Van Blerkom 1990; Hardy et al. 1993; Athayde Wirka et al. 2014; Vera-Rodriguez et al. 2015). Zygotes derived from intra-cytoplasmic sperm injection (ICSI) of a single sperm cell can transit from a normal two pronuclei (2PN) to a tri-pronucleated phase (3PN) (Lammers et al. 2014), indicating that endomitosis may cause multinucleation of zygotes. Interestingly, 2PN ICSI zygotes can divide directly into three or four blastomeres even in the absence of dispermy, suggesting the formation of a tripolar spindle through the introduction of a supernumerary centriole. The cellular and molecular components of these noncanonical zygotic divisions are not yet known; however, the combination of live-cell imaging techniques that detect aberrant cleavages with state-of-the-art single-cell RNA assays are starting to yield deeper insight into the molecular mechanisms (Vera-Rodriguez et al. 2015).
Heterogoneic divisions entailing gonomery, residual meiotic spindles, and noncanonical cytokinesis following normal fertilization, shed new insight (Fig. 6D) into the origin of human parthenogenetic chimeric cases described by Strain et al. (1995) and Yamazawa et al. (2010) and the androgenetic chimeras reported by Kaiser-Rogers et al. (2006) and Makrydimas et al. (2002). These human parthenogenetic and androgenetic chimeras developed in the absence of a fertilization error and contained two distinct cell lineages, a biparental and a uniparental maternal or paternal, respectively. The models evoked to explain their emergence (i.e., parthenogenetic activation of the oocyte) rely solely on the detection of the two cell lineages that were not selectively eliminated during early embryonic development (i.e., the parthenogenetic and the biparental cell lines). As a consequence, they are based only on DNA replication asymmetry between the parental nuclei without providing any insight into the mechanism underlying the allocation of one parental genome in a uniparental lineage.
This is the first study to demonstrate the segregation of parental genomes in chimeric and mixoploid lineages in cleavage-stage embryos. Therefore, despite the historical evidence that such events may occur in the human early embryo, studies using haplotyping methods are required to investigate the incidence and the biological significance of this phenomenon during preimplantation development. With the present study, we establish a valuable model organism for research on genomic instability in embryos conceived in vivo, which is unattainable for human conceptions. Interestingly, it has been shown that 25% of bovine blastocysts conceived in vivo were mixoploid, suggesting the phenomenon is not an artifact of in vitro fertilization and culture (Viuff et al. 2001).
In this study, we uncovered distinct blastomere lineages spontaneously emerging from (ab)normally fertilized zygotes (androgenetic, gynogenetic, and biparental) and propose that perturbations of fundamental zygotic programs—i.e., concurrent operation or residual meiotic spindles, loss of the gonomeric spindle pole integrity, endomitotic cycles—provide a novel conceptual framework for the origin of both parthenogenetic and androgenetic chimeras as well as mixoploid entities (Malan et al. 2006). The discovery of heterogoneic segregation adds to the remarkable genome plasticity observed during mammalian cleavage-stage development.
Methods
Selection of bovine gametes and embryos
Oocytes from eight Belgian Blue cows (Bos taurus) and semen from two Holstein-Friesian bulls (Bos taurus) were used for embryo production (BRP004 to BRP012 crosses) (Supplemental Fig. S1). Eleven embryos were isolated at day-2 post-insemination (pi), and 14 embryos were isolated on day-3 pi. Ovarian tissue from the donor cows (mothers) and semen from the two bulls (fathers) were used to extract bulk DNA (DNeasy Blood and Tissue kit, Qiagen). In addition, we obtained bulk DNA from the parents of the bulls (paternal grandparents). DNA from the entire expanded blastocysts (day-8 pi) of BRP010 and BRP011 crosses (Supplemental Fig. S1) was whole-genome amplified using the REPLI-g Single Cell Kit according to the manufacturer's protocol (Qiagen).
Bovine embryo in vitro production (IVP)
Bovine embryos were produced by routine in vitro methods (Wydooghe et al. 2014). Briefly, bovine ovaries were collected at the local slaughterhouse per donor and processed within 2 h. The ovaries were washed three times in warm physiological saline solution supplemented with kanamycin (25 mg/mL). Follicles between 2- and 8-mm diameter were punctured with an 18G needle and a 10 mL syringe. Cumulus Oocyte Complexes were collected using a stereomicroscope, washed in Hepes-TALP, and subsequently washed in 500 µL maturation medium, which consisted of modified bicarbonate-buffered TCM-199 supplemented with 50 µg/mL gentamycin and 20 ng/mL epidermal growth factor (EGF). Subsequently, maturation occurred per donor in 500 µL maturation medium in four-well plates (Nunc) for 22 h at 38.5°C in 5% CO2 in humidified air. For fertilization, semen of a single IVF tested bull was used following separation of frozen-thawed spermatozoa over a discontinuous Percoll gradient (45% and 90%; GE Healthcare Biosciences) at a final sperm concentration of 1 × 106 spermatozoa/mL. Fertilization was achieved by incubating the matured oocytes with spermatozoa for 21 h at 38.5°C in 5% CO2 in humidified air. The presumptive zygotes were transferred to synthetic oviductal fluid (SOF) supplemented with essential and nonessential amino acids (SOFaa), 0.4% BSA, and ITS (5 µg/mL insulin, 5 µg/mL transferrin, and 5 ng/mL selenium). In vitro culture (IVC) occurred in four-well dishes in 20 µL drops (1 per donor cow) covered with mineral oil. Embryos were incubated at 38.5°C in 5% CO2, 5% O2, and 90% N2.
Single blastomere isolation and whole-genome amplification
The embryos were treated with pronase to dissolve the zona pellucida (0.1% protease from S. griseus, P8811, Sigma-Aldrich, in TCM-199) and were subsequently washed in TCM-199 with 10% FBS followed by Ca+2/Mg+2-free PBS with 0.05% BSA to stimulate blastomere dissociation. Subsequently, the zona-free embryos were washed in Ca+2/ Mg+2-free PBS with 0.1% PVP (wash medium) and were transferred onto a petri dish for blastomere dissociation and tubing. Blastomere pick-up and tubing was performed with the use of a mouth-pipetting system using a 75 μm capillary. Briefly, each blastomere was washed three times in wash medium, and it was transferred into a 0.2-mL PCR tube containing either 4 μL of the transport medium (PBS, supplied as part of the REPLI-g Single Cell kit, Qiagen) or 1.5 μL alkaline lysis buffer (50 mM DTT and 200 mM KOH), depending on the downstream MDA method. The isolated blastomeres were placed immediately on dry ice and stored in −20°C for 30 min prior to whole-genome amplification (WGA). WGA was performed on the day of blastomere isolation. DNA from single blastomeres was whole-genome amplified using commercial MDA kits (REPLI-g Single Cell Kit, Qiagen; and GenomiPhiV2, GE Healthcare Biosciences). MDA amplification with GenomiPhiV2 was performed as described previously (Spits et al. 2006). MDA amplification of single blastomeres with the single-cell Repli-g kit was performed according to the manufacturer's instructions for the fast 3-h protocol. WGA products were purified using SPRI-beads (AMPure) at 0.8× total reaction volume.
SNP genotyping
Purified whole-genome amplified products were normalized to 50 ng/μL before downstream use for the Illumina Infinium HD assay super protocol according to the manufacturer's instructions (Illumina).
Single-cell and multicell parental or sibling DNA genotype calls, BAF, and logR values were obtained from the BovineHD BeadChip raw intensity data by application of the GenCall algorithm, which is embedded in Illumina's GenomeStudio software (http://support.illumina.com/array/array_software/genomestudio/downloads.html). Genotypes were called by setting the GenCall score at 0.75, based on the optimization steps described by Zamani Esteki et al. (2015). The raw logR- and BAF-values as well as discrete SNP genotype calls were fed to a modified version of siCHILD (Zamani Esteki et al. 2015) (see below).
Single-cell genome analysis of bovine cleavage-stage embryos
Genome-wide single-cell haplarithm profiles were deduced by using a modified version of the siCHILD algorithm (siCHILD-bovine). Briefly, siCHILD-bovine is a computational workflow, which consists of the steps described in the following sections.
Quality control (QC) of single cell SNP data
The acquired blastomere data were assessed as discussed previously (Zamani Esteki et al. 2015). Briefly, we performed a combination of unsupervised hierarchical clustering on the discrete SNP genotype calls and cumulative chromosome specific standard deviation on the logR values. Substandard samples were excluded from further analysis.
Genome haplarithmisis of single blastomeres
The process of haplarithmisis makes use of WGA-distorted SNP B allele frequency (BAF-) values of single cells and yields parental haplarithms. These haplarithm profiles blueprint the amount of haplotypes, and thus, the exact allelic ratio of the genomic regions. The entire process of haplarithmisis was described previously by Zamani Esteki et al. (2015). Briefly, the parental genotypes must be first phased. In this study, we used the paternal grandparents (option 1) or an offspring (expanded blastocyst or a good quality euploid single blastomere; option 2). Specifically, we applied option 1 for BRP004 to BRP009 crosses and option 2 for BRP010 to BRP012. Subsequently, single-cell SNP BAF values corresponding to specific combinations of phased parental genotypes are retrieved, of which some are mirrored against the 0.5 axis (Zamani Esteki et al. 2015). Consequently, these values are segmented by piecewise constant fitting PCF (gamma = 30) (Nilsen et al. 2012) and exhibited into paternal and maternal haplarithms (Zamani Esteki et al. 2015). Parity and complementarity/reciprocity features of haplarithmisis signify different genomic aberrations and unravel their parental and mechanistic origin (Zamani Esteki et al. 2015).
Single-cell haplotyping using discrete SNP-genotype calls
In parallel with haplarithmisis, we reconstructed genome-wide haplotypes of single blastomeres using discrete SNP genotype data. To this end, we applied a modified version of 1D median filtering (1D-MF) module of siCHILD (Zamani Esteki et al. 2015) by modifying the 1D-MF window size to 66.
Normalization of single-cell logR-value and copy-number profiling
The raw SNP-logR values are first smoothed using a moving average (window of 30 SNPs) and subsequently corrected for GC% bias by application of a Loess fit. The smoothed logR values are subsequently normalized toward a trimmed mean of the likely “normal” disomic chromosomes in the same cell. Initially, the disomic chromosomes are determined based on the parent-of-origin values (Voet et al. 2011a) and parental scoring criteria, as described previously (Zamani Esteki et al. 2015). These preliminary logR-values are consequently being corrected by haplarithm profiles to confirm copy-number balance. The normalized logR-values are integrated with haplarithm profiles for copy-number profiling. Subsequently, the normalized logR-values are segmented by PCF (gamma = 1000) (Nilsen et al. 2012). Copy-number aberrations are not called in loci where the logR-values are aberrant but are not corroborated by a haplarithm profile.
Data visualization and analyses
For illustration purposes, we applied siCHILD (Zamani Esteki et al. 2015), Circos (Krzywinski et al. 2009), and R (R Core Team 2015).
Data access
All SNP-array data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE76305.
Competing interest statement
M.Z.E., J.R.V., and T.V. are coinventors on a patent application ZL913096-PCT/EP2014/068315-WO/2015/028576, which is licensed to Cartagenia (Agilent Technologies).
Acknowledgments
Funding for this study is provided by the Agency for Innovation by Science and Technology (IWT) (TBM-090878 to J.R.V. and T.V.); the Research Foundation Flanders (FWO) (G.A093.11N to T.V. and J.R.V. and G.0392.14N to A.V.S. and J.R.V.); the European Union's Research and Innovation funding programme (FP7-PEOPLE-2012-IAPP SARM: 324509 to J.R.V., T.V., O.T, A.D., A.S., and A.K.); the University of Leuven (KU Leuven); and SymBioSys (PFV/10/016 to J.R.V. and T.V.). We thank S. Jackmaert and P. Van Damme for their technical assistance. We thank Professor Jan Traeger-Synodinos for a critical reading of the manuscript.
Author contributions: A.D., M.Z.E., T.V., and J.R.V. conceived the study. A.D., M.Z.E., A.V.S., T.V., and J.R.V. designed the experiments. M.Z.E. and T.V. developed haplarithmisis. A.D. and M.Z.E. performed data analysis. A.D., M.Z.E., M.C., and K.S. performed the experiments. O.T. and E.D. assisted with SNP-array technology. A.D. and M.Z.E. wrote the original draft of the manuscript. A.D., M.Z.E, A.V.S, A.K., A.S, T.V., and J.R.V. wrote and edited the final draft of the manuscript. J.R.V., T.V., A.V.S., A.K., and A.S. provided funding. T.V. and J.R.V. supervised the study.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.200527.115.
References
- Athayde Wirka K, Chen AA, Conaghan J, Ivani K, Gvakharia M, Behr B, Suraj V, Tan L, Shen S. 2014. Atypical embryo phenotypes identified by time-lapse microscopy: high prevalence and association with embryo development. Fertil Steril 101: 1637–1648.e1631–1635. [DOI] [PubMed] [Google Scholar]
- Boonen SE, Hoffmann AL, Donnai D, Tümer Z, Ravn K. 2011. Diploid/triploid mosaicism: a rare event or an under-diagnosed syndrome? Eur J Med Genet 54: 374–375. [DOI] [PubMed] [Google Scholar]
- Campbell IM, Yuan B, Robberecht C, Pfundt R, Szafranski P, McEntagart ME, Nagamani SC, Erez A, Bartnik M, Wisniowiecka-Kowalnik B, et al. 2014. Parental somatic mosaicism is underrecognized and influences recurrence risk of genomic disorders. Am J Hum Genet 95: 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez SL, Loewke KE, Han J, Moussavi F, Colls P, Munne S, Behr B, Reijo Pera RA. 2012. Dynamic blastomere behaviour reflects human embryo ploidy by the four-cell stage. Nat Commun 3: 1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford JH, Brown JK, Lew WY, Peters GB. 1986. Diploid complete hydatidiform mole, mosaic for normally fertilized cells and androgenetic homozygous cells. Case report. Br J Obstet Gynaecol 93: 1181–1186. [DOI] [PubMed] [Google Scholar]
- Giltay JC, Brunt T, Beemer FA, Wit JM, van Amstel HK, Pearson PL, Wijmenga C. 1998. Polymorphic detection of a parthenogenetic maternal and double paternal contribution to a 46,XX/46,XY hermaphrodite. Am J Hum Genet 62: 937–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovsky MD. 2003. Postzygotic diploidization of triploids as a source of unusual cases of mosaicism, chimerism and twinning. Hum Reprod 18: 236–242. [DOI] [PubMed] [Google Scholar]
- Hardy K, Winston RM, Handyside AH. 1993. Binucleate blastomeres in preimplantation human embryos in vitro: failure of cytokinesis during early cleavage. J Reprod Fertil 98: 549–558. [DOI] [PubMed] [Google Scholar]
- Iwasaki S, Nakahara T. 1990. Incidence of embryos with chromosomal anomalies in the inner cell mass among bovine blastocysts fertilized in vitro. Theriogenology 34: 683–690. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Gemelos G, Baner J, Ryan A, Cinnioglu C, Banjevic M, Ross R, Alper M, Barrett B, Frederick J, et al. 2010. Preclinical validation of a microarray method for full molecular karyotyping of blastomeres in a 24-h protocol. Hum Reprod 25: 1066–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser-Rogers KA, McFadden DE, Livasy CA, Dansereau J, Jiang R, Knops JF, Lefebvre L, Rao KW, Robinson WP. 2006. Androgenetic/biparental mosaicism causes placental mesenchymal dysplasia. J Med Genet 43: 187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura N. 2001. Fertilization and the first cleavage mitosis in insects. Dev Growth Differ 43: 343–349. [DOI] [PubMed] [Google Scholar]
- King WA. 2008. Chromosome variation in the embryos of domestic animals. Cytogenet Genome Res 120: 81–90. [DOI] [PubMed] [Google Scholar]
- Kola I, Trounson A, Dawson G, Rogers P. 1987. Tripronuclear human oocytes: altered cleavage patterns and subsequent karyotypic analysis of embryos. Biol Reprod 37: 395–401. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers J, Splingart C, Barrière P, Fréour T. 2014. Morphokinetic parameters of ICSI tripronucleated embryos observed using time lapse. Reprod Biomed Online 28: 658–660. [DOI] [PubMed] [Google Scholar]
- Lin HF, Wolfner MF. 1991. The Drosophila maternal-effect gene fs(1)Ya encodes a cell cycle-dependent nuclear envelope component required for embryonic mitosis. Cell 64: 49–62. [DOI] [PubMed] [Google Scholar]
- Macas E, Suchanek E, Grizelj V, Puharic I, Simunic V. 1988. Chromosomal preparations of human triploid zygotes and embryos fertilized in vitro. Eur J obstet Gynecol Reprod Biol 29: 299–304. [DOI] [PubMed] [Google Scholar]
- Maiato H, Logarinho E. 2014. Mitotic spindle multipolarity without centrosome amplification. Nat Cell Biol 16: 386–394. [DOI] [PubMed] [Google Scholar]
- Makrydimas G, Sebire NJ, Thornton SE, Zagorianakou N, Lolis D, Fisher RA. 2002. Complete hydatidiform mole and normal live birth: a novel case of confined placental mosaicism: case report. Hum Reprod 17: 2459–2463. [DOI] [PubMed] [Google Scholar]
- Malan V, Vekemans M, Turleau C. 2006. Chimera and other fertilization errors. Clin Genet 70: 363–373. [DOI] [PubMed] [Google Scholar]
- Mayer W, Smith A, Fundele R, Haaf T. 2000. Spatial separation of parental genomes in preimplantation mouse embryos. J Cell Biol 148: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertzanidou A, Wilton L, Cheng J, Spits C, Vanneste E, Moreau Y, Vermeesch JR, Sermon K. 2013. Microarray analysis reveals abnormal chromosomal complements in over 70% of 14 normally developing human embryos. Hum Reprod 28: 256–264. [DOI] [PubMed] [Google Scholar]
- Navara CS, First NL, Schatten G. 1994. Microtubule organization in the cow during fertilization, polyspermy, parthenogenesis, and nuclear transfer: the role of the sperm aster. Dev Biol 162: 29–40. [DOI] [PubMed] [Google Scholar]
- Nilsen G, Liestøl K, Van Loo P, Moen Vollan HK, Eide MB, Rueda OM, Chin SF, Russell R, Baumbusch LO, Caldas C, et al. 2012. Copynumber: efficient algorithms for single- and multi-track copy number segmentation. BMC Genomics 13: 591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieters MH, Dumoulin JC, Ignoul-Vanvuchelen RC, Bras M, Evers JL, Geraedts JP. 1992. Triploidy after in vitro fertilization: cytogenetic analysis of human zygotes and embryos. J Assist Reprod Genet 9: 68–76. [DOI] [PubMed] [Google Scholar]
- Plachot M, Crozet N. 1992. Fertilization abnormalities in human in-vitro fertilization. Hum Reprod 7(Suppl 1): 89–94. [DOI] [PubMed] [Google Scholar]
- Quigley DI, McDonald MT, Krishnamuthy V, Kishnani PS, Lee MM, Haqq AM, Goodman BK. 2005. Triploid mosaicism in a 45,X/69,XXY infant. Am J Med Genet Part A 138A: 171–174. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2015. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- Robinson WP, Lauzon JL, Innes AM, Lim K, Arsovska S, McFadden DE. 2007. Origin and outcome of pregnancies affected by androgenetic/biparental chimerism. Hum Reprod 22: 1114–1122. [DOI] [PubMed] [Google Scholar]
- Rosenbusch B, Schneider M, Sterzik K. 1997. The chromosomal constitution of multipronuclear zygotes resulting from in-vitro fertilization. Hum Reprod 12: 2257–2262. [DOI] [PubMed] [Google Scholar]
- Sathananthan AH, Ratnam SS, Ng SC, Tarin JJ, Gianaroli L, Trounson A. 1996. The sperm centriole: its inheritance, replication and perpetuation in early human embryos. Hum Reproduct 11: 345–356. [DOI] [PubMed] [Google Scholar]
- Sathananthan AH, Tatham B, Dharmawardena V, Grills B, Lewis I, Trounson A. 1997. Inheritance of sperm centrioles and centrosomes in bovine embryos. Arch Androl 38: 37–48. [DOI] [PubMed] [Google Scholar]
- Spits C, Le Caignec C, De Rycke M, Van Haute L, Van Steirteghem A, Liebaers I, Sermon K. 2006. Optimization and evaluation of single-cell whole-genome multiple displacement amplification. Hum Mutat 27: 496–503. [DOI] [PubMed] [Google Scholar]
- Staessen C, Van Steirteghem AC. 1997. The chromosomal constitution of embryos developing from abnormally fertilized oocytes after intracytoplasmic sperm injection and conventional in-vitro fertilization. Hum Reprod 12: 321–327. [DOI] [PubMed] [Google Scholar]
- Strain L, Warner JP, Johnston T, Bonthron DT. 1995. A human parthenogenetic chimaera. Nat Genet 11: 164–169. [DOI] [PubMed] [Google Scholar]
- Surti U, Hill LM, Dunn J, Prosen T, Hoffner L. 2005. Twin pregnancy with a chimeric androgenetic and biparental placenta in one twin displaying placental mesenchymal dysplasia phenotype. Prenat Diagn 25: 1048–1056. [DOI] [PubMed] [Google Scholar]
- Tram U, Fredrick K, Werren JH, Sullivan W. 2006. Paternal chromosome segregation during the first mitotic division determines Wolbachia-induced cytoplasmic incompatibility phenotype. J Cell Sci 119(Pt 17): 3655–3663. [DOI] [PubMed] [Google Scholar]
- Uchida IA, Freeman VC. 1985. Triploidy and chromosomes. Am J Obstet Gynecol 151: 65–69. [DOI] [PubMed] [Google Scholar]
- Van Blerkom J. 1990. Occurrence and developmental consequences of aberrant cellular organization in meiotically mature human oocytes after exogenous ovarian hyperstimulation. J Electron Microsc Tech 16: 324–346. [DOI] [PubMed] [Google Scholar]
- Van Blerkom J, Davis P, Alexander S. 2004. Occurrence of maternal and paternal spindles in unfertilized human oocytes: possible relationship to nucleation defects after silent fertilization. Reprod Biomed Online 8: 454–459. [DOI] [PubMed] [Google Scholar]
- van de Laar I, Rabelink G, Hochstenbach R, Tuerlings J, Hoogeboom J, Giltay J. 2002. Diploid/triploid mosaicism in dysmorphic patients. Clin Genet 62: 376–382. [DOI] [PubMed] [Google Scholar]
- Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C, Debrock S, Amyere M, Vikkula M, Schuit F, et al. 2009. Chromosome instability is common in human cleavage-stage embryos. Nat Med 15: 577–583. [DOI] [PubMed] [Google Scholar]
- Vera-Rodriguez M, Chavez SL, Rubio C, Pera RA, Simon C. 2015. Prediction model for aneuploidy in early human embryo development revealed by single-cell analysis. Nat Commun 6: 7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viuff D, Greve T, Avery B, Hyttel P, Brockhoff PB, Thomsen PD. 2000. Chromosome aberrations in in vitro-produced bovine embryos at days 2–5 post-insemination. Biol Reprod 63: 1143–1148. [DOI] [PubMed] [Google Scholar]
- Viuff D, Hendriksen PJ, Vos PL, Dieleman SJ, Bibby BM, Greve T, Hyttel P, Thomsen PD. 2001. Chromosomal abnormalities and developmental kinetics in in vivo-developed cattle embryos at days 2 to 5 after ovulation. Biol Reprod 65: 204–208. [DOI] [PubMed] [Google Scholar]
- Voet T, Vanneste E, Van der Aa N, Melotte C, Jackmaert S, Vandendael T, Declercq M, Debrock S, Fryns JP, Moreau Y, et al. 2011a. Breakage–fusion–bridge cycles leading to inv dup del occur in human cleavage stage embryos. Hum Mutat 32: 783–793. [DOI] [PubMed] [Google Scholar]
- Voet T, Vanneste E, Vermeesch JR. 2011b. The human cleavage stage embryo is a cradle of chromosomal rearrangements. Cytogenet Genome Res 133: 160–168. [DOI] [PubMed] [Google Scholar]
- Weaver DT, Fisher RA, Newlands ES, Paradinas FJ. 2000. Amniotic tissue in complete hydatidiform moles can be androgenetic. J Pathol 191: 67–70. [DOI] [PubMed] [Google Scholar]
- Wydooghe E, Heras S, Dewulf J, Piepers S, Van den Abbeel E, De Sutter P, Vandaele L, Van Soom A. 2014. Replacing serum in culture medium with albumin and insulin, transferrin and selenium is the key to successful bovine embryo development in individual culture. Reprod Fertil Dev 26: 717–724. [DOI] [PubMed] [Google Scholar]
- Xia XY, Wang WP, Li TF, Li WW, Wu QY, Li N, Zhang C, Gao HL, Li XJ, Cui YX. 2014. A parthenogenetic maternal and double paternal contribution to an ovotesticular disorder of sex development. Mol Cytogenet 7: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazawa K, Nakabayashi K, Kagami M, Sato T, Saitoh S, Horikawa R, Hizuka N, Ogata T. 2010. Parthenogenetic chimaerism/mosaicism with a Silver-Russell syndrome-like phenotype. J Med Genet 47: 782–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Kruskall MS, Yunis JJ, Knoll JH, Uhl L, Alosco S, Ohashi M, Clavijo O, Husain Z, Yunis EJ, et al. 2002. Disputed maternity leading to identification of tetragametic chimerism. N Engl J Med 346: 1545–1552. [DOI] [PubMed] [Google Scholar]
- Yunis EJ, Zuniga J, Romero V, Yunis EJ. 2007. Chimerism and tetragametic chimerism in humans: implications in autoimmunity, allorecognition and tolerance. Immunol Res 38: 213–236. [DOI] [PubMed] [Google Scholar]
- Zamani Esteki M, Dimitriadou E, Mateiu L, Melotte C, Van der Aa N, Kumar P, Das R, Theunis K, Cheng J, Legius E, et al. 2015. Concurrent whole-genome haplotyping and copy-number profiling of single cells. Am J Hum Genet 96: 894–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza MV, Surti U, Redline RW, Millie E, Chakravarti A, Hassold TJ. 2000. Parental origin and phenotype of triploidy in spontaneous abortions: predominance of diandry and association with the partial hydatidiform mole. Am J Hum Genet 66: 1807–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]