Abstract
Although over 35 different histone acetylation marks have been described, the overwhelming majority of regulatory genomics studies focus exclusively on H3K27ac and H3K9ac. In order to identify novel epigenomic traits of regulatory elements, we constructed a benchmark set of validated enhancers by performing 140 enhancer assays in human T cells. We tested 40 chromatin signatures on this unbiased enhancer set and identified H2BK20ac, a little-studied histone modification, as the most predictive mark of active enhancers. Notably, we detected a novel class of functionally distinct enhancers enriched in H2BK20ac but lacking H3K27ac, which was present in all examined cell lines and also in embryonic forebrain tissue. H2BK20ac was also unique in highlighting cell-type-specific promoters. In contrast, other acetylation marks were present in all active promoters, regardless of cell-type specificity. In stimulated microglial cells, H2BK20ac was more correlated with cell-state-specific expression changes than H3K27ac, with TGF-beta signaling decoupling the two acetylation marks at a subset of regulatory elements. In summary, our study reveals a previously unknown connection between histone acetylation and cell-type-specific gene regulation and indicates that H2BK20ac profiling can be used to uncover new dimensions of gene regulation.
A fundamental question in molecular biology is how chromatin modifications reflect the state of a cell. Over 100 histone modifications have been catalogued (Tan et al. 2011), but only a handful have been studied in depth for their effects on genome regulation (Barski et al. 2007; Wang et al. 2008; Bernstein et al. 2010; Hawkins et al. 2010; Boros 2012; Weiner et al. 2015). In particular, genome-scale analyses of histone acetylation have overwhelmingly focused on lysine 9 and lysine 27 on histone H3 (H3K9ac, H3K27ac), and these two marks have also been prioritized by international consortia, such as the NIH Roadmap Epigenomics Mapping Consortium (Bernstein et al. 2010) and ENCODE (The ENCODE Project Consortium 2012), as predictors of active enhancers. Other acetylation marks have occasionally been profiled using high-throughput methods (Wang et al. 2008; Hawkins et al. 2010; Ng et al. 2013), but little is known about the distinctions between them. It is likely that some of the ∼35 known histone acetylations could serve unique gene regulatory functions and play distinct roles in cellular processes (Agalioti et al. 2002; Ernst and Kellis 2010; Lasserre et al. 2013; Rajagopal et al. 2013).
In this study, we focus on histone marks at enhancers and promoters, since these are the two most abundant regulatory element classes in the human genome. The current paradigm is that enhancers exist in multiple poised or primed chromatin states characterized by various combinations of H2A.Z, H3K4me1, H3K4me2, and H3K27me3 (Rada-Iglesias et al. 2011; Loh et al. 2014) before they become active. A similar model holds for promoters, with H3K4me3 taking the place of H3K4me1 (Mikkelsen et al. 2007). It is believed that regulatory elements acquire histone acetylation when they transition from these “pre-active” states to an active state that drives gene expression (Mikkelsen et al. 2007; Rada-Iglesias et al. 2011; Calo and Wysocka 2013). However, little is known about the relative contributions of acetylation marks (except for H3K27ac) to this critical step in gene regulation and cell-state specification.
In order to address the above questions, we performed a comprehensive analysis of acetylation states at enhancers based on a novel, unbiased data set of enhancers and integration with Hi-C data. Surprisingly, we found that H2BK20ac, a little-studied histone acetylation mark, was the most predictive of enhancer activity. We therefore profiled this acetylation mark alongside more well-characterized histone modifications in multiple cell types and primary tissues. Our results revealed a diversity of acetylation signatures at active enhancers and also active promoters, with systematic differences in cell-type specificity, biological function, responsiveness to stimulus, and transcription factor (TF) recruitment between H2BK20ac and other more well-characterized acetylation marks.
Results
Histone acetylation signature of an unbiased set of enhancers
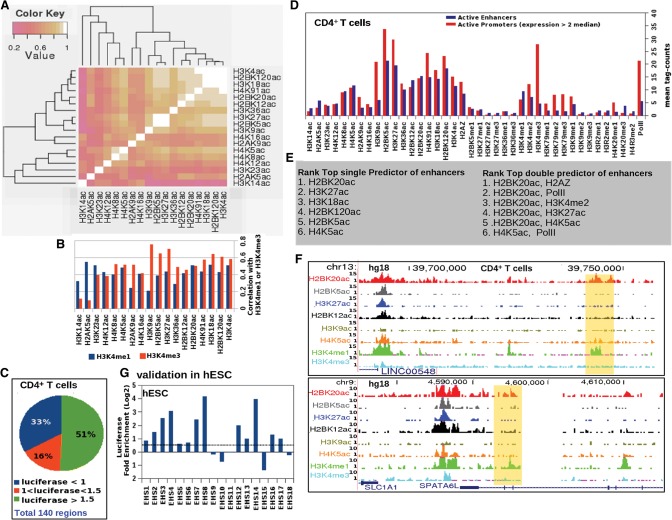
In order to identify similarities and differences between acetylation marks, we first examined 18 histone acetylation ChIP-seq profiles from CD4+ T cells (Fig. 1A; Wang et al. 2008). We clustered the 18 acetylations by their ChIP-seq tag count at open chromatin regions genome wide and noticed that they fell into distinct subgroups of co-occurrence. For example, H3K27ac, H2BK5ac, and H3K9ac (Fig. 1A) clustered as a group, with the former two showing the highest correlation. Although histone methylation data were not used in the clustering analysis, these three acetylations were also more strongly correlated with the promoter mark H3K4me3 than with the enhancer-enriched H3K4me1 mark (Fig. 1B). The tightest cluster consisted of H3K4ac, H2BK120ac, H3K18ac, and H4K91ac, which again showed greater correlation with H3K4me3 than with H3K4me1 (Fig. 1A,B). Yet another subgroup comprised H4K5ac, H4K8ac, and H4K12ac, which are known to be associated with transcriptional elongation (Hargreaves et al. 2009). These results suggest the existence of coherent subgroups within the set of histone acetylation marks, potentially reflecting distinct molecular mechanisms and functional roles as enhancers, promoters, and transcribed regions.
Figure 1.
Distinct chromatin signatures of active enhancers and promoters. (A) Heatmap of Pearson correlation coefficients between ChIP-seq signals for 18 histone acetylation marks at open chromatin sites in CD4+ T cells. (B) Bar graph of correlation between 18 acetylation marks with H3K4me1 and H3K4me3 ChIP-seq signals at open chromatin regions in CD4+ T cells. (C) Distribution of enhancer activity of tested open chromatin regions in luciferase assays. (D) Enrichment over genomic background of 40 ChIP-seq signals at active promoters and validated enhancers in CD4+ T cells. (E) Ranked list of top predictors of active enhancers (based on logistic regression). (F) Examples of validated CD4+ T cell enhancers (highlighted in yellow) marked by H2BK20ac but not by H3K27ac. (G) Enhancer assay results from testing 18 genomic regions marked by H2BK20ac in H1 human embryonic stem cells (hESCs). Candidate regions were chosen randomly from among the top 20,000 H2BK20ac ChIP-seq peaks in H1-hESCs.
We hypothesized, based on the above result, that histone acetylation marks could differ in their power to predict the genomic locations of active enhancers. Studies in vertebrates have frequently used indirect proxies such as EP300 binding or H3K4me1 to define enhancer regions. In order to more directly evaluate the chromatin signatures of enhancers, we generated an unbiased benchmark set of enhancers by performing reporter gene assays on 140 randomly chosen open chromatin regions in a CD4+ T cell line (Methods; Supplemental Table 1). Of the tested elements, 71 showed positive enhancer activity, 46 were negative, and 23 were discarded as ambiguous (Fig. 1C). We evaluated enrichment of the 18 histone acetylations at the validated enhancers and also included for completeness the ChIP-seq signals of 20 histone methylations, H2A.Z, and Pol II (Fig. 1D; Barski et al. 2007; Wang et al. 2008). Notably, H3K9ac, H2BK5ac, and H4K91ac showed a strong preference for active promoters over enhancers. As expected, H3K4me3 and Pol II were stronger at active promoters than at validated enhancers, while H3K4me1 showed the opposite trend. Note, however, that the chromatin signatures of enhancers and promoters showed substantial overlap; and as has previously been noted (Calo and Wysocka 2013), H3K4me3 and Pol II were also partially enriched at enhancers (Fig. 1D). Similarly, H3K4me1 showed some enrichment at active promoter regions. Most notably, H3K27ac showed higher enrichment at active promoters than at active enhancers.
To systematically assess the power of chromatin signals to predict active enhancers, we used machine learning on the 40 individual ChIP-seq data sets as well as on pairs (Fig. 1E). Surprisingly, the single most predictive enhancer mark was H2BK20ac, a covalent modification on the N-terminal tail of histone H2B, which has not been widely studied in the past. Moreover, every one of the top five pairs of ChIP-seq signals predictive for enhancer locations included H2BK20ac (Fig. 1E). Upon manual examination, we observed that certain validated enhancers were marked by H2BK20ac but not by H3K27ac, thus further supporting the distinction between different acetylation marks (Fig. 1F). To independently test the predictive power of H2BK20ac, we again used luciferase enhancer assays as above to test 18 genomic regions randomly chosen from among the top 20,000 H2BK20ac ChIP-seq peaks in human embryonic stem cells (H1-ESCs). We found that 72% of the tested elements showed enhancer activity (1.5-fold up-regulation) (Fig. 1G; Supplemental Table 2). Thus, H2BK20ac was strongly associated with enhancer function in both the tested cell types.
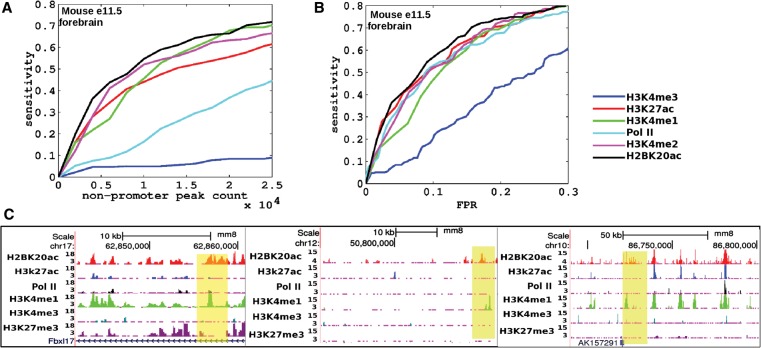
To test the association of H2BK20ac with tissue-specific enhancers in vivo, we performed ChIP-seq on five histone modifications (H3K27ac, H3K4me1, H3K4me2, H2BK20ac, and H3K27me3) and Pol II in the embryonic mouse forebrain. Forebrain samples were collected at embryonic day 11.5 (e11.5) so that chromatin profiles could be validated against the large number of enhancers reported to be active in this tissue at the VISTA Enhancer Browser (Visel et al. 2007). We found that the top-ranked forebrain ChIP-seq peaks for H2BK20ac were more likely to overlap the validated enhancer set than the corresponding peaks from other ChIP-seq data sets (Fig. 2A). We also estimated the specificity of enhancer predictions relative to the set of tested genomic regions that failed to act as nervous-system enhancers (Supplemental Methods). Again, H2BK20ac was observed to be the most predictive mark of forebrain enhancers (Fig. 2B). As above, we noticed that some of the validated forebrain enhancers showed H2BK20ac even in the absence of H3K27ac (Fig. 2C). Notably, the top H3K4me1 peaks were less predictive of forebrain enhancer function, perhaps due to the presence of this mark at inactive but poised enhancers (Rada-Iglesias et al. 2011; Calo and Wysocka 2013). To confirm this prediction, we examined three genomic regions marked by H3K4me1 and H3K27me3 but devoid of H2BK20ac. When assayed for reporter gene expression in transgenic mouse embryos at e11.5, none showed evidence of reproducible forebrain enhancer activity. We only observed infrequent forebrain expression as would be expected from occasional insertion of the transgene within forebrain-expressed loci (Supplemental Fig. 1). Based on these in vitro and in vivo results, we concluded that H2BK20ac was a hallmark of active enhancers distinct from H3K27ac.
Figure 2.
Power of H2BK20ac to predict active enhancers in complex tissues in vivo. (A) Sensitivity in detecting active enhancers in embryonic day 11.5 (e11.5) mouse forebrain (downloaded from VISTA Enhancer Browser) (Visel et al. 2007) as a function of number of peaks called. (B) Sensitivity versus false-positive rate (receiver-operator characteristic). The positive set comprised e11.5 forebrain enhancers, and the negative set comprised those that were tested but did not show activity in neural tissues at e11.5. (C) UCSC Genome Browser snapshots of three forebrain enhancers (from VISTA browser, highlighted in yellow) marked by H2BK20ac but not H3K27ac.
Genome-wide pattern of H2BK20ac at different enhancer classes
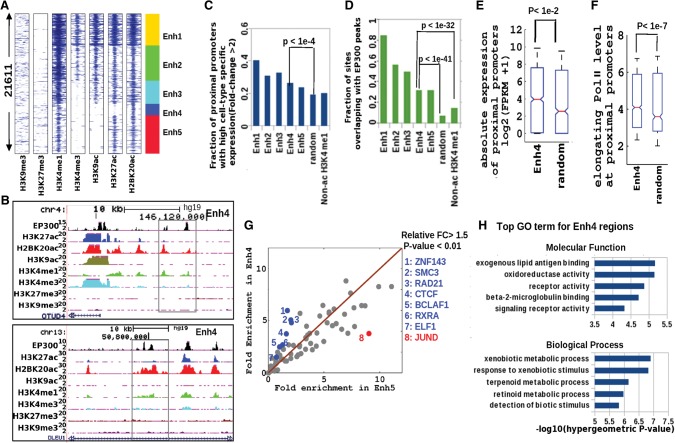
Encouraged by the differential enrichment of H2BK20ac and H3K27ac at validated enhancers, we analyzed the prevalence of these two marks at putative enhancers genome wide. For this, we performed H2BK20ac and H3K27ac ChIP-seq in GM12878 cells and also incorporated ENCODE ChIP-seq data on H3K9ac and multiple histone methylation marks. We predicted 21,611 enhancers in GM12878, based on the presence of H3K4me1 and at least one of H2BK20ac and H3K27ac (peak P-value <1 × 10−5). These predicted enhancer regions fell into five major chromatin classes (Fig. 3A,B). The first three classes (Enh1-3) were enriched for all three histone acetylations and appear to be strong enhancers based on EP300 binding and the cell-type-specific expression of their flanking genes (Fig. 3C,D). Notably, the Enh4 class was enriched for H2BK20ac but not H3K27ac or H3K9ac (Fig. 3A,B). Genes near Enh4 regions in GM12878 cells showed significantly higher absolute expression and also higher levels of elongating Pol II than genes flanking randomly chosen genomic regions (Fig. 3E,F). These features of Enh4 regions were observed even when we replaced our H3K27ac data set with one generated by ENCODE (data not shown). Enh4 regions overlapped EP300 binding sites (Fig. 3C) and showed a significant association with genes whose expression was cell-type specific (Fig. 3D). Despite the fact that Enh5 regions were marked by both H2BK20ac and H3K27ac, they did not appear to be stronger enhancers than Enh4 (Fig. 3C,D). The latter two classes (Enh4, Enh5) thus appear to represent two different chromatin states of moderate enhancers.
Figure 3.
A novel histone acetylation signature at putative distal enhancer regions in GM12878 cells. (A) Clustering of nonpromoter histone-acetylated regions (H2BK20ac or H3K27ac) based on ChIP-seq profiles of seven histone marks. Flanking regions’ width in spatial heatmap is 4 kb, and the number of Enh4 regions is ∼2000. (B) Genome Browser view of two regions from the Enh4 class (black boxes), showing H2BK20ac and EP300 binding in the absence of the two other acetylation marks. (C) Cell-type-specific expression of promoters proximal (closest within 100 kb) to enhancer regions of different classes compared to randomly chosen regions and also regions marked by H3K4me1 but not by histone acetylation (non-ac H3K4me1). (D) EP300 occupancy of the same sets of regions. (E) Absolute expression (FPKM) of promoters proximal to Enh4 regions relative to random regions. (F) Elongating (S2-phosphorylated) Pol II ChIP-seq signal at proximal promoters of the same regions. (G) Fold excess above genomic background of TF binding sites (ChIP-seq peaks) in Enh4 and Enh5 regions. (Blue dots) TFs enriched more in Enh4 than in Enh5 (P-value <0.01, FC > 1.5). (Red dots) TFs enriched more in Enh5 than in Enh4. (H) Top five enriched functional categories of genes proximal to Enh4 regions (GREAT gene ontology tool).
We repeated the above chromatin state analysis in IMR90 cells and detected the same five enhancer types as in GM12878 (Supplemental Fig. 2A). In this case, we exploited the availability of high-resolution Hi-C data (Jin et al. 2013) to link IMR90 enhancers to their target promoters and recapitulated the findings from GM12878 on cell-type specificity of Enh4 gene regulation (Supplemental Fig. 2A–D). It is likely that the validated enhancers highlighted above in Figure 1F and Figure 2C represent the Enh4 class in CD4+ T cells and mouse forebrain, respectively. We further used an independent algorithm, ChromHMM (Ernst and Kellis 2012), to detect chromatin states from ChIP-seq data on seven commonly used histone modifications plus H2BK20ac. In all three cell lines examined in this manner, ChromHMM detected a chromatin state that matched the Enh4 state (N15 in GM12878, IN6 in IMR90, and mN5 in mouse embryonic stem cells [mESCs]) (Supplemental Fig. 3). Thus, two independent classification procedures, applied to up to three cell types, reveal the existence of an enhancer chromatin state marked by H2BK20ac and H3K4me1, but not H3K27ac. Notably, when H2BK20ac was not included in the data provided to ChromHMM, enhancers with this chromatin state were annotated as inactive (Supplemental Fig. 3). Thus, H2BK20ac is essential for comprehensive annotation of active regulatory elements in the human genome.
To explore potential differences in molecular mechanism between Enh4 and other enhancer classes, we examined the prevalence of TF binding sites (The ENCODE Project Consortium 2012; Gerstein et al. 2012) within acetylated regions in GM12878 cells. Intriguingly, binding sites for seven TFs (ZNF143, SMC3, RAD21, CTCF, BCLAF1, RXRA, and ELF1) were more strongly enriched in Enh4 than in Enh5, relative to genomic background (Fig. 3G). In contrast, only one TF (JUND) showed significantly greater enrichment in Enh5. Notably, SMC3 and RAD21 are constituents of the cohesin complex, which has been shown to link active enhancers to promoters (Ing-Simmons et al. 2015), and CTCF frequently associates with such cohesin complexes (Sanyal et al. 2012). Similarly, in IMR90 cells, enhancers acetylated only on H2BK20 (Ienh4) showed greater enrichment for cohesin and CTCF binding than enhancers marked by H3K27ac as well as H2BK20ac (Ienh5) (Supplemental Fig. 2F). Thus, it is likely that Enh4 enhancers participate in cohesin-mediated looping interactions with their target promoters. Given that Enh4 regions showed differential TF binding, we hypothesized that they might also drive genes with unique functions. Using the GREAT tool (McLean et al. 2010), we discovered that Enh4 elements were enriched in loci associated with exogenous lipid antigen binding, xenobiotic metabolism, and xenobiotic response relative to the entire set of 21,611 acetylated regions (Fig. 3H). These enriched molecular function and biological process annotations indicate that Enh4 regions support some of the basic functions of B cells (Ross et al. 2011). Thus, Enh4 enhancers are enriched for cell-type-specific functions, perhaps due to the action of general enhancer-binding factors (cohesin complex) and B-cell TFs such as BCLAF1, RXRA, and ELF1. On a similar trend, genes proximal to Ienh4 regions in IMR90 cells were enriched for fibroblast-related functions (Supplemental Fig. 2G).
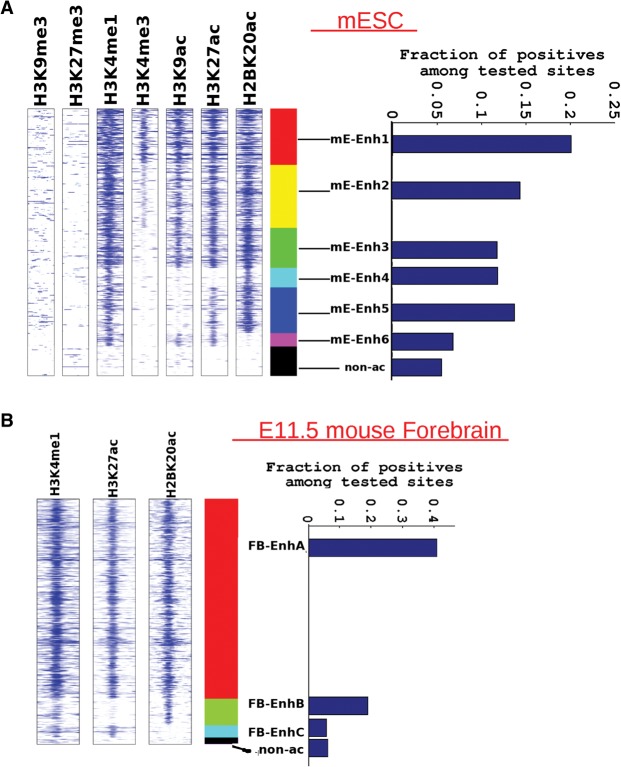
The above experiments indicate that H2BK20ac is a more reliable signature of active enhancers than H3K27ac, but they do not provide a direct head-to-head comparison between H2BK20ac-only and H3K27ac-only enhancers. Recently, Kwasnieski et al. (2014) used a massively parallel reporter gene assay (MPRA) to compare the enhancer activity of genomic regions in different chromatin states in K562 cells. We adopted a similar approach to compare H2BK20ac-only and H3K27ac-only enhancers. For this analysis, we exploited the availability of data from FIREWACh, an MPRA that was used to test ∼80,000 genomic fragments for enhancer activity in mESCs (Murtha et al. 2014). At the default P-value threshold for peak detection, DFilter detected a negligible number of H3K27ac-only enhancer peaks. We therefore lowered the threshold for H3K27ac and H2BK20ac peak calling to 1 × 10−4, which yielded an adequate number of such peaks (mE-Enh6 class) (Fig. 4A). We then categorized the genomic sequences tested in the FIREWACh assay (Murtha et al. 2014) according to their histone modification-based enhancer class and calculated the enhancer success rate for each class. Notably, sequences from the mE-Enh4 class, which were only marked by H2BK20ac, showed almost twice the enhancer activity of H3K27ac-only sequences (mE-Enh6). In fact, mE-Enh6 regions were only marginally more likely than random nonacetylated sequences to drive reporter gene activity. A similar trend was observed when we examined 1926 sequences tested for enhancer function in e11.5 mouse embryo using pronuclear injection of a construct containing the LacZ reporter gene (VISTA Enhancer Browser) (Fig. 4B; Visel et al. 2007). Thus, results from two large sets of enhancer assays indicate that H2BK20ac marks functionally active enhancers even in the absence of H3K27ac, whereas H3K27ac-only regions are not predictive of enhancer function.
Figure 4.
Evaluating regulatory activity of different enhancer classes in reporter gene assays in mESCs and e11.5 mouse forebrain. (A) Spatial heatmap of histone-acetylated regions genome wide, showing six enhancer classes in mESCs. Flanking regions’ width in spatial heatmap is 4 kb, and number of Enh4 regions is ∼1200. Each class represents a distinct combination of the seven examined histone marks. In this case, the ChIP-seq peak-calling threshold was lowered to 1 × 10−4 so as to detect the mE-Enh6 class, which was marked by H3K27ac in the absence of H2BK20ac (∼800 mEenh6 regions). Bar graph shows the success rate of tested regions from each enhancer class in the massively parallel FIREWACh enhancer assay (Murtha et al. 2014). Nonacetylated regions tested in the enhancer assay (non-ac) are shown as a control. The number of tested elements overlapping different enhancer classes is as follows: mE-Enh1:1603, mE-Enh2:1397, mE-Enh3:761, mE-Enh4:227, mE-Enh5:574, mE-Enh6:204. (B) Similar spatial heatmap showing three enhancer classes in e11.5 mouse forebrain, again with a peak-calling threshold of 1 × 10−4. There are ∼1700 FB-EnhB regions (H2BK20ac and H3K4me1) and ∼850 FB-EnhC sites (H3K27ac and H3K4me1). Bar graph shows the success rate of tested regions from each enhancer class in LacZ reporter gene assays in e11.5 mouse forebrain (VISTA Enhancer Browser). The control set is smaller in this case since only a few non-ac regions were tested in mouse embryos. The number of tested regions from each enhancer class is as follows: FB-EnhA:515, FB-EnhB:21, FB-EnhC:17.
Heterogeneity of histone acetylation marks at promoters
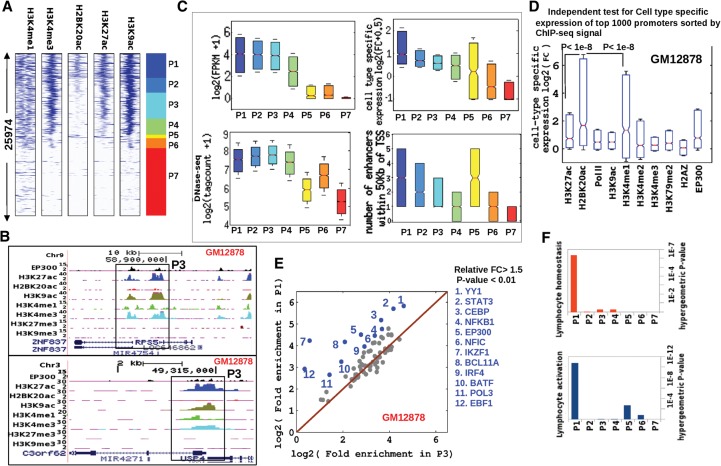
Inspired by the heterogeneity we observed at distal enhancers, we asked whether histone acetylation marks in promoter regions might also show distinct patterns and functions. To test this hypothesis, we clustered RefSeq transcription start sites by their histone modification profiles in GM12878 and thus identified seven distinct promoter classes (Fig. 5A,B). Promoters from the P1, P2, and P3 classes were the most active: They showed similarly high levels of H3K27ac, similar absolute gene expression, and also similar chromatin openness (Fig. 5A,C). P5 promoter regions were acetylated despite low H3K4me3 and low absolute expression. This class of promoter regions could contain enhancers proximal to inactive transcription start sites of non-target genes. Notably, H2BK20ac was strongly enriched only in the P1 class. P1 promoters also exhibited the greatest cell-type specificity and enrichment for flanking enhancers. To assess the cell-type specificity of chromatin signatures independently of one another, we also examined the top 1000 promoter regions for each mark in GM12878 (Fig. 5D). The top promoters marked by H2BK20ac displayed a striking increase in cell-type specificity relative to other marks, further supporting the clustering-based result. Top promoters for H3K4me1 also frequently showed cell-type-specific expression. However, a subset of H3K4me1 promoter regions also showed negative specificity; i.e., they were either inactive or down-regulated in GM12878. In contrast, top H3K27ac promoters showed relatively weak cell-type specificity. Consistently, in this lymphoblastoid cell line, genomic loci containing H2BK20ac-enriched promoters were the most likely to associate with susceptibility to autoimmune disorders, as shown by our analysis using mutations reported by genome-wide association studies (GWAS SNPs) (Supplemental Fig. 5A).
Figure 5.
Regulatory properties of active promoters with and without H2BK20ac in GM12878 cells. (A) Clustering of RefSeq promoters by profiles of five histone marks in GM12878 cells: seven major classes. (B) UCSC Genome Browser view of two P3-class promoter regions (black boxes) showing enrichment of H3K27ac but not H2BK20ac. (C) Absolute and cell-type-specific expression, DNase hypersensitivity, and number of flanking enhancers for each of the seven promoter classes in GM12878 cells. Cell-type-specific expression was quantified as fold change relative to the median of 12 cell types. (D) Cell-type-specific expression of top 1000 promoters for each ChIP-seq profile in GM12878 cells. (E) Fold excess above background of TF binding sites in P1 and P3 promoter regions. P7 promoter regions were used as the background set. (Blue dots) TFs enriched more in P1 than in P3 (P-value <0.01, FC > 1.5). (F) Enrichment of lymphocyte-specific functions among genes associated with the seven promoter classes.
As before, we examined the TF binding properties of H3K27ac-enriched promoter regions with and without co-occurrence of H2BK20ac (P1 vs. P3) (Fig. 5E). In total, we analyzed 78 GM12878 ChIP-seq data sets generated by ENCODE (The ENCODE Project Consortium 2012; Gerstein et al. 2012). Promoters marked by H2BK20ac (P1 class) were generally more enriched in TF binding than P3 promoters. While none of the individual TFs showed a preference for P3 promoters, 12 TFs were enriched in P1 regions, including well-known B-lymphocyte factors such as STAT3, NFKB1, NFIC, IKZF1, BCL11A, IRF4, EBF1, and BATF (Feng et al. 2004; Reynaud et al. 2008). Most strikingly, two master regulators of B-cell development, IKZF1 and EBF1, were over sevenfold enriched in P1 relative to background, despite showing almost no enrichment in P3. It is likely that the cell-type specificity of P1 promoters (Fig. 5C,D) is related to their greater propensity for binding B-cell TFs. The unique TF binding properties of P1 promoters potentially also explain their enrichment in biological functions specific to B lymphocytes (Fig. 5F).
To confirm the generality of our findings regarding H2BK20ac at promoters, we repeated the above analyses in mESCs. As before, we performed ChIP-seq on H2BK20ac and H3K27ac in these cells and also incorporated mESC RNA-seq and ChIP-seq data for other histone marks (Mikkelsen et al. 2007; Shen et al. 2012). Again, we found multiple distinct histone acetylation patterns in promoter regions (Supplemental Fig. 4A). mP1 promoters, which were maximally enriched in H2BK20ac, recapitulated all of the qualitative features of the P1 promoter class identified in GM12878, including cell-type specificity of gene expression and enrichment for ChIP-seq peaks of TFs associated with pluripotency of mESCs (Supplemental Fig. 4C–E; Chen et al. 2008; Percharde et al. 2012). Moreover, just as P1 promoters were enriched for B-cell-specific functions, mP1 promoters were maximally enriched for functions specific to mESCs (Supplemental Fig. 4F).
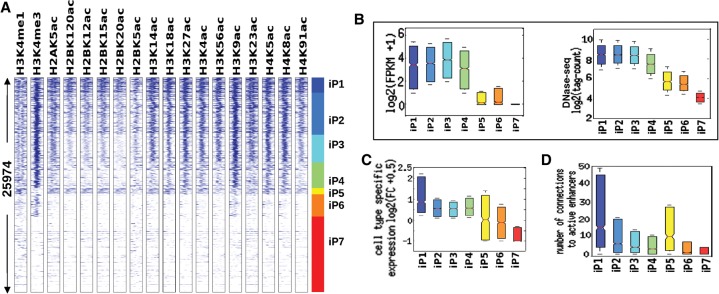
To rule out potential artifacts arising from the specifics of our ChIP-seq assays, we also examined published ChIP-seq data (GEO accession: GSE16256) on human ESCs generated in another laboratory using different antibodies and a different ChIP-seq protocol. Again, we observed that promoters enriched in H2BK20ac showed the strongest signal of cell-type specific gene expression, in comparison to 15 other histone acetylations and seven methylations (Supplemental Fig. 5B). We also used externally generated ChIP-seq data (GEO accession: GSE16256) to classify promoter histone modification states in IMR90 cells: 16 acetylation marks, plus H3K4me1 and H3K4me3. Again, we observed the same seven promoter classes as discovered in GM12878 and mESCs, and the iP1 promoter class, which was equivalent to P1 in GM12878 cells, again showed the greatest cell-type specificity (Fig. 6A,B). Notably, three of the other four H2B acetylation marks profiled in IMR90 cells (H2BK12ac, H2BK15ac and H2BK120ac) showed an enrichment pattern similar to H2BK20ac (Fig. 6A). This correlation between H2BK20ac and H2BK120ac at the enhancers has also been noted in another study (Rajagopal et al. 2014). Perhaps due to the overlap among acetylation marks on histone H2B, promoters highlighted by H2BK120ac were also highly cell-type specific (Supplemental Fig. 5B). In IMR90 cells, we were able to assign promoters to enhancers more accurately than in the other cell types, due to the availability of high-resolution Hi-C data on three-dimensional chromatin interactions (Jin et al. 2013). This increased the distinction between the H2B-acetylated promoter class (iP1) and other promoter classes in terms of connectivity to enhancer regions (Fig. 6B). More broadly, we found that distal promoters were more likely to form loops and contact each other if they possessed similar histone signatures, suggesting a mechanistic link between three-dimensional chromatin architecture and covalent modifications of histones (Supplemental Fig. 5C). Overall, promoter analysis across four different cell lines indicates that H2BK20 acetylation is a hallmark of cell-type-specific promoters.
Figure 6.
Regulatory properties of active promoters in IMR90 cells using 18 histone acetylation profiles and Hi-C data. (A) Clustering of RefSeq promoters by IMR90 histone modification profiles: iP1-iP7 classes identified de novo from this data set match P1–P7 classes in Figure 4. (B) Absolute expression and DNase hypersensitivity for each of the seven IMR90 promoter classes. (C) Cell-type-specific expression. (D) Number of enhancer–promoter connections (Hi-C) for each of the seven IMR90 promoter classes.
Histone acetylation dynamics of cell-state transitions
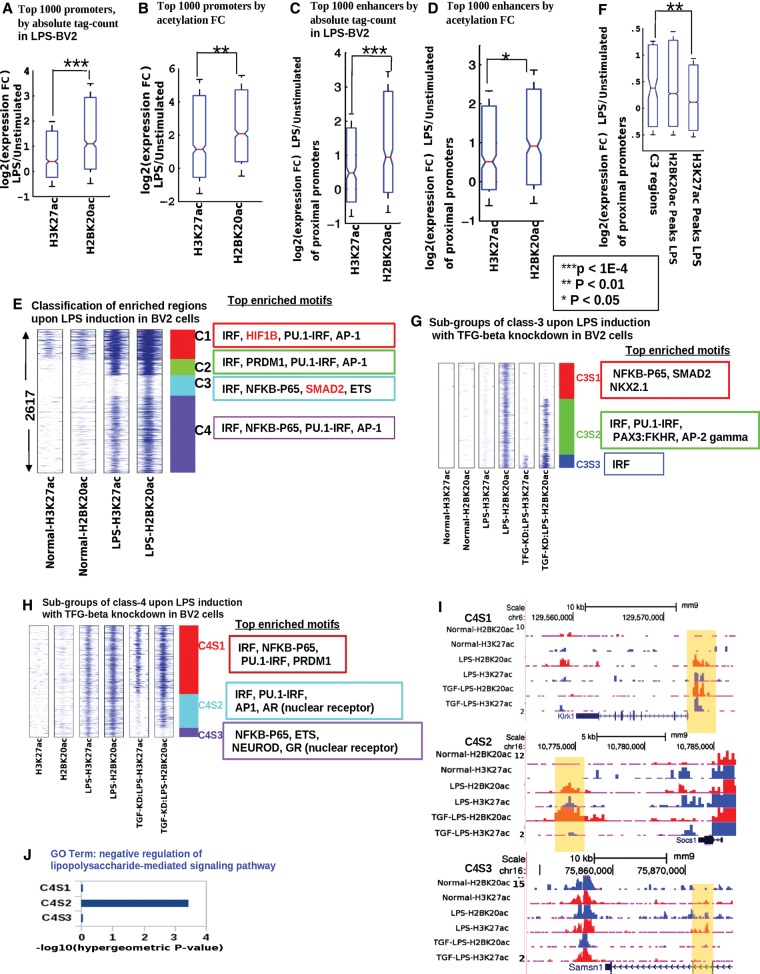
Given the high cell-type specificity of H2BK20ac, we hypothesized that this acetylation mark might also be cell-state specific, i.e., associated with dynamic changes in gene expression upon cell stimulation. To test this hypothesis, we examined the mouse microglial BV2 cell line, which shows inflammatory and apoptotic responses to stimulation by lipopolysaccharides (LPSs) present on the bacterial cell surface. Indeed, we found that promoters marked by H2BK20ac after LPS stimulation were up-regulated to a greater extent than promoters marked by H3K27ac (Fig. 7A). Instead of prioritizing promoters by their level of histone acetylation after LPS treatment, we ranked them by their acetylation fold change (LPS/unstimulated). Again, the top-ranked promoters for H2BK20ac fold change were more responsive to LPS than the corresponding promoters for H3K27ac (Fig. 7B). We observed the same trend when we compared H2BK20ac to H3K27ac at enhancer regions (Fig. 7C,D). Thus, in addition to being cell-type specific, H2BK20ac is also associated with dynamic expression changes during cell-state transitions.
Figure 7.
Differential acetylation dynamics during cell-state transition: decoupling of H2BK20ac and H3K27ac at a subset of regulatory regions. (A) Top 1000 promoters sorted by absolute ChIP-seq tag count (H2BK20ac, H3K27ac) in lipopolysaccharide (LPS)-stimulated BV2 cells (LPS-BV2): expression fold change upon stimulation. (B) Similar to A, promoters sorted by gain in ChIP-seq signal (fold change) upon LPS stimulation. (C) Top 1000 enhancers sorted by absolute ChIP-seq tag count (H2BK20ac, H3K27ac) in LPS-stimulated BV2 cells: expression fold change of proximal genes (nearest within 50 kb) after stimulation. (D) Similar to C, enhancers sorted by gain in ChIP-seq signal (fold change) upon stimulation. (E) Chromatin-based clustering of regulatory regions showing greater than fourfold increase in H2BK20ac or H3K27ac upon LPS stimulation of BV2 cells. Top four significantly enriched (P-value <0.01) motifs in each class are shown. (F) Fold change in expression for genes flanking (nearest within 50 kb) C3 regulatory regions, which are marked by increased H2BK20ac but not H3K27ac. Control sets: genes flanking all H2BK20ac or H3K27ac peaks in LPS-BV2. (G) Subclassification of C3 regulatory regions by histone acetylation patterns in BV2 cells stimulated by LPS after TGF-beta inhibition (TGF-KD:LPS). (H) Subclassification of C4 regulatory regions by histone acetylation patterns in TGF-KD:LPS cells. (I) Genome Browser views of sample regulatory regions from C4S1, C4S2, and C4S3 classes (highlighted by yellow). (J) Enrichment of LPS-response genes near regulatory regions belonging to the three subclasses of C4.
To investigate heterogeneity in histone acetylation dynamics, we examined the 2617 regulatory elements that showed at least fourfold increase in H2BK20ac or H3K27ac upon LPS stimulation in BV2 cells (Fig. 7E). Overall, most of these activated regulatory elements showed comparable increases in H2BK20ac and H3K27ac. Intriguingly however, one subgroup comprising 375 regulatory elements showed a clear asymmetry—they responded almost exclusively via H2BK20ac (C3 cluster) (Fig. 7E). The vast majority of these regions were promoter-distal (92%), and presumably functioned as enhancers. Consistently with this hypothesis, promoters flanking C3 regions showed increased expression after LPS stimulation (Fig. 7F).
To gain insights into molecular mechanisms of differential H2BK20ac and H3K27ac recruitment, we scanned regulatory elements from the four activated clusters for enrichment in TF binding motifs (Fig. 7F; Supplemental Methods). C3 regions were unique in showing enrichment for the TGF-beta effecter SMAD2 (Abutbul et al. 2012). In addition, we found that the TGF-beta signaling pathway, which has a well-known role (Suzumura et al. 1993; Abutbul et al. 2012) in modulating microglial LPS response, was the most enriched functional category among genes flanking C3 regions (Supplemental Fig. 6). We therefore hypothesized that TGF-beta signaling could be responsible for some of the observed differential histone acetylation in response to LPS. To test this prediction, we treated BV2 cells with the TGFBR1 inhibitor A83-01, which inhibits TGF-beta signaling (Loh et al. 2014), and repeated the LPS induction assay (TGF-KD:LPS BV2). As predicted, TGF-beta inhibition rendered a substantial fraction (33%) of C3 regions unresponsive to LPS (C3S1 subcluster) (Fig. 7G; Supplemental Fig. 6). This subcluster was also uniquely enriched for SMAD2 binding motifs (Fig. 7). Thus, TGF-beta signaling, presumably acting via SMAD2, recruits H2BK20ac to C3S1 regulatory regions independently of H3K27ac.
Although the 1406 regulatory regions belonging to the C4 class were not enriched for SMAD2 motifs, they nevertheless exhibited diverse histone acetylation responses to TGF-beta inhibition (Fig. 7H,I). Most notably, the C4S2 subclass, which originally responded to LPS with increases in both acetylation marks, lost H3K27ac after TGF-beta inhibition. These regulatory elements were proximal to genes (e.g., Socs1, Prdm1, Trib1, Tnfaip3) involved in negative regulation of LPS response, which is a known biological function of TGF-beta signaling (Fig. 7J; Supplemental Fig. 6; Suzumura et al. 1993). In contrast to C3 and C4 regulatory elements, C1 and C2 regions were not substantially affected by TGF-beta knockdown. Taken together, the above results indicate that TGF-beta decouples H2BK20ac and H3K27ac recruitment at >20% of the 2617 regulatory elements activated by LPS stimulation of microglial cells.
Discussion
We have used an unbiased training set of 140 assayed hypersensitive sites and 40 whole-genome chromatin profiles to define the histone signatures of active enhancers. Surprisingly, H2BK20ac, a little-studied mark, was the top predictor of active enhancers. This histone mark is generally not used to identify or characterize enhancers. The vast majority of previous studies focused instead on H3K9ac and H3K27ac, and the latter is the only acetylation mark included in the minimal reference epigenome by the NIH Roadmap Epigenomics Mapping Consortium (Bernstein et al. 2010) and the International Human Epigenome Consortium (http://ihec-epigenomes.org/).
Our results highlight substantial differences between H2BK20ac and H3K27ac at thousands of regulatory elements in multiple cell types. Specifically, we found a class of enhancers (Enh4) marked by H2BK20ac but little or no H3K27ac. Enh4 enhancers are enriched for EP300 and cohesin binding and loop to cell-type-specific promoters. They also have a substantially higher success rate in enhancer assays than non-promoter regions marked by H3K27ac in the absence of H2BK20ac. Moreover, in all four cell types examined, H2BK20ac-enriched promoter regions drove substantially greater cell-type-specific expression than top-ranked promoters for any other histone acetylation, perhaps due to binding of cell-type-specific TFs. Finally, promoter regions containing H2BK20ac were associated with cell-type-specific biological functions. In contrast, H3K27ac was present at all active promoter classes, regardless of cell-type specificity, which is consistent with the association of this histone mark with transcriptional elongation (Stasevich et al. 2014). Overall, these results indicate a unique, and hitherto unexplored, contribution of H2BK20ac to cell-type specific gene regulation and cell-type-specific biological functions at distal and proximal cis-regulatory elements.
Antibody specificity is a common concern in chromatin profiling studies, since nonspecific antibodies could confound signals from multiple histone modifications. In this study, we therefore used a monoclonal H2BK20ac antibody for ChIP-seq on mouse forebrain, GM12878 cells, mESCs, and BV2 cells, whose specificity has previously been exhaustively validated (Price et al. 2012). Our findings were also independently supported by analysis of CD4+ T cell, IMR90, and hESC ChIP-seq data from two other laboratories, based on a different H2BK20ac antibody that has also been rigorously tested for specificity (Wang et al. 2008). Thus, our conclusions are robust to artifacts from antibody cross-reactivity. Another potential concern is that the minimal promoter used in the enhancer assays could potentially be biased toward H2BK20ac-enriched enhancers. However, our conclusions in this regard are supported by four different minimal promoters (POU5F1 in hESCs, SV40 in Jurkat T cells, Hsp68 in the mouse embryo, and Fgf4 in mESCs) and three different reporter gene assay protocols (luciferase transfection, LacZ pronuclear microinjection, FIREWACh). Moreover, they are also supported by analysis of cell-type-specific expression at flanking genes, which is independent of reporter gene assays.
Yet another potential concern is that promoter DNA could be immunoprecipitated via crosslinking to H2BK20-acetylated histones at spatially proximal (looped) enhancers. This would then create artefactual H2BK20ac ChIP-seq signals at promoters. However, we note that H2BK20ac was enriched at active promoters even in ChIP-seq data generated without cross-linking (Wang et al. 2008). Thus, H2BK20 acetylation is likely to be a genuine property of a subset of active promoters, rather than a cross-linking artifact. One straightforward explanation for the observation of H2BK20ac at cell-type-specific promoters would be that H2BK20ac marks proximal enhancers lying within 2 kb of their transcription start sites. Another possibility is that promoter histones gain this acetylation mark via physical proximity to the enzymes that also acetylate looped enhancers.
In addition to cell-type-specific gene regulation, H2BK20ac also associates with dynamic changes in gene expression during transitions in cell state. In the BV2 microglial cell line, H2BK20ac showed significantly greater association with LPS-responsive genes than H3K27ac. The BV2-LPS system also allowed us to investigate potential molecular mechanisms of independent H2BK20ac and H3K27ac recruitment at cis-regulatory elements. Specifically, we found that TGF-beta signaling can decouple the two acetylation marks. One set of enhancers that gained only H2BK20ac upon LPS stimulation was reliant on TGF-beta signaling for activation, apparently mediated by SMAD2 (C3S1). Another set of enhancers gained both H3K27ac and H2BK20ac, but TGF-beta was only required for recruitment of H3K27ac to these regulatory elements (C4S2, associated with negative regulation of LPS response).
Previous analyses of “histone codes,” i.e., combinatorial patterns of histone marks with specific mechanistic and biological functions, relied primarily on patterns of histone methylation, complemented by one, or at most two, acetylation marks (Ernst et al. 2011; Shen et al. 2012; Loh et al. 2014; Kundaje et al. 2015). This approach was necessitated by the paucity of studies on specific histone acetylation marks that could provide additional gene regulatory information. Along the same lines, the current reference epigenome standard includes five histone methylation marks and only one acetylation mark (H3K27ac; http://ihec-epigenomes.org/research/reference-epigenome-standards). However, our results demonstrate clear heterogeneity in the genomic distribution, cell-type specificity, stimulus response, TF associations, and biological functions of H2BK20ac and H3K27ac. It will be important for future epigenomics studies to expand the set of chromatin profiles to include at least H3K27ac and H2BK20ac, and perhaps additional acetylation marks as well. For example, H2BK120ac also appears to correlate with cell-type-specific expression (Supplemental Fig. 5B). Broader acetylation profiling will be essential for comprehensively detecting enhancers, elucidating mechanisms of gene regulation, and characterizing responses to physiological and disease-related stimuli (epigenome-wide association studies).
Methods
Chromatin profile generation and analysis
We performed ChIP-seq on five histone modifications (H2BK20ac, H3K27ac, H3K4me1, H3K4me2, H3K27me3) and Pol II in the e11.5 mouse forebrain (Supplemental Methods). We also performed ChIP-seq on H2BK20ac and H3K27ac in mESCs, GM12878, and BV2 cells. Peaks were called in histone modification ChIP-seq data sets using DFilter (Kumar et al. 2013). TF ChIP-seq peak calls were downloaded from external sources (Chen et al. 2008; Gerstein et al. 2012) whenever available, and DFilter was used to call peaks in the remaining data sets. ChIP-seq tag counts were corrected for GC bias as previously described (del Rosario et al. 2015). ChIP-seq profiles around regulatory elements (Figs. 3A, 4A,B, 5A, 6A, 7E,G,H) were clustered as previously described (Ng et al. 2013).
Cell culture and treatment
H1-ESCs were cultured on Matrigel-coated plates using mTESR (Stemcell Technologies). Transfection of plasmid DNA for luciferase assay was performed using Fugene HD (Roche) according to the manufacturer's instructions. Jurkat T cells were grown in RPMI medium supplemented with 10% fetal bovine serum, 10 Mm HEPES, 100 units/mL penicillin, 100 µg/mL streptomycin. GM12878 cells were grown in RPMI 1640 medium (15% FBS, 2 mM L-glutamine) to approximately 1 million cells per milliliter and then collected for ChIP-seq or snap-frozen in liquid nitrogen and stored at −80°C. Cell culture and transfection feeder–free E14 mESCs were cultured at 37°C with 5% CO2. All cells were maintained on 0.1% gelatin-coated dishes in DMEM (Gibco), supplemented with 15% heat-inactivated ES FBS (Gibco), 0.055 mM β-mercaptoethanol (Gibco), 2 mM L-glutamine, 0.1 mM MEM nonessential amino acid, 5000 units per mL gentamycin, and 1000 units per mL LIF (Chemicon).
BV-2 cells (murine microglial cell line) were maintained in 75-cm2 culture flasks in Dulbecco's Modified Eagle's Medium (DMEM, Sigma, catalog no. 1152) supplemented with 10% fetal bovine serum (FBS, HyClone) and 1% antibiotic anti-mycotic solution (Sigma, catalog no. A5955), and cultured in 37°C in a humidified atmosphere of 5% CO2 and 95% air incubator. Cells were plated on a six-well plate at 8.0 × 105 per well and were subjected to different treatments the following day. Cells assigned for LPS treatment were incubated with LPS (1 μg/mL) for 3 h before cell collection. TGF-beta inhibitor A83-01 was added to the medium at a final concentration of 1 μM 1 h before LPS treatment. After 3 h, cells from each group were crosslinked for chromatin immunoprecipitation.
Statistical significance
While comparing different fractions, the statistical significance of difference was measured using a two-proportion z-test. Hence the P-values shown in Figures 3C,D,G, and 5E, Supplemental Figures 2C–F, 4E, and 5A were estimated using a two-proportion test. The P-values for boxplots shown in Figures 3E,F, 5D, 7A–D,F, Supplemental Figures 2B, 4D, and 5B were calculated using a Wilcoxon rank-sum test.
Luciferase reporter assays
For testing enhancer activities, the test sites were amplified from human genomic DNA; for H1ESC, the amplicons were cloned in the SalI site, downstream from the luciferase gene, of the pGL3-POU5F1 pp vector (a POU5F1 minimal promoter driving luciferase) using the Gateway System (Invitrogen). For Jurkat assays, amplicons were cloned on pGL4.23 vector downstream from the luciferase gene. A Renilla luciferase plasmid (pRL-SV40 from Promega) was cotransfected as an internal control. Cells were harvested 48 h after transfection, and the luciferase activities of the cell lysate were measured by using the Stop-Glow Dual-Luciferase reporter assay system (Promega).
LacZ in embryonic mouse
The LacZ reporter clones were constructed as previously described (del Rosario et al. 2014). PCR fragments corresponding to tested regions were cloned into the pENTR plasmid (Invitrogen) and transferred into the Gateway-compatible hsp68-LacZ reporter vector using LR recombination. The constructs were validated by Sanger sequencing. NotI digestion excises the vector backbone from the reporter construct. The enhancer-reporter fragment was gel-extracted fragment and used for pronuclear microinjection of mouse zygotes. Pronuclear microinjection of the DNA was performed by Cyagen Biosciences using standard procedures. Two rounds of injection were performed for each construct. Embryonic day 11.5 embryos were collected and stained for LacZ activity. The stained embryos were fixed with 4% paraformaldehyde overnight at 4°C and stored. Embryos were imaged by LeicaM205.
Identifying predictive histone modifications for active enhancers
Luciferase assays were used to measure the enhancer activity of 140 regions in CD4+ T cells. Since CD4+ T cells consist of Jurkat T cells, we used Jurkat T cells for testing enhancer activity in luciferase assays. For this purpose, regions were randomly chosen that had DNase hypersensitivity (DNase-seq peaks) (Boyle et al. 2008) in both CD4+ T cells and Jurkat T cells but did not lie on CpG islands or CTCF peaks (Barski et al. 2007).
Initial filtering was done to choose histone marks that could differentiate between active and inactive tested enhancers using ChIP-seq (39 histone marks and Pol II) from CD4+ T cells. We defined regions with luciferase activity above 1.5 as positives and regions with luciferase activity below one as negative. We compared the tag count of bins (200 bp) in regions of positive and negative using the Wilcoxon rank-sum test. The ChIP-seq data sets for which there was significant difference (Wilcoxon rank-sum P-value <0.01) between tag counts at enhancers and nonenhancers were chosen for further analysis. At this stage, we acquired 20 ChIP-seq data sets related to gene activation (Wang et al. 2008) that had a significant difference between tested enhancers and nonenhancers. Those marks included 14 histone acetylations (H2BK120ac, H2BK12ac, H2BK20ac, H2BK5ac, H3K18ac, H3K27ac, H3K36ac, H3K4ac, H3K9ac, H4K12ac, H4K16ac, H4K5ac, H4K8ac, H4K91ac), H2AZ, Pol II, and four histone methylations (H3K4me1, H3K4me2, H3K4me3, H3K9me1).
There could be overlapping occurrences and cross-talk between histone modifications. Hence we sought to find single or double best predictors of enhancers among the 20 ChIP-seq data sets that showed significant difference between tested enhancers and nonenhancers. For this purpose, in the second round, we used a regression model where positives were taken as enhancers (luciferase >1.5), and negatives were taken as regions which did not lie near promoters, CpG islands, or regions with DNase hypersensitivity or CAGE tag enrichment in CD4+ T cells (Carninci et al. 2006). At this stage, we tried to remove background artifacts in ChIP-seq profiles due to biases such as copy number variation (CNV) by convolving each ChIP-seq signal with a zero-mean filter (an approach used in DFilter) (Kumar et al. 2013). For each ChIP-seq data set, a zero-mean filter was made using the average profile of binned tag count around the center of DNase hypersensitive sites. After convolving with filter, for each enhancer we took the maximum score within 1 kb from the center of tested enhancers. We used a logistic regression model to find the best single and double predictor of active enhancers using our set of positives. We repeated the regression-based analysis 10 times while choosing negatives randomly for each iteration. Then the mean square errors from 10 iterations were added to get the final error in prediction. The ChIP-seq mark with the least error in prediction was considered the best predictor.
Data sources
For CD4+ T cells, ChIP-seq data were adapted from a previously published study (Barski et al. 2007; Wang et al. 2008). Histone modification data sets (except for H3K27ac and H2BK20ac) for GM12878 cells were adapted from ENCODE (GEO accession GSE26320) (Ernst et al. 2011). RNA-seq data for human cell lines were downloaded from ENCODE (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeCaltechRnaSeq/) and from other published sources (GEO accession GSE16190, GSE43070) (Chepelev et al. 2009; Jin et al. 2013). The ChIP-seq peaks of TFs were downloaded from ENCODE (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/). ChIP-seq and RNA-seq data of IMR90 and H1-hESC were downloaded from the NCBI GEO data set (GEO accession: GSE16256) submitted by Bing Ren's group. The table of Hi-C interactions in IMR90 used here was previously published (Jin et al. 2013).
RNA-seq data sets for several tissues and organs of mouse generated by Shen et al. (2012) were used for finding cell-type-specific expression in mESC (GEO accession: GSE29184). For histone modification of ChIP-seq data sets (except for H3K27ac and H2BK20ac) in mESCs, we relied on other published sources (GEO accession: GSE29184, GSE12241) (Mikkelsen et al. 2007; Shen et al. 2012).
Data access
The sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE72886.
Acknowledgments
This work was supported by funds from the Agency for Science Technology and Research, Singapore (A*STAR) and from grant 1R01MH094714-01 from the National Institute of Mental Health, USA.
Author contributions: S.P., H.H.N., and V.K. designed the study with assistance from N.A.R. and M.M. for the experimental components. V.K. performed data analysis. V.K. and S.P. interpreted the results and wrote the manuscript, with assistance from N.A.R. and M.M. N.A.R., M.M., B.E., and J.P. performed ChIP-seq and RNA-seq experiments. S.L., L.X., T.L., and H.M. performed or contributed to the in vitro enhancer assays. M.M., H.H.N., T.L., S.L., and N.A.R. designed and performed the in vivo assays.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.201038.115.
References
- Abutbul S, Shapiro J, Szaingurten-Solodkin I, Levy N, Carmy Y, Baron R, Jung S, Monsonego A. 2012. TGF-β signaling through SMAD2/3 induces the quiescent microglial phenotype within the CNS environment. Glia 60: 1160–1171. [DOI] [PubMed] [Google Scholar]
- Agalioti T, Chen G, Thanos D. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111: 381–392. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. 2010. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 28: 1045–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros IM. 2012. Histone modification in Drosophila. Brief Funct Genomics 11: 319–331. [DOI] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. 2008. High-resolution mapping and characterization of open chromatin across the genome. Cell 132: 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Wysocka J. 2013. Modification of enhancer chromatin: what, how, and why? Mol Cell 49: 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. 2006. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38: 626–635. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117. [DOI] [PubMed] [Google Scholar]
- Chepelev I, Wei G, Tang Q, Zhao K. 2009. Detection of single nucleotide variations in expressed exons of the human genome using RNA-seq. Nucleic Acids Res 37: e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rosario RC, Rayan NA, Prabhakar S. 2014. Noncoding origins of anthropoid traits and a new null model of transposon functionalization. Genome Res 24: 1469–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rosario RC, Poschmann J, Rouam SL, Png E, Khor CC, Hibberd ML, Prabhakar S. 2015. Sensitive detection of chromatin-altering polymorphisms reveals autoimmune disease mechanisms. Nat Methods 12: 458–464. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. 2010. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol 28: 817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. 2012. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 9: 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Cheng S, Pear WS, Liou HC. 2004. NF-kB inhibitor blocks B cell development at two checkpoints. Med Immunol 3: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. 2012. Architecture of the human regulatory network derived from ENCODE data. Nature 489: 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. 2009. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138: 129–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. 2010. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 6: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ing-Simmons E, Seitan VC, Faure AJ, Flicek P, Carroll T, Dekker J, Fisher AG, Lenhard B, Merkenschlager M. 2015. Spatial enhancer clustering and regulation of enhancer-proximal genes by cohesin. Genome Res 25: 504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. 2013. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503: 290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Muratani M, Rayan NA, Kraus P, Lufkin T, Ng HH, Prabhakar S. 2013. Uniform, optimal signal processing of mapped deep-sequencing data. Nat Biotechnol 31: 615–622. [DOI] [PubMed] [Google Scholar]
- Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwasnieski JC, Fiore C, Chaudhari HG, Cohen BA. 2014. High-throughput functional testing of ENCODE segmentation predictions. Genome Res 24: 1595–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasserre J, Chung HR, Vingron M. 2013. Finding associations among histone modifications using sparse partial correlation networks. PLoS Comput Biol 9: e1003168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh KM, Ang LT, Zhang J, Kumar V, Ang J, Auyeong JQ, Lee KL, Choo SH, Lim CY, Nichane M, et al. 2014. Efficient endoderm induction from human pluripotent stem cells by logically directing signals controlling lineage bifurcations. Cell Stem Cell 14: 237–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtha M, Tokcaer-Keskin Z, Tang Z, Strino F, Chen X, Wang Y, Xi X, Basilico C, Brown S, Bonneau R, et al. 2014. FIREWACh: high-throughput functional detection of transcriptional regulatory modules in mammalian cells. Nat Methods 11: 559–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng JH, Kumar V, Muratani M, Kraus P, Yeo JC, Yaw LP, Xue K, Lufkin T, Prabhakar S, Ng HH. 2013. In vivo epigenomic profiling of germ cells reveals germ cell molecular signatures. Dev Cell 24: 324–333. [DOI] [PubMed] [Google Scholar]
- Percharde M, Lavial F, Ng JH, Kumar V, Tomaz RA, Martin N, Yeo JC, Gil J, Prabhakar S, Ng HH, et al. 2012. Ncoa3 functions as an essential Esrrb coactivator to sustain embryonic stem cell self-renewal and reprogramming. Genes Dev 26: 2286–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JV, Tangsombatvisit S, Xu G, Yu J, Levy D, Baechler EC, Gozani O, Varma M, Utz PJ, Liu CL. 2012. On silico peptide microarrays for high-resolution mapping of antibody epitopes and diverse protein–protein interactions. Nat Med 18: 1434–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal N, Xie W, Li Y, Wagner U, Wang W, Stamatoyannopoulos J, Ernst J, Kellis M, Ren B. 2013. RFECS: a random-forest based algorithm for enhancer identification from chromatin state. PLoS Comput Biol 9: e1002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal N, Ernst J, Ray P, Wu J, Zhang M, Kellis M, Ren B. 2014. Distinct and predictive histone lysine acetylation patterns at promoters, enhancers, and gene bodies. G3 (Bethesda) 4: 2051–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud D, Demarco IA, Reddy KL, Schjerven H, Bertolino E, Chen Z, Smale ST, Winandy S, Singh H. 2008. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol 9: 927–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AC, Chen Q, Ma Y. 2011. Vitamin A and retinoic acid in the regulation of B-cell development and antibody production. Vitam Horm 86: 103–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. 2012. The long-range interaction landscape of gene promoters. Nature 489: 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasevich TJ, Hayashi-Takanaka Y, Sato Y, Maehara K, Ohkawa Y, Sakata-Sogawa K, Tokunaga M, Nagase T, Nozaki N, McNally JG, et al. 2014. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature 516: 272–275. [DOI] [PubMed] [Google Scholar]
- Suzumura A, Sawada M, Yamamoto H, Marunouchi T. 1993. Transforming growth factor-β suppresses activation and proliferation of microglia in vitro. J Immunol 151: 2150–2158. [PubMed] [Google Scholar]
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. 2011. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146: 1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. 2007. VISTA Enhancer Browser: a database of tissue-specific human enhancers. Nucleic Acids Res 35: D88–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A, Hsieh TH, Appleboim A, Chen HV, Rahat A, Amit I, Rando OJ, Friedman N. 2015. High-resolution chromatin dynamics during a yeast stress response. Mol Cell 58: 371–386. [DOI] [PMC free article] [PubMed] [Google Scholar]