SUMMARY
The identification of driver loci underlying arm-level somatic copy number alterations (SCNAs) in cancer has remained challenging and incomplete. Here we assess the relative impact and present a detailed landscape of arm-level SCNAs in 10985 patient samples across 33 cancer types from The Cancer Genome Atlas (TCGA). Further, using chromosome 9p loss in lower grade glioma (LGG) as a model, we employ a unique multi-tiered genomic dissection strategy using 540 patients from 3 independent LGG datasets to identify genetic loci that govern tumor aggressiveness and poor survival. This comprehensive approach uncovered several 9p loss-specific prognostic markers, validated existing ones, and re-defined the impact of CDKN2A loss in LGG.
INTRODUCTION
Cancer initiation and progression is a step-wise process that is characterized by the gradual accumulation of molecular events. Of these, somatic copy number alterations (SCNAs) are now well documented throughout human cancer. SCNAs comprise genetic losses or gains of varying size, ranging from only 1 kilobase (kb) to entire chromosome arms (Feuk et al., 2006). Importantly, focal SCNAs have yielded some of the earliest insight into mechanisms underlying malignant transformation, helping to identify loci that function as tumor suppressor genes (TSGs) or oncogenes (Zack et al., 2013). In contrast, less is known about the larger arm-level SCNAs despite occurring 30 times more frequently than focal SCNAs when adjusted for size (Beroukhim et al., 2010). These broad regions of gain or loss can include hundreds of genes, several of which are likely bona fide cancer genes (Weir et al., 2007). As a result, narrowing the list of potential “drivers” in large SCNAs has proven difficult, mainly due to limitations of traditional laboratory methods and conventional technologies (Beroukhim et al., 2010).
Recent advances in the field of genomics have helped shed light on some of cancer biology’s most difficult dilemmas (Green et al., 2011; Hoadley et al., 2014; Leiserson et al., 2014). Aided by multiple tiers of next-generation data and large patient cohorts, contemporary exploration of the genome via computational approaches allows an efficient means by which to reveal new aspects of tumor biology. Recent innovation in the field of genomics may allow a more thorough characterization of arm-level SCNAs, an incredibly common yet poorly understood aspect of cancer biology.
Large alterations have long been associated with poor outcomes, though underlying mechanisms and gene-specific biomarkers remain elusive (Baudis, 2007; Bown et al., 1999; Gross et al., 2014; Jen et al., 1994). For example, one of the most frequent arm-level SCNAs is chromosome 9p loss, which is linked to disease progression and worse survival in many types of cancer, including lower grade glioma (LGG; WHO grade II and III) (Idbaih et al., 2008; James et al., 1991; Wiltshire et al., 2004). Although known TSG CDKN2A is located on 9p, it is believed that other cancer genes exist that may be the target of the broad 9p deletion (Beatty et al., 1999; Bredel et al., 2005; Olopade et al., 1992; Pollock et al., 2001; Schmid et al., 2000; Ueki et al., 1994). The identity of these driver genes remains unknown, however. Additionally, we still lack a global portrait of arm-level SCNAs in cancer. Understanding the relative impact of these SCNAs across cancer and identifying underlying target genes is of vital importance to help uncover prognostic markers and potential therapeutic targets. We therefore set out to employ a multi-faceted genomic strategy to rank prognostic SCNAs in cancer as well as underlying driver loci.
RESULTS
Prognostic impact of arm-level SCNAs across cancer types
Here, we report a two-fold genomics approach to aid in clarifying the role of arm-level SCNAs in cancer (Figure 1A). First, using copy number and clinical data from 10,985 patient samples in 33 TCGA tumor datasets, we employed three statistical indices to determine the relative prognostic impact of all 82 arm-level SCNAs in cancer. Second, we implemented a multifaceted genomic analysis of the 9p loss event in LGG to identify specific genes involved in progression and overall survival (OS). We obtained all available clinical and molecular data from the TCGA (Cancer Genome Atlas Research et al., 2015) as of the Broad Firehose run on 21 August 2015. Six tiers of data – clinical variables, copy number, mRNA expression, miRNA expression, DNA methylation and somatic mutation – were analyzed for a total of 379 LGG cases. In addition, two independent datasets were used for validation, including the Repository for Molecular Brain Neoplasia Data (REMBRANDT) (Madhavan et al., 2009) and the MSKCC LGG dataset (Turcan et al., 2012), representing an additional 109 and 52 LGG cases, respectively.
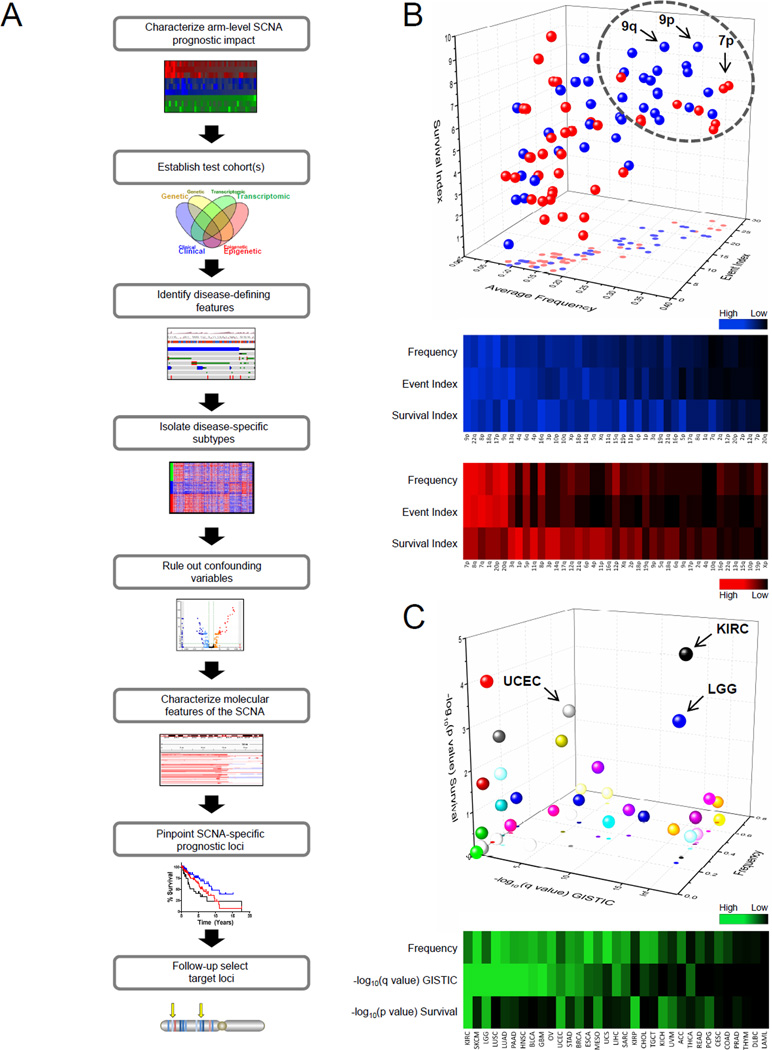
Figure 1. Prognostic impact of arm-level SCNAs in cancer.
(A) Schematic depicting arm-level SCNA dissection workflow to determine underlying driver loci.
(B) Three-dimensional (3D; top) and two-dimensional (2D; bottom) representation of individual arm-level SCNA across cancer as assessed by 3 parameters: average frequency, event index (total cancer types with GISTIC q<0.25), survival index (total cancer types with OS log-rank p<0.1). Blue, deletion; Red, amplification.
(C) 3D (top) and 2D (bottom) representation of frequency, GISTIC significance −log10(q value), and log-rank survival significance −log10(p value) for the 9p deletion event by cancer type.
SCNA, somatic copy number alteration; OS, overall survival; LGG, lower grade glioma; KIRC, kidney renal clear cell carcinoma; UCEC, uterine corpus endometrial carcinoma; Inf, infinite. High/Low scale corresponds to values within the 3 parameters individually.
In order to establish the prognostic associations of arm-level SCNA in cancer, arm-level copy number calls from GISTIC2.0 and corresponding survival data were obtained and analyzed. Specifically, the following three parameters were utilized in concert to determine overall prognostic impact of each arm-level SCNA in cancer: event frequency, GISTIC2.0 significance (q value), log-rank survival significance (p value) (Figure S1A).
Following a composite analysis of all three parameters, we characterized the relative prognostic impact of all arm-level SCNAs in cancer (Figures 1B, S1B and Table S1). Interestingly, chromosome 9p loss was ranked nearly highest in all three categories. In fact, across all types of cancer, 9p loss was the third most frequent arm-level deletion and eighth most frequent arm-level SCNA overall.
We further focused on the 9p loss event alone across 33 cancer types using a event-specific analysis of SCNA prognostic impact (Figure 1C). In fact, LGG had a high frequency of 9p loss (24%), highest event significance for 9p loss, and the third most significant 9p survival association. Additionally, the 9p loss event was among the top arm-level events within LGG when evaluated by cancer-specific analysis of prognostic impact (Figure S1C). These data suggest that 9p loss is one of the most substantial arm-level events in LGG, as well as across cancer. Therefore, 9p loss in LGG provided an ideal model to explore genomic approaches for identifying genetic loci responsible for arm-level SCNA-associated phenotypes in cancer.
Characterization of LGG test cohorts
The nature of genetic alterations on 9p and which underlying loci affect survival is an important question in cancer biology that remains controversial. To address this, we assembled data from the TCGA to form a 379 patient cohort that was complete for both clinical and molecular tiers of data (Figure S2A). GISTIC2.0 was used to determine focal and arm-level SCNA events – including 9p loss – across all patients in this cohort, and we found these data to be consistent with other published SCNAs in glioma (Figure S2B) (Beroukhim et al., 2007). For validation purposes, the REMBRANDT glioma database (n=109) and the MSKCC LGG dataset (n=52) were utilized (Table 1). In order to perform a genomic dissection of chromosome 9p in all three LGG cohorts, we first sought to validate the 9p survival phenotype, characterize tumor subtypes, identify any potential confounding variables, and confirm independent prognostic status for the 9p loss event.
Table 1.
Clinical characteristics of TCGA, REMBRANDT, and MSKCC LGG cohorts.
| TCGA (n=379) | REMBRANDT (n=109) | MSKCC (n=52) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of patients |
Median Survival (years) |
Log-rank (p value) |
No. of Patients |
Median Survival (years) |
Log-rank (p value) |
No. of Patients |
Median Survival (years) |
Log-rank (p value) |
||
| Gender | 0.435 | 0.22 | 0.684 | |||||||
| Male | 206 | 11.2 | 65 | 3.8 | 34 | 5.7 | ||||
| Female | 173 | 7.9 | 44 | 5.6 | 18 | 3.9 | ||||
| Race | 0.597 | 0.13 | 5.94E-13 | |||||||
| White | 357 | 7.9 | 70 | 4.2 | 41 | 4.7 | ||||
|
Black or African American |
14 | 3.7 | 3 | 2 | 0.111 | |||||
| Missing | 5 | 35 | 3 | 4 | 6.9 | |||||
| Asian | 2 | 1 | 0.9 | 4 | 2.9 | |||||
|
American Indian / Alaskan |
1 | 0 | 1 | |||||||
|
Year of diagnosis |
1.04E-03 | NA | 0.661 | |||||||
| post 2004 | 335 | 24 | 3.7 | |||||||
| pre 2004 | 44 | 5.2 | 28 | 5.7 | ||||||
|
Age (years) |
1.24E-12 | 7.45E-09 | 7.02E-03 | |||||||
| Younger than 55 | 297 | 9.5 | 75 | 5.6 | 37 | 6.4 | ||||
| Older than 55 | 82 | 2.5 | 34 | 1.1 | 15 | 2.2 | ||||
|
Tumor histology |
1.97E-02 | 0.96 | 0.182 | |||||||
| Astrocytoma | 137 | 5.2 | 74 | 3.7 | 19 | 6.4 | ||||
| Oligodendroglioma | 142 | 8 | 31 | 3.5 | 26 | 5.7 | ||||
| Mixed | 100 | 10.9 | 4 | 1.1 | 3 | 1.7 | ||||
| Other | 0 | 0 | 4 | 3.5 | ||||||
| Grade | 4.00E-06 | 5.10E-03 | 0.213 | |||||||
| II | 182 | 10.9 | 62 | 23 | ||||||
| III | 197 | 5.2 | 47 | 2 | 29 | 3.9 | ||||
|
Tumor location |
0.297 | NA | 0.583 | |||||||
| Frontal lobe | 237 | 8 | 23 | 4.7 | ||||||
| Temporal lobe | 100 | 7.9 | 11 | 6.4 | ||||||
| Occipital lobe | 3 | 4.3 | 3 | 6.9 | ||||||
| Parietal lobe | 31 | 5 | 2 | |||||||
| Cerebellum | 1 | 0 | ||||||||
| Other/NOS | 7 | 2 | 10 | 3.5 | ||||||
|
Extent of resection |
0.02 | NA | 0.483 | |||||||
| Gross total | 207 | 8 | 22 | 6.4 | ||||||
| Subtotal | 155 | 6.3 | 30 | 3.7 | ||||||
| Biopsy only | 6 | 0 | ||||||||
| Missing | 11 | 0 | ||||||||
NA, data not available.
NOS, not otherwise specified.
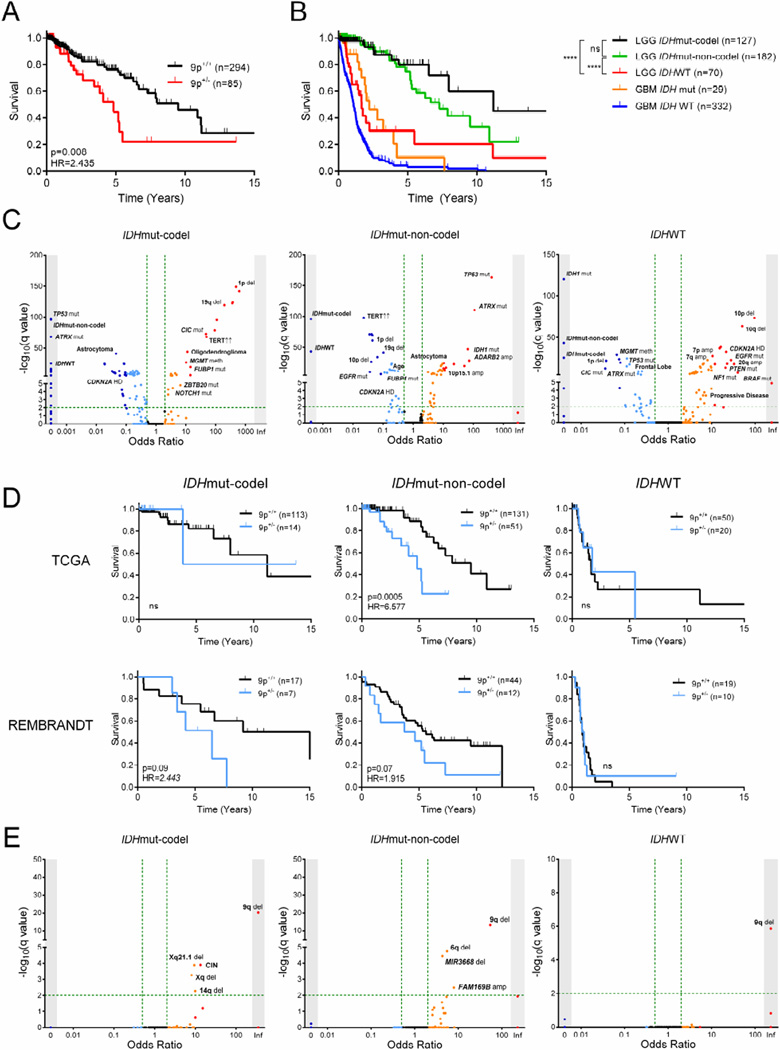
The frequency of 9p loss in TCGA, REMBRANDT, and MSKCC cohorts was 22.4%, 26.6%, and 15.4%, respectively, which is consistent with previous findings (Bello et al., 1994; James et al., 1991). The lower frequency of 9p loss in the MSKCC cohort may be due to smaller sample size and/or differences in cohort heterogeneity. Using a Kaplan-Meier log-rank test based on 9p status, we showed that 9p loss is significantly associated with worse OS in the TCGA, REMBRANDT, and MSKCC cohorts (Figures 2A and S2C).
Figure 2. Characterization of LGG test cohorts.
(A) Kaplan-Meier curve showing survival outcome for 9p loss in the TCGA LGG cohort.
(B) Kaplan-Meier curve showing survival outcome in TCGA LGG and GBM cohorts by subtype and IDH status, respectively. ****, p<0.0001; ns, not significant.
(C) Clinical and molecular variables in LGG and their association to LGG subtype. Fisher’s exact test −log10(q value) (y-axis) and odds ratio (x-axis) for association are shown. Arrows indicate increased mRNA expression.
(D) Kaplan-Meier curves showing survival outcome for 9p loss in both TCGA and REMBRANDT LGG cohorts by subtype.
(E) Clinical parameters and molecular events associated with 9p loss in the TCGA LGG cohort by subtype. Fisher’s exact test −log10(q value) (y-axis) and odds ratio (x-axis) for association are shown.
Mut, mutant; codel, 1p/19q codeletion; LGG, lower grade glioma; WT, wild-type; GBM, glioblastoma multiforme; HR, hazard ratio; del, deletion; amp, amplification; CIN, chromosomal instability; meth, promoter hypermethylation; HD, homozygous deletion.
In order to address the genetic diversity that characterizes LGG, we stratified patients into recently identified molecular subtypes that could potentially confound any subsequent analysis of survival outcomes. Using IDH mutation status and the presence of 1p/19q codeletion as proposed by Jiao and colleagues (Jiao et al., 2012), we separated the TCGA cohort into 3 molecular subtypes with distinct patterns of SCNAs (Figure S2D–S2G). All three subtypes showed marked differences in OS, with IDH wild-type (IDHWT) patients having survival outcomes similar to that of glioblastoma (GBM; Figure 2B). To uncover additional confounders underlying any subtype-specific analysis, we performed a large-scale associations test for LGG-specific clinical and molecular variables by subtype (Figure S2H). We confirmed the presence of several subtype-defining molecular characteristics – CIC and FUBP1 mutation in IDHmut-codel, ATRX and TP53 mutation in IDHmut-non-codel, 7p gain and 10p loss in IDHWT – as well as several unpublished associations of these 3 LGG subtypes (Figure 2C and Table S2) (Appin and Brat, 2014; Cancer Genome Atlas Research et al., 2015; Eckel-Passow et al., 2015). Importantly, these SCNA subtype associations allowed us to stratify subtypes in the REMBRANDT cohort, for which IDH mutation status is unavailable (Figure S2I and S2J). Similar to the TCGA cohort, LGG subtypes in both validation cohorts also corresponded to significant differences in OS (Figure S2K). This confirmed that any analysis of driver genes in LGG must consider the marked differences in OS and molecular signature between subtypes.
Notably, we observed differences in frequency of 9p loss in the TCGA cohort by subtype, occurring in 12.4%, 39.0%, and 28.6% of cases in IDHmut-codel, IDHmut-non-codel, and IDHWT, respectively, though no significant co-association with subtype was found via Fisher’s exact test. Further, in the TCGA cohort, 9p loss predicted significantly worse OS in IDHmut-non-codel gliomas, though its association with OS in IDHmut-codel tumors is less clear (Figure 2D). Within each LGG subtype, very few molecular alterations were found to significantly co-occur with 9p loss (Figure 2E and Table S3). Additionally, 9p loss alone, even in the absence of each co-occurring alteration, was sufficient to predict poor survival via log-rank test (Figure S2L and data not shown). All together, these results suggest that 9p loss may independently predict progression of LGG in IDH mutant tumors, at least when tested against 379 possible confounding variables.
Anatomy of the 9p commonly deleted region and gene-specific survival analysis
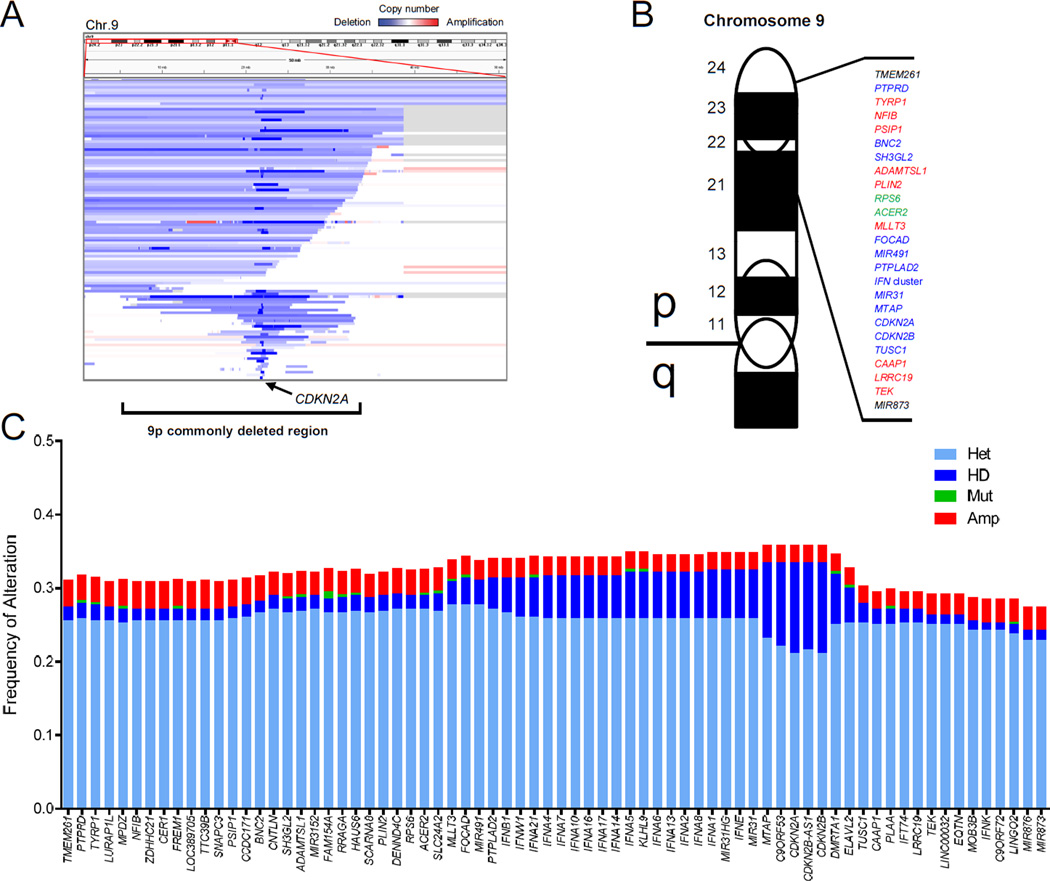
We defined a region on chromosome 9p that is lost in more than 90% of LGG patients with broad deletions on this arm (Figure S3A). The 9p commonly deleted region is characterized mostly by heterozygous loss, though one major focal homozygous deletion is observed at 9p21.3, which contains known TSGs CDKN2A and MTAP (Figures 3A and S2B). A total of 87 genetic loci are contained within the 9p commonly deleted region, including several which have been implicated in cancer through clinical and/or functional methods (Figure 3B and Table S4) (Kent et al., 2002).
Figure 3. Anatomy of the commonly deleted region on 9p in LGG.
(A) Segmentation map of somatic copy number alterations (SCNAs) on chromosome 9p in the TCGA LGG cohort.
(B) Diagram of select genes within the commonly deleted region on 9p ordered by chromosomal location. Known oncogenes (red), tumor suppressor genes (blue), and mixed role genes (green) are shown, along with uncharacterized loci (black).
(C) Overview of genetic alterations at 72 loci for which copy number data is available within the 9p commonly deleted region in the TCGA LGG cohort.
LGG, lower grade glioma; Het, heterozygous deletion; HD, homozygous deletion; Mut, mutation; Amp, amplification; Chr, chromosome; LGG, lower grade glioma.
To further dissect genetic alterations at 9p deleted loci, we extracted copy number status and sequencing mutation calls to identify additional genetic inactivation or second “hits” in the TCGA LGG cohort. Interestingly, very few genes were subject to homozygous deletion except for those within the focal peak at 9p21.3 (Figures 3C and S3B). Additionally, amplifications and nonsynonymous mutations were exceptionally rare. As such, it seemed unlikely that analysis of genetic alterations alone would allow us to pinpoint genetic loci involved in LGG progression on 9p.
Copy number and gene expression associations on 9p in LGG
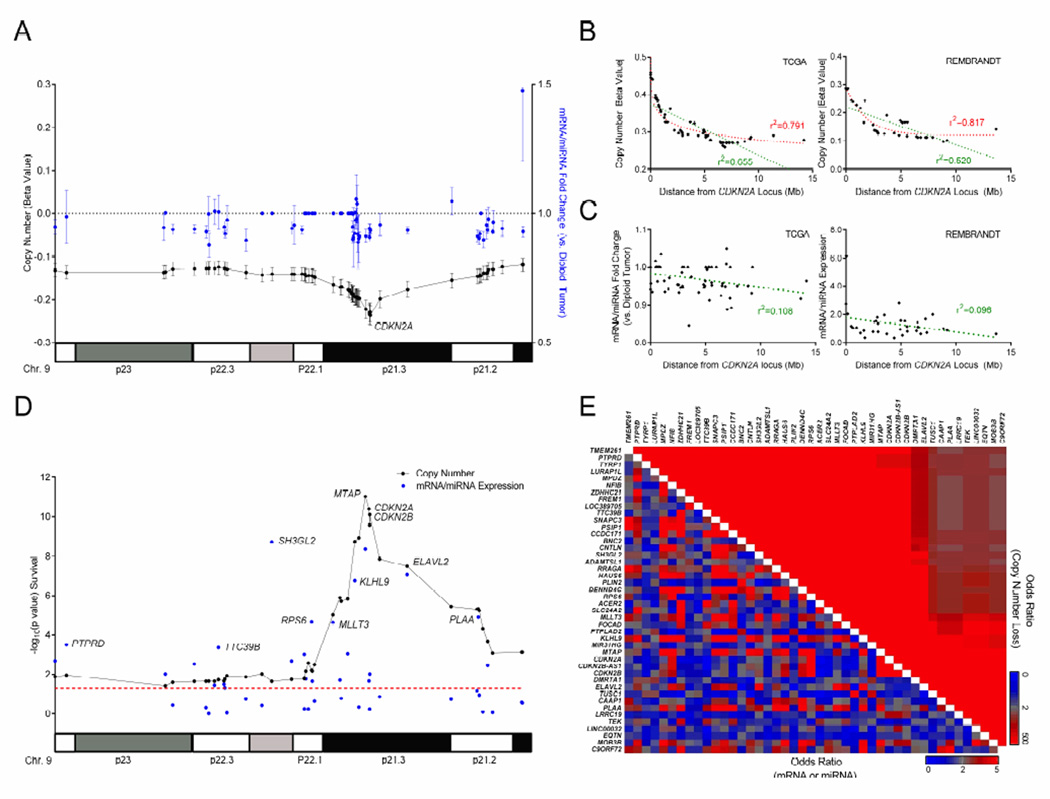
Using RNAseqV2 data from the TCGA, we showed widespread discrepancy between copy number status and gene expression at many loci within the 9p commonly deleted region (Figure 4A). Upon analysis of gene expression by zygosity, we found that copy number status did not uniformly predict gene expression levels. Specifically, several loci exhibited no change in expression following heterozygous deletion alone, while others such as CDKN2A even had elevated expression in this context (Figure S4A). Further, mRNA expression by genetic locus varied considerably across LGG subtypes (Figure S4B). In fact, when we examined copy number status of each gene in regard to distance from CDKN2A where homozygous deletions were most prominent, a strong correlation was observed in the entire LGG cohort and in most subtypes (Figures 4B and S4C). In contrast, mRNA/miRNA expression at each locus was poorly correlated to distance from CDKN2A (Figures 4C and S4C) as well as copy number status at each respective locus (Figure S4D). These findings are consistent with published data showing large SCNAs have variable effects on gene expression, which depends heavily on genomic location and genetic context (Henrichsen et al., 2009).
Figure 4. Copy number and gene expression associations on 9p in LGG.
(A) Plot showing mean copy number signal (beta value, black) and mean mRNA/miRNA fold change vs. diploid tumor (blue) at 44 loci contained within the 9p deletion in the TCGA LGG cohort. Error bars, ± SEM.
(B and C) Linear (green) and logarithmic (red) regression plots of copy number (B) or mRNA/miRNA fold change (C) vs. distance from the CDKN2A locus in the entire TCGA (left) and REMBRANDT (right) LGG cohorts.
(D) Copy number (black) and mRNA/miRNA expression (blue) −log10(p value) following Cox regression test for overall survival at 44 loci within the 9p commonly deleted region in the entire TCGA LGG cohort regardless of 9p copy number status. Red line, significance threshold (p<0.05).
(E) Heatmap depicting all pair-wise associations between loci in the 9p commonly deleted region by copy number loss and mRNA/miRNA status in the TCGA LGG cohort. Mb, megabase; Chr, chromosome.
See also Figure S4.
Historically, efforts to identify driver genes utilized copy number status, either via southern blotting or targeted semi-quantitative PCR, have resulted in a plethora of proposed prognostic loci along 9p (Caldas et al., 1994; Cheng et al., 1994; Stadler and Olopade, 1996). Furthermore, these studies were neither exhaustive nor complete for all gene loci within the 9p deleted region. In order to determine specific genetic loci driving poor survival in the 379 patient TCGA LGG cohort, we selected 44 of 87 loci that had measurable mRNA/miRNA expression, independent of 9p loss status, and performed a univariate Cox proportional hazards regression for each (Figure S4A and Table 2). Not surprisingly, copy number status was significantly correlated with worse OS at every locus whereas expression was much more variable across all LGG subtypes (Figures 4D and S4E). To better understand this phenomenon, we calculated odds ratios to test associations between all loci for both copy number and gene expression. Copy number status between every locus was found to significantly co-occur due to the overriding 9p loss event in absence of additional focal deletions (Figure 4E). Importantly, this trend toward co-occurrence was not observed with gene expression. These results suggest that any dissection of arm-level events must examine gene expression at least in addition to gene-specific copy number status, as the latter does not provide adequate resolution in the absence of additional genetic “hits.”
Table 2.
Survival associations by gene via Cox proportional hazard regression.
| Copy Number |
mRNA/miRNA expression |
|||
|---|---|---|---|---|
| Hazard Ratio (95% CI) | p value | Hazard Ratio (95% CI) | p value | |
| ACER2 | .343 (.158 – .744) | .007 | 1.486 (1.057 – 2.089) | .023 |
| ADAMTSL1 | .365 (.158 – .843) | .018 | 1.255 (1.086 – 1.450) | .002 |
| BNC2 | .357 (.158 – .808) | .013 | 1.167 (.933 – 1.460) | .177 |
| C9orf72 | .295 (.144 – .604) | .001 | 1.045 (.594 – 1.840) | .878 |
| CAAP1 | .209 (.107 – .409) | .000 | .617 (.368 – 1.034) | .067 |
| CCDC171 | .352 (.157 – .793) | .012 | 1.023 (.690 – 1.517) | .910 |
| CDKN2A | .144 (.081 – .255) | .000 | .829 (.706 – .974) | .022 |
| CDKN2B | .153 (.087 – .270) | .000 | .784 (.653 – .942) | .009 |
| CDKN2BAS1 | .156 (.087 – .277) | .000 | 1.095 (.802 – 1.495) | .567 |
| CNTLN | .343 (.153 – .769) | .009 | 1.250 (.769 – 2.031) | .368 |
| DENND4C | .314 (.148 – .668) | .003 | .852 (.499 – 1.457) | .559 |
| DMRTA1 | .171 (.093 – .314) | .000 | 1.281 (.925 – 1.775) | .135 |
| ELAVL2 | .170 (.091 – .319) | .000 | .665 (.573 – .772) | .000 |
| FOCAD | .185 (.094 – .366) | .000 | .564 (.348 – .914) | .020 |
| FREM1 | .384 (.169 – .874) | .023 | .999 (.845 – 1.182) | .993 |
| HAUS6 | .371 (.167 – .824) | .015 | 1.218 (.631 – 2.352) | .556 |
| IFT74 | .210 (.107 – .412) | .000 | .626 (.353 – 1.111) | .110 |
| KLHL9 | .148 (.079 – .275) | .000 | .413 (.296 – .575) | .000 |
| LOC389705 | .385 (.169 – .875) | .023 | .795 (.642 – .985) | .036 |
| LURAP1L | .414 (.180 – .953) | .038 | .655 (.476 – .902) | .009 |
| MIR31 | .148 (.080 – .273) | .000 | 1.057 (.914 – 1.222) | .453 |
| MIR491 | .191 (.097 – .378) | .000 | .859 (.694 – 1.063) | .163 |
| MIR873 | .270 (.126 – .578) | .001 | .931 (.818 – 1.060) | .281 |
| MIR876 | .270 (.126 – .578) | .001 | .906 (.762 – 1.077) | .263 |
| MLLT3 | .209 (.104 – .418) | .000 | .440 (.301 – .643) | .000 |
| MOB3B | .265 (.131 – .534) | .000 | .662 (.502 – .873) | .003 |
| MPDZ | .392 (.173 – .892) | .026 | .812 (.521 – 1.266) | .359 |
| MTAP | .128 (.071 – .231) | .000 | .390 (.285 – .534) | .000 |
| NFIB | .385 (.169 – .877) | .023 | .509 (.326 – .794) | .003 |
| PLAA | .208 (.107 – .408) | .000 | .321 (.192 – .534) | .000 |
| PLIN2 | .343 (.158 – .744) | .007 | 1.525 (1.078 – 2.157) | .017 |
| PSIP1 | .370 (.164 – .839) | .017 | .542 (.295 – .995) | .048 |
| PTPLAD2 | .194 (.099 – .378) | .000 | 1.524 (1.189 – 1.954) | .001 |
| PTPRD | .348 (.155 – .785) | .011 | .632 (.492 – .811) | .000 |
| RPS6 | .340 (.157 – .736) | .006 | .400 (.262 – .610) | .000 |
| RRAGA | .376 (.169 – .838) | .017 | .336 (.176 – .642) | .001 |
| SH3GL2 | .386 (.170 – .877) | .023 | .669 (.586 – .762) | .000 |
| SLC24A2 | .320 (.151 – .682) | .003 | 1.099 (.943 – 1.279) | .227 |
| SNAPC3 | .383 (.168 – .870) | .022 | .508 (.274 – .943) | .032 |
| TEK | .239 (.120 – .476) | .000 | .970 (.739 – 1.274) | .828 |
| TMEM261 | .336 (.142 – .796) | .013 | .320 (.155 – .659) | .002 |
| TTC39B | .377 (.167 – .852) | .019 | 1.533 (1.208 – 1.946) | .000 |
| TUSC1 | .209 (.108 – .406) | .000 | .848 (.667 – 1.080) | .181 |
| ZDHHC21 | .385 (.169 – .875) | .023 | .871 (.593 – 1.278) | .479 |
CI, confidence interval
Genes underlying 9p loss-dependent tumor aggressiveness in LGG
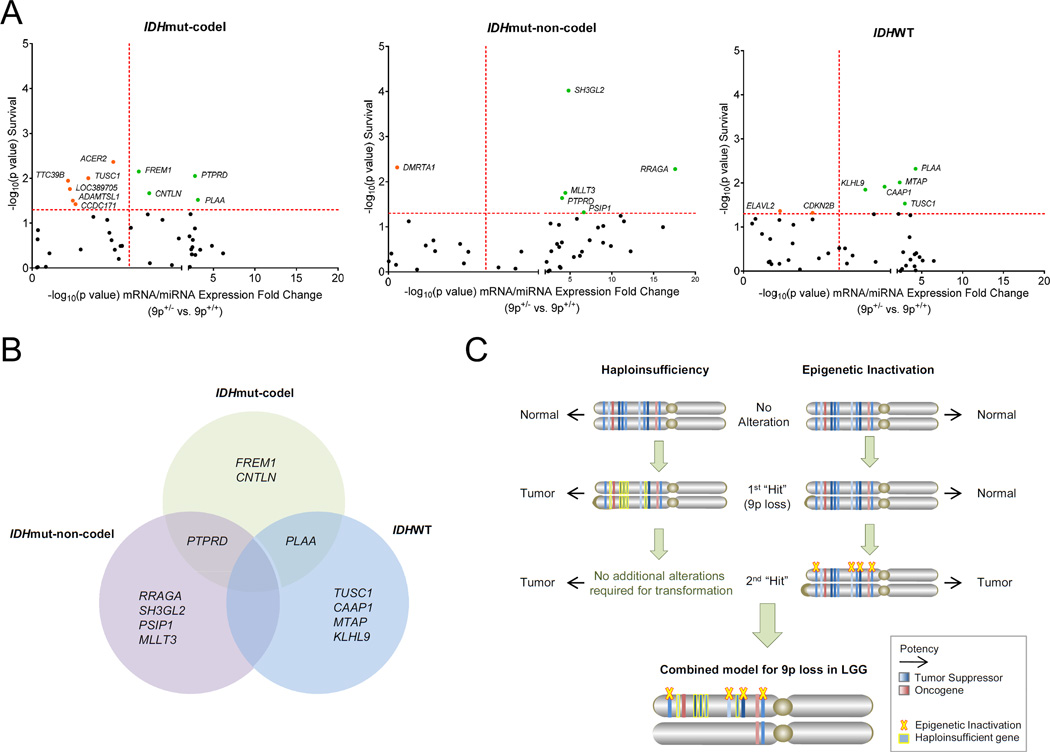
We next sought to determine which genes could predict worse OS in the context of 9p loss in our TCGA LGG cohort, as these would represent the most promising candidates responsible for the 9p survival phenotype in LGG (Figure S5). Student’s t-tests were performed for all 44 loci and average mRNA/miRNA fold change between 9p loss (9p+/−) and 9p diploid (9p+/+) was compared (Table S5). These significance values (−log10p) for 9p loss-driven expression changes were plotted against Cox survival significance values (−log10p) for each gene to reveal candidate 9p loss-targeted loci that drive progression in LGG (Figure 5A).
Figure 5. Genes targeted by 9p loss and their prognostic associations.
(A) Scatterplot showing −log10(p value) Cox survival and −log10(p value) expression fold change significance (9p+/− vs. 9p+/+) for loci within 9p commonly deleted region in the TCGA LGG cohort by subtype. Red dotted line, significance threshold (p<0.05); Green, 9p loss-dependent prognostic genes; Orange, 9p loss-independent prognostic genes.
(B) Genes with significantly decreased expression following 9p loss and poor overall survival from (A) are shown by LGG subtype.
(C) Schematic representation of the mechanisms underlying the 9p deletion phenotype in LGG.
LGG, lower grade glioma; mut, mutant; codel, 1p/19q codeletion.
In total, we identified 5 genes on 9p that likely contribute to tumor aggressiveness within LGG IDHmut-non-codel and IDHWT patients, along with 4 genes in IDHmut-codel patients (Figure 5B). Perhaps most striking was that only two candidates were identified as 9p loss-specific target genes in multiple LGG subtypes – PTPRD and PLAA. Still, several intriguing candidates emerged, including MTAP and the recently identified GBM TSG, KLHL9 (Chen et al., 2014; Schmid et al., 2000). Notably, no single candidate emerged in any of the LGG subtypes. This may be due to inactivation of several genes, likely by some combination of cumulative haploinsuffiency and additional epigenetic downregulation that provides the necessary second “hit(s)” (Figure 5C) (Davoli et al., 2013; McCarthy, 2012). These results suggest that a small number of specific genes on 9p are actually responsible for tumor aggressiveness of LGG and that the loci involved may be both cooperative and context dependent.
Characterization of CDKN2A as a prognostic marker in LGG
For many years, CDKN2A deletion has been associated with poor survival in LGG, leading to speculation that it is important for both initiation of high grade disease as well as tumor aggressiveness (Idbaih et al., 2008; Zhai and Yuan, 1998). In our 379 patient TCGA LGG cohort, CDKN2A downregulation was only weakly associated with survival outcome and expression did not predict outcome in any subtype-specific analysis (Figure 4A, Tables 2 and S5). Furthermore, CDKN2A did not emerge as a candidate to explain poor survival in the context of 9p loss (Figures 5A, 5B and Table S5). Based on these findings, we sought to clarify the role of CDKN2A in LGG.
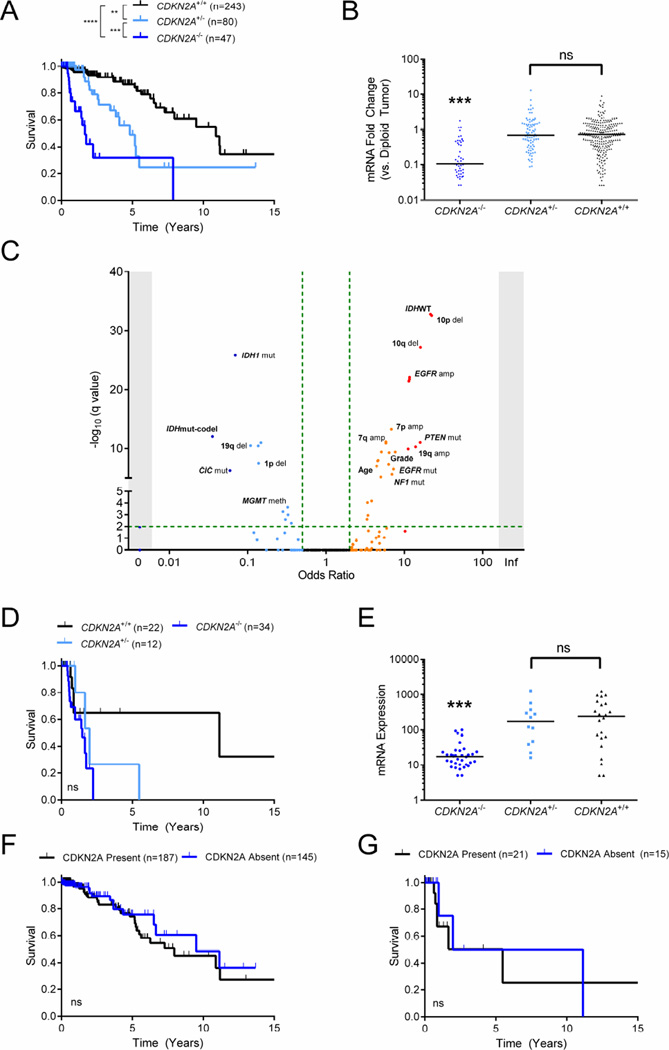
In agreement with prior publications (Bortolotto et al., 2000; James et al., 1999), patients with CDKN2A homozygous deletion (CDKN2A−/−) had significantly shorter survival in the TCGA and validation cohorts (Figures 6A and S6A). In addition, only in the TCGA LGG cohort did heterozygous loss at CDKN2A (CDKN2A+/−) predict worse OS, though only CDKN2A−/− and not CDKN2A+/− correlated with lower mRNA expression of the gene in all three cohorts (Figures 6B and S6B). This suggests that any change in survival in the context of CDKN2A heterozygous deletion is likely due to the confounding 9p loss phenotype.
Figure 6. Characterization of CDKN2A as a marker of prognosis in LGG.
(A and B) Kaplan-Meier curve showing survival outcomes (A) and mRNA fold change (B) in the TCGA LGG cohort based on CDKN2A deletion status. Horizontal bars in (B) depict median.
(C) Scatterplot showing any clinical and molecular associations with CDKN2A homozygous deletion in the TCGA LGG cohort. Fisher’s exact test −log10(q value) (y-axis) and odds ratio (x-axis) for association are shown.
(D and E) Kaplan-Meier curve showing survival outcomes (D) and mRNA fold change (E) in TCGA LGG IDHWT tumors according to the CDKN2A deletion status. Horizontal bars in (E) depict median.
(F and G) Kaplan-Meier curves showing survival outcome based on CDKN2A mRNA expression status in patients without CDKN2A homozygous deletion in the entire TCGA LGG cohort (F) and in IDHWT tumors (G).
LGG, lower grade glioma. amp, amplification; del, deletion; mut, mutation; meth, promoter hypermethylation; mut, mutant; codel, 1p/19q codeletion; WT, wild-type. **, p<0.01; ***, p<0.001; ****, p<0.0001; ns, not significant.
In order to reconcile the differences in OS between CDKN2A copy number status and expression, we performed a large associations test for 376 important clinical and molecular variables in the TCGA LGG cohort (Table S6). Notably, we found that CDKN2A−/− significantly co-occurred with both increased tumor grade and patient age. Additionally, CDKN2A−/− significantly associated with chromosome 7 gain, chromosome 10 loss, EGFR mutation, and PTEN mutation – the defining characteristics of IDHWT subtype tumors (Figure 6C). This association has been recently reported by others, as well (Brat and Group, 2014; Perry et al., 2014). Further, CDKN2A−/− was only observed in one patient in the IDHmut-codel subtype (0.9%) and few significant associations were found within a constrained analysis of IDHmut-non-codel and IDHWT tumors (Figure S6C, S6D and Table S6).
Since LGG IDHWT carries the worst prognosis of all 3 subtypes, we sought to clarify whether the worse survival observed in patients harboring CDKN2A homozygous deletion was a result of this association. We found that CDKN2A deletion status did not correlate with OS in either IDHWT or IDH mutant (IDHmut-codel and IDHmut-non-codel) patients in both TCGA and REMBRANDT LGG cohorts (Figures 6D, S6E and S6F). Although there was a trend toward worse survival following CDKN2A deletion of any type (CDKN2A−/− or CDKN2A+/−), decreased mRNA expression was only observed among samples with CDKN2A−/− (Figures 6E and S6E).
We then sought to determine if non-CDKN2A−/− tumors lacking CDKN2A expression were more likely to be associated with worse OS. First, we verified that CDKN2A mRNA expression correlated with protein expression in both glioma cell lines and in patient samples (Figure S6G and Table S7). Then, non-CDKN2A−/− tumors were categorized by the absence or presence of CDKN2A mRNA expression (Figure S6H). Absent CDKN2A mRNA expression did not associate with any change in OS across all subtypes (Figures 6F and S6I) or within IDHWT alone (Figures 6G and S6J). In fact, patients with absent CDKN2A protein expression (p16) had improved OS in the MSKCC cohort (Figure S6K). Further, in the setting of 9p loss, CDKN2A expression did not alter OS (Figure S6L). In fact, of all the molecular alterations that define IDHWT tumors, only chromosome 10 loss was linked to worse outcomes (Figure S6M and S6N). In light of these results, CDKN2A does not appear to promote tumor aggressiveness in LGG, though it is likely important for the initiation of IDHWT LGGs.
DISCUSSION
In this study, we present an analytical strategy to assess the relative prognostic impact of all arm-level events in pan-cancer SCNA cohort. This comprehensive landscape of arm-level gains/losses highlights the specific alterations associated most strongly with survival across human cancers and within each specific type. Importantly, this complete ranked list represents several attractive candidates for additional genomic and functional exploration. To provide a framework for subsequent analyses, we proceeded to perform a large-scale, multidimensional analysis of 9p loss in LGG to identify candidate driver genes linked to poor outcomes in this disease. Using several tiers of clinical and molecular data from 3 independent datasets, we analyzed 540 individual patient samples to clarify the roles of several known cancer genes and more poorly characterized genetic loci. Our data suggest that no single gene is responsible for the 9p loss phenotype in LGG and several likely act in concert to promote tumor aggressiveness in a context-dependent manner.
Although no single gene emerged as a driver of progression in all 3 LGG subtypes, PTPRD and PLAA influenced patient survival in 2 of 3 subtypes. There is extensive evidence already that PTPRD is a TSG in cancer, with widespread inactivation across many tumor types and the ability to slow tumor growth and invasion in vitro and in vivo (Funato et al., 2011; Veeriah et al., 2009). PLAA has been less extensively characterized, though there is some indication it may function as a TSG since it is capable of causing tumor regression in mouse models (Beatty et al., 1999; Goddard et al., 1998; Goddard et al., 1996). We also identified other intriguing 9p loss-specific candidates involved in progression, including the recently identified GBM TSG KLHL9 and much-debated TSG MTAP (Chen et al., 2014; Schmid et al., 2000). Both of these emerged in the LGG IDHWT, or GBM-like, tumors. Although we lacked sufficient power to further elucidate which combination of genes are driving tumor progression, the short list of genes discovered here provides attractive candidates for functional characterization. In addition, the identification of subtype-specific candidates allows the use of more accurate models and supports the possible implementation of these as biomarkers.
Perhaps most surprising was our discovery that CDKN2A inactivation did not promote tumor aggressiveness within any of the 3 LGG cohorts we examined. CDKN2A was among the first TSGs identified in glioma and has long been linked to poor outcomes in LGG (Bortolotto et al., 2000; James et al., 1999). We show, however, that only CDKN2A homozygous deletion – not expression – associates with poor OS and that this event almost exclusively occurs in IDHWT tumors, a subset of LGG with dismal prognosis (Yan et al., 2009). Further, within the IDHWT LGG subtype, CDKN2A loss is not a driver of disease progression based on our findings. This discovery has major practical implications, as it suggests that CDKN2A status cannot be used as an independent prognostic marker within LGG. Our data is also in line with the recent consensus that genes involved in cell cycle control – like CDKN2A – and those characterized in germline syndromes are most often involved in tumor initiation, not progression (Belinsky et al., 1998; Krepischi et al., 2012; Malumbres and Barbacid, 2001; Rocco and Sidransky, 2001). Although many have suggested that CDKN2A inactivation can lead to progression and worse survival in glioma (Idbaih et al., 2008; Zhai and Yuan, 1998), these early studies were unable to account for subtype-specific confounding alterations and deconvolute copy number status and transcriptional downregulation. Therefore, we believe this study adds clarity to the controversy surrounding CDKN2A in glioma progression. Additionally, these findings challenge existing beliefs that focal alterations alone can provide sufficient insight into underlying biology of superimposed arm-level SCNAs.
These results highlight the power of large datasets to shed light on existing dilemmas in cancer biology. Our approach of pairing clinical phenotypes (i.e. overall survival) with multiple statistical measures of genetic events (i.e. arm-level SCNAs) represents a powerful means by which to distinguish relevant alterations in a given disease. Furthermore, through next-generation data and robust computational analyses, dissecting the roles of genes underlying arm-level SCNAs is both possible and important in our quest to identify potential biomarkers and therapeutic targets in cancer.
EXPERIMENTAL PROCEDURES
Pan-cancer arm-level SCNA analysis
Arm-level SCNA TCGA data and associated clinical reports were downloaded from the Broad Firehose on 21 August 2015 from 33 different cancer types for which data were available. Processed SNP6.0 copy number data were available for a total of 10985 patients and clinical data available for 10594 patients. Copy number data were post-processed by GISTIC2.0 and arm-level SCNA calls were extracted, along with frequency and event significance (q value) by cancer type. Log-rank significance data (p value) were also extracted for each arm-level SCNA in all 33 TCGA cancer datasets. All data are available online (http://gdac.broadinstitute.org/).
Prognostic impact was assessed using the following three parameters: frequency of arm-level gain/loss, event significance (q values via GISTIC2.0), and event survival association (p values via log-rank test). Individual heatmaps for each parameter were constructed for all 82 arm-level SCNAs and 33 TCGA cancer datasets using obtained data. In the arm-level SCNA frequency heatmap, identification of events above background were defined as being 2 standard deviations above the average SCNA frequency within each TCGA dataset (i.e. cancer type).
To establish the relative impact of each arm-level SCNA across cancer, the following 3 parameters were utilized in both 3D and 2D visualizations in a composite analysis: averaged SCNA frequency across all 33 datasets (i.e. cancer types), event index, and survival index. Event index was defined as the number of independent TCGA datasets where an individual arm-level SCNA event was significantly gained or lost (q<0.25). Survival index was defined as the number of independent TCGA datasets where an arm-level SCNA was significantly associated with a change in overall survival on Kaplan-Meier log-rank test (p<0.1). Prognostic impact via event-specific and cancer-specific analyses utilized the following 3 parameters: frequency of arm-level gain/loss, −log10(q value) via GISTIC2.0, and −log10(p value) via log-rank test.
All three-dimensional (3D) representations were created using OriginPro v9.0. Two-dimensional (2D) representations were presented as a heatmap, ordered by linearly-transformed three-parameter composite sum, from highest to lowest.
TCGA LGG cohort
Data were assembled from the TCGA LGG dataset for 379 patients that had completed information for the following tiers: clinical, copy number, mRNA expression, miRNA expression, DNA methylation, and somatic mutation. All clinical, mutation, and normal brain mRNA/miRNA expression data was obtained directly from the TCGA data portal in November 2014 (https://tcga-data.nci.nih.gov/tcga/). Tumor-derived mRNA and miRNA expression data and level 3 SNP6.0 copy number segmentation was obtained from the Broad Firehose in October 2014 (http://gdac.broadinstitute.org/). Methylation data was obtained from the MSKCC cBio cancer genomics portal in October 2014 (http://cbioportal.org/) (Cerami et al., 2012).
Expression data in the 379 TCGA LGG patient cohort was derived from normalized RNAseqV2 RSEM read counts. Background noise was determined by comparing read counts of genes located on chromosome Y and median read count across the entire sample in women. Processing was performed by removing noise in the following manner: All read counts < 5 were considered 5 (baseline) for the purpose of subsequent analyses. Normalized read counts were used for all fold change calculations by comparing the experimental group’s median read count to median read count in diploid tumor samples. For miRNA expression analyses, unprocessed normalized miRNAseq data was used, as above for mRNA data. For all survival analyses, log2 transformed mRNA/miRNA data was used.
Level 3 exome data was used to identify mutation calls in the cohort. This matrix was run through MutSigCV to identify significantly mutated genes. Level 3 copy number segmentation was run through GISTIC2.0 to call SCNAs by patient and amplification/deletion peaks in the cohort. Clinical data and survival information was used from the TCGA without further processing. Only primary tumors were considered for analysis.
REMBRANDT LGG cohort
All clinical and mRNA expression data was downloaded directly from the REMBRANDT data portal (Madhavan et al., 2009). Copy number data was obtained directly from the NCI caArray data management system (www.array.nci.nih.gov/caarray). Only patients with complete data for all 3 tiers (109 patients total) were used for analysis.
Expression data derived from Affymetrix U133 plus 2.0 GeneChips was obtained directly from REMBRANDT (Madhavan et al., 2009). Raw data from Affymetrix GeneChip Mapping 100K array were analyzed and copy number segmentation constructed using Partek Genomics Suite v6.6. All SCNA calls were made using GISTIC2.0. Gender was not available for all patients, therefore copy number status was not determined for chromosome X.
MSKCC LGG cohort
Clinical, expression, and methylation data were obtained for 52 patients with LGG at MSKCC (GEO accession #GSE30336 and #GSE30338) (Turcan et al., 2012). Expression data was derived from Affymetrix U133 plus 2.0 GeneChips and raw intensity values were log2 transformed. Methylation data was derived from Illumina Infinium 450K Methylation Beadchip and were used to construct copy number segmentation for all 52 patient samples via the ChAMP package in R (Morris et al., 2014). Inclusion criteria for broad 9p loss were deletion beta value < −0.1 and deletion size > 5 Mb. Focal SCNAs were determined using GISTIC2.0.
GISTIC2.0
Within TCGA, REMBRANDT, and MSKCC cohorts, SCNA determination was made by processing copy number segmentation data through GISTIC2.0 (Mermel et al., 2011). For all analyses, GISTIC was run using the following parameters:
Amplification Threshold = 0.1
Deletion Threshold = 0.1
Cap Values = 1.5
Broad Length Cutoff = 0.7
Remove X-Chromosome = 0
Confidence Level = 0.99
Join Segment Size = 4
Arm Level Peel Off = 1
Maximum Sample Segments = 2000
Gene GISTIC = 1
Gene Pattern = 0
Arm-level SCNAs were defined as a region of amplification or deletion (GISTIC2.0 beta value > 0.1 or < −0.1, respectively) and occupying 0.7 or 70% of the chromosomal arm. Narrow peak gene loci were obtained from the following output tables: table_amp.conf_99 and table_del.conf_99.
Chromosome 9p commonly deleted region
Level 3 SNP6.0 copy number segmentation data was downloaded from the Broad Firehose on 15 July 2014 for all available patients in the TCGA LGG dataset. Deletion was defined as any beta value less than −0.2. Any deletion less than 5 Mb in size on chromosome 9p was not considered broad. All remaining 9p deletions were mapped to genomic coordinates, with the 9p commonly deleted region defined as the chromosomal start/end site which contained 90% or more of all broad deletions on 9p. Genetic loci contained within this deletion region were curated from the UCSC Genome Browser using human reference genome build hg19 (Kent et al., 2002).
Pearson correlation coefficient was used to determine association between mRNA/miRNA expression and copy number status, as well as genomic distance, for all normally distributed data. For copy number status vs. genomic distance, a logarithmic regression model was used to derive r2.
MutSigCV
To determine if any genes were significantly mutated in the 379 patient TCGA LGG cohort, level 3 mutation calls were downloaded directly from the TCGA data portal. These data were processed through MutSigCV (Lawrence et al., 2013) to identify significant mutation calls in LGG. Default settings were used for the analysis.
Survival analyses
Cox proportional hazards regression analysis was performed for 44 loci in the TCGA LGG cohort using IBM SPSS statistical software. Both copy number data and log-transformed expression data were used. These results were used to determine 9p loss-specific candidate loci.
For Kaplan-Meier survival analyses, patients without a date of death were censored. For each analysis, significance was calculated by log-rank test and the hazard ratio reported. For validation of 9p loss-specific candidates, the top/bottom 25% of patients were used for each gene analyzed. For determination of CDKN2A role in LGG, patients with amplifications at this locus were not analyzed. In TCGA, REMBRANDT, and MSKCC cohorts, CDKN2A mRNA expression threshold of “absent” vs. “present” was set at the 90th percentile of CDKN2A−/− tumors.
Statistical Analyses for clinical and molecular associations
In order to test for any molecular or clinical associations in regard to 9p loss, CDKN2A, or LGG subtype, we analyzed 7 distinct parameters within the 379 patient TCGA LGG cohort: clinical variables, arm-level amplification/deletion, significant gene amplifications/deletions, significant focal peaks, significant MutSigCV mutated genes, TERT upregulation, and MGMT promoter hypermethylation. 376 variables were tested in total. Significantly amplified or deleted genes, as well as focal peaks, were extracted from our GISTIC2.0 run on the TCGA cohort. Mutated genes were identified using MutSigCV and manual curation based on literature evidence. Fisher’s exact test p values and Bonferroni adjusted q values are reported along with odds ratios for each comparison. Statistical tests were performed using IBM SPSS.
Statistical testing for gene expression
Determination of mRNA/miRNA expression differences was made using a one-tailed Student’s t-test in MSKCC validation cohort only, otherwise all Student’s t-test used were two-tailed. All groups compared had similar variance and estimates of variation are shown in figures using standard error of mean (SEM). 9p loss-targeted loci were determined by comparing mRNA/miRNA expression in 9p loss patients (9p+/−) to average expression in 9p diploid (9p+/+) patients.
Establishment of LGG subtypes through copy number status
In order to cluster previously published LGG subtypes by copy number alone, we used results from association testing by subtype to test new combinations of SCNAs. In the 379 patient TCGA LGG cohort, using 1p/19q codeletion along with 10p, 7p and 7q, and CDKN2A status, we achieved > 90% accuracy. In order to test this on a validation set, we utilized 134 patients from the TCGA LGG dataset that were excluded from our 379 patient cohort but still had SNP6.0 copy number data and IDH mutation calls from whole exome sequencing. In this set, 95% accuracy was achieved using the new copy number criteria alone.
CDKN2A Protein Quantification
The immunohistochemical detection of p16 antibody (Roche, cat# 0669522100) was performed at Molecular Cytology Core Facility of Memorial Sloan Kettering Cancer Center using Discovery XT processor (Ventana Medical Systems). Sections were blocked for 30 minutes with Background Buster solution (Innovex). Antibody was applied and sections were incubated for 5 hours, followed by 60 minutes incubation with biotinylated goat anti-mouse IgG (Vector labs) or biotinylated horse anti-mouse IgG (Vector Labs) at 1:200 dilution. The detection was performed with DAB detection kit (Ventana Medical Systems) according to manufacturer instruction. Slides were counterstained with hematoxylin and coverslips mounted with Permount (Fisher Scientific). Staining was blindly scored by a clinical pathologist.
All cell lines were obtained from ATCC and cultured in the recommended growth conditions. Total cell extracts were obtained using CellLytic M protein lysis buffer (Sigma) and the protein concentration was determined via BCA assay (Thermo Scientific). Equal amounts of total protein were run at 120V on 12% Bis-Tris gels (Invitrogen) with MOPS running buffer (Invitrogen). Proteins were transferred onto PVDF membranes (Millipore) and blocked using 0.1% TBST containing 5% non-fat dry milk. The following antibodies were used: p14 ARF (Cell Signaling, 1:1000), p16 (Millipore, 1:1000), and actin (Sigma, 1:2000). Protein bands were visualized using enhanced chemiluminescence (Imagequant LAS 4000, GE Health Life Sciences) and quantified by densitometry (Imagequant TL, GE Health Life Sci).
Supplementary Material
SIGNIFICANCE.
Somatic copy number alterations (SCNAs) are frequent genetic events that promote tumor initiation and progression. Characterization of focal SCNAs has led to the identification of many cancer genes, yet the identity of potential drivers within broad SCNAs remains a critical unanswered question in cancer genetics. We describe an analytic strategy for elucidating underlying loci responsible for arm-level SCNA phenotypes. Implementing this methodology on the 9p loss event in lower grade glioma (LGG) revealed that CDKN2A inactivation is not associated with progression. Instead, other 9p loci represent potential drivers of progression and attractive targets for further study and therapeutic exploitation. These findings provide insight behind 9p loss in LGG and establish a framework for examining other arm-level SCNAs in cancer.
HIGHLIGHTS.
-
-
There exists a wide spectrum of arm-level SCNAs of variable prognostic “impact”
-
-
Arm-level dissection via copy number status alone frequently yields false positives
-
-
Only a minority of genetic loci influence powerful arm-level SCNA phenotypes
-
-
Several 9p loci, but not CDKN2A, are linked to survival outcomes in LGG
Roy et al. present a landscape of arm-level somatic copy number alterations (SCNA) across 33 cancer types and assess the relative impact. Using chromosome 9p loss in lower grade glioma as an example, they describe an analytic approach for elucidating underlying loci responsible for arm-level SCNA phenotypes.
Acknowledgments
The results published here are based upon data generated by the TCGA Research Network and REMBRANDT. This grant was supported in part by the National Institutes of Health (RO1-CA154767A1 and RO1 CA177828) (T.A.C.), The Geoffrey Beene Foundation (T.A.C.), the MSKCC Brain Tumor Center (T.A.C.), the Sontag Foundation (T.A.C.), and the Louis Gerstner Foundation (T.A.C.). D.M.R. was supported by the HHMI Medical Research Fellows Program and NIH MSTP grant T32GM007739. LAW was supported by The Canadian Institutes of Health Research PDF Award MFE-127325.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
T.A.C. and D.M.R. conceived the study. D.M.R. and L.A.W. designed the experiments. L.A.W. and W.W. performed the experiments. D.M.R, L.A.W., A.D., J.J.G., and P.B. conducted the bioinformatics analysis. D.M.R, L.A.W., T.A.C., A.D., J.T.H., J.J.G., and W.L. analyzed the data. D.M.R. and T.A.C. wrote the manuscript. All authors reviewed and edited the final manuscript.
REFERENCES
- Appin CL, Brat DJ. Molecular genetics of gliomas. Cancer journal. 2014;20:66–72. doi: 10.1097/PPO.0000000000000020. [DOI] [PubMed] [Google Scholar]
- Baudis M. Genomic imbalances in 5918 malignant epithelial tumors: an explorative meta-analysis of chromosomal CGH data. BMC cancer. 2007;7:226. doi: 10.1186/1471-2407-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty BG, Qi S, Pienkowska M, Herbrick JA, Scheidl T, Zhang ZM, Kola I, Scherer SW, Seth A. Chromosomal localization of phospholipase A2 activating protein, an Ets2 target gene, to 9p21. Genomics. 1999;62:529–532. doi: 10.1006/geno.1999.5999. [DOI] [PubMed] [Google Scholar]
- Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, Baylin SB, Herman JG. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:11891–11896. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello MJ, de Campos JM, Vaquero J, Kusak ME, Sarasa JL, Pestana A, Rey JA. Molecular and cytogenetic analysis of chromosome 9 deletions in 75 malignant gliomas. Genes, chromosomes & cancer. 1994;9:33–41. doi: 10.1002/gcc.2870090107. [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolotto S, Chiado-Piat L, Cavalla P, Bosone I, Chio A, Mauro A, Schiffer D. CDKN2A/p16 inactivation in the prognosis of oligodendrogliomas. International journal of cancer Journal international du cancer. 2000;88:554–557. doi: 10.1002/1097-0215(20001115)88:4<554::aid-ijc6>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Bown N, Cotterill S, Lastowska M, O’Neill S, Pearson AD, Plantaz D, Meddeb M, Danglot G, Brinkschmidt C, Christiansen H, et al. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. The New England journal of medicine. 1999;340:1954–1961. doi: 10.1056/NEJM199906243402504. [DOI] [PubMed] [Google Scholar]
- Brat DJ, Group TTLGGAW. INTEGRATIVE GENOMIC CHARACTERIZATION OF LOWER GRADE GLIOMAS. Neuro-oncology. 2014;16 [Google Scholar]
- Bredel M, Bredel C, Juric D, Harsh GR, Vogel H, Recht LD, Sikic BI. High-resolution genome-wide mapping of genetic alterations in human glial brain tumors. Cancer research. 2005;65:4088–4096. doi: 10.1158/0008-5472.CAN-04-4229. [DOI] [PubMed] [Google Scholar]
- Caldas C, Hahn SA, da Costa LT, Redston MS, Schutte M, Seymour AB, Weinstein CL, Hruban RH, Yeo CJ, Kern SE. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nature genetics. 1994;8:27–32. doi: 10.1038/ng0994-27. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N, Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, Rheinbay E, Miller CR, Vitucci M, et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. The New England journal of medicine. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Alvarez MJ, Talos F, Dhruv H, Rieckhof GE, Iyer A, Diefes KL, Aldape K, Berens M, Shen MM, Califano A. Identification of causal genetic drivers of human disease through systems-level analysis of regulatory networks. Cell. 2014;159:402–414. doi: 10.1016/j.cell.2014.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JQ, Jhanwar SC, Klein WM, Bell DW, Lee WC, Altomare DA, Nobori T, Olopade OI, Buckler AJ, Testa JR. p16 alterations and deletion mapping of 9p21-p22 in malignant mesothelioma. Cancer research. 1994;54:5547–5551. [PubMed] [Google Scholar]
- Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, Elledge SJ. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013;155:948–962. doi: 10.1016/j.cell.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, Pekmezci M, Rice T, Kosel ML, Smirnov IV, et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. The New England journal of medicine. 2015;372:2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nature reviews Genetics. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- Funato K, Yamazumi Y, Oda T, Akiyama T. Tyrosine phosphatase PTPRD suppresses colon cancer cell migration in coordination with CD44. Experimental and therapeutic medicine. 2011;2:457–463. doi: 10.3892/etm.2011.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard DH, Bomalaski JS, Clark MA. Phospholipase A2 activating protein induces tumor regression. Drug news & perspectives. 1998;11:104–110. doi: 10.1358/dnp.1998.11.2.863682. [DOI] [PubMed] [Google Scholar]
- Goddard DH, Bomalaski JS, Lipper S, Shorr RG, Clark MA. Phospholipase A2-mediated inflammation induces regression of malignant gliomas. Cancer letters. 1996;102:1–6. doi: 10.1016/0304-3835(96)04142-0. [DOI] [PubMed] [Google Scholar]
- Green ED, Guyer MS National Human Genome Research I. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470:204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- Gross AM, Orosco RK, Shen JP, Egloff AM, Carter H, Hofree M, Choueiri M, Coffey CS, Lippman SM, Hayes DN, et al. Multi-tiered genomic analysis of head and neck cancer ties TP53 mutation to 3p loss. Nature genetics. 2014;46:939–943. doi: 10.1038/ng.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrichsen CN, Vinckenbosch N, Zollner S, Chaignat E, Pradervand S, Schutz F, Ruedi M, Kaessmann H, Reymond A. Segmental copy number variation shapes tissue transcriptomes. Nature genetics. 2009;41:424–429. doi: 10.1038/ng.345. [DOI] [PubMed] [Google Scholar]
- Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, Leiserson MD, Niu B, McLellan MD, Uzunangelov V, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158:929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idbaih A, Carvalho Silva R, Criniere E, Marie Y, Carpentier C, Boisselier B, Taillibert S, Rousseau A, Mokhtari K, Ducray F, et al. Genomic changes in progression of low-grade gliomas. Journal of neuro-oncology. 2008;90:133–140. doi: 10.1007/s11060-008-9644-z. [DOI] [PubMed] [Google Scholar]
- James CD, Galanis E, Frederick L, Kimmel DW, Cunningham JM, Atherton-Skaff PJ, O’Fallon JR, Jenkins RB, Buckner JC, Hunter SB, et al. Tumor suppressor gene alterations in malignant gliomas: histopathological associations and prognostic evaluation. International journal of oncology. 1999;15:547–553. doi: 10.3892/ijo.15.3.547. [DOI] [PubMed] [Google Scholar]
- James CD, He J, Carlbom E, Nordenskjold M, Cavenee WK, Collins VP. Chromosome 9 deletion mapping reveals interferon alpha and interferon beta-1 gene deletions in human glial tumors. Cancer research. 1991;51:1684–1688. [PubMed] [Google Scholar]
- Jen J, Kim H, Piantadosi S, Liu ZF, Levitt RC, Sistonen P, Kinzler KW, Vogelstein B, Hamilton SR. Allelic loss of chromosome 18q and prognosis in colorectal cancer. The New England journal of medicine. 1994;331:213–221. doi: 10.1056/NEJM199407283310401. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Killela PJ, Reitman ZJ, Rasheed AB, Heaphy CM, de Wilde RF, Rodriguez FJ, Rosemberg S, Oba-Shinjo SM, Nagahashi Marie SK, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome research. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krepischi AC, Pearson PL, Rosenberg C. Germline copy number variations and cancer predisposition. Future oncology. 2012;8:441–450. doi: 10.2217/fon.12.34. [DOI] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson MD, Vandin F, Wu H, Dobson JR, Eldridge JV, Thomas JL, Papoutsaki A, Kim Y, Niu B, McLellan M, et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nature genetics. 2014 doi: 10.1038/ng.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K. Rembrandt: helping personalized medicine become a reality through integrative translational research. Molecular cancer research : MCR. 2009;7:157–167. doi: 10.1158/1541-7786.MCR-08-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. To cycle or not to cycle: a critical decision in cancer. Nature reviews Cancer. 2001;1:222–231. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- McCarthy N. Tumorigenesis. Better by half. Nature reviews Cancer. 2012;12:450–451. doi: 10.1038/nrc3307. [DOI] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome biology. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris TJ, Butcher LM, Feber A, Teschendorff AE, Chakravarthy AR, Wojdacz TK, Beck S. ChAMP: 450k Chip Analysis Methylation Pipeline. Bioinformatics. 2014;30:428–430. doi: 10.1093/bioinformatics/btt684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olopade OI, Jenkins RB, Ransom DT, Malik K, Pomykala H, Nobori T, Cowan JM, Rowley JD, Diaz MO. Molecular analysis of deletions of the short arm of chromosome 9 in human gliomas. Cancer research. 1992;52:2523–2529. [PubMed] [Google Scholar]
- Perry A, Reis GF, Pekmezci M, Hansen HM, Marshall RE, Rice T, Wiencke JF, Wrensch MR, Walsh KM, Perry A. CDKN2A LOSS IS ASSOCIATED WITH SHORTENED SURVIVAL IN INFILTRATING ASTROCYTOMAS BUT NOT OLIGODENDROGLIOMAS OR MIXED OLIGOASTROCYTOMAS. Neuro-oncology. 2014;16 [Google Scholar]
- Pollock PM, Welch J, Hayward NK. Evidence for three tumor suppressor loci on chromosome 9p involved in melanoma development. Cancer research. 2001;61:1154–1161. [PubMed] [Google Scholar]
- Rocco JW, Sidransky D. p16(MTS-1/CDKN2/INK4a) in cancer progression. Experimental cell research. 2001;264:42–55. doi: 10.1006/excr.2000.5149. [DOI] [PubMed] [Google Scholar]
- Schmid M, Sen M, Rosenbach MD, Carrera CJ, Friedman H, Carson DA. A methylthioadenosine phosphorylase (MTAP) fusion transcript identifies a new gene on chromosome 9p21 that is frequently deleted in cancer. Oncogene. 2000;19:5747–5754. doi: 10.1038/sj.onc.1203942. [DOI] [PubMed] [Google Scholar]
- Stadler WM, Olopade OI. The 9p21 region in bladder cancer cell lines: large homozygous deletion inactivate the CDKN2, CDKN2B and MTAP genes. Urological research. 1996;24:239–244. doi: 10.1007/BF00295899. [DOI] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki K, Rubio MP, Ramesh V, Correa KM, Rutter JL, von Deimling A, Buckler AJ, Gusella JF, Louis DN. MTS1/CDKN2 gene mutations are rare in primary human astrocytomas with allelic loss of chromosome 9p. Human molecular genetics. 1994;3:1841–1845. doi: 10.1093/hmg/3.10.1841. [DOI] [PubMed] [Google Scholar]
- Veeriah S, Brennan C, Meng S, Singh B, Fagin JA, Solit DB, Paty PB, Rohle D, Vivanco I, Chmielecki J, et al. The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9435–9440. doi: 10.1073/pnas.0900571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltshire RN, Herndon JE, 2nd, Lloyd A, Friedman HS, Bigner DD, Bigner SH, McLendon RE. Comparative genomic hybridization analysis of astrocytomas: prognostic and diagnostic implications. The Journal of molecular diagnostics : JMD. 2004;6:166–179. doi: 10.1016/S1525-1578(10)60507-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. IDH1 and IDH2 mutations in gliomas. The New England journal of medicine. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhang CZ, Wala J, Mermel CH, et al. Pan-cancer patterns of somatic copy number alteration. Nature genetics. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai G, Yuan X. Study of deletion of P16 gene in the progression of brain astrocytomas. Chinese Journal of Cancer Research. 1998;10:284–287. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.