Abstract
The morphological, biological, and molecular characteristics of Cryptosporidium avian genotype V are described, and the species name Cryptosporidium avium is proposed to reflect its specificity for birds under natural and experimental conditions. Oocysts of C. avium measured 5.30–6.90 μm (mean = 6.26 μm) × 4.30–5.50 μm (mean = 4.86 μm) with a length to width ratio of 1.29 (1.14–1.47). Oocysts of C. avium obtained from four naturally infected red-crowned parakeets (Cyanoramphus novaezealandiae) were infectious for 6-month-old budgerigars (Melopsittacus undulatus) and hens (Gallus gallus f. domestica). The prepatent periods in both susceptible bird species was 11 days post infection (DPI). The infection intensity of C. avium in budgerigars and hens was low, with a maximum intensity of 5,000 oocysts per gram of faeces. Oocysts of C. avium were microscopically detected at only 12–16 DPI in hens and 12 DPI in budgerigars, while PCR analyses revealed the presence of specific DNA in faecal samples from 11 to 30 DPI (the conclusion of the experiment). Cryptosporidium avium was not infectious for 8-week-old SCID and BALB/c mice (Mus musculus). Naturally or experimentally infected birds showed no clinical signs of cryptosporidiosis and no pathology was detected. Developmental stages of C. avium were detected in the ileum and caecum using scanning electron microscopy. Phylogenetic analyses based on small subunit rRNA, actin, and heat shock protein 70 gene sequences revealed that C. avium is genetically distinct from previously described Cryptosporidium species.
Keywords: Cryptosporidium avium, Morphology, Molecular analyses, Transmission studies, Cryptosporidium avian genotype V
1. Introduction
Cryptosporidium parasites belong to the phylum Apicomplexa and infect the gastrointestinal tract of a broad range of vertebrate species (Fayer 2010), causing the diarrheal disease cryptosporidiosis. Currently, around 30 species of Cryptosporidium infecting fish, amphibians, reptiles, birds, and mammals are considered to be valid (Kváč et al. 2014a; Liu et al. 2013; Qi et al. 2011). Of these, only three have specificity for birds:Cryptosporidium meleagridis, Cryptosporidium baileyi, and Cryptosporidium galli (Current et al. 1986; Ryan et al. 2003b; Slavin 1955). In addition, 11 Cryptosporidium genotypes have been described in more than 30 bird species worldwide, including avian I–V, goose genotypes I–IV, duck genotype, and Euroasian Woodcock genotype (Ryan 2010). Of these, only C. meleagridis is known to also infect humans (Alves et al. 2003; Cama et al. 2003; McLauchlin et al. 2000; Xiao and Ryan 2004). Although mammal-specific Cryptosporidium species and genotypes are rarely detected in birds, C. hominis, C. hominis-like, C. parvum, and muskrat genotype I have been reported in faecal samples from Canada geese (Branta canadensis) (Graczyk et al. 1998; Jellison et al. 2004, 2009; Zhou et al. 2004).
Natural cryptosporidiosis of birds caused by C. meleagridis and C. galli affects the gastrointestinal tract and manifests in different degrees of enteritis (Gharagozlou et al. 2006; Ryan et al. 2003b), whereas C. baileyi infects many sites, including conjunctiva, nasopharynx, trachea, bronchi, air sac, gut, bursa of Fabricius, kidneys, and urinary tract, and manifests in three clinical forms: respiratory disease, enteritis, and renal disease (Lindsay and Blagburn 1990). Usually only one form of the disease is present in an outbreak (Lindsay and Blagburn 1990). Also Cryptosporidium avian genotype III was reported as a possible cause of chronic vomiting in peach-faced lovebirds (Agapornis roseicollis) (Makino et al. 2010). Pathogenicity has not been described for other bird-derived Cryptosporidium genotypes (Ng et al. 2006).
The redescription of Cryptosporidium genotypes as new species requires morphometric studies of oocysts, genetic characterizations, and demonstration of host specificity (natural and, where possible, experimental) (Xiao et al. 2004). These data have thus far been lacking for Cryptosporidium genotypes from birds (Ng et al. 2006). The present study aimed to address this deficiency for Cryptosporidium avian genotype V, a genotype first reported in cockatiels (Nymphicus holandicus) in Japan (Abe and Makino 2010) and subsequently in many other bird hosts (Table 1). Based on the collective data from this and other studies, we conclude that Cryptosporidium avian V is genetically and biologically distinct from recognized Cryptosporidium species, and we propose that it be named Cryptosporidium avium.
Table 1.
Occurrence of Cryptosporidium avium n. sp. (previously known as avian genotype V) demonstrated on the basis of partial sequences of SSU, actin, and HSP70 in various bird hosts in the world.
| Host (Scientific name) | Location | Genes (GenBank accession number) | References |
|---|---|---|---|
| Cockatiel (Nymphicus holandicus) | Japan | SSU (AB471646); actin (AB471660); HSP70 (AB471665) | Abe and Makino (2010) |
| China | SSU (HM116381) | Qi et al. (2010) | |
| China | SSU (JQ246415); actin (JQ320301) | unpublished | |
| China | SSU (KM267556)* | Zhang et al. (2015) | |
| Chicken (Gallus gallus) | China | SSU (JX548299) | Wang et al. (2014) |
| Blue-fronted Amazon (Amazona aestiva) | Brazil | SSU (KJ487974) | Nakamura et al. (2014) |
| Major Mitchell's Cockatoo (Lophochroa leadbeateri) | USA | SSU (KP342400) | Curtiss et al. (2015) |
| Budgerigar (Melopsittacus undulates) | China | SSU (KM267556)* | Zhang et al. (2015) |
Identical GenBank accession number for sequence acquired from two different hosts cockatiel and budgerigar; SSU – small ribosomal subunit rRNA; HSP70 – 70 kDa heat shock protein
2. Materials and methods
2.1. Source of oocysts for studies
Oocysts of C. avium were originally isolated from faecal samples of four naturally infected adult red-crowned parakeets (Cyanoramphus novaezealandiae), which were caged by a private owner in České Budějovice (Czech Republic). Cryptosporidium avium oocysts from these red-crowned parakeets were pooled and used to infect a single 6-month-old hen (hen 1; Gallus gallus f. domestica). Oocysts from hen 1 were used to infect other animals (see 2.6.).
2.2. Parasitological examination and oocyst preparation
Animal faeces were screened for Cryptosporidium oocysts using faecal smears stained with aniline-carbolmethyl violet (ACMV) (Miláček and Vítovec 1985). Faecal specimens were collected daily and stored in a 2.5% potassium dichromate solution at 4–8°C.
Cryptosporidium oocysts originated from red-crowned parakeets and from hen 1 were purified using caesium chloride gradient centrifugation for morphometry analyses and transmission studies (Kilani and Sekla 1987). The viability of oocysts was examined using propidium iodide (PI) staining by a modified assay of Sauch et al. (1991). Briefly, examined oocysts were washed in distilled water (DW; 10,000 oocysts in 100 μl) and mixed with 1 μl of PI (1% solution, SIGMA). After 30 min of incubation at room temperature in the dark, the oocysts were washed twice with DW. Oocyst viability was examined using fluorescence microscopy (filter 420 nm, Olympus IX70). Oocysts with red fluorescence were considered to be dead, and those without fluorescence were considered viable.
2.3. Oocyst morphology
Cryptosporidium avium oocysts for morphology and morphometry analyses were examined using differential interference contrast (DIC) microscopy, brightfield microscopy following ACMV staining, and fluorescence microscopy following labelling with genus-specific FITC-conjugated antibodies (Cryptosporidium IF Test, Crypto cel, Medac) (Olympus IX70 microscope, filter 520 nm). Morphology and morphometry were determined using digital analysis of images (M.I.C. Quick Photo Pro v.3.0 soft-ware; Optical Service, Czech Republic) collected using a Camedia C 5060 WIDEZOOM 5.1 megapixel digital camera (Optical Service). A 20 μl aliquot containing ~10,000 purified oocysts was examined for each measurement. Length and width of oocysts (n = 100) were measured under DIC at 1000× magnification, and these were used to calculate the length-to-width ratio of each oocyst. As a control, the morphometry of C. baileyi (n=100) from a naturally infected adult Common Peafowl (Pavo cristatus) were measured by the same person using the same microscope. Photomicrographs of C. avium (avian genotype V) oocysts observed by DIC, ACMV and IFA were deposited as a phototype at the Institute of Parasitology, Biology Centre of the Czech Academy of Sciences, Czech Republic.
2.4. DNA extraction and molecular analyses
Total DNA was extracted from 200 mg of faeces, 10,000 purified oocysts, or 200 mg of tissue by bead disruption for 60 s at 5.5 m/s using 0.5 mm glass beads in a FastPrep®24 Instrument (MP Biomedicals, CA, USA). DNA was isolated and purified using a commercially available kit in accordance with the manufacturer's instructions (QIAamp® DNA Stool Mini Kit or DNeasy® Blood & Tissue Kit, Qiagen, Hilden, Germany). Purified DNA was stored at −20 °C prior to being used for PCR. A nested PCR approach was used to amplify a region of the SSU (~830 bp; Jiang et al. 2005; Xiao et al. 1999), actin (~1066 bp; Sulaiman et al. 2002), and HSP70 genes (~1950 bp; Sulaiman et al. 2000). Both primary and secondary PCR reactions were carried out in a volume of 50 μl; the primary reaction contained 2 μl of genomic DNA (or water as a negative control) and the secondary reaction contained 2 μl of the primary reaction as template. DNA of C. parvum and C. baileyi were used as positive controls. Secondary PCR products were detected by agarose gel (2%) electrophoresis, visualized by ethidium bromide staining, and extracted using QIAquick® Gel Extraction Kit (Qiagen). Purified secondary products were sequenced in both directions with an ABI 3130 genetic analyser (Applied Biosystems, Foster City, CA) using the secondary PCR primers and the BigDye1 Terminator V3.1 cycle sequencing kit (Applied Biosystems, Foster City, California) in 10 μl reactions.
2.5. Phylogenetic analyses
The nucleotide sequences of each gene obtained in this study were edited using the ChromasPro 1.7.5 software (Technelysium, Pty, Ltd.) and aligned with each other and with reference sequences from GenBank using MAFFT version 7 online server with automatic selection of alignment mode (http://mafft.cbrc.jp/alignment/software/). Phylogenetic analyses were performed and best DNA/Protein phylogeny models were selected using the MEGA6 software (Guindon and Gascuel 2003; Tamura et al. 2011). Phylogenetic trees were inferred by the maximum likelihood (ML) method, with the substitution model that best fit the alignment selected using the Bayesian information criterion. The Tamura 3-parameter model (Tamura 1992) was selected for SSU and HSP70 alignments, and the general time reversible model (Tavaré 1986) was selected for actin alignment. Bootstrap support for branching was based on 1000 replications. Phylograms were drawn using the MEGA6 and were manually adjusted using CorelDrawX7. Sequences of SSU, actin, and HSP70 derived in this study have been deposited in GenBank under accession numbers KU058875–KU058886.
2.6. Transmission studies
2.6.1. Animals
Three 8-week-old SCID mice (strain C.B-17), three 8-week-old BALB/c mice (Charles River, Germany), three 6-month-old hens (hen 2-4; Gallus gallus f. domestica), and three 6-month-old budgerigars (bud 1-3; Melopsittacus undulatus) were used for experimental infection studies. In addition, three animals from each host species/strain were used as negative control.
2.6.2. Experimental design
To prevent environmental contamination with oocysts, laboratory rodents were housed in plastic cages and supplied with a sterilized diet (TOP-VELAZ, Prague, Czech Republic) and sterilized water ad libitum. Hens and budgerigars were kept in species-appropriate birdcages with sterilized wood-chip bedding and without bedding, respectively, and were supplied with sterilized food and water ad libitum. Each animal was inoculated orally by stomach tube with 100,000 purified viable oocysts suspended in 200 μl of distilled water. Animals serving as negative controls were inoculated orally by stomach tube with 200 μl of distilled water. Faecal samples from all animals were screened daily for the presence of Cryptosporidium oocyst using ACMV staining and the presence of Cryptosporidium specific DNA was confirmed using nested PCR targeting the SSU gene. All experiments were terminated 30 days post infection (DPI). Infection intensity was reported as the number of oocysts per gram (OPG) of faeces as previously described by Kváč et al. (2007). In addition, faecal consistency and colour and general health status was examined daily. One Cryptosporidium avium positive animal from each host group was euthanized 20 DPI. Tissue specimens were processed for PCR detection, histology, and electron microscopy.
2.6.3. Histopathological examinations
The complete examination of all gastrointestinal organs was conducted at necropsy. Tissue specimens from the stomach, small intestine, and large intestine (the entire tract was divided into 1 cm sections) were sampled and processed for histology according to Kváč and Vítovec (2003) and for PCR analyses (see Section 2.3). Histology sections were stained with haematoxylin and eosin (HE), Wolbach's modified Giemsa stain, and genus-specific FITC conjugated monoclonal antibodies targeting Cryptosporidium oocyst wall antigens (Cryptosporidium IF Test, Crypto cel, Medac).
2.6.4. Scanning electron microscopy
Samples of intestinal tissue originating from a host confirmed to be infected with C. avium were fixed in freshly prepared 3% glutaraldehyde (v/v) in cacodylate buffer (0.1 M, pH 7.4) at 4°C and further processed for SEM as described in Valigurová et al. (2008). All samples were examined by JEOL JSM-7401F.
2.6.5. Animal care
Animal caretakers wore new disposable coveralls, shoe covers, and gloves every time they entered the experimental room. All wood-chip bedding, faeces, and disposable protective clothing were sealed in plastic bags, removed from the experimental room, and incinerated. All housing, feeding, and experimental procedures were conducted under protocols approved by the Institute of Parasitology, Biology Centre and Central Commission for Animal Welfare, Czech Republic (Protocols No. 071/2010 and 114/2013).
3. Results
In the present study, Cryptosporidium avium was detected in naturally infected red-crowned parakeets (Cyanoramphus novaezealandiae) (n=4), which continuously shed oocysts for more than 5 months.
3.1. Oocyst morphology
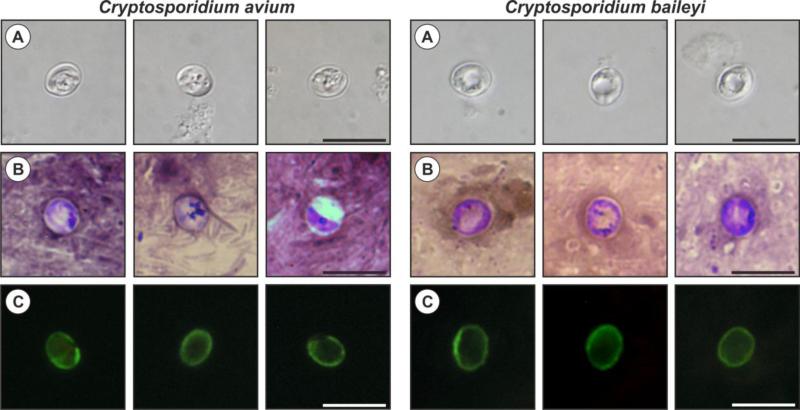
Oocysts of C. avium originated from naturally infected red-crowned parakeets were morphometrically identical to those recovered from experimentally infected hen no. 1, measuring 5.30–6.90 μm (mean = 6.26 μm) × 4.30–5.50 μm (mean = 4.86 μm) with a length to width ratio of 1.29 (1.14–1.47) (n = 100; Fig. 1a), and they were smaller than oocysts of C. baileyi, measuring 5.90–7.60 μm (mean = 6.90 μm) × 4.30–6.60 μm (mean = 5.50 μm) with a length to width ratio of 1.25 (1.06–1.43) (n = 100; Fig. 1). Oocysts in faecal smears showed typical Cryptosporidium ACMV staining characteristics (Fig. 1b). Fixed C. avium oocysts labelled with FITC conjugated anti-Cryptosporidium oocyst wall antibody and examined by fluorescence microscopy had typical apple green, halo-like fluorescence (Fig. 1c).
Figure 1.
Cryptosporidium avium and Cryptosporidium baileyi oocysts visualized in various preparations: (A) differential interference contrast microscopy, (B) aniline–carbol–methyl violet staining, and (C) labelled with anti-Cryptosporidium FITC-conjugated antibody. Bar = 10 μm.
3.2. Molecular characterization
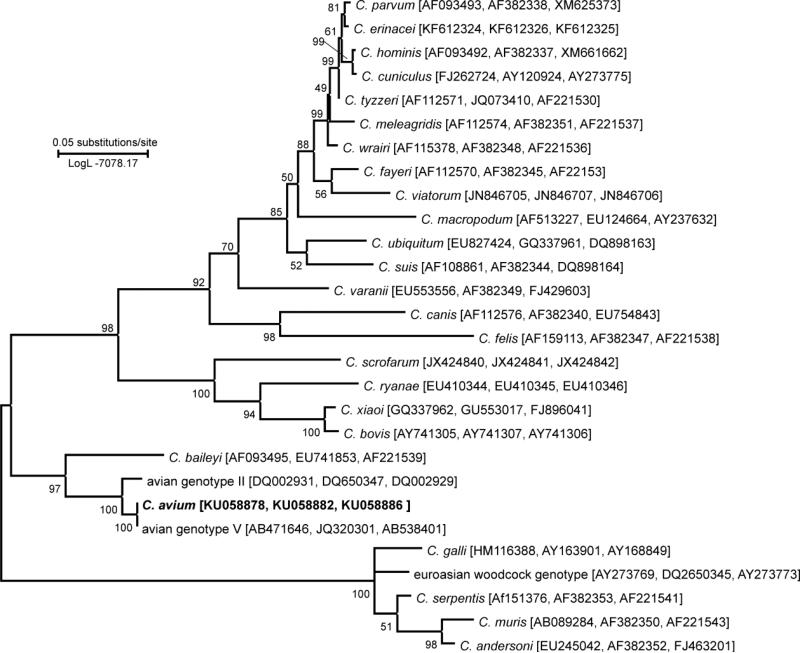
At the SSU locus, all isolates of C. avium (from naturally infected red-crowned parakeets and experimentally infected hens and budgerigars) shared 100% identity with each other and with Cryptosporidium avian genotype V obtained from cockatiels in Japan (AB471646, AB471647) and China (HM116381). At the actin locus, C. avium isolates from all experimentally susceptible hosts shared 100% identity with each other each and with the GenBank sequences of Cryptosporidium avian genotype V obtained from cockatiels in Japan (AB471660, AB471661) and China (JQ320301). At HSP70 locus, all sequences of C. avium isolates were identical to sequences obtained from a cockatiel (AB471665) and from a rosy-faced lovebird (Agapornis roseicollis; AB538401) in Japan. Maximum likelihood trees inferred from sequences of individual genes (data no shown) and concatenated SSU, actin, and HSP70 sequences (Fig. 2) showed that C. avium is most closely related to Cryptosporidium avian genotype II and also clusters with Cryptosporidium avian genotype I and C. baileyi.
Figure 2.
Phylogenetic relationships between Cryptosporidium avium and selected Cryptosporidium spp. as inferred by a maximum likelihood (ML) analysis of concatenated sequences constructed from partial DNA sequences of SSU, actin, and HSP70 loci (1234 base positions in the final dataset; model Tamura 3-parametr G+I). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates). Numbers at the nodes represent bootstrap values for the nodes gaining more than 50% support. Scale bar included in tree.
3.3. Experimental transmission studies
Oocyst used for experimental infections had >90% viability, determined by PI staining. Experimentally inoculated SCID and BALB/c mice did not produce detectable C. avium oocysts by microscopy or specific DNA by PCR in faecal samples within 30 DPI. No clinical signs of cryptosporidiosis were detected in any laboratory rodent. Histological and molecular examination of gastrointestinal tract tissue from these rodents did not reveal the presence of Cryptosporidium developmental stages or Cryptosporidium-specific DNA.
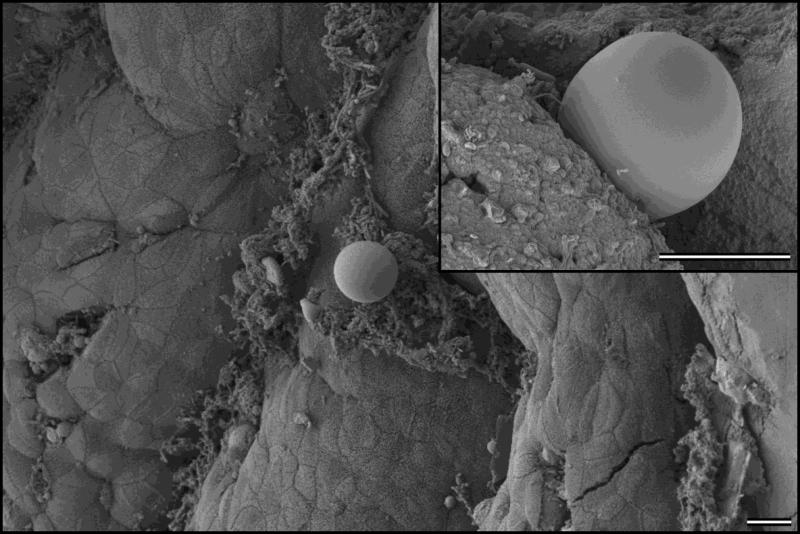
Cryptosporidium avium was fully infectious for all hens and budgerigars. Oocysts were microscopically detected by 12 DPI in both hens and budgerigars. Oocysts of C. avium were microscopically detected 12–16 DPI in hens and 12 DPI in budgerigars. The infection intensity of C. avium in hens and budgerigars was generally low – hens shed oocysts in range 2,000 to 5,000 OPG, while budgerigars did not shed more than 2,000 OPG. Specific DNA of C. avium was detected in faeces of both hens and budgerigars from 11 DPI and then intermittently until the end of the experiment. Infected birds showed no symptoms of the disease and hens and budgerigars necropsied at 20 DPI or 30 DPI showed no macroscopic signs of cryptosporidiosis. No developmental stages of C. avium were histologically observed in either hens or budgerigars. However, scanning electron microscopy revealed the presence of developmental stages of C. avium attached to the microvilli in the ileum and caecum of hens (Fig. 3) and budgerigars. No pathology-associated changes were observed.
Figure 3.
Scanning electron photomicrograph. Epithelium of ileum of a hen, sacrificed 20 days DPI, showing attached Cryptosporidium avium. Detail of the attached C. avium developmental stage is provided in the upper right corner. Bar = 10 μm.
3.4. Taxonomic summary
Cryptosporidium avium
Diagnosis: Oocysts are shed fully sporulated. Sporulated oocysts (n=100) measure 5.30–6.90 μm (mean = 6.26 μm) × 4.30–5.50 μm (mean = 4.86 μm) with a length to width ratio of 1.29 (1.14–1.47). Endogenous stages are unknown.
Type host: red-crowned parakeet (Cyanoramphus novaezealandiae)
Other natural hosts: rosy-faced lovebird (Agapornis roseicollis), Chicken (Gallus gallus), blue-fronted Amazon (Amazona aestiva), major Mitchell's cockatoo (Lophochroa leadbeateri), cockatiel (Nymphicus holandicus), budgerigar (Melopsittacus undulates) (Table 1)
Experimental hosts: hen (Gallus gallus domesticus), budgerigar (Melopsittacus undulatus)
Prepatent period: 11 DPI
Patent period: at least 30 DPI
Type locality: České Budějovice, Czech Republic
Other localities: Brazil, China, Japan, USA
Site of infection: ileum, ceacum (this study), kidney, ureter, and cloaca (Curtiss et al. 2015)
Material deposited: A phototype, description of oocysts, and DNA are deposited at the Institute of Parasitology, Biology Centre of the Academy of Sciences of the Czech Republic.
DNA sequences: Partial sequences of SSU, actin, and HSP70 genes were submitted to GenBank under the accession numbers KU058875–KU058886.
Etymology: The species name avium is derived from the Latin noun “avis” (meaning a bird) according to ICZN Article 11.9.1-3 as a plural in the genitive case, as it appears to be adapted to birds.
Morphological, genetic, and biological data support the establishment of Cryptosporidium avian genotype V as a new species. According to ICZN and criteria for naming species we propose the name Cryptosporidium avium.
4. Discussion
Avian-adapted Cryptosporidium species and genotypes appear to infect a broad range of bird species (Ryan 2010). This is supported by our finding that C. avium could be transmitted from parrots, which are in the order Psittaciformes, to hens, which are in the order Galliformes. It is therefore unsurprising that the host range of C. avium overlaps that of other avian-adapted Cryptosporidium, including the closely related avian genotype II (Abe and Makino 2010). In contrast to C. meleagridis, which has been reported in calves, pigs, rabbits, rats, mice, and humans (Akiyoshi et al. 2003; Cama et al. 2003; Darabus and Olariu 2003; Elwin et al. 2012; Huang et al. 2003; O'Donoghue 1995; Xiao and Ryan 2004) there is no evidence that C. avium infects non-avian hosts (present study; Kváč et al. 2014b).
Most birds infected with C. avium, including experimentally infected hens and budgerigars, showed no clinical signs of cryptosporidiosis (present study; Ng et al. 2006); however, a 7-yr-old Major Mitchell's cockatoo (Lophochroa leadbeateri) showed signs of lethargy, anorexia, and cloacal prolapse (Curtiss et al. 2015). Cryptosporidium avium was detected in the kidneys, ureter, and cloaca of the Major Mitchell's cockatoo, and developmental stages were found in the ileum and caecum in the present study. This broad tissue tropism is similar to the genetically related species and genotype, C. baileyi and avian genotype II (Nakamura and Meireles 2015).
Until now, the course of Cryptosporidium infection in birds has been described only for by C. meleagridis, C. baileyi, and C. galli (Current et al. 1986; Ryan et al. 2003b; Slavin 1955). We have shown that the prepatent period of C. avium (12 days) is significantly longer than that of C. meleagridis and C. baileyi (4–8 days; Hornok et al. 1998; Lindsay et al. 1988; Rhee et al. 1991; Tůmová et al. 2002) and shorter than that of C. galli (25 days, Pavlásek 2001). Differences in the prepatent period of Cryptosporidium species are not unusual, even for phylogenetically closely related species infecting the same host. For example, C. bovis and C. ryanae have a same host range (cattle) and share 98% sequence identity at the SSU locus, but C. ryanae has a shorter prepatent period (11 days) than C. bovis (16 days) (Fayer et al. 2005, 2008).
Although infected birds shed low numbers of C. avium oocysts, shedding continued for the duration of experimental infections (30 DPI), and naturally infected red-crowned parakeets continued to shed oocysts for at least 5 months. A several month-long natural infection was previously observed in various passerines naturally infected with the gastric species C. galli. The reported duration of C. baileyi and C. meleagridis infections ranges from 4 to 151 days and 4 to 21 days, respectively, depending on species and age of the host (Bermudez et al. 1988; Sreter et al. 1995; Tůmová et al. 2002; Woodmansee et al. 1988).
Cryptosporidium avium oocysts from this study (5.30–6.90 × 4.30–5.50 μm) are morphometrically indistinguishable from those of Cryptosporidium avian genotype V (5.0–6.6 × 4.1–5.2 μm, Qi et al. 2011), similar to those of Cryptosporidium avian genotype II (6.0–6.5 × 4.8–6.6 μm, Meireles et al. 2006; Ng et al. 2006; Qi et al. 2011) and C. baileyi (6.3 × 4.6 μm, Current et al. 1986), larger than those of C. meleagridis (5.0 × 4.3 μm, Slavin 1955), and smaller than those of C. galli (8.0–8.5 × 6.2–6.4 μm, Ryan et al. 2003b), Cryptosporidium avian III (7.5 × 6.3 μm, Meireles et al. 2006; Ng et al. 2006) and Euroasian woodcock genotype (8.5 × 6.4 μm, Ryan et al. 2003a).
Phylogenetic analyses based on SSU, actin, and HSP70 gene sequences showed that C. avium is genetically distinct from known species and is most closely related to C. bailey and Cryptosporidium avian genotypes I and II.
At the SSU locus, C. avium exhibits 1.70% and 0.28% genetic distance from avian genotypes I and II, respectively, and 2.27% genetic distance from C. baileyi. At the actin locus, the genetic distance from avian genotypes I and II is 10.8%, 1.86%, respectively, the genetic distance from C. baileyi is 11.04%. At the HSP70 locus, C. avium exhibits 4.49% and 12.92% genetic distance from avian genotype II and C. baileyi, respectively. These differences are comparable to genetic distances of currently accepted species. For example, at the SSU, actin, and HSP70 loci, the respective genetic distances between C. parvum and C. erinacei is 0.42%, 0.41%, and 0.72%; C. hominis and C. cuniculus is 1.11%, 0.37%, and 1.65%; and C. muris and C. andersoni is 0.70%, 3.54%, and 2.21% at SSU, actin, and HSP70 loci, respectively.
Acknowledgements
This study was funded by the Czech Science Foundation (15-01090S), and by National Institutes of Health project 2P20 RR015566. Sarah Menchaca was supported in part by a grant of the University of Arizona from the National Institutes of Health (T37 MD001427).
References
- Abe N, Makino I. Multilocus genotypic analysis of Cryptosporidium isolates from cockatiels, Japan. Parasitol Res. 2010;106:1491–1497. doi: 10.1007/s00436-010-1810-5. doi:10.1007/s00436-010-1810-5. [DOI] [PubMed] [Google Scholar]
- Akiyoshi DE, Dilo J, Pearson C, Chapman S, Tumwine J, Tzipori S. Characterization of Cryptosporidium meleagridis of human origin passaged through different host species. Infect Immun. 2003;71:1828–1832. doi: 10.1128/IAI.71.4.1828-1832.2003. doi:10.1128/IAI.71.4.1828-1832.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves M, Xiao LH, Sulaiman I, Lal AA, Matos O, Antunes F. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J Clin Microbiol. 2003;41:2744–2747. doi: 10.1128/JCM.41.6.2744-2747.2003. doi:10.1128/JCM.41.6.2744-2747.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez AJ, Ley DH, Levy MG, Ficken MD, Guy JS, Gerig TM. Intestinal and bursal cryptosporidiosis in turkeys following inoculation with Cryptosporidium sp. isolated from commercial poults. Avian Dis. 1988;32:445–450. doi:10.2307/1590910. [PubMed] [Google Scholar]
- Cama VA, Bern C, Sulaiman IM, Gilman RH, Ticona E, Vivar A, Kawai V, Vargas D, Zhou L, Xiao LH. Cryptosporidium species and genotypes in HIV-positive patients in Lima, Peru. J Eukaryot Microbiol. 2003;50:531–533. doi: 10.1111/j.1550-7408.2003.tb00620.x. doi:10.1111/j.1550-7408.2003.tb00620.x. [DOI] [PubMed] [Google Scholar]
- Current WL, Upton SJ, Haynes TB. The life cycle of Cryptosporidium baileyi n. sp. (Apicomplexa, Cryptosporidiidae) infecting chickens. J Protozool. 1986;33:289–296. doi: 10.1111/j.1550-7408.1986.tb05608.x. doi:10.1111/j.1550-7408.1986.tb05608.x. [DOI] [PubMed] [Google Scholar]
- Curtiss JB, Leone AM, Wellehan JF, Jr., Emerson JA, Howerth EW, Farina LL. Renal and Cloacal Cryptosporidiosis (Cryptosporidium Avian Genotype V) in a Major Mitchell's Cockatoo (Lophochroa Leadbeateri). J Zoo Wildl Med. 2015;46:934–937. doi: 10.1638/2015-0025.1. doi:10.1638/2015-0025.1. [DOI] [PubMed] [Google Scholar]
- Darabus G, Olariu R. The homologous and interspecies transmission of Cryptosporidium parvum and Cryptosporidium meleagridis. Pol J Vet Sci. 2003;6:225–228. doi:10.1371/journal.pone.0054857. [PubMed] [Google Scholar]
- Elwin K, Robinson G, Hadfield SJ, Fairclough HV, Iturriza-Gomara M, Chalmers RM. A comparison of two approaches to extracting Cryptosporidium DNA from human stools as measured by a real-time PCR assay. J Microbiol Methods. 2012;89:38–40. doi: 10.1016/j.mimet.2012.02.006. doi:10.1016/j.mimet.2012.02.006. [DOI] [PubMed] [Google Scholar]
- Fayer R. Taxonomy and species delimitation in Cryptosporidium. Exp Parasitol. 2010;124:90–97. doi: 10.1016/j.exppara.2009.03.005. doi:10.1016/j.exppara.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Fayer R, Santín M, Trout JM. Cryptosporidium ryanae n. sp. (Apicomplexa: Cryptosporidiidae) in cattle (Bos taurus). Vet Parasitol. 2008;156:191–198. doi: 10.1016/j.vetpar.2008.05.024. doi:10.1016/j.vetpar.2008.05.024. [DOI] [PubMed] [Google Scholar]
- Fayer R, Santín M, Xiao L. Cryptosporidium bovis n. sp. (Apicomplexa: Cryptosporidiidae) in cattle (Bos taurus). J Parasitol. 2005;91:624–629. doi: 10.1645/GE-3435. doi:10.1645/GE-3435. [DOI] [PubMed] [Google Scholar]
- Gharagozlou MJ, Dezfoulian O, Rahbari S, Bokaie S, Jahanzad I, Razavi AN. Intestinal cryptosporidiosis in turkeys in Iran. J Vet Med A Physiol Pathol Clin Med. 2006;53:282–285. doi: 10.1111/j.1439-0442.2006.00843.x. doi:10.1111/j.1439-0442.2006.00843.x. [DOI] [PubMed] [Google Scholar]
- Graczyk TK, Fayer R, Trout JM, Lewis EJ, Farley CA, Sulaiman I, Lal AA. Giardia sp. cysts and infectious Cryptosporidium parvum oocysts in the feces of migratory Canada geese (Branta canadensis). Appl Environ Microbiol. 1998;64:2736–2738. doi: 10.1128/aem.64.7.2736-2738.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. doi:10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hornok S, Bitay Z, Szell Z, Varga I. Assessment of maternal immunity to Cryptosporidium baileyi in chickens. Vet Parasitol. 1998;79:203–212. doi: 10.1016/s0304-4017(98)00170-8. doi:10.1016/S0304-4017(98)00170-8. [DOI] [PubMed] [Google Scholar]
- Huang K, Akiyoshi DE, Feng X, Tzipori S. Development of patent infection in immunosuppressed C57Bl/6 mice with a single Cryptosporidium meleagridis oocyst. J Parasitol. 2003;89:620–622. doi: 10.1645/0022-3395(2003)089[0620:DOPIII]2.0.CO;2. doi:10.1645/0022-3395(2003)089[0620:dopiii]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Jellison KL, Distel DL, Hemond HF, Schauer DB. Phylogenetic analysis of the hypervariable region of the 18S rRNA gene of Cryptosporidium oocysts in feces of Canada geese (Branta canadensis): evidence for five novel genotypes. Appl Environ Microbiol. 2004;70:452–458. doi: 10.1128/AEM.70.1.452-458.2004. doi:10.1128/AEM.70.1.452-458.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellison KL, Lynch AE, Ziemann JM. Source tracking identifies deer and geese as vectors of human-infectious Cryptosporidium genotypes in an urban/suburban watershed. Environ Sci Technol. 2009;43:4267–4272. doi: 10.1021/es900081m. doi:10.1021/es900081m. [DOI] [PubMed] [Google Scholar]
- Jiang J, Alderisio KA, Xiao L. Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl Environ Microbiol. 2005;71:4446–4454. doi: 10.1128/AEM.71.8.4446-4454.2005. doi:10.1128/AEM.71.8.4446-4454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilani RT, Sekla L. Purification of Cryptosporidium oocysts and sporozoites by cesium chloride and Percoll gradients. Am J Trop Med Hyg. 1987;36:505–508. doi: 10.4269/ajtmh.1987.36.505. [DOI] [PubMed] [Google Scholar]
- Kváč M, Hofmannová L, Hlásková L, Květoňová D, Vítovec J, McEvoy J, Sak B. Cryptosporidium erinacei n. sp. (Apicomplexa: Cryptosporidiidae) in hedgehogs. Vet Parasitol. 2014a;201:9–17. doi: 10.1016/j.vetpar.2014.01.014. doi:10.1016/j.vetpar.2014.01.014. [DOI] [PubMed] [Google Scholar]
- Kváč M, McEvoy J, Stenger B, Clark M. Cryptosporidiosis in other vertebrates. In: Cacciò SM, Widmer G, editors. Cryptosporidium: Parasite and Disease. 1st edn. Springer; Wien: 2014b. pp. 237–326. [Google Scholar]
- Kváč M, Ondráčková Z, Květoňová D, Sak B, Vítovec J. Infectivity and pathogenicity of Cryptosporidium andersoni to a novel host, southern multimammate mouse (Mastomys coucha). Vet Parasitol. 2007;143:229–233. doi: 10.1016/j.vetpar.2006.08.031. doi:10.1016/j.vetpar.2006.08.031. [DOI] [PubMed] [Google Scholar]
- Kváč M, Vítovec J. Prevalence and pathogenicity of Cryptosporidium andersoni in one herd of beef cattle. J Vet Med B. 2003;50:451–457. doi: 10.1046/j.0931-1793.2003.00701.x. doi:10.1046/j.0931-1793.2003.00701.x. [DOI] [PubMed] [Google Scholar]
- Lindsay DS, Blagburn BL. Cryptosporidiosis in Birds. In: Dubey JP, Speer CA, Fayer R, editors. Cryptosporidiosis in Man and Animals. Boca Raton, FL. 1st edn. CRC Press; 1990. pp. 133–148. [Google Scholar]
- Lindsay DS, Blagburn BL, Sundermann CA, Giambrone JJ. Effect of broiler chicken age on susceptibility to experimentally induced Cryptosporidium baileyi infection. Am J Vet Res. 1988;49:1412–1414. [PubMed] [Google Scholar]
- Liu X, He T, Zhong Z, Zhang H, Wang R, Dong H, Wang C, Li D, Deng J, Peng G, Zhang L. A new genotype of Cryptosporidium from giant panda (Ailuropoda melanoleuca) in China. Parasitol Int. 2013;62:454–458. doi: 10.1016/j.parint.2013.06.004. doi:10.1016/j.parint.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Makino I, Abe N, Reavill DR. Cryptosporidium avian genotype III as a possible causative agent of chronic vomiting in peach-faced lovebirds (Agapornis roseicollis). Avian Dis. 2010;54:1102–1107. doi: 10.1637/9227-123009-Case.1. doi:10.1637/9227-123009-Case.1. [DOI] [PubMed] [Google Scholar]
- McLauchlin J, Amar C, Pedraza-Diaz S, Nichols GL. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: results of genotyping Cryptosporidium spp. in 1,705 fecal samples from humans and 105 fecal samples from livestock animals. J Clin Microbiol. 2000;38:3984–3990. doi: 10.1128/jcm.38.11.3984-3990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meireles MV, Soares RM, dos Santos MM, Gennari SM. Biological studies and molecular characterization of a Cryptosporidium isolate from ostriches (Struthio camelus). J Parasitol. 2006;92:623–626. doi: 10.1645/0022-3395(2006)92[623:BSAMCO]2.0.CO;2. doi:10.1645/0022-3395(2006)92[623:bsamco]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Miláček P, Vítovec J. Differential staining of cryptosporidia by aniline-carbol-methyl violet and tartrazine in smears from feces and scrapings of intestinal mucosa. Folia Parasitol. 1985;32:50. [PubMed] [Google Scholar]
- Nakamura AA, Homem CG, da Silva AM, Meireles MV. Diagnosis of gastric cryptosporidiosis in birds using a duplex real-time PCR assay. Vet Parasitol. 2014;205:7–13. doi: 10.1016/j.vetpar.2014.07.033. doi:10.1016/j.vetpar.2014.07.033. [DOI] [PubMed] [Google Scholar]
- Nakamura AA, Meireles MV. Cryptosporidium infections in birds--a review. Rev Bras Parasitol Vet. 2015;24:253–267. doi: 10.1590/S1984-29612015063. doi:10.1590/S1984-29612015063. [DOI] [PubMed] [Google Scholar]
- Ng J, Pavlásek I, Ryan U. Identification of novel Cryptosporidium genotypes from avian hosts. Appl Environ Microbiol. 2006;72:7548–7553. doi: 10.1128/AEM.01352-06. doi:10.1128/AEM.01352-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donoghue PJ. Cryptosporidium and cryptosporidiosis in man and animals. Int J Parasitol. 1995;25:139–195. doi: 10.1016/0020-7519(94)e0059-v. doi:10.1016/0020-7519(94)E0059-V. [DOI] [PubMed] [Google Scholar]
- Pavlásek I. Findings of cryptosporidia in the stomach of hens and of exotic and wild birds. Veterinářství (Czech) 2001;51:103–108. [Google Scholar]
- Qi M, Wang R, Ning C, Li X, Zhang L, Jian F, Sun Y, Xiao L. Cryptosporidium spp. in pet birds: genetic diversity and potential public health significance. Exp Parasitol. 2011;128:336–340. doi: 10.1016/j.exppara.2011.04.003. doi:10.1016/j.exppara.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Rhee JK, Seu YS, Park BK. Isolation and identification of Cryptosporidium from various animals in Korea. III. Identification of Cryptosporidium baileyi from Korean chicken. Korean J Parasitol. 1991;29:315–324. doi: 10.3347/kjp.1991.29.4.315. doi:10.3347/kjp.1991.29.4.315. [DOI] [PubMed] [Google Scholar]
- Ryan U. Cryptosporidium in birds, fish and amphibians. Exp Parasitol. 2010;124:113–120. doi: 10.1016/j.exppara.2009.02.002. doi:10.1016/j.exppara.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Ryan U, Xiao L, Read C, Zhou L, Lal AA, Pavlásek I. Identification of novel Cryptosporidium genotypes from the Czech Republic. Appl Environ Microbiol. 2003a;69:4302–4307. doi: 10.1128/AEM.69.7.4302-4307.2003. doi:10.1128/AEM.69.7.4302-4307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan UM, Xiao L, Read C, Sulaiman IM, Monis P, Lal AA, Fayer R, Pavlásek I. A redescription of Cryptosporidium galli Pavlásek, 1999 (Apicomplexa : Cryptosporidiidae) from birds. J Parasitol. 2003b;89:809–813. doi: 10.1645/GE-74RI. doi: 10.1645/GE-74RI. [DOI] [PubMed] [Google Scholar]
- Sauch JF, Flanigan D, Galvin ML, Berman D, Jakubowski W. Propidium iodide as an indicator of Giardia cyst viability. Appl Environ Microbiol. 1991;57:3243–3247. doi: 10.1128/aem.57.11.3243-3247.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavin D. Cryptosporidium meleagridis (sp. nov.). J Comp Pathol. 1955;65:262–266. doi: 10.1016/s0368-1742(55)80025-2. [DOI] [PubMed] [Google Scholar]
- Sreter T, Varga I, Bekesi L. Age-dependent resistance to Cryptosporidium baileyi infection in chickens. J Parasitol. 1995;81:827–829. doi:10.2307/3283992. [PubMed] [Google Scholar]
- Sulaiman IM, Lal AA, Xiao LH. Molecular phylogeny and evolutionary relationships of Cryptosporidium parasites at the actin locus. J Parasitol. 2002;88:388–394. doi: 10.1645/0022-3395(2002)088[0388:MPAERO]2.0.CO;2. doi:10.1645/0022-3395(2002)088[0388:MPAERO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Sulaiman IM, Morgan UM, Thompson RC, Lal AA, Xiao L. Phylogenetic relationships of Cryptosporidium parasites based on the 70-kilodalton heat shock protein (HSP70) gene. Appl Environ Microbiol. 2000;66:2385–2391. doi: 10.1128/aem.66.6.2385-2391.2000. doi:10.1128/AEM.66.6.2385-2391.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol Biol Evol. 1992;9:678–687. doi: 10.1093/oxfordjournals.molbev.a040752. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. doi:10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavaré S. Some probabilistic and statistical problems in the analysis of DNA sequences. In: Miura RM, editor. Some Mathematical Questions in Biology: DNA Sequence Analysis (Lectures on Mathematics in the Life Sciences) New York. American Mathematical Society; 1986. pp. 57–86. [Google Scholar]
- Tůmová E, Skřivan M, Marounek M, Pavlásek I, Ledvinka Z. Performance and oocyst shedding in broiler chickens orally infected with Cryptosporidium baileyi and Cryptosporidium meleagridis. Avian Dis. 2002;46:203–207. doi: 10.1637/0005-2086(2002)046[0203:PAOSIB]2.0.CO;2. doi:10.1637/0005-2086(2002)046[0203:PAOSIB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Valigurová A, Jirku M, Koudela B, Gelnar M, Modrý D, Šlapeta J. Cryptosporidia: epicellular parasites embraced by the host cell membrane. Int J Parasitol. 2008;38:913–922. doi: 10.1016/j.ijpara.2007.11.003. doi:10.1016/j.ijpara.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Wang LM, Xue X, Li JQ, Zhou QJ, Yu YC, Du AF. Cryptosporidiosis in broiler chickens in Zhejiang Province, China: molecular characterization of oocysts detected in fecal samples. Parasite. 2014;21 doi: 10.1051/parasite/2014035. doi:10.1051/Parasite/2014035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodmansee DB, Pavlásek I, Pohlenz JF, Moon HW. Subclinical cryptosporidiosis of turkeys in Iowa. J Parasitol. 1988;74:898–900. doi:10.2307/3282279. [PubMed] [Google Scholar]
- Xiao L, Fayer R, Ryan U, Upton SJ. Cryptosporidium taxonomy: recent advances and implications for public health. Clin Microbiol Rev. 2004;17:72–97. doi: 10.1128/CMR.17.1.72-97.2004. doi:10.1128/CMR.17.1.72-97.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Morgan UM, Limor J, Escalante A, Arrowood M, Shulaw W, Thompson RC, Fayer R, Lal AA. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl Environ Microbiol. 1999;65:3386–3391. doi: 10.1128/aem.65.8.3386-3391.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Ryan UM. Cryptosporidiosis: an update in molecular epidemiology. Curr Opin Infect Dis. 2004;17:483–490. doi: 10.1097/00001432-200410000-00014. doi:10.1097/00001432-200410000-00014. [DOI] [PubMed] [Google Scholar]
- Zhang XX, Zhang NZ, Zhao GH, Zhao Q, Zhu XQ. Prevalence and Genotyping of Cryptosporidium Infection in Pet Parrots in North China. Biomed Research International. 2015 doi: 10.1155/2015/549798. doi:10.1155/2015/549798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Kassa H, Tischler ML, Xiao L. Host-adapted Cryptosporidium spp. in Canada geese (Branta canadensis). Appl Environ Microbiol. 2004;70:4211–4215. doi: 10.1128/AEM.70.7.4211-4215.2004. doi:10.1128/aem.70.7.4211-4215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]