Abstract
Using the ATP-independent transacylase CapW required for the biosynthesis of capuramycin-type antibiotics, we developed a biocatalytic approach for the synthesis of 43 analogues via a one-step aminolysis reaction from the methyl ester precursor as the acyl donor and various nonnative amines as acyl acceptors. Further examination of the donor substrate scope for CapW revealed this enzyme can also catalyze a direct transamidation reaction using the major capuramycin congener as a semisynthetic precursor. Biological activity tests revealed that a few of the new capuramycin analogues have significantly improved antibiotic activity against Mycobacterium smegmatis MC2 155 and Mycobacterium tuberculosis H37Rv. Furthermore, most of the analogues are able to be covalently modified by the phosphotransferase CapP/Cpr17 involved in self resistance, providing critical insight for future studies regarding clinical development of the capuramycin antimycobacterial antibiotics.
Introduction
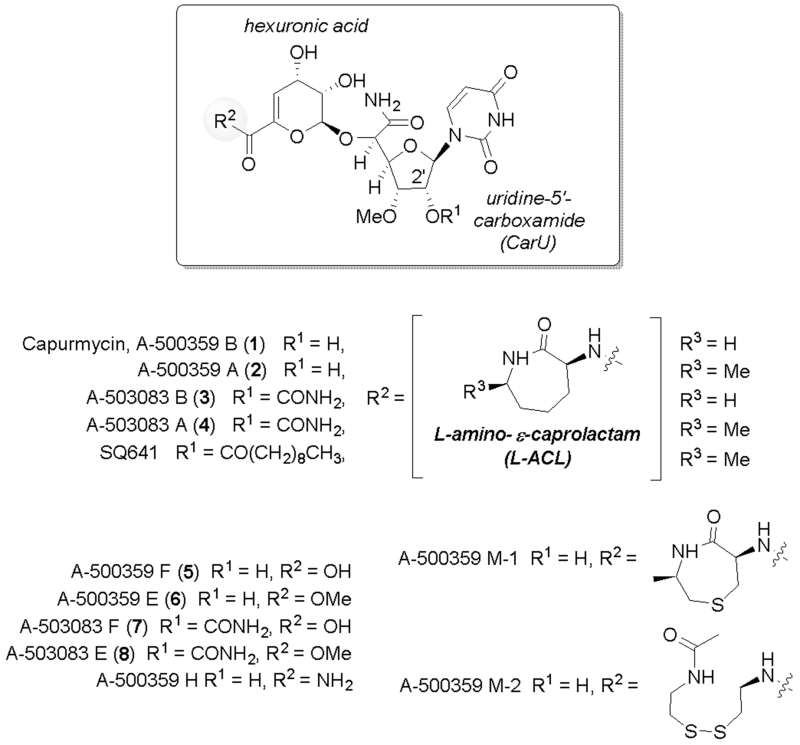
Capuramycin was discovered in the 1980s based on its antibacterial activity.1 The structure consists of three components, a uridine-5′-carboxamide (CarU) nucleoside core, a rare unsaturated hexuronic acid, and an l-amino-β-caprolactam (l-ACL) (Fig. 1).2 Since 2003 several capuramycin-related compounds have been discovered from different strains of actinomycetes.3-5 These compounds, which includes a series of congeners termed A-500359s from Streptomyces griseus SANK 60196 (exemplified by 1 and 2) and the 2′-O-carbamoylated variants A-503083s from Streptomyces sp. SANK 62799 (3 and 4), were initially identified as inhibitors of bacterial translocase I (MraY), an essential enzyme for bacterial cell wall biosynthesis.4-6 Although narrow in spectrum, bioactivity studies with 2 and a few acylated semisynthetic derivatives have revealed their potential utility as anti-tuberculosis (TB) antibiotics.7,8 For example, SQ641 (Fig. 1), a 2′-O-monoacylated lead candidate discovered by Sankyo and now under development by Sequella (Rockville, MD), has shown several advantageous attributes including in vitro activity against multiple-drug-resistant strains of Mycobacterium tuberculosis, the primary causative agent of TB; high efficacy in a murine model of TB; rapid kill time in vitro and in vivo; bactericidal activity, and no intrinsic toxicity in mice.9-11
Fig. 1.
Structures of capuramycin, representative capuramycin-related congeners isolated from two different strains, and a semisynthetic variant of 2.
Fermentation of the 1 and 2 producer in media supplemented with 2-aminoethyl-l-cysteine (AEC), an inhibitor of aspartokinase required for the biosynthesis of l-Lys in actinomycetes, resulted in the discovery of the de-l-ACL derivatives represented by A-500359 F (5; ~6 mg/L culture) and A-500359 E (6; 2.9 mg/L), the latter the methyl ester of 5.12 Three additional capuramycins (termed A-500359 M-1, M-2, and H) were discovered upon feeding AEC that were produced with higher titres than 6 (~4 mg/L, 6 mg/L, and 3.2 mg/L, respectively). Structural elucidation of A-500359 M-1 revealed that AEC itself was cyclized, methylated, and incorporated in place of the l-ACL; A-500359 M-2 contained an N-acetylcystamine, and A-500359 H consisted of a terminal carboxamide (Fig. 1). Despite the relatively low yields (compared with 472 mg/L for 1 and 2 combined from nonsupplemented media), 6 has been used as a semisynthetic precursor to prepare a library of l-ACL-substituted capuramycins.13 The synthesis utilized either a single-step substitution with excess amine and heat or a four-step procedure with protection, saponification, aminolysis, and deprotection. In contrast to the 1 and 2 producer, the de-l-ACL variants A-503083 F (7) and A-503083 E (8) are produced during standard fermentations (~3 mg/L and 1 mg/L, respectively, compared to 5 mg/L for 3 and 4, combined).4 Comparable semisynthetic efforts to modify these carbamoylated capuramycins have not been reported.
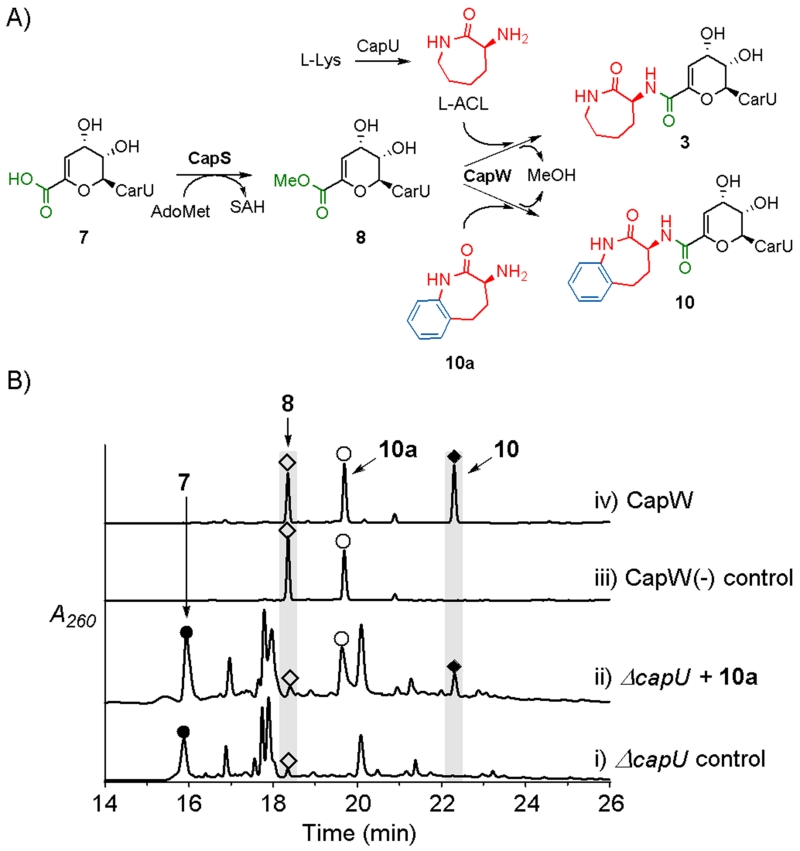
We previously identified the biosynthetic process for formation and the attachment of the l-ACL, which proceeds from 7 to 3. A key discovery was the coupling enzyme CapW, an N-transacylase that catalyzes the attachment of l-ACL to 8 with concomitant formation of methanol and 3 (Fig. 2A).14 The formation of this amide bond was demonstrated to follow esterification of 7 to give 8, a reaction that is catalyzed by an S-adenosyl-l-methione-dependent carboxymethyltransferase, CapS. In contrast to this unusual ATP-independent transacylation strategy, l-ACL was more recently shown to be formed from l-Lys in a relatively standard ATP-dependent reaction catalyzed by a nonribosomal peptide synthetase (NRPS) CapU.15 Importantly, a ΔcapU mutant strain was prepared during these functional characterization studies, which led to a strain unable to produce 3 and 4 and produced almost exclusively 7 at relatively high titers (17 mg/L vs. 2.9 mg/L for 6).15
Fig. 2.
Biocatalytic synthesis of capuramycin and analogues. A) Role of CapS and CapW in capuramycin biosynthesis and CapW-catalyzed incorporation of alternative acyl acceptors exemplified with 10a. B) In vivo feeding experiments with the ΔcapU mutant strain and in vitro enzyme assay with recombinant CapW. The HPLC traces include i) metabolite extract from the ΔcapU mutant strain; ii) extract from the ΔcapU mutant strain grown in media supplemented with 10a; iii) control reaction omitting CapW; iv) reaction with 8, 10a, and CapW.
Herein we take advantage of the existing biosynthetic knowledge and the availability of a ΔcapU mutant strain that is unable to generate l-ACL to establish chemoenzymatic and mutasynthetic approaches for generating new, l-ACL-substituted capuramyucin analogues. The results establish the utility and limitations of CapW as a biocatalytic tool for introducing novel amines into the capuramycin scaffold. Significantly, the development of a biocatalytic platform revealed some conceptual appealing advantages that aided in streamlining the semisynthetic process. Finally, a select set of compounds were analysed for antimycobacterial activity as well as their likelihood to be covalently modified by a self-resistance phosphotransferase.
Results
Establishment of mutasynthesis and chemoenzymatic platforms
Compound (S)-3-amino-4,5-dihydro-1H-benzo[b]azepin-2(3H)-one (10a), a structural analogue of l-ACL, was initially fed to the ΔcapU mutant strain. When compared to the control that produces 7 as the major congener (Fig. 2B, trace i), extract from media supplemented with 10a yielded a new peak with a UV/VIS spectrum suggestive of a capuramycin-like metabolite (Fig. 2B, trace ii). In parallel with the in vivo biotransformation, 8, which was synthetically prepared from 7 that was purified from the ΔcapU mutant strain, was reacted with 10a using recombinant CapW to reveal a new peak with an identical retention time (Fig. 2B, traces iii and iv). The HPLC-purified product was subjected to HRMS to reveal an [M + H]+ ion of m/z = 661.2015, consistent with the molecular formula of C28H32N6O13 for the expected N-transacylated product 10 (Fig. 2A). The structure of 10 was further supported by 1H-NMR spectroscopic analysis (ESI†)
Scope of amine-containing acyl acceptors for CapW
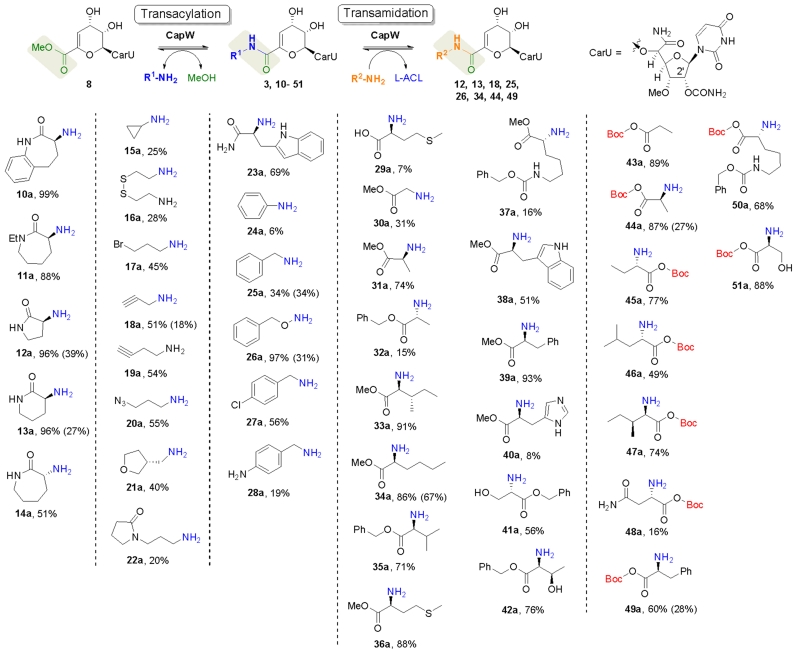
A diverse structural range of commercial acyl acceptors were next tested with CapW using the in vitro assay, which was preferred during the screening phase due to its relative simplicity. The amine-containing acyl acceptors incorporated by CapW are shown in Fig. 3 along with product yields; amine-containing compounds not recognized by CapW are shown in Fig. S1, ESI†. To summarize N-ethyl-l-ACL (11a) and smaller α-lactam rings (12a, 13a) were readily incorporated with high yields, whereas rings lacking the lactam functionality (for example, carbocycles, heterocycles, and lactones) were not incorporated by CapW. Contrary to expectation, d-ACL (14a) was incorporated, yet the reaction was accompanied by a significant amount of hydrolysis to form the carboxylic acid 7. Linear aliphatic amines were also identified as good substrates giving decent yields, which included aliphatic amines with monofunctional groups like bromine (17a), alkynyl (18a and 19a), and azide (20a). Excluding aniline (24a), which gave a relatively low incorporation, substituted arylamines were not utilized as acyl acceptors. In contrast benzylamine and variants with different aryl substituents (25a-28a) were substrates for CapW and afforded the respective capuramycin analogues with modest yields.
Fig. 3.
Biocatalytic approach to new capuramycins and structures of acyl acceptors that are incorporated by CapW in vitro. The transacylation and transamidation yields (the latter in parenthesis) were determined based on HPLC/LC-MS analysis after an 8 hr CapW-catalyzed reaction. Product structures are illustrated in the supplementary information.
Amino acid incorporation
To differentiate our approach and the generated analogues from other synthetic efforts, we aimed to incorporate α-amino acids with a single-step reaction. However, a survey of the proteinogenic α-amino acids revealed CapW was inactive except with l-methionine (29a). In general free carboxylates—regardless of the regiochemistry—were not utilized as substrates. Nonetheless, the use of ester-protected α-amino acids bypassed this selectivity, and several new capuramycin analogues were obtained with good yields (30a-40a). Unexpectedly and in contrast to the methyl and benzyl esters, the t-butyl esters (43a-51a) were hydrolyzed during the transformation to reform the free carboxylate following incorporation. The BOC protection strategy, therefore, made it possible to incorporate ‘unprotected’ amino acids in a single step.
Relative activity of CapW
To gain a better understanding regarding the specificity of CapW with respect to the acyl acceptor, amines with reasonable turnover (yield of 50 % or greater) were tested under initial velocity conditions to estimate the relative activity (Fig. S2, ESI†). When compared to the natural substrate l-ACL, an approximate 2-fold higher specific activity for CapW was observed with substrates benzyloxyamine (26a) and 4-chlorophenyl methylamine (27a). Contrastingly, CapW was less active with all other substrates, having specific activities ranging from 0.3-0.9 relative to reactions with l-ACL. From the cumulative activity data and considering the substrates not utilized by CapW, it appears that the activity of CapW is primarily influenced by the nucleophilicity of the amine, and bulky groups and/or free carboxylates are detrimental to turnover.
Single-step transamidation
In the presence of excess glycerol, CapW was previously shown to convert 3 to the glyceryl ester analogue of 8.14 Follow-up reactions that substituted glycerol with methanol confirmed the reversibility. With this in mind, we interrogated the possibility of preparing capuramycin analogues via direct transamidation starting from 3. Using in vitro conditions to thermodynamically drive product formation, eight representative amine-containing acyl acceptors were incorporated at the expense of l-ACL. Furthermore, these reactions were independent upon the inclusion of methanol. Although the yields for transamidation starting from 3 were generally less than transacylation starting from 8 (Fig. 3), the results demonstrate the feasibility of employing a single-step transamidation strategy to generate differentially amidated capuramycins.
Anti-mycobacterial activity
Representative analogues were prepared on a large scale using CapW and minimally characterized by HR-MS and 1H-NMR spectroscopy. To assess the antimycobacterial activity as well documented for 1 and 2,8,9 these new analogues were initially screened against the lab strain Mycobacterium smegmatis MC2 155 using 3, isoniazid, and rifampin, the last two of which are first-line anti-TB drugs, as positive controls (Table 1). Several of the compounds retained comparable activity to the parent 3 (MIC 60 μM) with 10, 11 and 26 have a significantly better MIC (8-fold improvement). Comparison of 31 (MIC = 32 μM), which contains the methyl ester of l-Ala, with 46 (MIC > 200 μM), which contains the free acid of l-Ala, is consistent with a detrimental effect of a carboxylate functionality as previously noted for 7. A subset of these compounds were also tested against Mycobacterium tuberculosis H37Rv, and similar to before, compounds 10, 11, and 26 had relatively good anti-TB activity that was comparable to 2 yet slightly worse than the preclinical lead SQ641.9
Table 1.
Antimycobacterial activity of representative analogues.
| Com. | MIC b | Com. | MIC | Com. | MIC |
|---|---|---|---|---|---|
| INH a | 137 (0.9) c | 12 | 16 | 34 | 15 |
| RIF | 1.5 (0.007) | 13 | 63 (>200) | 36 | 58 |
| 2 | 11 (14) | ||||
| 3 | 61 | 14 | 61 (100) | 44 | >200 |
| 8 | 73 | 19 | 34 (200) | 49 | 29 (57) |
| SQ641 | (~1) | ||||
| 10 | 7.1 (13) | 26 | 7.7 (15) | 52 | >200 |
| 11 | 7.3 (14) | 31 | 32 (>200) | 53 | >200 |
INH, isoniazid; RIF, rifampicin;
Miminum inhibitory concentration (μM) against Mycobacterium smegmatis MC2 155;
The number in paranthesis is the MIC against Mycobacterium tuberculosis H37Rv.
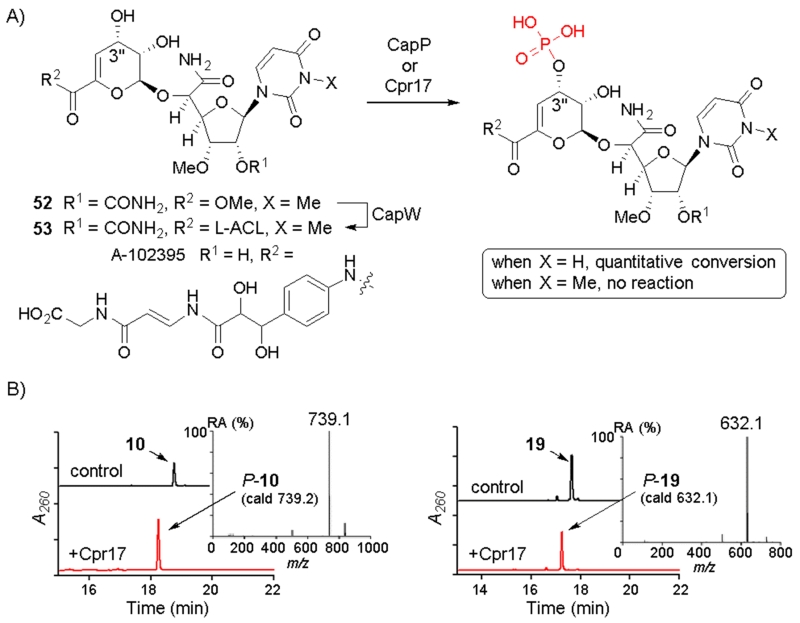
During the preparation of the methyl ester 8 starting from 7, a second, minor product (52) was observed with a mass corresponding to a dimethylated product. Increasing the methylating reagent from 1.2 to 2.4 eq yielded 52 as the major product (Fig. 4). The 13C chemical shift (27.54 ppm) for the additional methyl group was consistent with N3-methylation of the uracil base, which was subsequently confirmed by MS-MS analysis. This new 8 analogue was reacted with l-ACL using CapW, and a new product 53 was obtained with 92% yield. Spectroscopic analysis was consistent with the structure of 53 as N-3-methyl-3. Both 52 and 53 were inactive against M. smegmatis MC2 155 (Table 1), demonstrating the significance of the uracil base for antimycobacterial activity.
Fig. 4.
Enzymatic phosphorylation of 3 analogues. A) The reaction catalysed by CapP and Cpr17 (encoded in the 3 and A-102395 biosynthetic gene clusters, respectively). B) HPLC traces for representative 3 analogues following the reaction with Cpr17. The inset shows the mass spectrum in negative ionisation mode for the monophosphorylated product.
Enzymatic phosphorylation of 3 analogues
The C-3″ hydroxyl of 3 and the capuramycin-type antibiotic A-102395 (Fig. 4A) were previously shown to undergo enzymatic phosphorylation by CapP and the orthologous enzyme Cpr17, respectively, as a mechanism of self-resistance.16,17 Both enzymes were also demonstrated to phosphorylate the noncognate capuramycin, suggesting lack of specificity regarding the l-ACL/polyamide component.17 Several of the CapW-generated compounds were investigated as potential substrates for the phosphotransferase Cpr17. All of the analogues where converted to a new product that yielded a mass consistent with the monophosphorylated product (Fig. 4B and Fig. S3 ESI†). In contrast 53 was not a substrate for Cpr17, revealing that the uracil base is important for substrate recognition and phosphorylation.
Discussion
Two complementary biocatalytic strategies (chemoenzymatic and mutasynthetic) for the production of l-ACL-substituted capuramycins have been developed. The implementation of these strategies was initially inspired by the discovery of M-1 and M-2, which revealed the biosynthetic machinery involved in l-ACL incorporation was able to accommodate alternative amine-containing substrates as acyl acceptors.12 CapW, which was previously identified as the enzyme catalyst responsible for this N-acylation,14 was shown here to efficiently incorporate a variety of amine-containing substrates using mild, aqueous conditions and without the need for protection/deprotection of the hydroxyl groups within the acyl donor 8. Unlike past semisynthetic efforts that relied on the precursor 6,13 which is produced in relatively low yields as the fifth most abundant congener when feeding the wild-type strain with AEC, our biocatalytic strategy took advantage of a recently reported ΔcapV mutant strain that produces predominantly the de-l-ACL capuramycin 7 with a relatively superior yield (17 mg/L compared to 2.9 mg/L for 6).15 Not only did this provide a means to obtain ample precursor for the chemoenzymatic synthesis, it also enabled the establishment of a mutasynthetic platform, whose proof of concept was demonstrated with 10a. Such a fermentation-based platform is anticipated to be critical for future scale-up efforts.
During this study, we identified and took advantage of two inherent properties of CapW that make this enzyme particularly useful as a biocatalyst and might serve as a model for other mechanistically related enzymes. First and foremost was the discovery that CapW can catalyze a direct transamidation under the highly controlled in vitro setting. The first hint of this possiblity was realized upon functional assignment of CapW, when it was observed that the glyceryl ester was generated upon incubation of 3 with a large excess of glycerol and CapW.14 Prior biochemical characterization of CapW also revealed a Ser residue is critical for activity, suggesting that catalysis occurs through the formation of an acyl-enzyme intermediate similar to Class C β-lactamases and the evolutionarily related penicillin binding proteins.18 Two distinct and mechanistically comparable penicillin binding proteins have recently been demonstrated to catalyze an intermolecular transamidation that was exploited to introduce alternative amines including fluorescent probes in place of the terminal d-Ala of peptidoglycan.19,20 Similarly, all of the substrates tested here with CapW were incorporated in place of the naturally occurring l-ACL, and it is hypothesized that this thermodynamically driven reaction proceeds through the reversible formation of an acyl-enzyme intermediate. The ability of CapW to catalyze direct amine exchange is particularly significant when considering that 1 or 3 are the congeners with the highest yield from the respective wild-type stains, thus making them desirable semisynthetic precursors. In addition to reversibility, the second, and in this case unexpected, property of CapW was its moonlighting yet substrate specific hydrolytic activity for the t-butyl protection group following the transacylation reaction. This enabled a level of control for incorporating amino acids with or without protected carboxylates in a single step.
From a structure-activity relationship perspective, this work has reinforced prior conclusions from semi- and recent total synthetic efforts that l-ACL substitution can lead to capuramycins with improved antimycobacterial activity and/or better pharmaceutical properties.7-11,13,21 What was not known prior to this effort, however, was whether substituting for the l-ACL had any effect on the activity of the hexuronic acid phosphotransferase, which we previously established is a mechanism of self-resistance by the producing strain.16,17 A similar phenomenon is found within aminoglycoside producing strains that often encode for phosphotransferases to covalently modify and inactivate the self-made antibacterial product, and it has been established that identical mechanisms—often acquired through horizontal gene transfer—are employed by pathogens to render these antibiotics inactive.22 This resistance problem has been partially remedied by semisynthetic modification of an aminoglycoside at a distant site that prevents binding and catalysis by the covalently modifying enzyme.23 Hence, we were interested in testing whether the l-ACL substitutions had any effect on phosphorylation. However, all of the derivatives were readily converted to the monophosphorylated product, suggesting that the phosphotransferase has little to no specificity for this component. In stark contrast, N3-methylation of the uracil, which did not affect CapW catalysis, yielded a capuramycin that was unable to be phosphorylated. Although the N3-methylated variant was inactive in the antimycobacterial assays, it does suggest that the rational design of uracil-substituted variants may be a reasonable strategy to generate an antimycobacterial capuramycin that is less prone to inactivation by phosphorylation and hence resistance.
Conclusions
A total of 43 capuramycin analogues were prepared using the substrate permissive enzyme CapW. In addition to CapW’s ability to catalyse O→N transacylation, this enzyme was demonstrated to directly substitute the l-ACL via an N→N transamidation. Furthermore, CapW was revealed to hydrolyze t-butyl protected acids but not other protecting groups. Some of the new analogues had comparable or better antimycobacterial activity compared to the parent yet—with the exception of 53—the new analogues were prone to covalent inactivation by a phosphotransferase involved in self resistance.
Experimental section
Chemicals, reagents, and standards
All chemicals, reagents, buffers, and salts were purchased from Sigma-Aldrich and Alfa-Aesar unless noted. Protected amino acids were from Chem-Impex International, L-(−)-α-amino-ε-caprolactam (L-ACL) hydrochloride was from Fluka, 3-aminopyrrolidin-2-one hydrochloride and 3-aminopiperidin-2-one hydrochloride were from AAT Pharmaceutical, (S)-a-amino-e-N-ethyl-caprolactam was from J&W, and 3-amino-2,3,4,5-tetrahydro-1H-1-benzazepin-2-one was from Astatechl Inc. Compounds 3 and 7 were isolated from Streptomyces sp. SANK 62799 as previously described.14
Instrumentation
Analytic HPLC was performed with one of three systems: a Waters Alliance 2695 separation module (Milford, MA) equipped with a Waters 2998 diode array detector and an analytical Apollo C-18 column (250 mm × 4.6 mm, 5 μm) or a Dionex Ultimate 3000 Focused separation module (Bannockburn, IL) equipped with a DAD-3000(RS) and MWD-3000(RS) diode array detector and an Acclaim 120 C-18 column (4.6 mm × 100 mm, 3 μm) or an Agilent 1200 Series Quaternary LC system and an Eclipse XDB-C18 column (150mm × 4.6 mm, 5 μm, 80Å) equipped with an Agilent 6120 Quadrupole MSD mass spectrometer (Agilent Technologies, Santa Clara, CA). Semi-preparative HPLC was performed with a Waters 600 controller and pump (Milford, MA) equipped with a 996 diode array detector, 717plus autosampler, and an Apollo C-18 column (250 mm × 10 mm, 5 μm) purchased from Grace (Deerfield, IL). LC-electrospray ionization (ESI)-mass spectroscopy (MS) was performed using an Agilent 6120 Quadrupole MSD mass spectrometer (Agilent Technologies, Santa Clara, CA) equipped with an Agilent 1200 Series Quaternary LC system and an Eclipse XDB-C18 column (150mm × 4.6 mm, 5 μm, 80Å). High resolution (HR)-MS was performed using a Bruker BioTOF II, and NMR data were collected using a Varian Unity Inova 400 MHz spectrometer (Varian, Inc., Palo Alto, CA).
Synthetic and biocatalytic procedures
Synthesis of A-503083 E (8) and A-503083 G (52)
To a magnetically stirred solution of 7 (100.4 mg, 0.2 mmol) in anhydrous methanol (0.4 mL) under anaerobic conditions at 0 °C, trimethylsilyl diazomethane solution in 2.0 M hexanes (0.24 mmol for 8, 0.48 mmol for 52) was added dropwise. After stirring until completion based on LC-MS (30 min), the reaction was quenched with 0.1 mL of distilled water and the solvent removed under vacuum. The product, containing predominantly 8 (85 % yield) or 52 (94 % yield), was used without subsequent purification for CapW-catalyzed reactions. 52: 1H NMR (400 MHz, D2O) δ 7.66 (d, J=8.1 Hz, 1H), 6.18-6.13 (m, 1H), 5.95-5.88 (m, 2H), 5.31 (d, J=3.5 Hz, 1H), 5.13 (t, J=4.9 Hz, 1H), 4.68 (d, J=2.2 Hz, 1H), 4.53-4.46 (m, 2H), 4.14 (dd, J=5.4, 2.5 Hz, 1H), 4.03 (t, J=5.2 Hz, 1H), 3.77 (s, 3H), 3.36 (s, 3H), 3.21 (s, 3H). 13C NMR (100 MHz, D2O) δ 172.63, 165.07, 163.48, 157.02, 151.46, 139.44, 139.06, 113.97, 101.55, 99.51, 88.91, 82.00, 77.94, 76.38, 73.86, 64.76, 61.67, 58.34, 52.96, 27.54. HRMS (ESI): C20H26N4O13+H+, Calc: 531.1569, Found: 531.1571.
CapW-catalyzed transacylation of 8 with different acyl acceptors
Reactions (100 μL) consisted of 25 mM potassium phosphate (pH 8.0), 2 mM 8, 10 mM amine-containing substrate and 37.5 μM CapW at 30 °C for the 8 h. Following removal of the protein by ultrafiltration, reactions with 10a-13a were analyzed using reverse-phase chromatography using the Waters HPLC system. A series of linear gradients was developed from 0.1 % TFA in 2.5 % acetonitrile (A) to 0.1 % TFA in 90 % acetonitrile (B) in the following manner (beginning time and ending time with linear increase to % B): 0-5 min, 0 % B; 5-10 min, 0-50 % B; 10-15 min, 50-100 % B; 15-18 min, 100 % B, and 18-20 min, 100-0 % B. The flow rate was kept constant at 1 mL/min, and elution was monitored at 260 nm. The Dionex Ultimate 3000 HPLC system was used for analysis of reactions with 15a - 17a, and 21a-51a with the same linear gradient program as above. The Agilent 1200 Series Quaternary LC-MS was used for analysis of reactions with compounds 14a, 18a-20a, and reactions with l-ACL and 52 substituted for 8. The linear gradient was developed from 0.1 % formic acid in DI water (C) to 0.1 % formic acid in 100 % acetonitrile (D) in the following manner (beginning time and ending time with linear increase to % D): 0-18 min, 5-95 % D; and 18-20 min, 95-5 % D. The flow rate was kept constant at 0.4 mL/min, and elution was monitored at 260 nm.
CapW-catalyzed transamidation of 3 with different acyl acceptors
Reactions (100 μL) consisted of 25 mM potassium phosphate (pH 8.0), 2 mM 3, 10 mM amine-containing substrates, and 37.5 μM CapW at 30 °C for the 8 hours. Following removal of the protein by ultrafiltration, the reaction components were analyzed using the Agilent 1200 Series Quaternary LC-MS system as described in the previous section.
Large-scale isolation of CapW products
10 - 13, 53
A large-scale reaction (1 mL) consisted of 25 mM potassium phosphate (pH 8.0), 2 mM 8, 10 mM amine-containing substrate, and 37.5 μM CapW. Following incubation for 12 h at 30 °C, protein was removed by ultrafiltration and the product was purified by HPLC using a C-18 reverse-phase semi-preparative column. A series of linear gradients was developed from A to B in the following manner (beginning time and ending time with linear increase to % B): 0-2 min, 0 % B; 2-18 min, 0-90 % B; 18-22 min, 90 % B; 22-24 min, 90-0 % B; and 24-26 min, 0 % B. The flow rate was kept constant at 3.5 mL/min, and elution was monitored at 260 nm.
14, 19, 26, 31, 33, 34, 36, 44, 49
A large-scale reaction (200 μl) consisted of 25 mM potassium phosphate (pH 8.0), 2 mM 8, 10 mM amine-containing substrates, and 37.5 μM CapW. Following incubation for 16 h at 30 °C, protein was removed by ultrafiltration and the product was purified by HPLC using semi-preparative column as described above.
Bioactivity against M. smegmatis MC2 155 and M. tuberculosis H37Rv
MIC values were determined using the micro-dilution method as previously reported.24 M. smegmatis MC2 155 was tested in Mueller-Hinton broth. Cultures were grown for 24-48 h before visual inspection and addition of alamar blue (5 μL of 5 mg/mL). Determination of the MIC value was evaluated when the growth control color change was complete (an additional 24-48 h later). MIC values against M. tuberculosis H37Rv were determined under Contract No. HHSN272201100009I with the Division of Microbiology and Infectious Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services.
Supplementary Material
Acknowledgements
This work was supported by a National Institute of Health grant AI087849 (S.V.L.) and UL1TR000117 (S.G.-T. and S.V.L.), a National Science Foundation CAREER award MCB 114927 (S.G-T.), and a National Natural Science Foundation of China grant 81261120417, 81321004, and 81273414 (Z.Y.)
Footnotes
Electronic Supplementary Information (ESI) available: [Analytical analyses of new compounds and supplementary figures].
Notes and references
- 1.Yamaguchi H, Sato S, Yoshida S, Takada K, Itoh M, Seto H, Otake N. J. Antibiot. 1986;39:1047–1053. doi: 10.7164/antibiotics.39.1047. [DOI] [PubMed] [Google Scholar]
- 2.Seto H, Otake N, Sato S, Yamaguchi H, Takada K, Itoh M, Masayoshi L, Lu HSM, Clardy J. Tetrahedron Lett. 1988;29:2343–2346. [Google Scholar]
- 3.Muramatsu Y, Muramatsu A, Ohnuki T, Ishii MM, Kizuka M, Enokita R, Tsutsumi S, Arai M, Ogawa Y, Suzuki T, Takatsu T, Inukai M. J. Antibiot. 2003;56:243–252. doi: 10.7164/antibiotics.56.243. [DOI] [PubMed] [Google Scholar]
- 4.Muramatsu Y, Ohnuki T, Ishii M, Kizuka MM, Enokita R, Miyakoshi S, Takatsu T, Inukai M. J. Antibiot. 2004;57:639–646. doi: 10.7164/antibiotics.57.639. [DOI] [PubMed] [Google Scholar]
- 5.Murakami R, Fujita Y, Kizuka M, Kagawa T, Muramatsu Y, Miyakoshi S, Takatsu T, Inukai M. J. Antibiot. 2007;60:690–695. doi: 10.1038/ja.2007.88. [DOI] [PubMed] [Google Scholar]
- 6.Muramatsu Y, Ishii MM, Inukai M. J. Antibiot. 2003;56:253–258. doi: 10.7164/antibiotics.56.253. [DOI] [PubMed] [Google Scholar]
- 7.Hotoda H, Daigo M, Furukawa M, Murayama K, Hasegawa CA, Kaneko M, Muramatsu Y, Ishii MM, Miyakoshi S, Takatsu T, Inukai M, Kakuta M, Abe T, Fukuoka T, Utsui Y, Ohya S. Bioorg. Med. Chem. Lett. 2003;13:2833–2836. doi: 10.1016/s0960-894x(03)00597-3. [DOI] [PubMed] [Google Scholar]
- 8.Koga T, Fukuoka T, Doi N, Harasaki T, Inoue H, Totoda H, Kakuta M, Muramatsu Y, Yamamura N, Hoshi M, Hirota T. J. Antimicrob. Chemother. 2004;54:755–760. doi: 10.1093/jac/dkh417. [DOI] [PubMed] [Google Scholar]
- 9.Reddy VM, Einck L, Nacy CA. Antimicrob. Agents Chemother. 2008;52:719–721. doi: 10.1128/AAC.01469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nikonenko BV, Reddy VM, Protopopova M, Bogatcheva E, Einck L, Nacy CA. Antimicrob. Agents Chemother. 2009;53:3138–3139. doi: 10.1128/AAC.00366-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reddy VM, Bogatcheva E, Einck L, Nacy CA. Drug Delivery Lett. 2011;1:150–158. [Google Scholar]
- 12.Muramatsu Y, Miyakoshi S, Ogawa Y, Ohnuki T, Ishii MM, Arai M, Takatsu T, Inukai M. J. Antibiot. 2003;56:259–267. doi: 10.7164/antibiotics.56.259. [DOI] [PubMed] [Google Scholar]
- 13.Hotoda H, Furukawa M, Daigo M, Murayama K, Kaneko M, Muramatsu Y, Miyazawa Ishii M, Miyakoshi S, Takatsu T, Inukai M, Katuta M, Abe T, Harasaki T, Fukuoka T, Utsui Y, Ohya S. Bioorg. Med. Chem. Lett. 2003;13:2829–2832. doi: 10.1016/s0960-894x(03)00596-1. [DOI] [PubMed] [Google Scholar]
- 14.Funabashi M, Yang Z, Nonaka K, Hosobuchi M, Fujita Y, Shibata T, Chi X, Van Lanen SG. Nat. Chem. Biol. 2010;6:581–586. doi: 10.1038/nchembio.393. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Jin Y, Cui Z, Nonaka K, Baba S, Funabashi M, Yang Z, Van Lanen SG. ChemBioChem. 2016 doi: 10.1002/cbic.201500701. doi:10.1002/cbic.201500701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Z, Funabashi M, Nonaka K, Hosobuchi M, Shibata T, Pahari P, Van Lanen SG. J. Biol. Chem. 2010;285:12899–12905. doi: 10.1074/jbc.M110.104141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai W, Goswami A, Yang Z, Liu X, Green KD, Barnard-Britson S, Baba S, Funabashi M, Nonaka K, Sunkara M, Morris AJ, Spork AP, Ducho C, Garneau-Tsodikova S, Thorson JS, Van Lanen SG. J. Biol. Chem. 2015;290:13710–13724. doi: 10.1074/jbc.M115.646414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher JF, Meroueh SO, Mobashery S. Chem. Rev. 2005;105:395–424. doi: 10.1021/cr030102i. [DOI] [PubMed] [Google Scholar]
- 19.Lebar MD, May JM, Meeske AJ, Leiman SA, Lupoli TJ, Tsukamoto H, Losick R, Rudner DZ, Walker S, Kahne D. J. Am. Chem. Soc. 2014;136:10874–10877. doi: 10.1021/ja505668f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiao Y, Lebar MD, Schirner K, Schaefer K, Tsukamoto H, Kahne D, Walker S. J. Am. Chem. Soc. 2014;136:14678–14681. doi: 10.1021/ja508147s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siricilla S, Mitachi K, Wan B, Franzblau SG, Kurosu M. J. Antibiot. 2015;68:271–278. doi: 10.1038/ja.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magnet S, Blanchard JS. Chem. Rev. 2005;105:477–498. doi: 10.1021/cr0301088. [DOI] [PubMed] [Google Scholar]
- 23.Huang F, Haydock SF, Mironenko T, Spiteller D, Li Y, Spencer JB. Org. Biomol. Chem. 2005;3:1410–1418. doi: 10.1039/b501199j. [DOI] [PubMed] [Google Scholar]
- 24.Gajadeera C, Willby MJ, Green KD, Shaul P, Fridman M, Garneau-Tsodikova S, Posey J, Tsodikov EOV. J. Antibiot. 2015;68:153–157. doi: 10.1038/ja.2014.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.