Synopsis
The peroxiredoxins are a ubiquitous family of proteins involved in protection against oxidative stress through the detoxification of cellular peroxides. In addition the typical 2-cys peroxiredoxins function in signalling of peroxide stress and as molecular chaperones, functions that are influenced by oligomeric state. Of the human peroxiredoxins, Prx IV is unique in possessing an N-terminal signal peptide believed to allow secretion from the cell. Here we present a characterisation of Prx IV in human cells demonstrating that it is actually retained within the endoplasmic reticulum. Stable knockdown of Prx IV expression led to detrimental effects upon viability of human HT1080 cells following treatment with exogenous hydrogen peroxide. However, these effects were not consistent with a dose-dependent correlation between Prx IV expression and peroxide tolerance. Moreover, modulation of Prx IV expression showed no obvious effect on ER-associated stress, redox conditions or hydrogen peroxide turnover. Subsequent investigation demonstrated Prx IV to form complex structures within the ER consistent with the formation of homodecamers. Furthermore, Prx IV oligomeric interactions are stabilised by additional non-catalytic disulphide bonds indicative of a primary role other than peroxide elimination.
Keywords: peroxiredoxin IV, hydrogen peroxide, reactive oxygen species, molecular chaperone, peroxidatic cysteine, oxidative stress
Introduction
Exposure to reactive oxygen species (ROS) has long been recognised as a major problem to aerobic organisms and numerous biological mechanisms exist which facilitate removal of ROS within cells. Imbalances between these protective processes and those generating ROS have been implicated in pathogenesis of cancers and important degenerative disorders including Alzheimer’s and Parkinson’s diseases [1].
ROS also impart significant benefits. For example, hydrogen peroxide (H2O2) has importance as a cytotoxic agent during microbial engulfment by phagocytic immune cells [2]. Such cells generate H2O2 catalytically from NADPH oxidase-derived superoxide anions (O2-) and there is increasing evidence that a wide range of mammalian cytokines and growth factors also stimulate H2O2 production via NADPH oxidases for second messenger signalling purposes [3]. Despite its defined roles H2O2 remains a reactive molecule that can directly modify lipids, proteins and nucleic acids. This is particularly relevant given that substantial H2O2 is generated as a by-product of metabolic processes such as electron transport ‘leakage’ releasing O2- from the mitochondria [4]. Such reactions highlight the importance of peroxide detoxification pathways.
Peroxide degradation can occur via several routes including direct reaction with glutathione, breakdown by catalase (free H2O2) or glutathione peroxidases (H2O2 and lipid hydroperoxides), or reaction with vitamins and other non-enzymatic antioxidants [1]. In addition, the peroxiredoxin family of proteins has combined peroxidase activity with other functions, including the ability to communicate peroxide stress in the cell.
Mammals possess six peroxiredoxin isoforms [5, 6] with cellular locations including the cytosol (Prx I, II, VI), nucleus (Prx I) mitochondria (Prx III, VI), peroxisomes (Prx V) and potentially secreted (Prx IV). Each is characterised by a redox-active ‘peroxidatic’ cysteine residue that attacks peroxides, becoming oxidised to a cysteine sulfenic acid in the process [7–9]. The best characterised peroxiredoxin family members are the typical 2-cys peroxiredoxins (including Prx I, II, III and IV in humans) which contain an additional ‘resolving’ cysteine near the C-terminus. Typical 2-cys peroxiredoxins function as obligate homodimers and following peroxide elimination the peroxidatic cysteine-sulphenic acid reacts with the resolving cysteine of its partner to form a stable intermolecular disulphide [7, 10]. This disulphide may be reduced by a cell-specific disulphide reductase regenerating the active site thiols.
Most 2-cys peroxiredoxins undergo further fluid transition from dimers to toroid decamers and back again [11–14]. This process optimises active site arrangement for efficient catalysis, with active site disulphide formation subsequently destabilising the complex. Several studies have shown that hyperoxidation of the peroxidatic cysteine to a sulphinic acid (SO2H) derivative can occur in high peroxide concentrations [15, 16] which in turn stabilises peroxiredoxin decamers by preventing formation of the resolving disulphide [12]. In mouse lung cells this was demonstrated to lead to aggregates of Prx II decamers whose appearance and subsequent breakdown correlated with arrest and eventual resumption of cell cycle [17]. Hence Prx II oligomeric state may be used as a monitor of cytosolic H2O2. Further oligomeric state-dependent peroxiredoxin functions have been implicated, most notably a chaperone activity associated with high molecular weight fractions of Saccharomyces cerevisiae cytosolic peroxiredoxins following stress [18] and with decameric human Prx I in vitro [19]. In the latter case the decamer is covalently stabilised by non-catalytic disulphides preventing dimer-decamer transitions. This rigidity appears to reduce peroxidase activity and increase prevalence of chaperone activity [19].
Prx IV is the least well characterised of the human 2-cys peroxiredoxins and is unique in possessing an N-terminal secretory signal. Despite being identified a decade ago some confusion exists as to the true nature of Prx IV in mammalian cells. Prx IV has been described as both a cytosolic protein attenuating activity of NF-κB [20] and as a secreted protein activating NF-κB [21]. Later studies investigating rat Prx IV concluded it was secreted and bound at the cell surface following transient over-expression in African green monkey cells [22, 23]. The only consistent finding between these studies was the ability of Prx IV to act as a peroxidase in vitro. Consequently many questions remain unanswered regarding the size, sub-cellular location and physiological relevance of Prx IV.
The possibility of Prx IV traversing the secretory pathway is intriguing, particularly given that oxidative protein folding in the endoplasmic reticulum (ER) has been recently proposed as another significant source of H2O2 within the cell [24, 25]. H2O2 may be generated through electron transfer to molecular oxygen by Ero1 proteins during the oxidation of protein disulphide isomerase (PDI) [26, 27]. Such situations may not be limited to the ER as oxidative protein folding by other means may continue through the secretory pathway into the Golgi apparatus and perhaps beyond [28]. Given the delicate redox balance required for native disulphide bond formation, we hypothesise that localised mechanisms might exist for the detection and removal of ROS generated during the oxidative folding process. Prx IV provides an attractive candidate given the described bifunctionality for other human Prx family members. To this end we performed an investigation of Prx IV in vivo, in which endogenous Prx IV was demonstrated to be both translocated to and retained within the ER of human cells. Cell lines were created in which Prx IV was stably over-expressed or knocked-down and the influence of altered expression upon hydrogen peroxide turnover and ER homeostasis were investigated. Subsequent structural analyses of Prx IV complexes formed within cells provided insight into the behaviour of Prx IV in comparison with the previously characterised human 2-cys peroxiredoxins.
Experimental
Chemicals and reagents
All reagents were acquired from Sigma-Aldrich (Dorset, UK) and enzymes from Promega (Southampton, UK) unless otherwise stated.
Antibodies
A rabbit polyclonal antibody to Prx IV was purchased from Lab Frontier (Seoul, Korea) whilst rabbit polyclonal antibody to Alix was kindly donated by Phil Woodman (University of Manchester). Mouse monoclonal antibodies recognising α-tubulin and the KDEL ER-retrieval motif have been described previously [29, 30] and were generous gifts from Keith Gull (University of Oxford) and Stephen Fuller (University of Heidelberg) respectively. A goat polyclonal specific to the N-terminal region of BiP was purchased from Santa Cruz Biotechnology inc. (Santa Cruz, CA, USA), whilst antibodies to the ER oxidoreductases PDI, ERp57 and ERp72 have been described previously [31].
Transcription and translation in vitro
A clone encoding human Prx IV in pOTB7 was obtained from Geneservice Ltd. (Cambridge, UK) and in vitro transcription and translation performed essentially as described previously [32]. DNA was linearised with Xho I and transcribed using SP6 polymerase. Transcript was translated using rabbit reticulocyte lysate (Flexi-lysate, Promega, USA) with semi-permeabilised (SP) cells added as required. Proteinase K treatment of SP cells was performed for 25 min. on ice ± 1% v/v Triton X-100, using 0.2 mg/ml proteinase K in the presence of 10 mM CaCl2, and terminated by 1mM phenylmethylsulphonyl fluoride (PMSF). When added, SP cells were isolated by centrifugation and resuspended in SDS-PAGE sample buffer (31.25 mM Tris-HCL pH 6.8, 2% w/v SDS, 5% v/v glycerol, 0.01% w/v bromophenol blue). Otherwise reactions were mixed directly with SDS-PAGE sample buffer.
Electrophoresis and Western blotting
Samples for SDS-PAGE were resuspended in SDS-sample buffer and heated to 100°C for 5 min. For reducing conditions, dithiothreitol (DTT) was added to 50 mM. Gels containing radioactive samples were fixed in 10% v/v acetic acid and 10% v/v methanol, dried and exposed to Kodak Biomax MR film (GRI, Essex, UK). For Western blotting, gels were transferred to nitrocellulose and blocked using 3% milk in TTBS (10 mM Tris, 150 mM NaCl, pH 7.5, 0.1% Tween-20). Primary antibody incubations were performed for 1 hour at room temperature with 3% milk. As secondary antibodies polyclonal goat anti-rabbit, rabbit anti-goat and rabbit anti-mouse immunoglobulins – each conjugated to horseradish peroxidase - were obtained from Dako (Ely, UK). Secondary antibodies were diluted 1:2000 in TTBS and incubation performed at room temperature for 1 hour. Products were visualised using enhanced chemiluminescent substrate (Perbio, Northumberland, UK) and Fuji Super RX film (Fujifilm UK, Bedford, UK).
Sub-cellular fractionation
HT1080 human fibrosarcoma cells were suspended in buffer A (50mM Tris-HCl, 0.25 M sucrose, 25 mM KCl, 0.5 mM MgCl2, 1 mM EDTA) at 2 x 107 cells/ml and disrupted using a ball bearing homogeniser with 10 µm clearance. Insoluble debris and nuclear material was removed at 500 × g and post-nuclear supernatant centrifuged at 150,000 × g to pellet organelle membranes. Membranes were resuspended in buffer A and treated with proteinase K, when required, as described above.
Pulse-chase analysis
107 sub-confluent HT1080 cells were deprived of essential amino acids for 30 min., incubated with radioactive methionine/cysteine protein labelling mix (50 µCi/ml, NEN, Boston, MA, USA) for a further 30 min., and then medium was replaced with DMEM + 10% FCS. At required times, cells and media were separated and cells were lysed using IP buffer (50 mM Tris-HCl, 150 mM NaCl, 2 mM EDTA, 0.5 mM PMSF, 1% v/v Triton X-100). Insoluble material was removed by centrifugation at 10,000 × g for 1 min. and lysates were mixed with SDS to 1% w/v, boiled for 3 min. and diluted ten-fold with lysis buffer. Pre-incubation with protein-A Sepharose for 30 minutes preceded incubation with protein-A Sepharose and anti-Prx IV for 16 hours at 4°C. Beads were washed three times with 100 volumes of lysis buffer and resuspended in SDS-PAGE sample buffer. Immunoprecipitation was repeated for whole media samples.
Immunofluorescence
Immunofluorescence was performed as described previously [33]. Anti-Prx IV was detected by an Alexa Fluor 594 anti-rabbit antibody, whereas anti-KDEL was detected by an Alexa Fluor 448 anti-mouse antibody. Cells were visualised on an Olympus BX60 upright microscope at × 40 magnification.
Creation of stable cell lines
Human Prx IV was excised from the pOTB7 vector using Bam HI and Xho I and ligated with pcDNA3.1/Hygro(+) (Invitrogen, Paisley, UK). The final construct was linearised with Ssp I for transfection. HuSH Vectors encoding shRNA for Prx IV knockdown were obtained from Origene (Rockville, MD, USA) and linearised with Sca I. All constructs were transfected into sub-confluent HT1080 cells using Fugene8 (Roche, Indiannapolis, IN, USA). Stable transfectants were selected using 250 µg/ml hygromycin B or 1 µg/ml puromycin. After 14 days growth, colonies were selected and screened for Prx IV expression by Western blotting.
Analysis of XBP1 splicing
Total RNA was isolated from cells using TRI-reagent according to the manufacturer’s instructions. Following DNase treatment, cDNA was prepared from 5µg RNA using Bioscript reverse transcriptase (Bioline, London, UK). XBP1 mRNA was amplified from 1 µg cDNA with primers corresponding to XBP1 nucleotides 450-469 and 671-690 (Eurogentech, Southampton, UK). The resulting PCR products were digested using Pst I, separated by electrophoresis through 2% agarose, stained with 1 µg/ml ethidium bromide and visualised using a UVItec UVIDoc gel documentation system.
Crystal violet viability assay
Adherent cells were supplemented with hydrogen peroxide and incubated for 24 hours. Detached cells were removed by PBS washing and remaining cells fixed in methanol at −20°C for 5 min. Nuclei were stained with 0.2% w/v crystal violet for 5 min. and washed thrice with PBS. Stain was solubilised in 0.1% w/v SDS, diluted five-fold and absorbance recorded at λ = 540 nm.
Hydrogen peroxide turnover assay
Adherent cells were trypsinised and resuspended in PBS or SP cells prepared and suspended in KHM buffer (110mM KOAc, 20mM HEPES, 2mM MgOAc). Hydrogen peroxide was supplemented to 20 µM final concentration and cells removed by centrifugation at appropriate times. Remaining hydrogen peroxide concentration was determined using Amplex Red reagent (Invitrogen, Paisley, UK) in accordance with the manufacturer’s instructions.
Determination of oxidoreductase redox states
Redox states were determined exactly as described previously [31].
Sucrose gradient fractionation
107 cells were lysed by vortexing in SG buffer (5 mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM PMSF, 0.1% NP-40) following treatment with 25 mM NEM. Cleared lysates were applied to a continuous gradient of 9 parts 5% w/v sucrose into 9 parts 25% w/v sucrose, laid over 2 parts 50% w/v sucrose in SG buffer. Gradients were centrifuged for 16 hours at 285,000 × g, 4°C, and then separated into ten equal-volume fractions. Proteins were precipitated with 5% trichloracetic acid and resuspended in SDS-PAGE sample buffer. For denaturing gradients, cleared lysates were supplemented with 1% w/v SDS and boiled for 5 min. prior to fractionation. Gradients contained 0.1% SDS. For denaturing & reducing gradient, NEM was added after boiling in presence of 1% SDS and 10 mM DTT.
Site-directed mutagenesis of Prx IV
Mutagenesis was performed based on the method of Hemsley et al. [34] using pcDNA3.1/hygro(+); Prx IV as a template and primers encoding alanine in place of cysteine residues. Parental DNA was removed by Dpn I digestion prior to transformation into E. coli. Mutant constructs were transiently expressed for 24 hours in HT1080 cells following transfection of 5 × 106 cells with 5 µg DNA using Lipofectamine 2000 (Invitrogen, UK).
Results
Prx IV is co-translationally translocated to the human ER
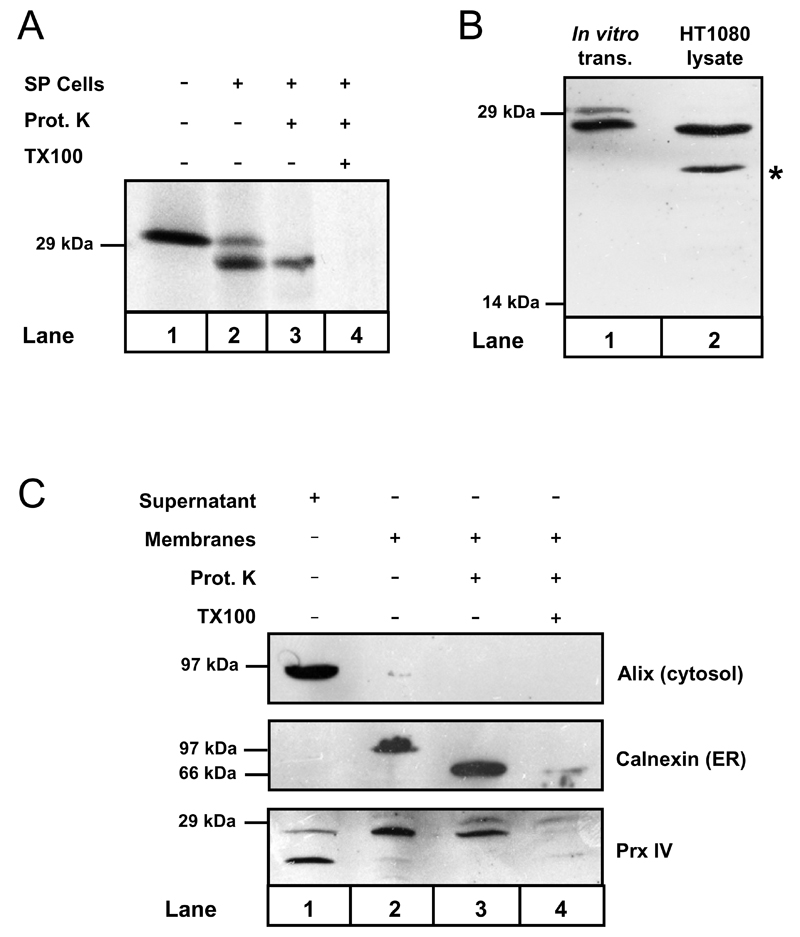
Prx IV was initially estimated to possess a secretory signal peptide consisting of 79 amino acids [20], leading to mature Prx IV with a predicted molecular weight of 22 kDa. In contrast, our predictive analysis using the SignalP 3.0 program [35] indicated signal peptide cleavage to occur between residues 37 and 38 generating a 27 kDa product. To clarify this issue, cDNA encoding the full length protein was translated in vitro in the presence and absence of semi-permeabilised (SP) HT1080 human fibrosarcoma cells (Fig.1A). In addition to the pre-protein (upper band), a second species with enhanced mobility predominated when translated in the presence of SP cells (compare lanes 1 and 2). This was consistent with co-translational translocation of Prx IV to the ER and signal peptide cleavage. The cleaved product displayed apparent molecular weight of 27 kDa, whilst protection of this fragment from proteinase K digestion confirmed that translocation was complete (lane 3).
Figure 1. Prx IV is co-translationally translocated to the ER in vitro and in vivo.
A. Autoradiograph showing Prx IV mRNA translated in vitro using 35S-labelled methionine & cysteine. Translation was performed in the presence or absence of semi-permeabilised HT1080 cells (SP cells) as indicated. SP cells were harvested and treated with proteinase K with or without Triton X-100 detergent (TX100) as required. B. Prx IV was translated in vitro plus SP cells. Translation products were compared with HT1080 whole-cell lysate by Western blotting using antibody to Prx IV. C. HT1080 cells were homogenised and the post-nuclear supernatant separated by ultracentrifugation. The resulting supernatant and organelle membrane fractions were probed by Western blotting using antibodies to cytosolic and ER proteins along with anti-Prx IV. Membrane samples were also treated with proteinase K in the presence or absence of TX100 as indicated.
A comparison between Prx IV translated in vitro and HT1080 whole cell lysates clearly demonstrated that the translocated product in SP cells corresponded with the prevalent form of Prx IV in vivo (Fig. 1B). Pre-Prx IV was not detected in whole cells though a smaller immuno-reactive species of 22-24 kDa was apparent (marked by *). This species was judged as cytosolic by virtue of its co-localisation with the cytosolic marker ALG2-interacting-protein X (Alix) following HT1080 homogenisation and preparation of organelle membranes (Fig. 1C, lane 1). In contrast, the predominant form of Prx IV co-localised mainly with calnexin in the organellar fraction (lane 2, representing the membranes of the secretory pathway) and like the calnexin lumenal domain was again protected from proteinase K digestion (lane 3). Thus, human Prx IV exists primarily as an ER-translocated peptide of approximately 27 kDa.
The most probable explanation for the presence of the cytosolic species is that it is an artefact of antibody cross-reactivity during Western blotting. This is based upon the observation that its levels remained unaffected in cells stably over-expressing Prx IV and also in cell lines with up to 90% stable knockdown of Prx IV expression (see Fig. 3A). Furthermore, unlike ER translocated Prx IV, this species could not be immunoprecipitated from cell lysate (Fig. 2A). Nonetheless, the possibility remains that it may constitute some hitherto unknown Prx IV splice variant.
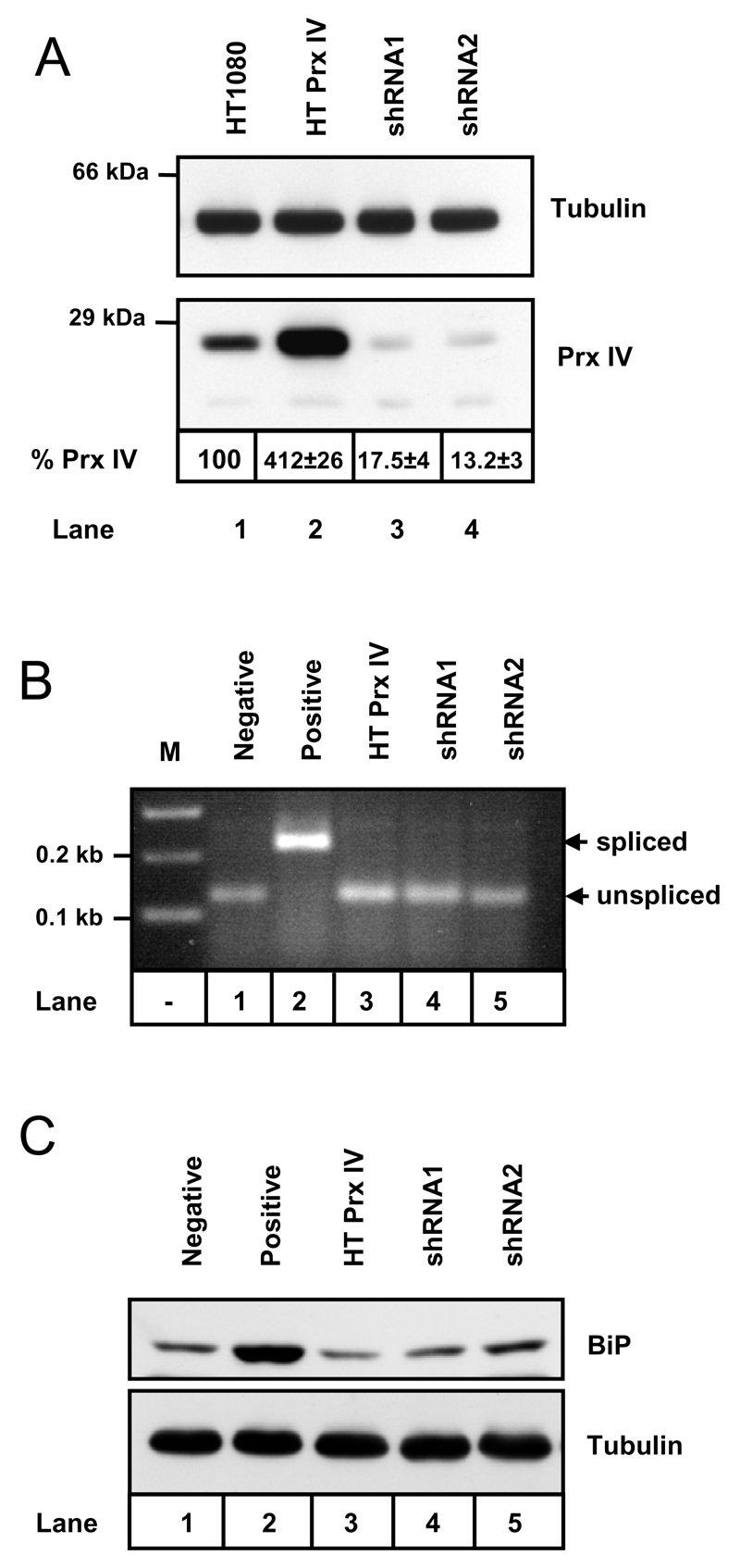
Figure 3. Stable alteration of Prx IV expression does not induce ER stress.
A. Western blot analysis of whole-cell lysates prepared from HT1080 cells and HT1080s engineered to stably overexpress (HT Prx IV) and under-express (shRNA1 and shRNA2) Prx IV. Prx IV expression levels (% Prx IV) relative to the parent cells are indicated ± standard deviation (n = 4). Anti-tubulin blot serves as a loading control. B. Agarose gel stained with ethidium bromide and imaged under UV light to visualise Pst I digests of XBP1 cDNA prepared from cell lines utilised in A. Negative control is prepared from the untreated HT1080s, positive control = HT1080s treated for 2 hours with 10 mM DTT. C. Western blot analysis of whole cell lysates to examine expression levels of BiP (with tubulin loading control). HT1080 lysate again serves as negative control and HT1080s treated for 12 hours with 10 µg/ml tunicamycin provide a positive control for UPR induction.
Figure 2. Prx IV is not secreted and remains ER associated.
A. HT1080 cells over-expressing Prx IV were pulsed with 35S-labelled methionine & cysteine for 30 min then chased for 5 hours with fresh medium. Cells and media were separated at the indicated times and radioactive Prx IV immunoprecipitated from each sample before being visualised by SDS-PAGE and autoradiography. B. Fluorescence microscopy of HT1080 cells following 5 hours incubation with 0.5 mM cycloheximide. Panels show a representative cell with nuclear staining (DAPI), immuno-staining of proteins containing the KDEL ER retrieval motif (anti-KDEL) and immuno-staining of Prx IV along with composite image.
Prx IV is retained in the ER
To evaluate secretion of Prx IV into the extracellular environment, pulse-chase time-courses were performed for HT1080 cells (not shown) and HT1080s transfected to over-express Prx IV (Fig. 2A). Transfection was stable rather than transient, ensuring Prx IV expression was consistent and physiologically tolerable. In each case the labelled pool of Prx IV was immunoprecipitated exclusively from cell lysates throughout the following five hours (lanes 1-3); with none detected in culture supernatant samples (lanes 4-6). In addition, Western blot analyses of protein precipitates from HT1080, HeLa, HEK 293 and Hep G2 culture supernatants showed no detectable secretion of Prx IV (not shown).
As Prx IV clearly remains associated with cells, an experiment was conducted to determine its final cellular localisation. Endogenous Prx IV was visualised by immunofluorescence microscopy of HT1080 cells following cycloheximide treatment to inhibit de novo protein synthesis (Fig. 2B). Interestingly Prx IV remained co-localised with ER markers for a full five hours after cycloheximide addition, mirroring the distribution seen in untreated cells and in cells over-expressing Prx IV (not shown). The data indicate that native Prx IV is ER-retained in human cells even when over-produced, despite lacking a recognised retention/retrieval motif.
Altering Prx IV expression does not induce ER stress
To investigate function of Prx IV in the ER, cell lines were created in which Prx IV expression was stably modified (Fig. 3A). In addition to the aforementioned cell line over-expressing Prx IV (lane 2), two HT1080 cell lines were established displaying stable expression of shRNA directed against the Prx IV coding sequence (lanes 3 & 4). Densitometry performed for multiple Western blot exposures, within the linear range of chemiluminescent substrate response, allowed quantification of Prx IV knockdowns as indicated (Fig. 3A).
To determine if modulated Prx IV expression resulted in ER stress induction, components of the unfolded protein response (UPR) were examined in each of the cell lines created. Xbp1 is a transcription factor involved in expression of UPR induced genes following accumulation of protein aggregates within the ER [36]. Production of active Xbp1 requires IRE1–dependent mRNA splicing, which in turn eliminates a Pst I restriction enzyme recognition site from the mRNA sequence. Xbp1 splicing was induced in HT1080 cells by incubation with DTT (Fig. 2B, lane 2). For each of our cell lines however, the status of XBP1 mRNA reflected that seen for the HT1080 parent cells (compare lanes 3-5 and lane 1) indicating no induction of UPR. This was confirmed by Western blotting of whole cell lysates (Fig. 2C) using antibodies to BiP - an ER chaperone up-regulated during UPR [37, 38].
In addition to investigating the influence of Prx IV expression upon the UPR, levels of Prx IV itself were evaluated under conditions of UPR induction. Prx IV expression in HT1080 cells was unaffected by 12 hour incubation with DTT (2 mM), tunicamycin (10 µg/ml) or thapsigargin (2 µM)), indicating that it does not respond to ER stress (data not shown).
Prx IV knockdown does not alter ER redox balance
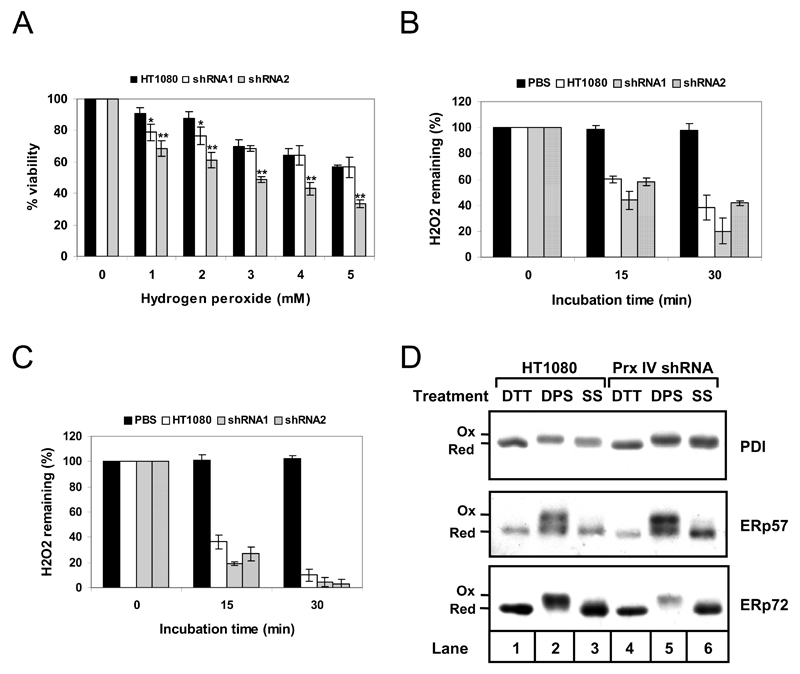
We hypothesised that if Prx IV has important peroxidase activity, knockdown of Prx IV expression would limit the ability of cells to remove H2O2 from the ER and increase susceptibility of cells to H2O2-induced cell death. To test the latter, our Prx IV knockdown cell lines were treated with increasing concentrations of H2O2 and viability recorded after a 24 hour incubation period (Fig. 4A). Compared to the HT1080 parent, shRNA2 cells displayed reduced survival at all H2O2 concentrations tested (compare black and grey bars). This differed from the response of the shRNA1 cell line which, whilst demonstrating some attenuation of viability at 1-2 mM H2O2, matched that of the parent at higher concentrations (black versus white bars). Disparity in response of the knockdown cell lines may possibly reflect the respective levels at which Prx IV is expressed in each.
Figure 4. Prx IV knockdown affects viability but not hydrogen peroxide turnover or ER redox balance.
A. Viability profiles for HT1080s (black bars) and Prx IV knockdown cell lines, shRNA1 (white) and shRNA2 (grey), 24 hours after addition of indicated H2O2 concentrations to culture medium. Viability for each was recorded using crystal violet staining and spectrophotometry, with % viability at each concentration calculated relative to untreated cells. Differences in viability for each treatment were assessed for statistical significance using two-tailed, unpaired Student’s t-tests with unequal variance assumed; * Statistically significant with P<0.05, **P<0.005. B. Decomposition of H2O2 during incubation of 20 µM final concentration in PBS (black bars, negative control), and in 5 x 105 cells/ml suspensions of HT1080s (white), shRNA1 (grey) and shRNA2 (hatched). Samples were harvested at the indicated times, cells removed and supernatant H2O2 concentration determined using Amplex Red detection. Remaining H2O2 is expressed as % of that determined for the corresponding time = 0 sample. C. As for B except suspensions consist of semi-permeabilised cells at 106 cells/ml in KHM buffer instead of PBS. For A, B and C, data represent mean averages of three replicates with error bars indicating standard deviation. D. Western blot analysis of whole cell lysates from HT1080 cells (lanes 1-3) and Prx IV knockdown cell line shRNA2 (lanes 4-6). Lysates were prepared from cells following treatment with 10 mM DTT, 1 mM DPS or at steady state (SS). Consecutive modifications with NEM, TCEP and AMS preceded analysis using antibodies to the indicated ER oxidoreductases.
To establish whether the attenuated viability could be a manifestation of reduced peroxidase activity, Prx IV knockdown cell lines were evaluated for their ability to decompose exogenously supplied H2O2. Surprisingly, both shRNA1 and shRNA2 cell lines were able to remove H2O2 at least as efficiently as unmodified HT1080s (Fig. 4B). Similar results were obtained using SP cells (lacking any cytosol) in place of intact cells (Fig. 4C), confirming that the results observed were not due to cytosolic factors masking any underlying ER activity.
As we could detect no decrease in peroxide turnover following Prx IV knockdown, an approach was taken to examine more general effects upon the ER redox environment. Several ER-resident oxidoreductases are known to exist in a reduced steady state in HT1080 cells, the maintenance of which can be influenced by altering redox conditions [31]. We therefore postulated that if Prx IV was a major ER peroxidase, substantially reducing its expression could result in a more oxidising environment within the ER. As markers we investigated the redox states of PDI, ERp57 and ERp72 in the Prx IV knockdown cell lines. In both shRNA1 (not shown) and shRNA2 cells (Fig. 4D), each oxidoreductase was in a reduced state exactly mirroring that of the HT1080 control cells (compare lanes 3 and 6). The redox states of these enzymes therefore appear unaffected by reduced Prx IV expression, indicating no gross change in ER redox homeostasis.
Prx IV forms oligomeric complexes containing non-catalytic disulphide bonds in vivo
As alluded to previously, human Prx I and Prx II display distinct oligomeric behaviours that potentially influence function [19]. Whilst each forms a penta-dimeric decamer via hydrophobic interactions, Prx I decamers may be stabilised by additional intermolecular disulphide bonds at the dimer-dimer interface. Relative to Prx II, Prx I displays reduced peroxidase activity and increased propensity to inhibit thermal aggregation of malate dehydrogenase in vitro [19]. Substitution of the cys-83 residue with alanine however, preventing formation of the inter-dimer disulphide, resulted in elevated and diminished Prx I peroxidase and chaperone activities respectively. Thus, increased oligomeric stability potentially diverts typical 2-cys peroxiredoxins from operating principally as peroxidases. As we observed no detectable peroxidase activity attributable to Prx IV in the preceding experiments, we investigated whether there could be a structural basis for these findings similar to those responsible for disparities between Prx I and Prx II.
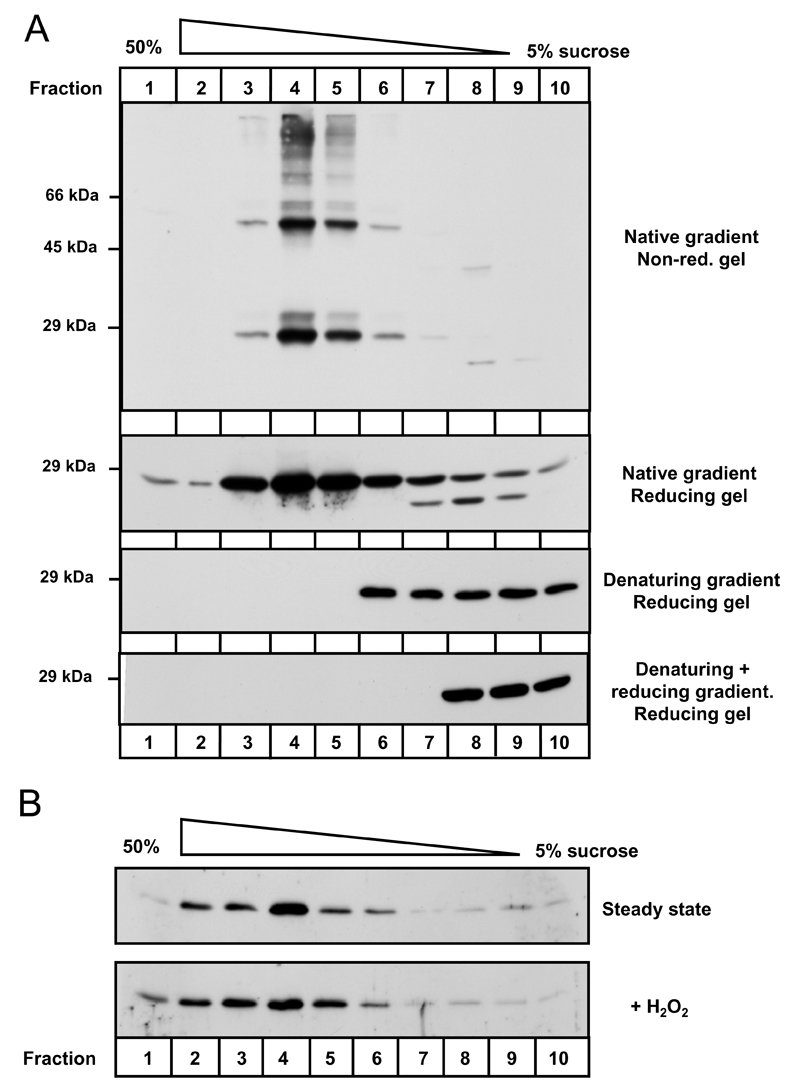
A crystal structure for a Prx IV fragment comprising amino acid residues 84-271 has recently been submitted to the RSCB Protein Data bank (Pilka et al. unpublished data, 2007. PDB ID: 2pn8). The structure confirmed that human Prx IV purified from E. coli forms decamers characteristic of typical 2-cys peroxiredoxins in vitro. To establish if this occurs in vivo, HT1080 cells over-expressing Prx IV were alkylated to preserve protein thiol-disulphide status. Cell lysates were subsequently fractionated through a sucrose gradient to separate native protein complexes principally on the basis of molecular weight. Prx IV predominantly sedimented towards the high-sucrose end of the gradient (Fig. 5A, top two panels, lanes 4-5), as opposed to the fractionation patterns obtained for a mixture of dimeric and monomeric Prx IV (denaturing gradient), or monomeric Prx IV alone (denaturing + reducing gradient). Comparable results were obtained for endogenous Prx IV in HT1080 cells under reducing conditions though expression levels limited non-reducing examination (data not shown). These data indicate that Prx IV does exist in an oligomeric state in human cells.
Figure 5. Prx IV forms oligomeric complexes in the ER.
A. NEM-treated lysates prepared from HT1080s over-expressing Prx IV were subjected to sucrose gradient fractionation. Lysates were fractionated under mild detergent (native) or denaturing conditions. An additional sample was fully reduced by treatment with 20 mM DTT and alkylation with NEM prior to denaturing gradient separation (denaturing + reducing). Following fractionation and TCA precipitation, SDS-PAGE was performed under non-reducing or reducing conditions (as indicated) and anti-Prx IV Western blotting undertaken. B. Organelle membranes were prepared from Prx IV over-expressing HT1080 cells and alkylated with NEM immediately (steady state) or following 15 minutes incubation with 5 mM H2O2. Contents were subsequently fractionated through sucrose gradients and TCA precipitated. Anti-Prx IV Western blotting analysis was carried out following SDS-PAGE under reducing conditions.
In addition to whole-cell extracts, protease treated organelle membranes were subjected to sucrose gradient analyses confirming that Prx IV oligomeric complexes formed within the ER (Fig. 5B). Moreover, incubation of membranes with H2O2, at concentrations substantially greater than those previously determined to cause ‘stacking’ of Prx II decamers [17], led to negligible effects upon the fractionation profile for Prx IV.
Prx IV oligomers contain two prevalent species of apparent molecular weight 27 kDa and 54 kDa (Fig. 5A, top panel), corresponding to Prx IV monomers and disulphide linked homo-dimers respectively. Intriguingly, mutation of the catalytic cysteine residues to alanine did not abolish the formation of Prx IV dimers (Fig. 6B, lanes 2-4), indicating that additional intermolecular disulphide bonds could form independently of the active sites.
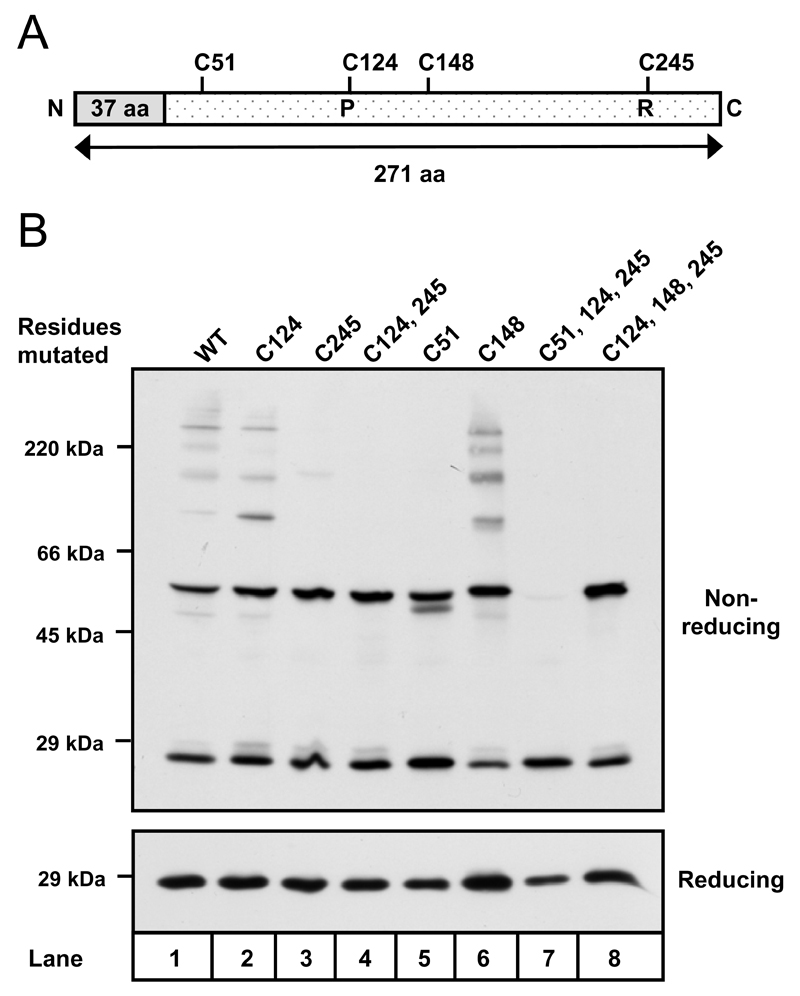
Figure 6. Cysteine 51 is required to form a non-catalytic disulphide in Prx IV.
A. Linear representation of Prx IV indicating signal peptide (grey) and positions of cysteine residues. Peroxidatic (P) and resolving (R) cysteines are highlighted. B. Anti-Prx IV Western blots of HT1080 cell lysates following transient expression of cDNA encoding Prx IV native sequence (WT), or Prx IV with indicated cysteine residues mutated to alanine. Free thiols were alkylated with NEM prior to lysis.
Prx IV contains two non-catalytic cysteine residues at positions 51 and 148 in the primary structure (Fig. 6A). To evaluate the involvement of each in disulphide bond formation, cysteine to alanine mutants were created for cys-51 and cys-148 and expressed in HT1080 cells (Fig. 6B, lanes 5-8). Mutation of cys-148 - individually and in combination with the peroxidatic and resolving cysteines - had no detectable effect upon Prx IV dimerisation (lanes 7-8). The same was true of a cys-51 mutant in which active site disulphides were still intact (lane 5). However, mutation of cys-51 in conjunction with the catalytic residues led to a total depletion of covalently linked dimers. We therefore conclude that cys-51 facilitates formation of an inter-molecular disulphide bond independently of the Prx IV active site cysteine residues.
Discussion
The finding that Prx IV resides within the human ER clarifies an issue that has remained clouded for the last ten years. Our observation of a second anti-Prx IV immuno-reactive species within the cytosol may help to explain previous disagreement regarding the cellular localisation of Prx IV. Similarly, the detection of this product within human cell lysates may shed some light regarding the original prediction of such a large signal peptide [20]. Previously it has been suggested that the larger form of Prx IV detected in cells could be unprocessed pre-protein, anchored to membranes via the uncleaved hydrophobic sequence [39]. Here however, we clearly show that pre-Prx IV is not present at any substantial level in human cells and that the larger, predominant form is mature ER-localised Prx IV. Furthermore, whilst the implied Prx IV signal peptide may seem unusually long (37 residues) no obvious anomalies were visible within the amino acid sequence. All positions within the first 34 residues scored poorly as potential cleavage sites during SignalP 3.0 analysis, with the only potential alternative presented as possible cleavage occurring between positions 34 and 35.
The mechanism by which Prx IV is retained within the ER remains undetermined as no KDEL-type retrieval motif is present within the primary structure. This situation is not without precedent however. Human Ero1 proteins lack an ER retrieval motif yet remain within the ER. Recently this has been indicated to occur through competitive interactions in the ER lumen with both PDI (containing a KDEL motif) and ERp44 (RDEL) in a largely thiol-dependent fashion [40].
Prx IV may also have a requirement for thiol-mediated interaction with ER proteins. Recycling of the peroxidatic cysteine, by reduction of the resolving disulphide, can occur in the presence of both thioredoxin and glutathione in vitro [23]. Whilst glutathione is found within the ER, thioredoxin is cytosolic. Numerous thioredoxin-like proteins exist within the ER however, including the protein disulphide isomerase family members. It is therefore possible that Prx IV may be retained in the ER via interactions akin to those observed for the Ero1 family. If so, identification of partners involved in redox turnover of Prx IV may in turn provide insight into the maintenance of its ER localisation.
In conjunction with the established literature regarding typical 2-cys peroxiredoxin behaviour in vitro, the data presented here provide a new perspective for speculation of Prx IV function in vivo. Based on the available crystal data both for Prx IV and other closely related proteins [11, 12, 41, 42], it seems highly likely that the oligomeric complexes observed in the ER of HT1080 cells correspond to toroid Prx IV decamers. Furthermore, it is clear that Prx IV intermolecular interactions can be stabilised by disulphide bonds formed independently of the peroxidatic and resolving cysteines. The missing piece to this puzzle remains the position of cys-51 mediated disulphides within the Prx IV quaternary structure. The current crystal structure for Prx IV provides no clarification on this matter as cys-51 is absent from the crystallised fragment. Moreover, we can not assume that these bonds are analogous to those at the Prx I dimer-dimer interface as sequence alignments indicate that the cys-83 residue of Prx I does not correspond with cys-51 of Prx IV [22]. Comparisons instead suggest cys-148 of Prx IV as the residue most closely matching cys-83 of Prx I. Here however, we demonstrate that cys-148 is not required for disulphide bonding between Prx IV monomers. Consequently, we cannot currently discount the possibility that cys-51 disulphide bonds exist intra-dimerically, strengthening this particular interaction of Prx IV.
Irrespective of whether non-catalytic disulphides form between or within Prx IV dimers, it is clear that either situation may interfere with oligomeric transitions and therefore the peroxidatic cycle of Prx IV. Fluid conversion from dimeric > decameric > covalent-dimeric states are important for the efficient peroxidase activity of peroxiredoxins such as Prx II. This activity can clearly be manipulated by modulation of Prx I disulphide bonding capability at residue 83, though given the cytosolic/nuclear localisation attributed to Prx I it remains uncertain how prevalent this stabilising disulphide may be in vivo. Prx IV however, exists in an environment in which formation of disulphide bonds is positively encouraged. We would therefore predict the arrangement of Prx IV into disulphide-stabilised structures to be highly favoured, reflected in our inability to detect any significant peroxidase effects associated with Prx IV expression.
Despite no obvious defect in peroxidase activity, knockdown of Prx IV expression clearly caused some compromise in survival of HT1080 cells following H2O2 exposure. However, the differential responses of the two cell lines were not wholly consistent with a dose-dependent increase in H2O2 susceptibility relative to Prx IV expression level. Whilst Prx IV may not contribute significantly towards H2O2 elimination, the effects witnessed following Prx IV knockdown may be attributable to its presence helping to alleviate the after-effects of oxidative insult. As touched upon previously, an alternative role already prescribed to typical 2-cys peroxiredoxins is that of a molecular chaperone. Cytoplasmic peroxiredoxins from both yeast and humans have been demonstrated to inhibit thermal aggregation of substrates in vitro and to enhance heat shock resistance in vivo [18, 19, 43]. Yeast TSA1 has also been shown to prevent aggregation of ribosomal proteins following reductive stress [44]. The prospect of Prx IV displaying such activity within the mammalian ER is indeed attractive, particularly given that protein folding within the compartment is coupled with additional complexities such as introduction of native disulphide bonds. The possibility still remains however, that Prx IV function may revolve around its active site residues. Whilst considered primarily as a peroxidatic mechanism, the dynamic redox status of peroxiredoxin active sites may necessitate thiol-dependent interaction with other proteins. Peroxiredoxins may therefore be considered as oxidoreductases of said proteins in their own right. Consequently, identification of interacting partners may provide the key to determining Prx IV function, and may yet indicate a role for Prx IV in thiol-disulphide exchange processes within the human ER.
Acknowledgements
This work was supported by grants from the Wellcome Trust (ref. #74081) and BBSRC (ref. D00769). In addition we wish to acknowledge the generosity of Prof. Phil Woodman and Dr Lisa Swanton (both University of Manchester), Prof. Keith Gull (University of Oxford) and Prof. Stephen Fuller (European Molecular Biology Laboratory Heidelberg) for their contribution of reagents and antibodies.
Abbreviations
The abbreviations used are:
- ROS
reactive oxygen species
- Prx
peroxiredoxin
- ER
endoplasmic reticulum
- SP
semi-permeabilised
- TX100
triton X-100
- Alix
ALG2-interacting protein X
- UPR
unfolded protein response
- DTT
dithiothreitol
- DPS
2,2’-dithiodipyridine
- NEM
N-ethyl maleimide
- TCEP
Tris[2-carboxyethyl] phosphine
- AMS
4-acetamido-4´-maleimidyl-stilbene-2,2´-disulphonic acid
- shRNA
short hairpin RNA
- cys
cysteine
References
- 1.Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 2.El-Benna J, Dang PM, Gougerot-Pocidalo MA, Elbim C. Phagocyte NADPH oxidase: a multicomponent enzyme essential for host defenses. Arch Immunol Ther Exp (Warsz) 2005;53:199–206. [PubMed] [Google Scholar]
- 3.Veal EA, Day AM, Morgan BA. Hydrogen peroxide sensing and signaling. Mol Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 4.Muller FL, Liu Y, Van Remmen H. Complex III releases superoxide to both sides of the inner mitochondrial membrane. J Biol Chem. 2004;279:49064–49073. doi: 10.1074/jbc.M407715200. [DOI] [PubMed] [Google Scholar]
- 5.Wood ZA, Schroder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 6.Hofmann B, Hecht HJ, Flohe L. Peroxiredoxins. Biol Chem. 2002;383:347–364. doi: 10.1515/BC.2002.040. [DOI] [PubMed] [Google Scholar]
- 7.Ellis HR, Poole LB. Roles for the two cysteine residues of AhpC in catalysis of peroxide reduction by alkyl hydroperoxide reductase from Salmonella typhimurium. Biochemistry. 1997;36:13349–13356. doi: 10.1021/bi9713658. [DOI] [PubMed] [Google Scholar]
- 8.Choi HJ, Kang SW, Yang CH, Rhee SG, Ryu SE. Crystal structure of a novel human peroxidase enzyme at 2.0 A resolution. Nat Struct Biol. 1998;5:400–406. doi: 10.1038/nsb0598-400. [DOI] [PubMed] [Google Scholar]
- 9.Montemartini M, Kalisz HM, Hecht HJ, Steinert P, Flohe L. Activation of active-site cysteine residues in the peroxiredoxin-type tryparedoxin peroxidase of Crithidia fasciculata. Eur J Biochem. 1999;264:516–524. doi: 10.1046/j.1432-1327.1999.00656.x. [DOI] [PubMed] [Google Scholar]
- 10.Hirotsu S, Abe Y, Okada K, Nagahara N, Hori H, Nishino T, Hakoshima T. Crystal structure of a multifunctional 2-Cys peroxiredoxin heme-binding protein 23 kDa/proliferation-associated gene product. Proc Natl Acad Sci U S A. 1999;96:12333–12338. doi: 10.1073/pnas.96.22.12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alphey MS, Bond CS, Tetaud E, Fairlamb AH, Hunter WN. The structure of reduced tryparedoxin peroxidase reveals a decamer and insight into reactivity of 2Cys-peroxiredoxins. J Mol Biol. 2000;300:903–916. doi: 10.1006/jmbi.2000.3881. [DOI] [PubMed] [Google Scholar]
- 12.Schroder E, Littlechild JA, Lebedev AA, Errington N, Vagin AA, Isupov MN. Crystal structure of decameric 2-Cys peroxiredoxin from human erythrocytes at 1.7 A resolution. Structure. 2000;8:605–615. doi: 10.1016/s0969-2126(00)00147-7. [DOI] [PubMed] [Google Scholar]
- 13.Chauhan R, Mande SC. Characterization of the Mycobacterium tuberculosis H37Rv alkyl hydroperoxidase AhpC points to the importance of ionic interactions in oligomerization and activity. Biochem J. 2001;354:209–215. doi: 10.1042/0264-6021:3540209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood ZA, Poole LB, Hantgan RR, Karplus PA. Dimers to doughnuts: redox-sensitive oligomerization of 2-cysteine peroxiredoxins. Biochemistry. 2002;41:5493–5504. doi: 10.1021/bi012173m. [DOI] [PubMed] [Google Scholar]
- 15.Rabilloud T, Heller M, Gasnier F, Luche S, Rey C, Aebersold R, Benahmed M, Louisot P, Lunardi J. Proteomics analysis of cellular response to oxidative stress. Evidence for in vivo overoxidation of peroxiredoxins at their active site. J Biol Chem. 2002;277:19396–19401. doi: 10.1074/jbc.M106585200. [DOI] [PubMed] [Google Scholar]
- 16.Wagner E, Luche S, Penna L, Chevallet M, Van Dorsselaer A, Leize-Wagner E, Rabilloud T. A method for detection of overoxidation of cysteines: peroxiredoxins are oxidized in vivo at the active-site cysteine during oxidative stress. Biochem J. 2002;366:777–785. doi: 10.1042/BJ20020525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phalen TJ, Weirather K, Deming PB, Anathy V, Howe AK, van der Vliet A, Jonsson TJ, Poole LB, Heintz NH. Oxidation state governs structural transitions in peroxiredoxin II that correlate with cell cycle arrest and recovery. J Cell Biol. 2006;175:779–789. doi: 10.1083/jcb.200606005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jang HH, Lee KO, Chi YH, Jung BG, Park SK, Park JH, Lee JR, Lee SS, Moon JC, Yun JW, Choi YO, et al. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 2004;117:625–635. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Lee W, Choi KS, Riddell J, Ip C, Ghosh D, Park JH, Park YM. Human peroxiredoxin 1 and 2 are not duplicate proteins: the unique presence of CYS83 in Prx1 underscores the structural and functional differences between Prx1 and Prx2. J Biol Chem. 2007;282:22011–22022. doi: 10.1074/jbc.M610330200. [DOI] [PubMed] [Google Scholar]
- 20.Jin DY, Chae HZ, Rhee SG, Jeang KT. Regulatory role for a novel human thioredoxin peroxidase in NF-kappaB activation. J Biol Chem. 1997;272:30952–30961. doi: 10.1074/jbc.272.49.30952. [DOI] [PubMed] [Google Scholar]
- 21.Haridas V, Ni J, Meager A, Su J, Yu GL, Zhai Y, Kyaw H, Akama KT, Hu J, Van Eldik LJ, Aggarwal BB. TRANK, a novel cytokine that activates NF-kappa B and c-Jun N-terminal kinase. J Immunol. 1998;161:1–6. [PubMed] [Google Scholar]
- 22.Matsumoto A, Okado A, Fujii T, Fujii J, Egashira M, Niikawa N, Taniguchi N. Cloning of the peroxiredoxin gene family in rats and characterization of the fourth member. FEBS Lett. 1999;443:246–250. doi: 10.1016/s0014-5793(98)01736-0. [DOI] [PubMed] [Google Scholar]
- 23.Okado-Matsumoto A, Matsumoto A, Fujii J, Taniguchi N. Peroxiredoxin IV is a secretable protein with heparin-binding properties under reduced conditions. J Biochem (Tokyo) 2000;127:493–501. doi: 10.1093/oxfordjournals.jbchem.a022632. [DOI] [PubMed] [Google Scholar]
- 24.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 25.Tu BP, Weissman JS. Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol. 2004;164:341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tu BP, Weissman JS. The FAD- and O(2)-dependent reaction cycle of Ero1-mediated oxidative protein folding in the endoplasmic reticulum. Mol Cell. 2002;10:983–994. doi: 10.1016/s1097-2765(02)00696-2. [DOI] [PubMed] [Google Scholar]
- 27.Sevier CS, Cuozzo JW, Vala A, Aslund F, Kaiser CA. A flavoprotein oxidase defines a new endoplasmic reticulum pathway for biosynthetic disulphide bond formation. Nat Cell Biol. 2001;3:874–882. doi: 10.1038/ncb1001-874. [DOI] [PubMed] [Google Scholar]
- 28.Chakravarthi S, Jessop CE, Willer M, Stirling CJ, Bulleid NJ. Intracellular catalysis of disulfide bond formation by the human sulfhydryl oxidase, QSOX1. Biochem J. 2007;404:403–411. doi: 10.1042/BJ20061510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woods A, Sherwin T, Sasse R, MacRae TH, Baines AJ, Gull K. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J Cell Sci. 1989;93(Pt 3):491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- 30.Vaux D, Tooze J, Fuller S. Identification by anti-idiotype antibodies of an intracellular membrane protein that recognizes a mammalian endoplasmic reticulum retention signal. Nature. 1990;345:495–502. doi: 10.1038/345495a0. [DOI] [PubMed] [Google Scholar]
- 31.Jessop CE, Bulleid NJ. Glutathione directly reduces an oxidoreductase in the endoplasmic reticulum of mammalian cells. J Biol Chem. 2004;279:55341–55347. doi: 10.1074/jbc.M411409200. [DOI] [PubMed] [Google Scholar]
- 32.Wilson R, Allen AJ, Oliver J, Brookman JL, High S, Bulleid NJ. The translocation, folding, assembly and redox-dependent degradation of secretory and membrane proteins in semi-permeabilized mammalian cells. Biochem J. 1995;307(Pt 3):679–687. doi: 10.1042/bj3070679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jessop CE, Chakravarthi S, Garbi N, Hammerling GJ, Lovell S, Bulleid NJ. ERp57 is essential for efficient folding of glycoproteins sharing common structural domains. Embo J. 2007;26:28–40. doi: 10.1038/sj.emboj.7601505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hemsley A, Arnheim N, Toney MD, Cortopassi G, Galas DJ. A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Res. 1989;17:6545–6551. doi: 10.1093/nar/17.16.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 36.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 37.Kohno K, Normington K, Sambrook J, Gething MJ, Mori K. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol Cell Biol. 1993;13:877–890. doi: 10.1128/mcb.13.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 39.Fujii J, Ikeda Y. Advances in our understanding of peroxiredoxin, a multifunctional, mammalian redox protein. Redox Rep. 2002;7:123–130. doi: 10.1179/135100002125000352. [DOI] [PubMed] [Google Scholar]
- 40.Otsu M, Bertoli G, Fagioli C, Guerini-Rocco E, Nerini-Molteni S, Ruffato E, Sitia R. Dynamic retention of Ero1alpha and Ero1beta in the endoplasmic reticulum by interactions with PDI and ERp44. Antioxid Redox Signal. 2006;8:274–282. doi: 10.1089/ars.2006.8.274. [DOI] [PubMed] [Google Scholar]
- 41.Mizohata E, Sakai H, Fusatomi E, Terada T, Murayama K, Shirouzu M, Yokoyama S. Crystal structure of an archaeal peroxiredoxin from the aerobic hyperthermophilic crenarchaeon Aeropyrum pernix K1. J Mol Biol. 2005;354:317–329. doi: 10.1016/j.jmb.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 42.Papinutto E, Windle HJ, Cendron L, Battistutta R, Kelleher D, Zanotti G. Crystal structure of alkyl hydroperoxide-reductase (AhpC) from Helicobacter pylori. Biochim Biophys Acta. 2005;1753:240–246. doi: 10.1016/j.bbapap.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 43.Moon JC, Hah YS, Kim WY, Jung BG, Jang HH, Lee JR, Kim SY, Lee YM, Jeon MG, Kim CW, Cho MJ, et al. Oxidative stress-dependent structural and functional switching of a human 2-Cys peroxiredoxin isotype II that enhances HeLa cell resistance to H2O2-induced cell death. J Biol Chem. 2005;280:28775–28784. doi: 10.1074/jbc.M505362200. [DOI] [PubMed] [Google Scholar]
- 44.Rand JD, Grant CM. The thioredoxin system protects ribosomes against stress-induced aggregation. Mol Biol Cell. 2006;17:387–401. doi: 10.1091/mbc.E05-06-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]