Abstract
The DNA-binding protein PRDM9 has a critical role in specifying meiotic recombination hotspots in mice and apes, but appears to be absent from other vertebrate species, including birds. To study the evolution and determinants of recombination in species lacking PRDM9, we inferred fine-scale genetic maps from population resequencing data for two bird species, the zebra finch Taeniopygia guttata and the long-tailed finch Poephila acuticauda. We find that both species have hotspots, which are enriched near functional genomic elements. Unlike in mice and apes, the two species share most hotspots, with conservation seemingly extending over tens of millions of years. These observations suggest that in the absence of PRDM9, recombination targets functional features that both enable access to the genome and constrain its evolution.
Meiotic recombination is a ubiquitous and fundamental genetic process that shapes variation in populations, yet our understanding of its underlying mechanisms is based on a handful of model organisms, scattered throughout the tree of life. One pattern shared among most sexually reproducing species is that meiotic recombination tends to occur in short segments of 100s to 1000s of base pairs, termed “recombination hotspots” (1). In apes and mice, the location of hotspots is largely determined by PRDM9, a zinc-finger protein that binds to specific motifs in the genome during meiotic prophase and generates H3K4me3 marks, eventually leading to double-strand breaks (DSBs) and both crossover and non-crossover resolutions (2–5). In mammals, the zinc-finger domain of PRDM9 evolves quickly, with evidence of positive selection on residues in contact with DNA (2, 6), and as a result, there is rapid turnover of hotspot locations across species, subspecies, and populations (7–10).
Although PRDM9 plays a pivotal role in controlling recombination localization in mice and apes, many species lacking PRDM9 nonetheless have hotspots (6). An artificial example is provided by mice knockouts for PRDM9. Despite being sterile, they make similar numbers of DSBs as wild-type mice, and their recombination hotspots appear to default to residual H3K4me3 mark locations, notably at promoters (10). A natural but puzzling example is provided by canids, which carry premature stop codons in PRDM9 yet are able to recombine and remain fertile (11, 12). Like in mouse PRDM9 knockouts, in dogs and in other species without PRDM9 such as the yeast Saccharomyces cerevisae and the plant Arabidopsis thaliana, hotspots tend to occur at promoters or other regions with promoter-like features (11, 13, 14). In yet other taxa without PRDM9, including Drosophila species (15), honeybees (16), and Caenorhabditis elegans (17), short, intense recombination hotspots appear to be absent altogether.
To further explore how the absence of PRDM9 shapes the fine-scale recombination landscape and impacts its evolution, we turn to birds, because an analysis of the chicken genome suggested that it may not have PRDM9 (6). We first confirmed the absence of PRDM9 across reptiles by querying the genomes of 48 species of birds, three species of crocodilians, two species of turtles, and one species of lizard for PRDM9 (18), finding that only the turtle genomes contain putative orthologs with all three PRDM9 domains (Fig. S1). We also found no expression of any PRDM9-like transcripts in RNAseq data from testes tissue of the zebra finch (Taeniopygia guttata) (18). Given the likely absence of PRDM9 in birds, we ask: is recombination nonetheless concentrated in hotspots in these species? If so, how quickly do the hotspots evolve? Where does recombination tend to occur in the genome? To address these questions, we generated whole-genome resequencing data for wild populations from two bird species and inferred fine-scale genetic maps from patterns of linkage disequilibrium.
Inferring fine-scale recombination maps
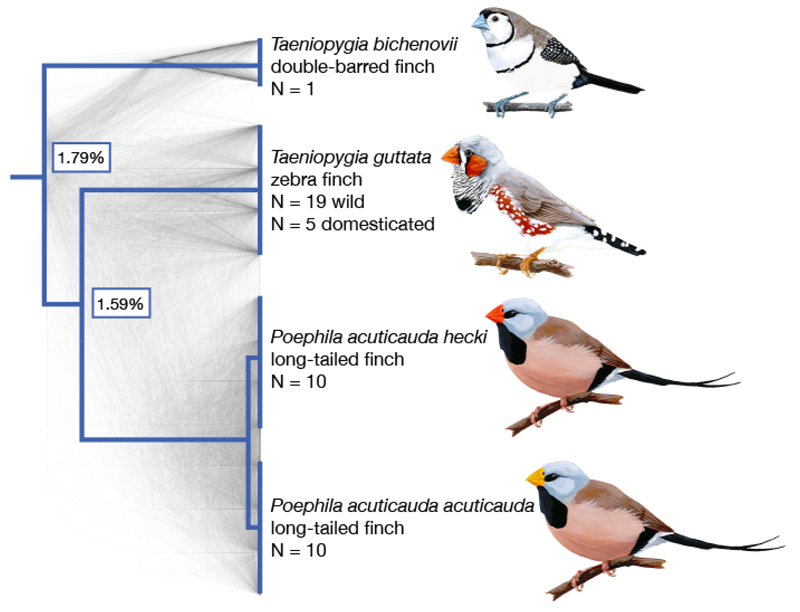
We sampled three species of finch in the family Estrildidae: zebra finch (Taeniopygia guttata; n=19 wild, unrelated birds and n=5 from a domesticated, nuclear family), long-tailed finch (Poephila acuticauda; n=20, including 10 of each of two, similar subspecies with average autosomal FST = 0.039), and, for use as an outgroup, double-barred finch (Taeniopygia bichenovii; n=1) [Fig. 1, Table S1; (18)]. Despite extensive incomplete lineage sorting between the species, they do not appear to have diverged with gene flow (Fig. S2). Moreover, nucleotide divergence among the three finch species is similar to that of human, chimpanzee and gorilla, providing a well-matched comparison to apes (8, 9) (Fig. 1).
Figure 1.
Species tree for the finch species in this study. Species sampled were double-barred finch (Taeniopygia bichenovii), zebra finch (T. guttata), and the two long-tailed finch subspecies (Poephila acuticauda hecki and P. a. acuticauda). Tree rooted with medium ground finch and collared flycatcher (Geospiza fortis and Ficedula albicollis; full phylogeny shown in Fig. 4). Shown in gray are 1000 gene trees, which were used to infer the species tree (18). The pairwise divergence between species is indicated at nodes, as measured by the genome-wide average across autosomes. Images of birds from Wikimedia Commons.
We mapped reads from all individuals to the zebra finch reference genome [1 Gb assembled across 34 chromosomes; (19)] and generated de novo SNP calls for all three species. After filtering for quality, we identified 44.6 million SNPs in zebra finch, 26.2 million SNPs in long-tailed finch, and 3.0 million SNPs in double-barred finch (Table S2). These SNP numbers correspond to autosomal nucleotide diversity of π=0.82% and θw =1.37% in zebra finch and π=0.55% and θw=0.73% in long-tailed finch, approximately ten times higher than estimates in apes (20). Assuming a mutation rate per base pair per generation of 7 × 10−10 (18), these diversity levels suggest a long-term effective population size (Ne) of 4.8 × 106 and 2.5 × 106 in the zebra finch and long-tailed finch, respectively. Thus, these two species have much larger Ne than most other species for which there exist fine-scale recombination maps, with Ne more reflective of biodiversity at large (Fig. S3).
Next, we inferred haplotypes for zebra finch and long-tailed finch using a linkage-disequilibrium approach that incorporated phase-informative reads and family phasing. From the haplotypes, we estimated fine-scale recombination maps using LDhelmet, which works well for species with higher nucleotide diversity (15). The resulting maps estimated median recombination rates in the zebra finch and long-tailed finch genomes as ρ = 26.2/kb and 14.0/kb, respectively, which corresponds to a median rate of 0.14 cM/Mb in both species (18). Simulations indicated that we had limited power to identify hotspots in regions with high recombination rates (Fig. S4), so we restricted our analyses to the 18 largest chromosomes in the reference genome (930 Mb; 91% of the assembled genome). For these 18 chromosomes, our results accord well with recombination maps inferred from a more limited pedigree-based study of zebra finch (21), with a correlation of 0.90 for rates estimated at the 5 Mb scale (Fig. S5), providing confidence in our rate inferences.
Hotspots and their evolution
To identify hotspots in the genome, we operationally define them as regions that are at least 2 kb in length, have at least 5-fold the background recombination rate as estimated across the 80 kb of sequence surrounding the region, and are statistically supported as hotspots by a likelihood ratio test (18). This approach yielded 3949 hotspots in zebra finch and 4933 hotspots in long-tailed finch (Figs. 2, S6, S7), with one hotspot detected on average every 215 and 179 kb in the two species, respectively. Both the lower density of hotspots in zebra finch than in long-tailed finch and the lower density of hotspots in the finches compared to humans is consistent with simulations that indicate decreased power to detect hotspots when the background population recombination rate is higher (18, Figs. S4, S8). Importantly, the hotspots were detected after aggressively filtering our SNP datasets and show no evidence of having higher phasing error rates than the rest of the genome (Table S3, S4, Fig. S9).
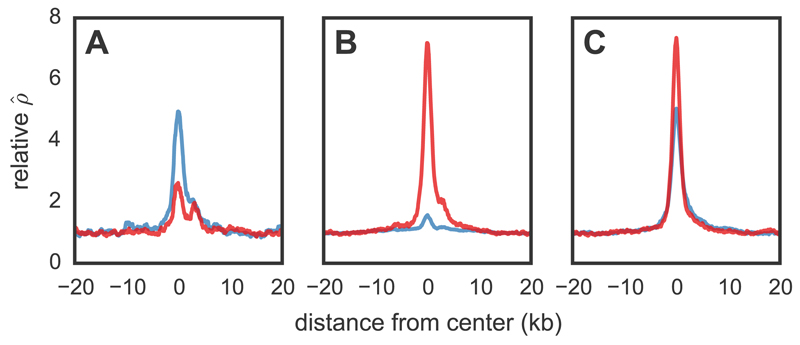
Figure 2.
Recombination rates across hotspots in zebra finch and long-tailed finch. Average relative recombination rate (/bp divided by the background of 20 kb on either side of the hotspot) across (A) hotspots detected only in zebra finch (n=1075; shown in blue), (B) those detected only in long-tailed finch (middle; n=2059; shown in red), and (C) those inferred as shared in the two species (right; n=2874). Shared hotspots are those whose midpoints occur within 3 kb of each other. The orientation of hotspots is with respect to the genomic sequence.
Considering hotspots as shared if their midpoints occur within 3 kb of each other, 73% of zebra finch hotspots (2874 of 3949 hotspots) were detected as shared between the two species (Fig. S10) when only 4.4% were expected to overlap by chance (Figs. S10, 11); similar results were obtained under different criteria for hotspot sharing (Table S5). The true fraction of shared hotspots between zebra finch and long-tailed finch is likely higher than observed, because we do not have complete power (Fig. S4) and simulations suggest that we are unlikely to detect spurious cases of hotspot sharing (18). On the other hand, the observed levels of sharing are somewhat lower than expected compared to a model in which all hotspots are identical in the two species (Fig. S12).
This conservation of hotspots contrasts sharply with comparative analyses in apes and mice, where, even across populations with modest levels of genetic differentiation, there is no hotspot sharing (8–10). In fact, if we apply the same criterion for hotspot sharing to humans and chimpanzees, only 10.5% of chimpanzee hotspots overlap with human hotspots when a 7.2% overlap is expected by chance (Fig. S11).
To provide further support for the validity of the inferred hotspots, we tested if they show evidence for GC-biased gene conversion (gBGC), measured as higher expected equilibrium levels of GC content (GC*; 18). Because evidence for gBGC in birds is somewhat indirect (22), we first looked for support for gBGC at broad genomic scales, finding a positive relationship between recombination rate and GC* (Fig. 3A-B). Narrowing our focus to the regions surrounding hotspots, we observe that hotspots exhibit peaked GC* relative to both flanking sequence and coldspots (i.e., regions without peaks in recombination) matched for the same overall GC and CpG content (Fig. 4A-B). A similar phenomenon is seen in intra-species variation data: at hotspots but not matched coldspots, derived alleles segregate at a higher frequency at AT to GC polymorphisms than at GC to AT polymorphisms (Fig. S13). Thus, two independent signatures of recombination—patterns of linkage disequilibrium and of base composition— converge in demonstrating that finches have recombination hotspots and that these are conserved over much larger time scales than in apes and mice (8–10).
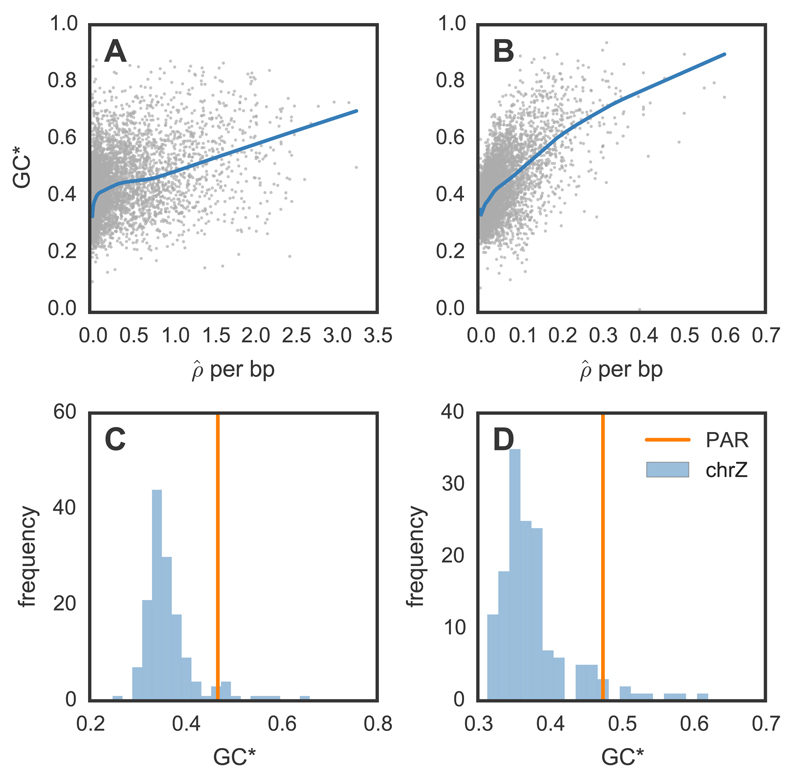
Figure 3.
Equilibrium GC content and broad-scale recombination rates in zebra finch (A, C) and long-tailed finch (B, D). (A-B) Relationship between equilibrium GC content [GC*; 18]) and /bp for zebra finch and long-tailed finch across all autosomal chromosomes. Both GC* and were calculated across 50 kb windows with LOESS curves shown for span of 0.2. (C-D) GC* and the pseudoautosomal region (PAR). The histogram shows GC* for chromosome Z across 500 kb windows; GC* for the 450 kb PAR shown by the vertical line.
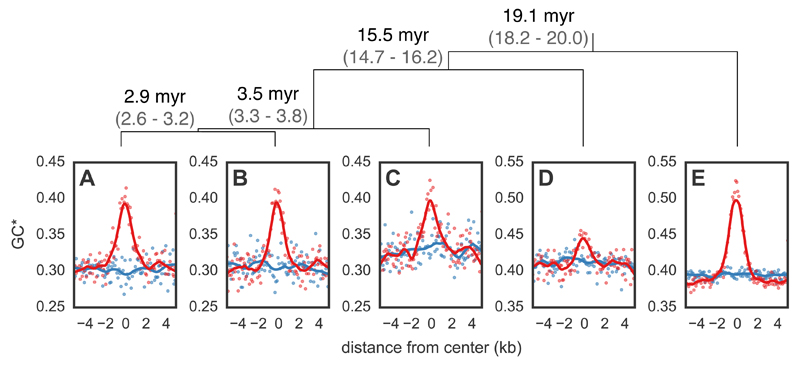
Figure 4.
Expected equilibrium GC content (GC*) around hotspots and matched coldspots for five bird species. Points (hotspots shown in red and coldspots in blue) represent GC* estimated from the lineage-specific substitutions aggregated in 100 bp bins from the center of all hotspots in (A) zebra finch and (B) long-tailed finch. GC* for (C) double-barred finch, (D) medium ground finch, and (E) collared flycatcher was calculated around hotspots identified as shared between zebra finch and long-tailed finch. LOESS curves are shown for a span of 0.2. The orientation of hotspots is with respect to the genomic sequence. Species tree (18) shown with estimated divergence times in millions of years (myr) and its 95% Highest Posterior Density in gray; top.
After observing the pattern of gBGC at hotspots in zebra finch and long-tailed finch, we tested how far conservation of hotspot locations extends across the avian phylogeny by additionally considering the genomes of double-barred finch (an estimated ~3.5 million years [myr] from zebra finch (18)), medium ground finch Geospiza fortis (~15.5 myr from zebra finch (23)), and collared flycatcher Ficedula albicollis (~19.1 myr from zebra finch (24)). Because we only had a single diploid genome from these species, we tested for hotspot conservation indirectly by determining if these species had peaks in GC* at the hotspot locations inferred as shared between zebra finch and long-tailed finch. We find localized GC* peaks at hotspots in all three species (Fig. 4C-E), suggesting that the conservation of hotspots extends across tens of millions of years of evolution. Intriguingly, these findings echo those obtained from four species of Saccharomyces yeast, which show nearly complete conservation of hotspot locations and intensities across species 15 myr diverged (25). Almost all hotspots in Saccharomyces yeast occur at promoters, which are evolutionarily stable, suggesting that how hotspot locations are specified influences how they evolve (12, 26).
The localization of hotspots in the genome
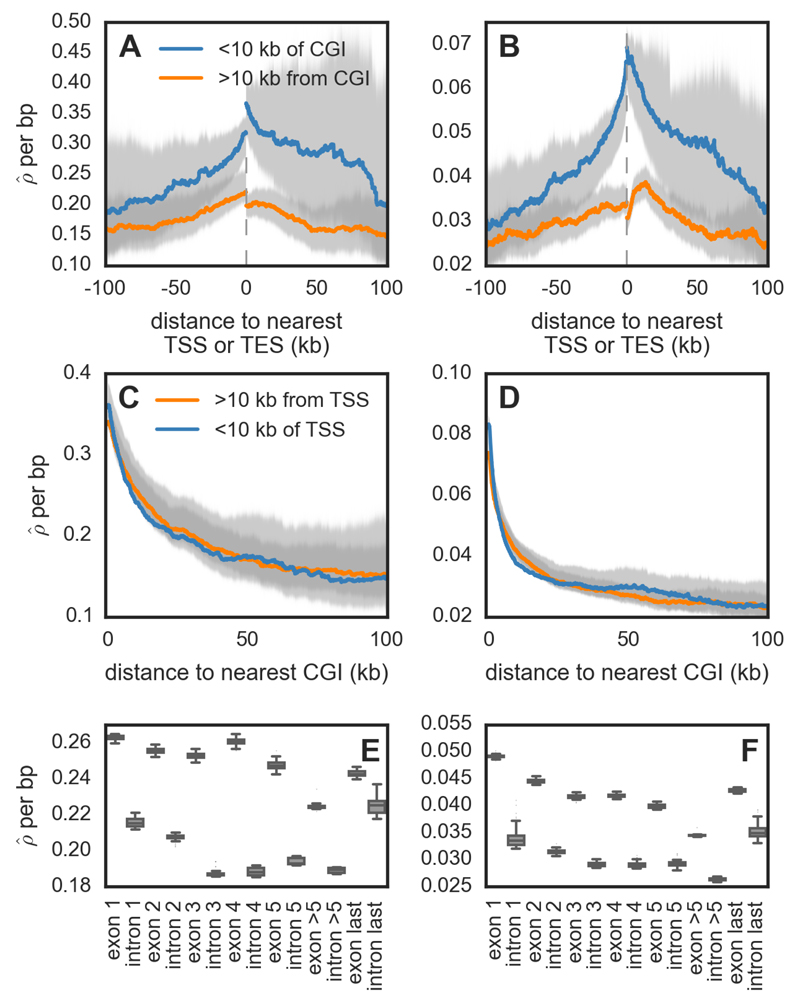
Hotspots in zebra finch and long-tailed finch are enriched near transcription start sites (TSSs), transcription stop sites (TESs) and CpG islands (CGIs), with close to half of all hotspots occurring within 3 kb of one of these features (~17% occur within 3 kb of both an annotated TSS and CGI, 3% within 3 kb of both a TES and CGI, and ~26% within 3 kb of a CGI only; Fig. S14). In particular, the hotspots near CGIs are more likely to be shared between species and exhibit stronger evidence for gBGC compared to hotspots distant from CGIs (Fig. S15), providing further support for the importance of these elements in the targeting of recombination.Consistent with the findings about hotspots, recombination rates are nearly two-fold higher near annotated TSSs and TESs (Fig. 5A-B). This pattern appears to be driven mainly by their co-localization with CGIs (Fig. 5A-B, Fig. S16): rates near CGIs are more than three-fold higher with only a small further increase if they are near a TSS or a TES (Fig. 5C-D; Fig. S17).
Figure 5.
Recombination rates across genomic features for zebra finch (A, C, E) and long-tailed finch (B, D, F). (A-B) Estimated recombination rates (/bp) around annotated transcription start sites (TSSs) and end sites (TESs), conditional on whether they are within 10 kb of a CpG island (CGI) or not. The gray dotted line represents the location of the gene, and the distances are shown accounting for the 5' → 3' orientation of genes. (C-D) shown as a function of distance to nearest CGI, conditional on whether the CGI is within 10 kb of an annotated TSS or not. See Fig. S17 for the pattern of CGIs relative to TESs. For figures A – D, uncertainty in rate estimates (shown in gray) was estimated by drawing 100 bootstrap samples and recalculating means. (E-F) within exons and introns for genes that have ≥5 exons (n=7,131). See Fig. S28 for simulation results that suggest the inference of higher background in exons does not reflect differences in diversity levels between exons and introns.
A positive association between proximity to the TSS and recombination rate has been previously reported in a number of species without PRDM9, including S. cerevisiae, the monkey-flower Mimulus guttatus, dogs, and A. thaliana (11, 13, 14, 27) and an association between TES and recombination rate has been shown in A. thaliana (14). In turn, the link between CGIs and recombination rates has been found both in species without PRDM9 including dogs (11) and, albeit more weakly, in species with PRDM9 including humans and chimpanzees (9). Moreover, the relationship between distance to CGIs and recombination rate remains significant after controlling for expression levels in zebra finch testes (Spearman’s r=-0.11; p=4.32 ×10−27, Fig. S18). This increase in recombination rates near TSSs, TESs and CGIs supports a model in which, particularly in the absence of PRDM9 binding specificity, recombination is concentrated at functional elements that are accessible to the recombination machinery. Indeed, all three coincide with destabilization of nearby nucleosome occupancy (28, 29) and both TSSs and CGIs serve as sites of transcription initiation (30). An intriguing implication is that the structure of linkage disequilibrium may differ systematically between species with and without PRDM9, with tighter coupling between regulatory and exonic variants in species with PRDM9.
Under a model in which the recombination machinery tends to target accessible genomic elements, we would not necessarily expect to see enrichment of specific binding motifs associated with hotspot activity. Accordingly then, when we test for motifs enriched in hotspots relative to coldspots, the top motifs in both species are a string of As, which are also enriched in A. thaliana and yeast hotspots and which may be nucleosome depleted or facilitate nucleosome removal (13, 31) (Fig. S19). We also find a number of additional motifs that are GC-rich and perhaps indicative of CGIs.
At even finer resolution, recombination rates are higher in exonic than intronic regions, which has also been seen in A. thaliana (14), dogs (11), and M. guttatus (27) and higher towards the ends of the gene than in the middle (Fig. 5E-F). One possibility for these patterns is that DSBs preferentially initiate in exons near the TSS and TES and their resolution occurs in intervening exons and introns. The specific mechanism by which DSBs would preferentially initiate in exons is unknown, but the pattern is consistent with an important role for chromatin marks that distinguish exons from introns (28).
Contrasting tempos of broad- and fine scale recombination rate evolution
Median recombination rates across and within chromosomes vary over nearly six orders of magnitude (Figs. S8, S20), creating a heterogeneous landscape of broad-scale recombination rates across the genome, with regions of elevated recombination near telomeres and large intervening deserts (as found in zebra finch pedigree data; (21)). Indeed, most of the recombination events in zebra finch and long-tailed finch occur in a narrow portion of the genome, with 82% and 70% of events localized to 20% of the genome in zebra finch and long-tailed finch, respectively (Fig. S21). Notably, recombination rates for the Z sex chromosome are two orders of magnitude lower than the most similarly sized autosome, chromosome 1A, even after accounting for the lack of recombination in females (Fig. S8, (21)). Although cytological data indicate that both zebra finch and long-tailed finch harbor a pericentric inversion polymorphism over much of chromosome Z (32, 33), such an inversion is unlikely to explain this extreme a difference (18).
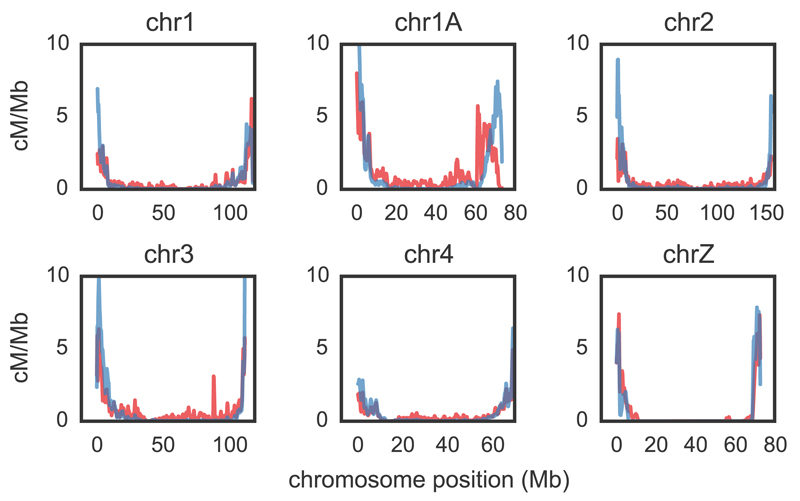
Between zebra finch and long-tailed finch, broad-scale rates are highly similar, with genome-wide correlations of 0.82 and 0.86 at the 10 kb and 1 Mb scales, respectively (Figs. 6, S20). Despite this broad-scale concordance, we infer that some genomic regions between the two species have very different rates of recombination (Fig. S22) and find tentative support for some of these changes in the derived allele frequency spectra (Fig. S23). Moreover, at a greater evolutionary distance, broad-scale patterns differ markedly; the collared flycatcher (~19 myr diverged) has a relatively homogeneous recombination landscape compared to zebra finch and long-tailed finch (24). This evolution of broad-scale rates is particularly notable because, in many species, shifts in broad-scale recombination patterns can be explained almost entirely by chromosomal rearrangements, shifts in karyotypes, and changes in chromosome lengths (9, 34, 35). However, there is no obvious pattern by which chromosomal rearrangements drive differences in recombination rates between zebra finch and long-tailed finch (Fig. S22), and, despite harboring a number of small inversions between them, collared flycatcher and zebra finch have similar karyotypes and syntenic genomes (24). That broad-scale recombination patterns have changed across the same phylogenetic breadth for which we see hotspot conservation suggests two non-exclusive possibilities that merit further investigation: either the heats or locations of some hotspots have evolved, or rates have changed in regions that fall outside of our operational definition of hotspots.
Figure 6.
Comparative recombination rates for zebra finch and long-tailed finch. Zebra finch rates shown in red; long-tailed finch in blue. Estimated rates [cM/Mb; obtained from /bp (18)] are shown as rolling means calculated across 100 kb windows. We show here the five largest autosomal chromosomes and chromosome Z; see Fig. S20 for all chromosomes. Rate estimates for chromosome Z should be taken with caution for both biological and technical reasons (see 18 for more information).
The impact of recombination on the genome
Given the marked variation in recombination rates across the genome, we consider the consequences for genome evolution. First, we note that increased recombination rates drive increasing GC content in the genome, presumably via gBGC, and we see this phenomenon both at the genome-wide scale (Fig. 3A-B) and the scale of hotspots (Fig. 4). An extreme example is provided by the pseudoautosomal region (PAR), which we identified on an unassembled scaffold from chromosome Z using estimates of coverage in males and females. We confirmed the PAR by inferring homology to PARs identified in medium ground finch and collared flycatcher (Fig. S24). The PAR is short—estimated to be just 450 kb—and is subject to an obligate crossover in every female meiosis (36); as such, it has very high recombination rates. The consequence is visible in the high GC* for the PAR, which exceeds estimates of GC* across most of the rest of chromosome Z in both species (Fig. 3C-D).
Further, as has been reported for many other organisms, including chickens (37–39), our results suggest that recombination is positively correlated with levels of nucleotide diversity, particularly on the Z (Fig. S25–27). This observation is consistent with widespread effects of linked selection in these species (40).
Conclusion
Finches lack PRDM9 yet nonetheless harbor hotspots, with recombination concentrated at functional elements (TESs, TSSs, and CGIs) that likely denote greater accessibility to the cellular recombination machinery. In sharp contrast to apes and mice, the hotspot locations are conserved among species several millions of years diverged and likely over tens of millions of years. These results suggest that the genetic architecture of recombination influences the rate at which hotspots evolve. Whereas the binding specificity of PRDM9 drives rapid turnover, the reliance on accessible, functional genomic features leads to stasis. This hypothesis accords with recent results in yeast, in which recombination is concentrated at promoters, and hotspots are stable in intensity and location over tens of millions of years (25). To further investigate how deeply this stasis extends and explore the taxonomic generality of these findings, the approaches illustrated here can be applied to other sequenced bird species (41) and beyond. In doing so, we will begin to better understand why species differ so drastically in their specification of hotspots and, in particular, why a subset rely on PRDM9.
Supplementary Materials
Materials and Methods
Figures S1-S35
Tables S1-S6
References S42-S94
One-Sentence Summary.
We show that the fine-scale recombination landscape is stable across tens of millions of years in birds, in sharp contrast to what is seen in primates and mice.
Acknowledgements
This project was started when MP was a Howard Hughes Medical Institute Early Career Scientist and was funded, in part, by Wellcome Trust grants 086786/Z/08/Z to O.V. and 090532/Z/09/Z to the Wellcome Trust Centre for Human Genetics. We thank B. de Massy, C. Grey, S. Myers, T. Price, M. Schumer, J. Wall, A. Williams and J. Willis for helpful discussions and/or comments on the manuscript; K. Argoud and P. Piazza at the Genomics Core at the Wellcome Trust Centre for Human Genetics for assistance with lab work; and M. T. Gilbert for sharing the zebra finch gene annotations in advance of publication. We thank I. Lam and S. Keeney for sharing their unpublished manuscript with us, and S. Keeney for many helpful discussions. BAM alignment files for genomic data and filtered variant call files (VCFs) for zebra finch, long-tailed finch, and double-barred finch are available at http://www.ebi.ac.uk/ena/data/view/PRJEB10586. Sequence reads for RNAseq experiments in zebra finch are available at http://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA295077. Masked genome files, the reconstructed ancestral genome, and recombination maps for both species are available at DataDryad: doi:10.5061/dryad.fd24j. All scripts and an electronic lab notebook for this work are available at https://github.com/singhal/postdoc and https://github.com/singhal/labnotebook, respectively.
References and Notes
- 1.de Massy B. Annual review of genetics. 2013;47:563. doi: 10.1146/annurev-genet-110711-155423. [DOI] [PubMed] [Google Scholar]
- 2.Myers S, et al. Science. 2010;327:876. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg IL, et al. Nature genetics. 2010;42:859. doi: 10.1038/ng.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baudat F, et al. Science. 2010;327:836. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams AL, et al. eLife. 2015;4:e04637. [Google Scholar]
- 6.Oliver PL, et al. PLoS Genet. 2009;5:e1000753. doi: 10.1371/journal.pgen.1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinch AG, et al. Nature. 2011;476:170. doi: 10.1038/nature10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevison LS, et al. bioRxiv. 2015:013755. [Google Scholar]
- 9.Auton A, et al. Science. 2012;336:193. doi: 10.1126/science.1216872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Nature. 2012;485:642. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Auton A, et al. PLoS Genetics. 2013;9 [Google Scholar]
- 12.Axelsson E, et al. Genome research. 2012;22:51. doi: 10.1101/gr.124123.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan J, et al. Cell. 2011;144:719. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi K, et al. Nature genetics. 2013;45:1327. doi: 10.1038/ng.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan AH, Jenkins PA, Song YS. PLoS genetics. 2012;8:e1003090. doi: 10.1371/journal.pgen.1003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wallberg A, Gle´min S, Webster MT. PLoS Genetics. 2015;11 doi: 10.1371/journal.pgen.1005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaur T, Rockman MV. Genetics. 2014;196:137. doi: 10.1534/genetics.113.158857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Materials and methods are available as supplementary materials on Science Online.
- 19.Warren WC, et al. Nature. 2010;464:757. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leffler EM, et al. PLoS Biol. 2012;10:e1001388. doi: 10.1371/journal.pbio.1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Backstrom N, et al. Genome research. 2010;20:485. doi: 10.1101/gr.101410.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber CC, Boussau B, Romiguier J, Jarvis ED, Ellegren H. Genome biology. 2014;15:549. doi: 10.1186/s13059-014-0549-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang G, Parker P, Li B, Li H, Wang J. GigaScience. 2012;1:13. [Google Scholar]
- 24.Kawakami T, et al. Molecular ecology. 2014;23:4035. doi: 10.1111/mec.12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lam I, Keeney S. bioRxiv. 2015:023176. [Google Scholar]
- 26.Nicolas A, Treco D, Schultes NP, Szostak JW. Nature. 1989;338:35. doi: 10.1038/338035a0. [DOI] [PubMed] [Google Scholar]
- 27.Hellsten U, et al. Proceedings of the National Academy of Sciences. 2013;110:19478. doi: 10.1073/pnas.1319032110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones PA. Nature Reviews Genetics. 2012;13:484. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 29.Kaplan N, et al. Nature. 2009;458:362. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deaton AM, Bird A. Genes & development. 2011;25:1010. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wijnker E, et al. Elife. 2013;2:e01426. doi: 10.7554/eLife.01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christidis L. Genetica. 1986;71:81. [Google Scholar]
- 33.Itoh Y, Kampf K, Balakrishnan CN, Arnold AP. Chromosoma. 2011;120:255. doi: 10.1007/s00412-010-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dumont BL, Payseur BA. Evolution. 2008;62:276. doi: 10.1111/j.1558-5646.2007.00278.x. [DOI] [PubMed] [Google Scholar]
- 35.Jensen-Seaman MI, et al. Genome research. 2004;14:528. doi: 10.1101/gr.1970304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otto SP, et al. Trends in Genetics. 2011;27:358. doi: 10.1016/j.tig.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 37.Nachman MW. TRENDS in Genetics. 2001;17:481. doi: 10.1016/s0168-9525(01)02409-x. [DOI] [PubMed] [Google Scholar]
- 38.Begun DJ, Aquadro CF. Nature. 1992;356:519. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- 39.Mugal CF, Arndt PF, Ellegren H. Molecular Biology and Evolution. 2013;30:1700. doi: 10.1093/molbev/mst067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charlesworth B, Morgan M, Charlesworth D. Genetics. 1993;134:1289. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang G, et al. Science. 2014;346:1311. doi: 10.1126/science.1251385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griffith SC, Pryke SR, Mariette M. Emu. 2009;108:311. [Google Scholar]
- 43.Rollins LA, Svedin N, Pryke SR, Griffith SC. Ecology and evolution. 2012;2:1208. doi: 10.1002/ece3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pesole G, et al. Gene. 2001;276:73. doi: 10.1016/s0378-1119(01)00674-6. [DOI] [PubMed] [Google Scholar]
- 45.Nylander J. Mraic.pl. 2004 https://github.com/nylander/MrAIC
- 46.Guindon S, et al. Systematic biology. 2010;59:307. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 47.Liu L, Yu L, Pearl DK, Edwards SV. Systematic Biology. 2009;58:468. doi: 10.1093/sysbio/syp031. [DOI] [PubMed] [Google Scholar]
- 48.Bouckaert R, et al. PLoS Comput Biol. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agate R, Scott B, Haripal B, Lois C, Nottebohm F. PNAS. 2009;106:17963. doi: 10.1073/pnas.0909139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hooper DM, Price TD. Evolution. 2015;69:890. doi: 10.1111/evo.12633. [DOI] [PubMed] [Google Scholar]
- 51.Price TD, et al. Nature. 2014;509:222. doi: 10.1038/nature13272. [DOI] [PubMed] [Google Scholar]
- 52.Jetz W, Thomas G, Joy J, Hartmann K, Mooers A. Nature. 2012;491:444. doi: 10.1038/nature11631. [DOI] [PubMed] [Google Scholar]
- 53.Jarvis ED, et al. Science. 2014;346:1320. [Google Scholar]
- 54.Pickrell JK, Pritchard JK. PLoS Genet. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Nature. 2009;461:489. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.St John JA, et al. Genome biology. 2012;13:415. doi: 10.1186/gb-2012-13-1-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Camacho C, et al. BMC bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Birney E, Clamp M, Durbin R. Genome research. 2004;14:988. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baudat F, Imai Y, de Massy B. Nature Reviews Genetics. 2013;14:794. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- 60.Kent WJ. Genome research. 2002;12:656. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quinlan AR, Hall IM. Bioinformatics. 2010;26:841. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McKenna A, et al. Genome research. 2010;20:1297. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou Q, et al. Science. 2014;346:1246338. doi: 10.1126/science.1246338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smeds L, et al. Nature communications. 2014;5 doi: 10.1038/ncomms6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harris RS. Improved pairwise alignment of genomic DNA. ProQuest; 2007. [Google Scholar]
- 66.Lunter G, Goodson M. Genome research. 2011;21:936. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H. arXiv preprint arXiv: 1303.3997. 2013 [Google Scholar]
- 68.Iqbal Z, Caccamo M, Turner I, Flicek P, McVean G. Nature genetics. 2012;44:226. doi: 10.1038/ng.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H, et al. Bioinformatics. 2009;25:2078. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.1000 Genomes Project Consortium. Nature. 2010;467:1061. [Google Scholar]
- 71.Purcell S, et al. The American Journal of Human Genetics. 2007;81:559. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pritchard JK, Przeworski M. The American Journal of Human Genetics. 2001;69:1. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rands CM, et al. Bmc Genomics. 2013;14:95. doi: 10.1186/1471-2164-14-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Langmead B, Salzberg SL. Nature methods. 2012;9:357. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Watterson G. Theoretical population biology. 1975;7:256. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 76.Tajima F. Genetics. 1989;123:597. doi: 10.1093/genetics/123.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Balakrishnan CN, Edwards SV. Genetics. 2009;181:645. doi: 10.1534/genetics.108.094250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Williams AL, Housman DE, Rinard MC, Gifford DK. Genome biology. 2010;11:R108. doi: 10.1186/gb-2010-11-10-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Delaneau O, Howie B, Cox AJ, Zagury JF, Marchini J. AJHG. 2013;93:687. doi: 10.1016/j.ajhg.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stephens M, Donnelly P. The American Journal of Human Genetics. 2003;73:1162. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Coop G, Przeworski M. Nature Reviews Genetics. 2007;8:23. doi: 10.1038/nrg1947. [DOI] [PubMed] [Google Scholar]
- 82.Choi K, Henderson IR. The Plant Journal. 2015 doi: 10.1111/tpj.12870. [DOI] [PubMed] [Google Scholar]
- 83.Auton A, McVean G. Genome research. 2007;17:1219. doi: 10.1101/gr.6386707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen GK, Marjoram P, Wall JD. Genome research. 2009;19:136. doi: 10.1101/gr.083634.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fearnhead P. Bioinformatics. 2006;22:3061. doi: 10.1093/bioinformatics/btl540. [DOI] [PubMed] [Google Scholar]
- 86.I. H. Consortium et al. Nature. 2005;437:1299. [Google Scholar]
- 87.Ellegren H, et al. Nature. 2012;491:756. doi: 10.1038/nature11584. [DOI] [PubMed] [Google Scholar]
- 88.Sueoka N. PNAS. 1962;48:582. doi: 10.1073/pnas.48.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meunier J, Duret L. Molecular Biology and Evolution. 2004;21:984. doi: 10.1093/molbev/msh070. [DOI] [PubMed] [Google Scholar]
- 90.Bailey TL, Johnson J, Grant CE, Noble WS. Nucleic acids research. 2015:gkv416. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Flicek P, et al. Nucleic acids research. 2011:gkr991. [Google Scholar]
- 92.Roberts A, Pachter L. Nature methods. 2013;10:71. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rausch T, et al. Bioinformatics. 2012;28:i333. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pratto F, et al. Science. 2014;346:1256442. doi: 10.1126/science.1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.