Abstract
BACKGROUND
The CpG island methylator phenotype (CIMP) is a promising biomarker for irinotecan/5-fluorouracil/leucovorin chemotherapy in stage III colon cancer. In the present study, we evaluated whether CIMP is a prognostic biomarker for standard-of-care oxaliplatin-based adjuvant therapy.
MATERIALS AND METHODS
The HE6C/05 trial randomized 441 stage II/III colorectal adenocarcinoma patients to adjuvant XELOX vs. mFOLFOX6. The primary and secondary objectives were disease-free and overall survival, respectively. CIMP status was determined using the DNA methylation status of CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1. Cox models assessed the association of CIMP with survival.
RESULTS
Twenty-eight (9.6%) of the 293 available tumors were CIMP-positive. In univariate Cox regression analysis, no significant differences in survival were observed between individuals with CIMP-positive vs. negative tumors. CIMP-positive tumors were more likely to be right-sided and BRAF-mutant (chi-square, p<0.001). In the multivariate model, TNM-stage II (vs. stage III) was associated with reduced risk for relapse (HR=0.25, 95% CI 0.11-0.55, Wald’s p<0.001), while colon primary located on the left side and earlier TNM-stage were associated with reduced risk for death (HR=0.48, 95% CI 0.28-0.81, p=0.006 and HR=0.22, 95% CI 0.10-0.49, p<0.001, respectively).
CONCLUSION
In this exploratory analysis, CIMP does not appear to be a prognostic biomarker in oxaliplatin-treated patients with resected colorectal cancer.
Keywords: Methylation, CIMP, prognosis, biomarker, adjuvant
INTRODUCTION
Colorectal cancer (CRC) treatment is currently guided by the tumor, nodal, and/or metastatic involvement (TNM-stage).1 However, biomarkers are greatly needed to help guide therapy and improve the accuracy of prognostic determinations for individual patients. Prior experience has demonstrated that treatment options and their associated biomarkers can be stage-specific, such as KRAS as a predictive biomarker for anti-EGFR therapy in metastatic CRC.2 However, other biomarkers, such as BRAF, can be informative for the prognosis of early- and advanced-stage cancers.3 Thus, prospective biomarkers must be evaluated in varied clinical contexts to best determine their appropriate use in clinical care.
Adding further complexity, CRC is increasingly being recognized as a heterogeneous entity that arises from multiple biologic pathways.4 The three most recognized CRC pathways are chromosomal instability (microsatellite stable), microsatellite instable (MSI), and CpG Island Methylator Phenotype (CIMP). Found in ~10-20% of CRCs, CIMP is characterized by an exceptionally high frequency of aberrantly methylated CpG islands across the genome. Occurring more often in women and older individuals, CIMP-positive CRCs are typically proximal, high-grade, mismatch repair (MMR)-deficient, TP53-wildtype and BRAF-mutant.5-11 Somatic MMR-deficiency due to MLH1 promoter hypermethylation is found in a subset of CIMP CRCs.4
Somatic molecular variations present in the CRCs are also relevant to predicting outcomes after a CRC diagnosis.12 CIMP is an emerging biologically plausible biomarker in CRC because of its distinct pattern of aberrantly methylated genes and gene mutations. Early studies using 5-fluorouracil (5-FU) suggested no association between CIMP and prognosis.10, 13-15 However, recent analyses in the context of doublet chemotherapy noted conflicting results whether individuals with CIMP-positive cancers have worse survival.11, 16 We sought to further evaluate CIMP as a prognostic biomarker within the context of modern oxaliplatin-based adjuvant chemotherapy.
MATERIALS AND METHODS
Patient selection
The Hellenic Cooperative Oncology Group HE6C/05 trial randomized individuals with high-risk stage II or stage III CRC to 6 months of adjuvant oxaliplatin and either 5-FU/leucovorin (LV) (mFOLFOX6) or capecitabine (XELOX).17 Patients with rectal cancer additionally received adjuvant capecitabine-based chemoradiotherapy (50Gy). High-risk stage II was defined as any of: high histological grade, lymphovascular/perineural invasion, mucinous component, T4-stage, extramural vein invasion, symptomatic bowel obstruction/perforation at diagnosis, or <12 lymph nodes removed.
Enrollment occurred between November 2005 and January 2008, with last follow-up in November 2013. The primary endpoint was 3-year disease-free survival (DFS); 3-year overall survival (OS) was a secondary endpoint. The trial closed early due to poor accrual; no difference in survival was noted between treatment arms.
Of 441 patients enrolled on this study, 408 were eligible for treatment and 293 had primary tumor DNA and RNA available. All patients provided informed consent prior to participation in the trial; optionally a separate informed consent was obtained for providing biological material for research purposes. The clinical protocol was approved by the Institutional Review Boards of the participating institutions and by the National Organization for Medicines; the trial was included in the Australian New Zealand Clinical Trials Registry (Registration Number: ANZCTR 12610000509066).
Patient Chemotherapy Treatment (HE6C/05 trial)
As recently published,17 mFOLFOX6 treatment consisted of oxaliplatin 85 mg/m2, LV 200 mg/m2 over 2 hours, bolus 5-FU 400 mg/m2 intravenously (IV) all on day 1 followed by 5-FU 2400 mg/m2 over 46-hour continuous infusion. The mFOLFOX6 cycle was repeated every 14 days for a total of 12 cycles. The XELOX group received oxaliplatin 130 mg/m2 IV on day 1 and capecitabine (Xeloda®) 1000 mg/m2 twice daily for days 1-14. The XELOX cycle was repeated every 21 days for a total of 8 cycles.
Patients with rectal primaries additionally received 50 Gray of adjuvant radiotherapy (46 Gray to the pelvic area and 4 Gray boost to the tumor) concomitantly with capecitabine 825 mg/m2 twice daily on the days of radiotherapy. Two cycles of chemotherapy, administered according to the assigned treatment arm, were given prior to chemoradiotherapy. The remaining 10 cycles of mFOLFOX6 or 6 cycles for XELOX were administered after the completion of chemoradiotherapy.
DNA and RNA Extraction
DNA processing and analysis was performed as described previously in a non-CLIA-approved research laboratory at the Fred Hutchinson Cancer Research Center (PI: Grady).11 DNA and total RNA was extracted from 1.0 mm TMA cores (approximately 15-18 tumor tissue cores for each sample), using a standardized isolation method based on silica-coated magnetic beads (VERSANT Tissue Preparation Reagents, Siemens Healthcare Diagnostics, Tarrytown, NY).18
Sodium Bisulfite Conversion and Sample Preparation
Sodium bisulfite conversion of approximately 500 ng genomic DNA was performed using the EZ DNA Methylation Kit (Zymo Research), with final eluted volume of 10μL. The converted DNA was diluted 1:50 for MethyLight analysis. Methylated and unmethylated control DNA (Epitect Methylated/Unmethylated human DNA; QIAGEN, Valencia, CA) was diluted 1:10. A total of 10 μl diluted DNA was used per polymerase chain reaction (PCR) reaction. Additionally, serial dilutions of the methylated control DNA (QIAGEN) were included on each PCR assay plate for standard curve generation.
MethyLight Analysis of CIMP Markers
Treated genomic DNA was analyzed by MethyLight using a Bio-Rad CFX96 Real-Time System (Bio-Rad, Hercules, CA). Real-time PCR amplification was performed using Bio-Rad Hard-Shell Thin-Wall 96-Well Skirted PCR plates with Microseal ‘B’ Adhesive Seals, using the same primer/probe sequences as previously described.11 A 20 μL reaction mixture (0.5 μl primers (10uM), 0.02μl probe, 10 μl iTaq universal probe supermix, 10 μL bisulfite-converted DNA) was cycled as: 95°C for 3 minutes, followed by 48 cycles of 95°C for 5 second s and 60°C for 1 minute.
Determination of CIMP status
Methylation of CpG islands was evaluated for a validated five-marker panel: CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1.7, 8 A percentage of methylated reference of ≥4% was used to define the threshold of methylation 19. “CIMP-positive” was defined as methylation of ≥3/5 methylated loci and “CIMP-negative” as methylation of <3 markers.
BRAF, KRAS, CD3, CD8 and MMR assays
Tumor molecular analysis was performed as previously described.20 KRAS mutations (exons 2, 3 and 4) and BRAF V600E mutations (exon 15) were assessed with dd-sequencing on nested PCR amplicons, at the Laboratory of Molecular Oncology, Hellenic Foundation for Cancer Research, Aristotle University of Thessaloniki School of Medicine, Thessaloniki, Greece, as previously described in detail. One-step qRT-PCR was used for the assessment of CD3 and CD8 mRNA expression (STRATIFYER Molecular Pathology GmbH, Cologne, Germany).20 IHC for the four MMR proteins (MLH1, MSH2, MSH6, and PMS2) was carried out at the Laboratory of Molecular Oncology, Hellenic Foundation for Cancer Research (Aristotle University of Thessaloniki School of Medicine, Thessaloniki, Greece).
Statistical Methods
Continuous variables were presented as median with the corresponding range and categorical variables as frequency with the respective percentages. Chi-Square or Fisher’s exact tests and the non-parametric Mann-Whitney test were used for comparing patient and tumor characteristics. SAS software was used for all statistical analyses (Windows v9.3, SAS Institute Inc., Cary, NC).
Survival was determined from date of randomization to date of death or last contact (OS) and/or to documented first recurrence (DFS), whichever occurred first. Surviving patients were censored at the date of last contact. Time-to-event distributions were estimated using Kaplan-Meier curves and compared using log-rank tests. Cox models were used to assess the relationship of OS/DFS with various clinical and histological variables, with significance level α=0.05 for estimating hazard ratios (HR) and 95% confidence intervals (95% CI).
Multivariate Cox regression analyses applying the Bald backwards procedure were performed using: TNM-stage (II/III), KRAS status (wildtype/mutant), BRAF status (wildtype/mutant), primary site (right/left), MMR status (deficient/proficient), CD3/CD8 mRNA expression (high/low), treatment arm (mFOLFOX6/XELOX), and CIMP (positive/negative). The significance threshold for keeping a variable in the final model was set at α=0.15. No adjustments for multiple comparisons were done.
RESULTS
Twenty-eight (9.6%) of the 293 patients in HE6C/05 with available tumor DNA were CIMP-positive. Compared to CIMP-negative CRCs, CIMP-positive CRCs were more likely to be right-sided (chi-square, p<0.001), BRAF-mutant (p<0.001), MMR-deficient (p=0.007), and have a low CD3/CD8 tumor T-cell mRNA expression (p=0.015) (Table 1). There was no difference in gender predominance or KRAS mutation status.
TABLE 1.
Comparison of patients with CIMP-positive and CIMP-negative tumors.
|
CIMP-Negative
(n=264) N (%) |
CIMP-Positive
(n=28) N (%) |
P-value* | ||
|---|---|---|---|---|
| Age | <65 | 160 (60.6) | 14 (50.0) | 0.28 |
| ≥65 | 104 (39.4) | 14 (50.0) | ||
| Gender | Female | 119 (45.1) | 11 (39.3) | 0.56 |
| Male | 145 (54.9) | 17 (61.7) | ||
| TNM stage | II | 83 (32.4) | 13 (50.0) | 0.07 |
| III | 173 (67.6) | 13 (50.0) | ||
| Primary type | Colon | 192 (73.3) | 24 (85.7) | 0.16 |
| Rectum | 70 (26.7) | 4 (14.3) | ||
| Primary side | Left | 192 (73.3) | 9 (32.1) | <0.001 |
| Right | 70 (26.7) | 19 (67.9) | ||
| Treatment group | mFOLFOX6 | 128 (48.5) | 16 (57.1) | 0.38 |
| XELOX | 136 (51.5) | 12 (42.9) | ||
| BRAF status | Mutated | 5 (1.9) | 10 (35.7) | <0.001 |
| KRAS status | Mutated | 109 (41.6) | 9 (32.1) | 0.33 |
| MMR status | Deficient | 23 (8.8) | 7 (25.0) | 0.007 |
|
CD3/CD8
mRNA expression |
Low | 170 (68.3) | 11 (44.0) | 0.015 |
TNM = tumor node metastasis staging; MMR = mismatch repair.
Age category by two-sample t-test for differences in means. All other categories compared with Chi-square tests of homogeneity. Significant p-values are shown in bold.
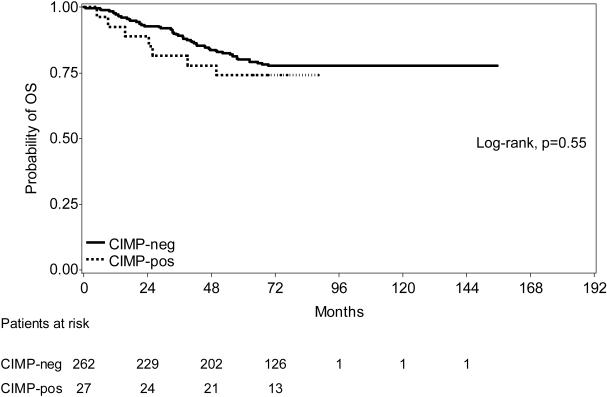
In univariate Cox regression analysis, there was no difference in OS by CIMP status (Figure 1). Comparing individuals with CIMP-positive and CIMP-negative tumors, individual clinical, molecular, and immunologic characteristics were analyzed (Table 2). However, no individual factor reached statistical significance for an effect on OS. DFS results were similar (Table 3); none of the variables reached statistical significance.
FIGURE 1. Overall survival of CIMP-positive and CIMP-negative patients.
There was no difference in overall survival (OS) in individuals with CIMP-positive versus CIMP-negative tumors at a median follow-up of 74.5 months.
TABLE 2.
Univariate Cox regression analysis for the effect of CIMP on overall survival.
| Parameter | Category |
HR (95% CI)
CIMP-positive vs. CIMP-negative |
P-value |
|---|---|---|---|
| CIMP | 1.27 (0.58, 2.80) | 0.55 | |
| TNM-stage | II | 1.13 (0.14, 9.37) | 0.91 |
| III | 1.45 (0.58, 3.66) | 0.43 | |
| Primary site | Right | 0.91 (0.34, 2.41) | 0.84 |
| Left | 1.23 (0.30, 5.13) | 0.77 | |
| Treatment arm | mFOLFOX6 | 1.25 (0.44, 3.57) | 0.84 |
| XELOX | 1.30 (0.39, 4.28) | 0.67 | |
| BRAF status | Wild-type | 1.11 (0.40, 3.08) | 0.83 |
| Mutated | 0.77 (0.13, 4.61) | 0.77 | |
| KRAS status | Wild-type | 1.14 (0.40, 3.24) | 0.81 |
| Mutated | 1.59 (0.48, 5.28) | 0.45 | |
| MMR status | Deficient | 0.84 (0.09, 7.50) | 0.87 |
| Proficient | 1.46 (0.62, 3.39) | 0.39 | |
| CD3/CD8 mRNA expression |
Low | 1.49 (0.46, 4.84) | 0.51 |
| High | 1.02 (0.30, 3.52) | 0.97 |
TNM = tumor node metastasis staging; MMR = mismatch repair.
TABLE 3.
Univariate Cox regression analysis for the effect of CIMP on disease-free survival.
| Parameter | Category |
HR (95% CI)
CIMP-positive vs. CIMP-negative |
P-value |
|---|---|---|---|
| CIMP | 0.75 (0.27, 2.07) | 0.58 | |
| TNM-stage | II | 0.92 (0.11, 7.44) | 0.93 |
| III | 0.94 (0.29, 3.04) | 0.92 | |
| Primary site | Right | 0.50 (0.11, 2.21) | 0.36 |
| Left | 1.11 (0.27, 4.62) | 0.88 | |
| Treatment arm | mFOLFOX6 | 0.63 (0.15, 2.65) | 0.53 |
| XELOX | 0.89 (0.21, 3.77) | 0.88 | |
| BRAF status | Wild-type | 0.57 (0.14, 2.33) | 0.43 |
| Mutated | 0.76 (0.07, 8.47) | 0.83 | |
| KRAS status | Wild-type | 0.62 (0.15, 2.64) | 0.52 |
| Mutated | 1.08 (0.26, 4.56) | 0.92 | |
| MMR status | Deficient | 0 (n/a) | 1.00 |
| Proficient | 1.01 (0.36, 2.81) | 0.98 | |
| CD3/CD8 mRNA expression |
Low | 0.92 (0.22, 3.83) | 0.91 |
| High | 0.80 (0.18, 3.53) | 0.76 |
TNM = tumor node metastasis staging; MMR = mismatch repair, n/a = not applicable.
In multivariate analysis, 237 patients had all relevant data available for the calculation of the risk for relapse and 260 patients for the risk for death. TNM-stage II (vs. stage III) was associated with reduced risk for relapse (HR=0.25, 95% CI 0.11-0.55, Wald’s p<0.001). In addition, colon primary located on the left side and earlier TNM-stage were associated with reduced risk for death (HR=0.48, 95% CI 0.28-0.81, p=0.006 and HR=0.22, 95% CI 0.10-0.49, p<0.001, respectively) (Table 4).
TABLE 4.
Significant results from the multivariate Cox regression analysis of CIMP and other factors on survival.
| Parameter | # of patients | # of events | HR (95% CI) | Wald’s p-value |
|
|---|---|---|---|---|---|
| DFS |
TNM-Stage
II vs. III |
96 vs. 186 | 12 vs. 68 | 0.25 (0.11, 0.55) | <0.001 |
| OS |
TNM-Stage
II vs. III |
96 vs. 186 | 7 vs. 52 | 0.22 (0.10, 0.49) | <0.001 |
|
Primary site
Left vs. Right |
197 vs. 85 | 35 vs. 24 | 0.48 (0.28, 0.81) | 0.006 |
DFS = disease-free survival; OS = overall survival; TNM = tumor node metastasis staging.
DISCUSSION
In this exploratory analysis of stage II/III CRC patients in a prospective, randomized trial of adjuvant mFOLFOX6 versus XELOX, there was no difference in survival between patients with CIMP-positive and CIMP-negative tumors. There was also no difference in the association of CIMP status and survival by treatment arm. DFS and OS results were similar.
Individuals with CIMP-positive CRCs exhibited similar features to what has been previously published, including a higher proportion of proximal tumors and tumors with BRAF mutations and deficient MMR.5-11 Unlike prior reports, no association with age or gender was noted in our analysis. A non-significant lower frequency of KRAS mutations was observed, as has been noted in previous analysis.11
The percent of individuals in the HE6C/05 trial who had CIMP-positive tumors was lower than has been reported in other studies.10, 13-16 This may be due to regional variability, as frequencies of 8-33% have been reported, while an international collaborative study noted a frequency of 18%.12 Variability in reported frequencies may also be related to differences in CIMP gene panels and definitions. In this analysis, we elected to use a validated 5-marker panel and categorized CRCs as CIMP-positive or CIMP-negative (rather than CIMP-high, -low, or -negative) for consistency with recent analyses.7, 8, 11, 12
Our prior analysis within CALGB (Alliance) 89803 demonstrated that CIMP was a stronger prognostic feature than MSI, a surrogate for MMR-deficiency.11, 21 The difference in observed exploratory outcomes may be related to the population differences (Greece vs. North America), baseline characteristics of the patients (e.g. inclusion of colon and rectal cancer), or differences in the specific features of the treatment regimens (e.g. the older IFL regimen of CALGB/Alliance 89803 has been replaced by FOLFIRI).
Han, et al also evaluated CIMP as a prospective biomarker in a Korean trial of adjuvant FOLFOX.16 Only 7.8% of individuals were CIMP-positive in this trial and no association between CIMP and DFS was observed. Compared to the Korean trial, HE6C/05 allowed for radiation of rectal primaries (consistent with guideline-recommended practice), had slightly different high-risk stage II criteria, included more stage III patients, and used updated oxaliplatin regimens.22 Additionally, we used the validated 5-marker panel with binary classification of CIMP as positive or negative,7, 8 while the Korean trial used an 8-marker panel and three-tier classification of CIMP. Despite these differences, both analyses noted no association between CIMP and survival among individuals receiving adjuvant oxaliplatin-based chemotherapy.
CONCLUSIONS
In conclusion, in this exploratory analysis of CIMP as a prognostic biomarker in stage II/III CRC patients treated with adjuvant oxaliplatin-based chemotherapy, we noted no association between CIMP and DFS or OS. As there was no 5-FU/capecitabine-only arm, CIMP could not be assessed as a predictive marker for benefit from oxaliplatin therapy. No predictive benefit for XELOX versus mFOLFOX6 was observed. Given the relatively small number of CIMP-positive CRCs in this study, the role of CIMP as a prognostic biomarker or a predictive biomarker for oxaliplatin benefit should be further investigated.
CLINICAL PRACTICE POINTS.
Biomarkers are greatly needed to inform and individualize treatment decisions. The CpG Island Methylator Phenotype (CIMP) is a key biologic pathway in colorectal cancer and has been suggested as a prognostic biomarker. We sought to validate prior findings that CIMP is a prognostic and predictive biomarker among non-metastatic colorectal cancer patients. In this analysis, that frequency of CIMP cancers was somewhat lower (9.6%) than has been described for other populations. We noted no association between CIMP and disease-free or overall survival. Further studies will need to focus on which populations CIMP is a relevant biomarker and whether CIMP can be used to inform chemotherapy treatment decisions.
MICROABSTRACT.
CpG Island Methylator Phenotype (CIMP) status was explored as a potential biomarker in 293 tumor specimens from stage II-III HE6C/05 patients receiving either mFOLFOX6 or XELOX adjuvant chemotherapy. Twenty-eight (9.6%) were CIMP-positive, but no association with disease-free or overall survival was noted. In this exploratory analysis, CIMP does not appear to be a prognostic biomarker in oxaliplatin-treated patients with resected colorectal cancer.
ACKNOWLEDGEMENTS
Research reported in this manuscript was supported by Seattle Cancer Care Alliance Development Funds (SS); National Institutes of Health (NIH) awards R01CA115513, P30CA15704, U01CA152756, U54CA143862, and P01CA077852 (WMG); Burroughs Wellcome Fund Translational Research Award for Clinician Scientist (WMG). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
ABBREVIATIONS
- CIMP
CpG Island Methylator Phenotype
- CRC
Colorectal cancer
- mFOLFOX6
modified FOLFOX-6 (5-fluorouracil, leucovorin, oxaliplatin combination chemotherapy)
- MMR
Mismatch repair
- MSI
Microsatellite instability
- XELOX
Capecitabine, oxaliplatin combination chemotherapy
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The findings in this manuscript were previously presented at the 2015 Gastrointestinal Cancer Symposium (January 2015; San Francisco, CA, USA).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1. Referenced with permission from The NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines(r)) for Colon Cancer V.2.2015. (c) National Comprehensive Cancer Network, Inc 2015. All rights reserved. Accessed March 11, 2015. To view the most recent and complete version of the guideline, go online to www.nccn.org. NATIONAL COMPREHENSIVE CANCER NETWORK(r), NCCN(r), NCCN GUIDELINES(r), and all other NCCN Content are trademarks owned by the National Comprehensive Cancer Network, Inc. NCCN Colon Cancer 2015.
- 2.De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. The lancet oncology. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 3.Lochhead P, Kuchiba A, Imamura Y, et al. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. Journal of the National Cancer Institute. 2013;105:1151–1156. doi: 10.1093/jnci/djt173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hawkins N, Norrie M, Cheong K, et al. CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology. 2002;122:1376–1387. doi: 10.1053/gast.2002.32997. [DOI] [PubMed] [Google Scholar]
- 7.Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. The Journal of molecular diagnostics : JMD. 2007;9:305–314. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nature genetics. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 9.van Rijnsoever M, Grieu F, Elsaleh H, Joseph D, Iacopetta B. Characterisation of colorectal cancers showing hypermethylation at multiple CpG islands. Gut. 2002;51:797–802. doi: 10.1136/gut.51.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Min BH, Bae JM, Lee EJ, et al. The CpG island methylator phenotype may confer a survival benefit in patients with stage II or III colorectal carcinomas receiving fluoropyrimidine-based adjuvant chemotherapy. BMC cancer. 2011;11:344. doi: 10.1186/1471-2407-11-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shiovitz S, Bertagnolli MM, Renfro LA, et al. CpG Island Methylator Phenotype Is Associated With Response to Adjuvant Irinotecan-Based Therapy for Stage III Colon Cancer. Gastroenterology. 2014;147:637–645. doi: 10.1053/j.gastro.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phipps AI, Limburg PJ, Baron JA, et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology. 2015;148:77–87. doi: 10.1053/j.gastro.2014.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Rijnsoever M, Elsaleh H, Joseph D, McCaul K, Iacopetta B. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2003;9:2898–2903. [PubMed] [Google Scholar]
- 14.Shen L, Catalano PJ, Benson AB, 3rd, O'Dwyer P, Hamilton SR, Issa JP. Association between DNA methylation and shortened survival in patients with advanced colorectal cancer treated with 5-fluorouracil based chemotherapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:6093–6098. doi: 10.1158/1078-0432.CCR-07-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jover R, Nguyen TP, Perez-Carbonell L, et al. 5-Fluorouracil adjuvant chemotherapy does not increase survival in patients with CpG island methylator phenotype colorectal cancer. Gastroenterology. 2011;140:1174–1181. doi: 10.1053/j.gastro.2010.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han SW, Lee HJ, Bae JM, et al. Methylation and microsatellite status and recurrence following adjuvant FOLFOX in colorectal cancer. International journal of cancer. Journal international du cancer. 2013;132:2209–2216. doi: 10.1002/ijc.27888. [DOI] [PubMed] [Google Scholar]
- 17.Pectasides D, Karavasilis V, Papaxoinis G, et al. Randomized phase III clinical trial comparing the combination of capecitabine and oxaliplatin (CAPOX) with the combination of 5-fluorouracil, leucovorin and oxaliplatin (modified FOLFOX6) as adjuvant therapy in patients with operated high-risk stage II or stage III colorectal cancer. BMC cancer. 2015;15:384. doi: 10.1186/s12885-015-1406-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bohmann K, Hennig G, Rogel U, et al. RNA extraction from archival formalin- fixed paraffin-embedded tissue: a comparison of manual, semiautomated, and fully automated purification methods. Clinical chemistry. 2009;55:1719–1727. doi: 10.1373/clinchem.2008.122572. [DOI] [PubMed] [Google Scholar]
- 19.Ogino S, Kawasaki T, Brahmandam M, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. The Journal of molecular diagnostics : JMD. 2006;8:209–217. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pentheroudakis G, Raptou G, Kotoula V, et al. Immune response gene expression in colorectal cancer carries distinct prognostic implications according to tissue, stage and site: a prospective retrospective translational study in the context of a hellenic cooperative oncology group randomised trial. PloS one. 2015;10:e0124612. doi: 10.1371/journal.pone.0124612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertagnolli MM, Niedzwiecki D, Compton CC, et al. Microsatellite instability predicts improved response to adjuvant therapy with irinotecan, fluorouracil, and leucovorin in stage III colon cancer: Cancer and Leukemia Group B Protocol 89803. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:1814–1821. doi: 10.1200/JCO.2008.18.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Referenced with permission from The NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines(r)) for Rectal Cancer V.2.2015. (c) National Comprehensive Cancer Network, Inc 2015. All rights reserved. Accessed March 11, 2015. To view the most recent and complete version of the guideline, go online to www.nccn.org. NATIONAL COMPREHENSIVE CANCER NETWORK(r), NCCN(r), NCCN GUIDELINES(r), and all other NCCN Content are trademarks owned by the National Comprehensive Cancer Network, Inc. NCCN Rectal Cancer 2015.