Abstract
Drug exposure during critical periods of development is known to have lasting effects, increasing one's risk for developing mental health disorders. Emerging evidence has also indicated the possibility for drug exposure to even impact subsequent generations. Our previous work demonstrated that adolescent exposure to Δ9-tetrahydrocannabinol (THC), the main psychoactive component of marijuana (Cannabis sativa), in a Long-Evans rat model affects reward-related behavior and gene regulation in the subsequent (F1) generation unexposed to the drug. Questions, however, remained regarding potential epigenetic consequences. In the current study, using the same rat model, we employed Enhanced Reduced Representation Bisulfite Sequencing to interrogate the epigenome of the nucleus accumbens, a key brain area involved in reward processing. This analysis compared 16 animals with parental THC exposure and 16 without to characterize relevant systems-level changes in DNA methylation. We identified 1027 differentially methylated regions (DMRs) associated with parental THC exposure in F1 adults, each represented by multiple CpGs. These DMRs fell predominantly within introns, exons, and intergenic intervals, while showing a significant depletion in gene promoters. From these, we identified a network of DMR-associated genes involved in glutamatergic synaptic regulation, which also exhibited altered mRNA expression in the nucleus accumbens. These data provide novel insight into drug-related cross-generational epigenetic effects, and serve as a useful resource for investigators to explore novel neurobiological systems underlying drug abuse vulnerability.
Introduction
Marijuana (Cannabis sativa) is the most commonly used illicit drug by teenagers and young adults in Western countries, with significant controversies regarding its immediate and long-term effects (SAMHSA, 2011; Ammerman, 2014). Alarmingly, marijuana use now exceeds that of tobacco in youths (Johnston et al, 2012). Recent movements regarding the legalization of Cannabis for medical or recreational purposes have sparked intense debates, yet controversies surrounding marijuana regarding medical, political, and societal issues have largely lacked scientific input (Hopfer, 2014). Despite the public perception that Cannabis is a relatively safe drug, there is now evidence that its abuse can lead to dependence, especially in individuals with vulnerable genetic backgrounds, as well as with neuropsychiatric abnormalities such as anxiety, depression or psychosis (Hurd et al, 2014).
The main psychoactive component of Cannabis, Δ9-tetrahydrocannabinol (THC), directly binds to cannabinoid receptor type 1 receptors (CB1R), which are expressed in neuronal circuits that have a critical role in the pathophysiology of addiction-related psychiatric phenotypes (Koob and Volkow, 2010; Mathur and Lovinger, 2012). Recently, we reported multiple lines of evidence that implied a previously unknown ‘cross-generational gateway' state in the offspring of THC-exposed parents (Szutorisz et al, 2014). Adult F1 offspring that were themselves not exposed to THC displayed increased heroin self-administration behavior, changes in the expression of CB1R, dopamine, and glutamatergic receptor genes in the striatum, and altered striatal synaptic plasticity. However, the molecular mechanisms by which parental THC exposure affects gene regulation in the offspring brain remain unknown.
Epigenetic processes are thought to underlie cross-generational effects of environmental insults, including those related to drugs of abuse (Szutorisz et al, 2014; Vassoler and Sadri-Vakili, 2014). Of the known epigenetic modifications, variation in DNA methylation has been shown both to be mediated by environmental influences and to frequently persist through multiple generations in various organisms (Lange and Schneider, 2010; Becker and Weigel, 2012; Heard and Martienssen, 2014). DNA methylation is central to many key biological processes, is known to colocalize with functional chromatin features (eg, DNaseI hypersensitivity and histone modifications), and has demonstrated effects on gene regulation via interactions with transcription factors and splicing machinery (Cedar and Bergman, 2009; Gutierrez-Arcelus et al, 2015; Hu et al, 2013). With relevance to behavioral and physiological alterations previously described in our model (Szutorisz et al, 2014), DNA methylation has also been widely implicated in normal brain development (Wilson and Sengoku, 2013; Spiers et al, 2015) and psychiatric phenotypes such as anxiety, depression, autism, schizophrenia, bipolar disorder, and addiction (Feng and Nestler, 2013; Schmitt et al, 2014; Tuesta and Zhang, 2014).
We hypothesized that that epigenetic disturbances involving DNA methylation are associated with parental THC exposure in the subsequent generation, and that these changes affect gene pathways with relevance to previously characterized behavioral impairments (Szutorisz et al, 2014). To test this, we performed genome-wide DNA methylation analysis using Enhanced Reduced Representation Bisulfite Sequencing (ERRBS) in the nucleus accumbens (NAc)—the ventral subregion of the striatum most linked to addiction vulnerability—of adult F1 offspring. Being the largest study of its kind to date, these data provide novel insight into potential drug-related cross-generational epigenetic effects.
Materials and Methods
Drugs
Δ9-THC (50 mg/ml in ethanol solution) was evaporated under nitrogen gas, dissolved in 0.9% NaCl (saline) containing 0.3% Tween 80 to a concentration of 0.75 mg/ml (Dinieri and Hurd, 2012). Control ‘vehicle' (VEH) solution was saline containing 0.3% Tween 80.
Animals and Cross-Generational THC Paradigm
The cross-generational THC animal model has been described in detail previously (Szutorisz et al, 2014). Briefly, adolescent male and female Long-Evan rats were administered THC (1.5 mg/kg every third day from postnatal day 28–49) or VEH. Animals were mated as adults when no detectable THC was present and their pups were raised by drug-naïve mothers (see Supplementary Materials and Materials and Methods for more details).
Dissection of NAc Tissue, Sample Preparation, and ERRBS Pipeline
Samples were processed following procedures outlined in Supplementary Figure S1. Bilateral NAc tissue was dissected (15-gauge punch), from frozen brains of 32 F1 rats (PND~62) from adult offspring of parents with repeated exposure to either THC or VEH during adolescence (Szutorisz et al, 2014). Eight females and eight males from both the THC- and VEH-exposed lines were derived from five to six different mothers and fathers in each group. Genomic DNA was extracted (DNeasy Blood & Tissue Kit; Qiagen, Valencia, CA, USA) and ~250 ng DNA/sample was processed for ERRBS following established protocols (Akalin et al, 2012a). ERRBS sequencing libraries were constructed and sequenced on a HiSeq2000 (Weill Cornell genomics core facility, New York, NY). As per Alkalin et al (2012a), reads were aligned to an in silico bisulfite-converted version of the rn4 reference genome assembly using Bismark (Krueger and Andrews, 2011), allowing 2 bp mismatches per read, and discarding reads mapping to multiple locations.
Data Processing and Statistical Analysis
Following read mapping, methylation levels were estimated on a per CpG basis by taking the fraction of reads containing methylated non-bisulfite-converted C nucleotides over the sum of all reads mapping to a given CpG, and multiplying this fraction by 100 (ie, CpG percent methylation=(count C)/(count C+count T) × 100). All processing of CpG read count files, sample quality control, and initial statistical analyses were carried out using the package methylKit (Akalin et al, 2012b), implemented in R v3.0 (www.R-project.org).
CpGs with either fewer than 30 mapped reads or read counts >99th percentile (to exclude sites with potential PCR clonal amplification bias) were removed. CpG read depths across the 32 samples were normalized using the ‘normalizeCoverage' function in methylKit (Alkalin et al, 2012b). CpGs not represented in all samples and those mapping to sex chromosomes were not considered, leaving a total of 776 220 CpGs genome wide. Using this filtered and normalized set of CpGs, principal component analysis (PCA) was applied to identify potential sample outliers. Following outlier removal, we discarded CpGs at which the range in observed CpG methylation values (ie, maximum observed methylation value − minimum observed methylation value) within either group was ≥30% (n=208 914). At the remaining autosomal CpGs (n=567 306), logistic regression was used to test for differential methylation between THC and VEH groups. Observed P-values were adjusted using the Benjamini–Hochberg false discovery rate (FDR) method (Benjamini and Hochberg, 1995).
A novel sliding-window method, written in PERL, was used to identify clusters of neighboring CpGs exhibiting concordant changes in methylation associated with cross-generational THC exposure (termed differentially methylated regions, DMRs; Supplementary Figure S2). DMRs were defined as regions of the genome containing at least three neighboring CpGs within a 500-bp interval; we required the presence of a minimum of three statistically significant CpGs (q<0.01) with a concordant (either hypo- or hypermethylated) mean methylation difference >2% between THC and VEH groups, representing at least 50% of CpGs within a given window/DMR.
DMR/CpG Annotation and Downstream Analyses
Coordinates for Refseq gene annotations (rn4), CpG islands (‘AL' UCSC track)(Gardiner-Garden and Frommer, 1987), and common repeats (RepeatMasker; see Supplementary Materials and Materials and Methods for more details) (Jurka, 2000; Smit et al, 1996–2010) were downloaded from the UCSC genome browser (http://genome.ucsc.edu/) (Kent et al., 2002). The overlap of CpGs with annotated genomic features were assessed using BedTools version 2.1 (Quinlan and Hall, 2011). Refseq gene annotations were segregated into four categories: exonic (within coding or 3′UTR sequence), intronic, promoter (±2 kbp of transcription start site (TSS)) and intergenic (outside of exons, introns, and promoters). Likewise, CpG island annotations were classified as one of four categories: CpG islands (CpGi); CpGi shores (±2 kbp from CpGi); CpGi shelves (±2 kbp from CpGi shores); and sea (outside of all other CpGi annotations). In cases where CpGs fell within more than one CpGi annotation, islands took precedence over shores and shelves, and shores took precedence over shelves. For both gene and CpGi feature overlaps, the distribution of DMR-CpGs was assessed using the χ2-test to compare against the background list of CpGs that included all tested sites (n=567 306; referred to as ‘All' below).
Enrichment for gene ontology (GO) terms was assessed using GOrilla (Eden et al, 2007; Eden et al, 2009). Refseq genes overlapped by significant DMRs (promoters or introns/exons; n=492) were compared with a background list of Refseq genes (n=11 389) overlapped by all interrogated CpGs. Enrichment of DMR-associated genes for interactions with a list of protein–protein interaction hubs (PPIhubs) was estimated using the Fisher's exact test implemented in the program List2Networks (Lachmann and Ma'ayan, 2010), based on physical protein–protein interaction networks cataloged from several mammalian data sets (Berger et al, 2007); PPIhubs were defined as proteins with >24 known interaction partners (Berger et al, 2007; Lachmann and Ma'ayan, 2010). PPIhub enrichments for DMR-associated genes were compared to those obtained from separate analyses using two gene sets of interest. Rat genome-annotated genes under GO terms ‘regulation of synaptic plasticity' (GO:0048167; n=146) and ‘glutamatergic synaptic transmission' (GO:0035249; n=79) were obtained from the AmiGO 2 database (http://amigo.geneontology.org/amigo). PPI gene network images were generated using genemania (Warde-Farley et al, 2010) based on PPI interactions from databases utilized by Lists2Networks (Berger et al, 2007; Lachmann and Ma'ayan, 2010).
Comparisons with known imprinted genes were conducted using lists compiled from rat, mouse, and human (www.geneimprint.com), considering gene IDs and aliases with status listed as ‘Imprinted' or ‘Predicted'.
Quantitative Reverse Transcription PCR (qRT-PCR) Analysis
mRNA levels for 13 genes were measured in the NAc of a separate cohort of THC and VEH animals (n=8 males and 8 females/group). The data were analyzed using the ΔΔCT method (Livak and Schmittgen, 2001), and statistical significance was assessed using the Welch two-sample t-test. More details are described in the Supplementary Information.
Results
Differentially Methylated Regions in the NAc Are Associated With Cross-Generational Effects of Adolescent THC Exposure in Adult F1 Offspring
To characterize cross-generational epigenetic signatures associated with adolescent Cannabis use, we compared genome-wide DNA methylation levels in the NAc of two groups of unrelated F1 Long-Evans rats born with F0 individuals that had either been exposed to THC or VEH during adolescence. From each ERRBS library (n=32), we generated a mean of 196 062 679 reads (50 bp single-end). An average of 93% of bases per library had quality (Q) scores >30, and >2.2 million CpGs were interrogated per individual genome (mean=3 454 502). Raw methylation files have been deposited in GEO (GSE69984). On the basis of PCA using autosomal CpG methylation profiles, two individuals were identified as outliers and were excluded from subsequent analyses (Supplementary Figure S3).
We first applied two important filtering criteria in an attempt to increase the reliability of data being analyzed at a given CpG and ultimately limit the occurrence of false-positives. First, we required high sequencing read depth per CpG per sample, filtering out loci with lower than 30-fold coverage, as the accuracy of methylation estimates at sites with lower read coverage are compromized by random sampling variation. For example, at 30-fold coverage, a single read at a given CpG accounts for 3% of the methylation estimate, in contrast to a site with 10-fold coverage at which a single read accounts for 10% of the estimate. In our data set, 758 204 autosomal CpGs met this requirement across all individuals, with an average read depth per CpG of 224 reads (min=49.5; max=585.7).
To further reduce the number of low-confidence CpGs, we also excluded sites exhibiting high levels of within group variation (range≥30%), representing approximately one quarter of sites in our data set. Individual inspection of these sites revealed characteristics of underlying genetic variation and technical noise (Supplementary Figure S4), both of which can contribute to inflated between-group differences and spurious statistical associations.
Differential methylation at the remaining 567 306 CpGs was first assessed using logistic regression. Although instances of methylation differences at single CpGs influencing changes in gene expression have been demonstrated, from a statistical perspective, given the burden of multiple testing, it can be difficult to assess the significance and reliability of variation observed at CpGs on a per site basis. To circumvent such issues, much recent work has focused on the development of methods to assess concurrent changes in methylation at neighboring CpGs (Bock, 2010). Furthermore, it has been shown that methylation levels are strongly correlated at CpGs that are closer in genomic space (Huynh et al, 2014). Thus, in this study we used a sliding-window approach (Supplementary Figure S2) to characterize a robust set of DMRs from our data set containing multiple statistically significant CpGs.
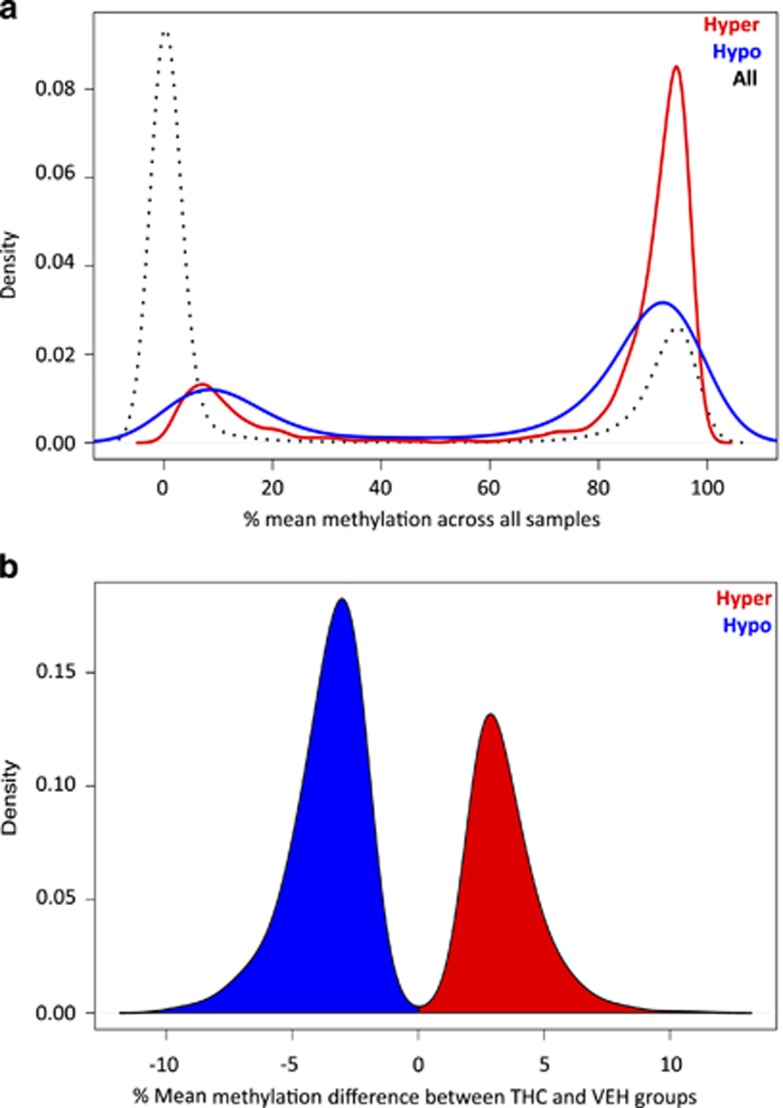
Using this method, we identified 406 hypermethylated and 621 hypomethylated DMRs in offspring with parental THC exposure, including 5611 CpGs, 3758 (66.9%) of which were independently significant by logistic regression (q<0.01; Supplementary Table S1). The distribution of population-wide methylation levels at CpGs in our data set was bimodal, and the majority of CpGs had low levels of methylation, <20% (Figure 1a). However, CpGs within DMRs were on average more highly methylated compared with the background set of CpGs, especially CpGs within hypermethylated DMRs (Figure 1a). DMR size ranged from 15 to 824 bp (mean=191.29; Supplementary Figure S5a), and the number of CpGs per DMR ranged from 3 to 37 (mean=5.46), with an average CpG density of 0.046 (Supplementary Figure S5b). Importantly, read depths for CpGs within DMRs (mean=231.28; min=61.87; max=552.40) were similar to those observed genome-wide, indicating that random error resulting from reduced read depth did not underlie the methylation differences observed at these sites.
Figure 1.
Global features of CpGs associated with cross-generational effects of THC exposure. (a) Density of CpGs plotted against the estimated mean methylation values between 0 and 100% (calculated across all THC and VEH animals) from both the background CpG list (black; n=567,306), and the hypermethylated (red) and hypomethylated (blue) DMRs; CpGs within DMRs have higher methylation levels on average. (b) Density of significant CpGs (q<0.01) from hyper- and hypomethylated DMRs (n=3,758) plotted against the difference in mean methylation values observed between THC and VEH groups, with hypermethylated changes (between-group difference>0%) shown in red, and hypomethylated changes shown in blue. DMRs, differentially methylated regions; THC, tetrahydrocannabinol; VEH, control vehicle
Overall, the methylation changes in the THC group were modest within most DMRs, with a bias toward hypomethylated changes (Figure 1b). The mean absolute change in methylation was ~3.6%, and the largest difference observed was ~11.3%. A list of the top 30 DMRs according to methylation differences are shown in Table 1. Despite the overall modest differences observed, on a per locus basis, neighboring CpGs within DMRs exhibited concordant changes in DNA methylation between groups (Figure 2). Each DMR shown in Figure 3 contains at least three significant CpGs (q<0.01), with absolute mean methylation differences ranging between 2.5% and 11.3%.
Table 1. Summary Data for top 30 DMRs Ranked by Mean Methylation Difference Between THC- and VEH-Exposed Lines.
| Position | DMR State | Size (Bp) | Number of CpGs | Significant CpGs | q valuea | Mean methylation difference | Max methylation difference | Gene |
|---|---|---|---|---|---|---|---|---|
| chr6:137806059-137806196 | Hyper | 137 | 4 | 3 (Hyper); 1 (Hypo) | < 2.2 × 10-16 | 4.82 | 11.30 | Mta1 (body) |
| chr19:53187556-53188054 | Hyper | 498 | 5 | 3 (Hyper); 1 (Hypo) | < 2.2 × 10-16 | 4.89 | 10.76 | Cdh15 (body) |
| chr3:17539229-17539721 | Hyper | 492 | 6 | 3 (Hyper); 1 (Hypo) | 1.42E-10 | 3.59 | 10.47 | Crb2 (promoter) |
| chr2:181392219-181392458 | Hypo | 239 | 9 | 6 (Hypo) | < 2.2 × 10-16 | −5.18 | −9.90 | intergenic |
| chr19:14461470-14461625 | Hypo | 155 | 4 | 4 (Hypo) | < 2.2 × 10-16 | −4.82 | −9.88 | Sez6l (body) |
| chr1:191363083-191363553 | Hyper | 470 | 3 | 3 (Hyper) | 1.49E-13 | 7.30 | 9.86 | intergenic |
| chr10:110006960-110007027 | Hypo | 67 | 3 | 3 (Hypo) | 5.22E-12 | -6.35 | -9.80 | Pcyt2 (body) |
| chr3:77115409-77115629 | Hyper | 220 | 3 | 3 (Hyper) | 7.91E-14 | 6.14 | 9.66 | intergenic |
| chr6:123810978-123811047 | Hypo | 69 | 3 | 3 (Hypo) | 8.19E-09 | −6.68 | −9.53 | intergenic |
| chr5:138731660-138731719 | Hypo | 59 | 4 | 4 (Hypo) | 4.20E-14 | −5.59 | −9.52 | Ptprf (body) |
| chr17:22355192-22355394 | Hypo | 202 | 4 | 3 (Hypo) | 4.20E-14 | −6.34 | −9.45 | intergenic |
| chr7:126694282-126694366 | Hypo | 84 | 3 | 3 (Hypo) | < 2.2 x 10-16 | −5.89 | −9.42 | intergenic |
| chr2:189652504-189652547 | Hypo | 43 | 4 | 3 (Hypo) | 6.71E-10 | −5.52 | −9.41 | intergenic |
| chr3:9850543-9850579 | Hyper | 36 | 4 | 3 (Hyper) | 1.26E-10 | 7.72 | 9.19 | RGD1311084 (body) |
| chr7:10033550-10033723 | Hypo | 173 | 4 | 3 (Hypo) | < 2.2 x 10-16 | −5.20 | −9.09 | Pias4 (body) |
| chr8:113967545-113967591 | Hypo | 46 | 3 | 3 (Hypo) | 1.17E-09 | −6.25 | −9.06 | Uqcrc1 (body) |
| chr4:175748047-175748127 | Hypo | 80 | 5 | 3 (Hypo) | 1.27E-10 | −5.04 | −8.99 | Igbp1b (promoter) |
| chr2:31418558-31418707 | Hypo | 149 | 6 | 4 (Hypo) | < 2.2 × 10-16 | −6.23 | −8.95 | Marveld2 (body) |
| chr5:142274098-142274244 | Hypo | 146 | 9 | 6 (Hypo) | < 2.2 × 10-16 | −4.56 | −8.89 | Mfsd2a (promoter) |
| chr16:24106684-24107019 | Hypo | 335 | 6 | 3 (Hypo) | < 2.2 × 10-16 | −6.32 | −8.84 | intergenic |
| chr1:84217357-84217486 | Hypo | 129 | 6 | 3 (Hypo) | < 2.2 x 10-16 | −5.69 | −8.83 | intergenic |
| chr2:240105866-240106285 | Hyper | 419 | 6 | 3 (Hyper) | < 2.2 × 10-16 | 5.05 | 8.66 | intergenic |
| chr18:35059059-35059101 | Hypo | 42 | 3 | 3 (Hypo) | < 2.2 × 10-16 | −7.24 | −8.63 | intergenic |
| chr14:87083145-87083356 | Hypo | 211 | 4 | 3 (Hypo) | 8.26E-08 | −5.64 | −8.53 | Ogdh (body) |
| chr10:89290919-89290957 | Hyper | 38 | 4 | 3 (Hyper) | 2.45E-13 | 5.84 | 8.52 | Hap1 (body) |
| chr3:118268340-118268622 | Hyper | 282 | 5 | 3 (Hyper) | 2.24E-10 | 6.43 | 8.50 | Lzts3 (promoter) |
| chr12:33604601-33604840 | Hypo | 239 | 6 | 3 (Hypo) | 3.62E-11 | −5.10 | −8.43 | Arl6ip4 (promoter) |
| chr17:11771962-11772121 | Hypo | 159 | 3 | 3 (Hypo) | 1.35E-12 | −5.69 | −8.40 | Ntrk2 (body) |
| chr3:158735564-158735714 | Hypo | 150 | 8 | 4 (Hypo); 1 (Hyper) | 2.44E-11 | −4.05 | −8.30 | intergenic |
| chr3:53850869-53850908 | Hypo | 39 | 4 | 4 (Hypo) | 1.52E-12 | −4.89 | −8.28 | intergenic |
FDR corrected P-value from logistic regression for most significant CpG in DMR.
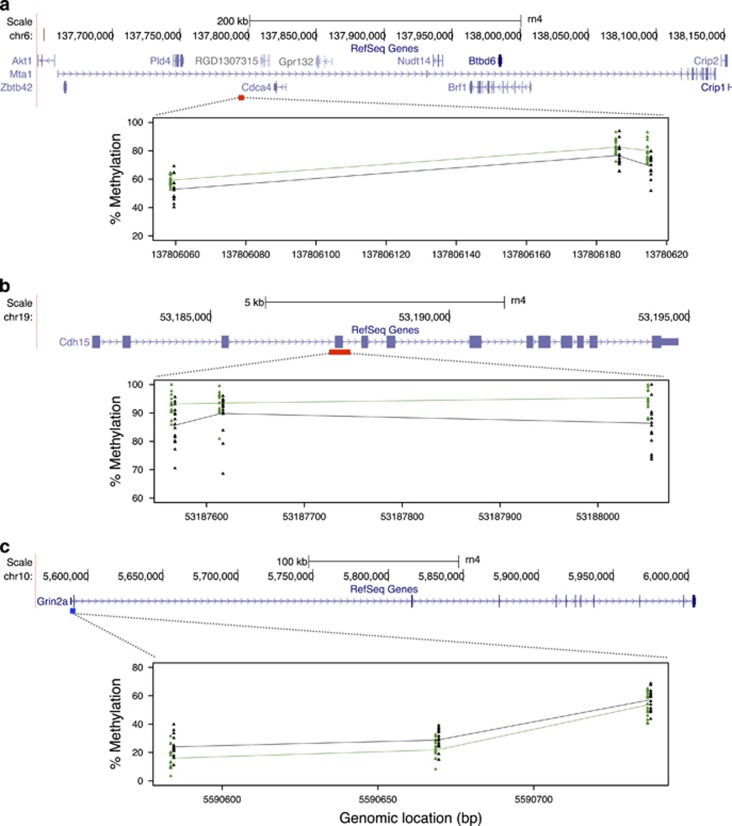
Figure 2.
Plots demonstrating consistent differences in mean methylation between THC and VEH groups across significant CpGs within: (a) a hypermethylated DMR within an intron of Mta1, showing only the three significant hypermethylated CpGs within the DMR; (b) a hypermethylated DMR identified spanning exon 3 of Cdh15, showing only the three significant hypermethylated CpGs within the region; and (c) a hypomethylated DMR residing in exon 1 of Grin2a, ~2.5 kbp downstream of the transcript start site. In each panel, a schematic of Refseq gene annotations for each loci are shown, with exons depicted as purple blocks. A scale bar and bp positions in the rat rn4 genome assembly are provided. The locations of each hyper- and hypomethylated DMRs are indicated under each gene by red and blue boxes, respectively. DMR plots are shown below each gene. Within each plot, clusters of colored triangles represent methylation values at a given CpG across all individuals (THC, green; VEH, black). The mean methylation values for THC and VEH groups are represented by solid green (THC) and black (VEH) lines, connecting each CpG position. Approximate CpG bp positions (rn4) are indicated on the x axis. DMR, differentially methylated region; THC, tetrahydrocannabinol; VEH, control vehicle.
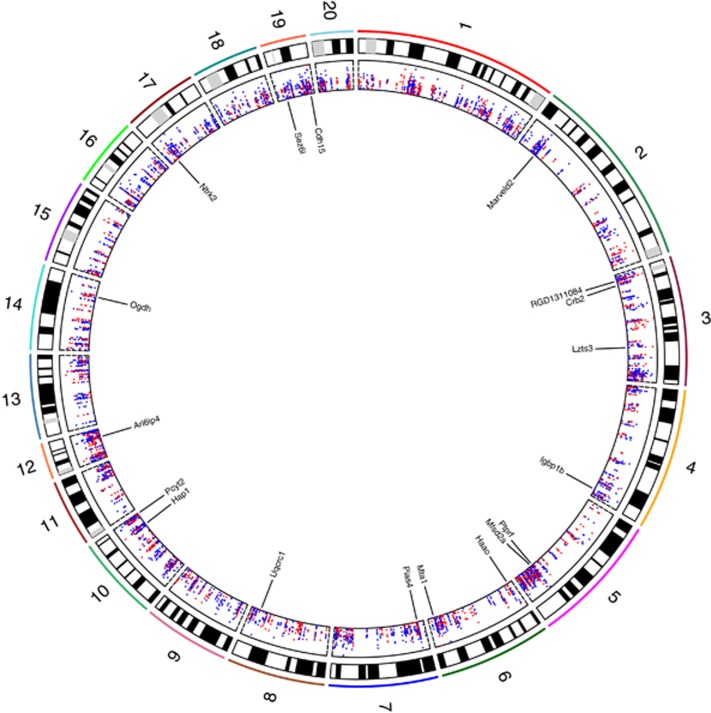
Figure 3.
A circos plot showing the positions of all CpGs (n=5611) within significant DMRs across autosomes (chr1–20). Absolute mean methylation differences between THC and VEH groups for each hypo- and hypermethylated CpG are shown in the inner most panel (scale, 0–12%) as blue and red dots, respectively. The locations and names of 18 Refseq genes overlapped by the top 30 DMRs (Table 1) are indicated. The plot was generated using Rcircos (Zhang et al, 2013). DMR, differentially methylated region; THC, tetrahydrocannabinol; VEH, control vehicle.
DMRs Are Preferentially Located in Gene Bodies, and Downstream of Transcription Start Sites
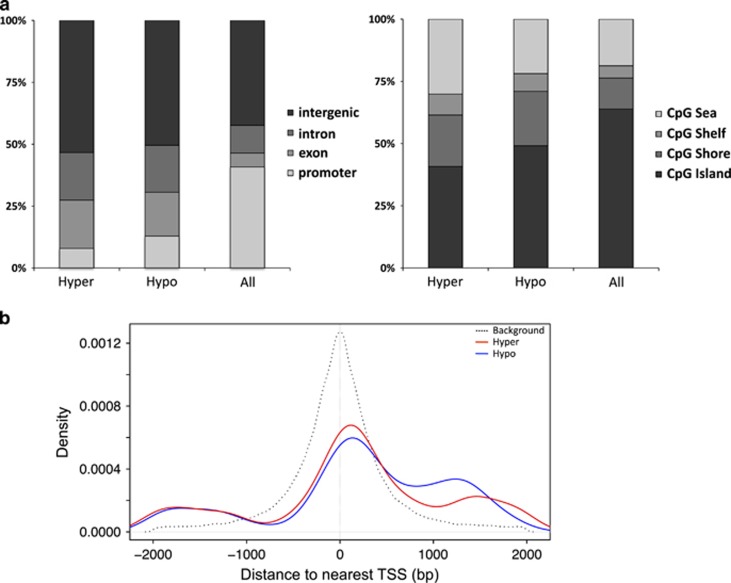
To gain potential insight into functional and mechanistic characteristics of DMRs, we assessed the overlap of DMR-CpGs with various genomic feature annotations from the rn4 genome. A genome-wide map of the DMRs identified in our study is plotted in Figure 3, including the locations of 30 genes associated with DMRs/CpGs exhibiting the largest differences in methylation between THC- and VEH-exposed groups (as per Table 1). The overlap distributions of hyper- and hypomethylated DMR-CpGs were relatively similar with respect to gene-associated annotations; however, each was significantly different from the distribution observed for all tested CpGs (‘All' P<2.2 × 10−16, both comparisons; Figure 4a). Both hyper- and hypomethylated DMR-CpGs were enriched approximately twofold in exonic and intronic regions (gene bodies) compared with background, which likely explains the bias of DMR-CpGs toward higher levels of methylation (Figure 2a)(Jones, 2012). Although we observed a reduction in gene promoter regions compared with the background CpG set (Figure 4a), promoter-associated DMR-CpGs were preferentially located downstream of the TSS compared with background promoter CpGs (Figure 4b); CpGs positioned downstream of the TSS have been shown to cause transcriptional silencing (Brenet et al, 2011). Similar to gene-related genomic features, hyper- and hypomethylated DMR-CpGs also showed different distribution patterns with respect to CpGi annotations compared with background (P<2.2 × 10−16, both comparisons; Figure 4a). Specifically, we observed a depletion of DMR-CpGs in CpGi's, with concomitant increases in the other three categories compared with background.
Figure 4.
Localization of CpGs with respect to exon/intron, promoter/TSS, and CpG island annotations in the rn4 genome. (a) The proportions of CpGs in hyper- and hypomethylated DMR (‘Hyper', ‘Hypo' n=5,611) and background sets (‘All' n=567,306) overlapping Refseq gene (left panel) and CpG island annotations (right panel). (b) Comparison of genomic distances (bp) from Refseq gene TSSs of promoter-associated CpGs within DMRs (n=612) and the background set (n=235 524), represented by red/blue and black lines; DMR-CpGs were preferentially located downstream of TSSs.DMR, differentially methylated region; TSS, transcription start site.
Imprinted genes and several genomic repeat classes are known to escape the developmental reprogramming of DNA methylation (Guibert et al, 2012). Given the potential biological relevance of these genomic features to our study question, we tested whether DMRs were preferentially associated with SINE, LINE, and LTR repeat elements, as well as imprinted loci. Neither analysis revealed significant enrichments (Supplementary Figures S6 and S7). Only six imprinted genes (Abcc9, Begain, Chst8, Dlgap2, Gata3, and Kcnq1) were present in our list of DMRs, representing 1.2% of all DMR-associated genes, which was not significantly different than that expected by chance (χ2, P>0.05).
DMRs Reside Within Genes Associated with Biological Functions Related to Neurotransmission and Synaptic Plasticity
In total, 513 DMRs (hypermethylated, 196; hypomethylated, 317) overlapped promoters or exons/introns of 492 annotated RefSeq genes. To gain insight into the broader functional roles of these DMR-associated genes, we assessed their potential enrichment in GO terms associated with biological processes, molecular functions, and cellular components; we combined genes associated with hyper- and hypomethylated DMRs to increase power in the analysis. The top 5 GO term enrichments and genes for each category are listed in Table 2 (see also Supplementary Table S2), highlighting functional involvement of DMR-associated genes in cell membrane function, animal behavior, synaptic organization, receptor activity, and also including proteins localized to cellular components of neurons and synapses. Genes contributing to GO enrichments were equally represented by hyper- and hypomethylated effects. The DMR state for each gene is indicated in Table 2. GO analysis results partitioned by hyper- and hypomethylated DMRs are also provided in Supplementary Tables S3 and S4.
Table 2. Top 5 DMR-Associated Gene GO Enrichments for Biological Processes, Molecular Functions, and Cellular Components.
| GO term | Description | P-value | Enrichment | Number of genes | Gene IDs |
|---|---|---|---|---|---|
| Biological process | |||||
| GO:0042391 | Regulation of membrane potential | 1.69E-7 | 3.02 | 28 | Cacna1a, Clic1, Grik3, Kcna5, Kcnma1, Kcnq2, Nedd4l, Nlgn2, Oprd1, Piezo1, Popdc2, Shank1, Prkce, Atp1a1, Atxn1, Gpr39, Grik5, Grin2a, Hcn3, Kcnh1, Kcnj10, Kcnn4, Kcnq1, P2rx7, Prkcz, Scn5a, Scn8a, Shank3 |
| GO:0050808 | Synapse organization | 2.22E-05 | 3.93 | 13 | Cacna1a, Lrrc4, Map1b, Nlgn2, Shank1, App, F2r, Farp1, Ntrk2, Sez6l, Sez6l2, Shank3, Unc13a |
| GO:0008344 | Adult locomotory behavior | 3.29E-05 | 3.79 | 13 | Cacna1a, Kcnma1, Nlgn2, Oprd1, Otog, Park2, App, Atxn1, Kcnj10, Npc1, Scn8a, Sez6l, Sez6l2 |
| GO:0030534 | Adult behavior | 3.57E-05 | 3.08 | 17 | Cacna1a, Kcnma1, Nlgn2, Oprd1, Otog, Park2, Shank1, App, Atxn1, Gpr39, Kcnj10, Npc1, Scn8a, Sez6l, Sez6l2, Shank3, Unc79 |
| GO:0021700 | Developmental maturation | 7.32E-05 | 3.03 | 16 | Cacna1a, Kcnma1, Map1b, Park2, Pcsk4, Shank1, Anks1a, App, Gata3, Lhx6, Ren, Runx3, Sez6l, Sez6l2, Sox10, Unc13a |
| Molecular function | |||||
| GO:0097110 | Scaffold protein binding | 1.02E-06 | 6.89 | 10 | Dlgap3, Kcna5, Shank1, Grin2a, Ikbkb, Kcnq1, Nos3, P2rx7, Scn5a, Shank3 |
| GO:0030165 | PDZ domain binding | 2.53E-04 | 3.31 | 12 | Dlgap3, Lnx2, Lphn2, Lrp2, Lzts3, Nos1ap, Park2, Sstr2, Tbc1d10a, Grik5, Lnx1, Sntb1 |
| GO:0003779 | Actin binding | 3.17E-04 | 2.25 | 22 | Coro7, Epb41l3, Kcnma1, Map1b, Myo1c, Park2, Spata32, Syne3, Actn1, Add3, Afap1, Hip1r, Myh8, Nos3, Parva, Pknox2, Plec, Prkce, Shank3, Shroom3, Sptbn1, Wipf1 |
| GO:0004872 | Receptor activity | 4.41E-04 | 1.64 | 49 | Abcc9, Bai1, Cx3cr1, Epha10, Ephb3, Gpr157, Grid1, Grik3, Itga11, Itpr3, Lphn2, Lrp2, Niacr1, Nlgn2, Nr0b2, Nrp2, Oprd1, Sema5b, Sstr2, Casr, Cckbr, Cd2, Cd5l, Chrm1, Disp1, Enpp3, Epha2, Ephb2, Esrrg, F2r, Gpr158, Gpr39, Grik5, Grin2a, Hcrtr2, Il27ra, Ildr1, Itpr2, Kcnh1, Npc1, Nr1h3, Nr4a1, Ntrk2, Olr1111, P2rx7, Paqr4, Ptprf, Robo4, Tbxa2r |
| GO:0046872 | Metal ion binding | 4.84E-04 | 1.31 | 123 | Adamts18, Adcy6, Adprm, Alpl, Bnc2, Cacna1a, Cdh15, Clstn1, Col1a2, Crb2, Cyp20a1, Efcab4a, Ikzf3, Itpr3, Kcnma1, Klf5, Lnx2, Lrp1, Lrp2, Mgmt, Mmp17, Nbr1, Nrp2, Padi2, Padi6, Park2, Pde6c, Pklr, Rbm20, Shh, Tph1, Xpnpep1, Zbtb5, Zcchc9, Zfp295, Zfp644, Actn1, Adamts2, Agap1, Agap3, App, Atp1a1, B4galt6, Bhmt2, Casr, Cblb, Cdh11, Cdh13, Cdh22, Ciz1, Col11a1, Cxxc1, Cyp2c22, Dpf3, Dpys, Efhd2, Enpp3, Esrrg, F8, Fat1, Fgd2, Fgd3, Fto, Galnt10, Gata3, Gem, Gli2, Gpd2, Grin2a, Haao, Isl2, Itpr2, Kalrn, Klf9, Lhx6, Lhx9, Lias, Lims2, Lnx1, Lpcat2, MAST1, Mta1, Necab2, Nos3, Nr1h3, Nr4a1, Ogdh, Osr2, P2rx7, Pcdh20, Pcdhga1, Pcdhga10, Pde2a, Pde6b, Pde9a, Pgm5, Pias4, Pim1, Pitpnm2, Prdm2, Prkce, Prkcz, RGD1561909, Rhbdl1, Rnf165, Rnf41, Rph3a, Scn8a, Shank3, Smoc1, Taf3, Traf3, Unc13a, Upb1, Uqcrc1, Usp21, Usp5, Zfp111, Zfp36, Zfp423, Zfp592, Zfp64, Zfp870 |
| Cellular component | |||||
| GO:0044459 | Plasma membrane part | 1.67E-07 | 1.70 | 92 | Abcc9, Adcy6, Ahnak, Cacna1a, Cdh15, Clstn1, Cx3cr1, Dlgap2, Dlgap3, Epb41l3, Epha10, Ephb3, Grid1, Grik3, Itga11, Itpr3, Kcna5, Kcnma1, Kcnq2, Lrp1, Lrp2, Lrrc4, Lzts3, Mttp, Myo1c, Nlgn2, Oprd1, Park2, Ptk7, Shank1, Slc4a8, Sstr2, Syk, Amigo2, Anks1b, Ap2s1, Apbb1, Atp1a1, C1qtnf1, Casr, Cd2, Cdh13, Chrm1, Cnnm2, Epha2, Ephb2, F2r, Farp1, Gem, Gng7, Grik5, Grin2a, Hcn3, Hcrtr2, Ikbkb, Itga8, Jak3, Kcnh1, Kcnj10, Kcnn4, Kcnq1, Ldlrap1, Marveld2, Mfsd2a, Myof, Nos3, Npc1, Ntng2, Ntrk2, P2rx7, Pde2a, Pgm5, Plec, Prkcz, Prr7, RT1-M6-2, Robo4, Scn5a, Scn8a, Scrib, Shank3, Shroom3, Sirpa, Slc4a4, Slc6a13, Sptbn1, Srcin1, Tbc1d5, Tmem123, Traf3, Unc13a, Vwf |
| GO:0045211 | Postsynaptic membrane | 2.13E-07 | 3.43 | 23 | Clstn1, Dlgap2, Dlgap3, Grid1, Grik3, Kcnma1, Lrrc4, Lzts3, Nlgn2, Oprd1, Park2, Shank1, Anks1b, Apbb1, Chrm1, F2r, Grik5, Grin2a, Ntrk2, Prr7, Scrib, Shank3, Srcin1 |
| GO:0097458 | Neuron part | 2.64E-07 | 1.83 | 72 | Begain, Cacna1a, Camk2b, Cx3cr1, Dlgap2, Dlgap3, Epb41l3, Grid1, Grik3, Hap1, Itpr3, Kcnma1, Kcnn1, Kcnq2, Lphn2, Lrp1, Lrrc4, Lzts3, Map1b, Nlgn2, Nrp2, Oprd1, Park2, Shank1, Shh, Slc4a8, Sstr2, Tph1, Aatk, Actn1, Add3, Anks1a, Anks1b, Apbb1, App, Atp1a1, Casr, Cdh13, Chrm1, Dpf3, Ephb2, Farp1, Grik5, Grin2a, Gtf2i, Itga8, Itpr2, Kalrn, Kcnn4, Kif5c, Ldlrap1, Nov, Ntng2, Ntrk2, P2rx7, Pde2a, Pde6b, Pde9a, Prkcz, Ptprf, Rph3a, Samd4a, Scn8a, Scrib, Sez6l, Sez6l2, Shank3, Smarcd1, Sptbn1, Srcin1, Ston2, Unc13a |
| GO:0030054 | Cell junction | 5.99E-08 | 1.88 | 64 | Ahnak, Cd151, Cdh13, Clstn1, Dlgap2, Dlgap3, Epb41l3, Grid1, Grik3, Hap1, Ifi30, Itga11, Kcna5, Lcp2, Lrp1, Lrrc4, Lzts3, Map1b, Nlgn2, Park2, Rfc1, Shank1, Svop, Actn1, Add3, Afap1, Anks1b, App, Atp1a1, Ccnd1, Cd2, Chrm1, Clmp, Epha2, Farp1, Fat1, Grik5, Grin2a, Irf2, Itga8, Kazn, Lims2, Marveld2, Misp, Myh8, Nphp4, P2rx7, Pard6b, Parva, Pgm5, Plec, Prkcz, Prr7, Ptk7, Rph3a, Samd4a, Scn5a, Scrib, Shank3, Shroom3, Sntb1, Srcin1, Ston2, Unc13a, Ywhab |
| GO:0097060 | Synaptic membrane | 6.27E-07 | 3.05 | 25 | Clstn1, Dlgap2, Dlgap3, Grid1, Grik3, Kcnma1, Lrrc4, Lzts3, Nlgn2, Oprd1, Park2, Shank1, Anks1b, Apbb1, Chrm1, F2r, Grik5, Grin2a, Ntrk2, Pde2a, Prr7, Scrib, Shank3, Srcin1, Unc13a |
Bold, genes in Table 1; Underlined, hypomethylated genes
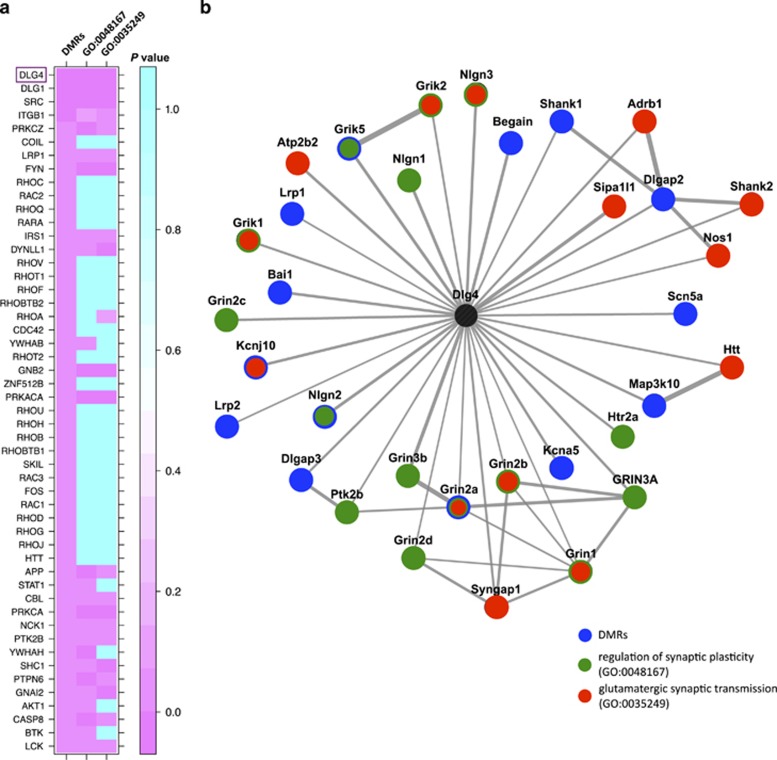
Building on the GO analysis, we further explored the relationship of DMR-associated genes and additional gene sets of interest associated with biological processes that we would expect to be altered in this model. Specifically, it is well established and is in line with our previous observations that glutamatergic-related genes such as glutamate receptors (GluRs) play important roles in mediating synaptic plasticity and transmission, with impacts on behavior including those involved in addiction (Szutorisz et al, 2014). For example, we have previously shown that F1 adult rats with parental THC exposure show electrophysiological impairments related to dysregulation of synaptic plasticity (Szutorisz et al, 2014). Consistent with these phenotypes, many DMRs overlapped genes encoding regulators of synaptic plasticity, including GluRs and kainite receptors (Grin2a, Grik3, and Grik5), G-protein-coupled receptors (GPCRs; Gpr39, Gpr157, and Gpr158), pre- and postsynaptic ion channels (Cacna1a, Kcna5, Kcnma1, Kcnq2, Kcnh1, Kcnn1, Kcnm, Kcnj10, Kcnn4, Kcnq1, Hcn3, Scn5a, and Scn8a) and scaffolding proteins (Table 2). Thus, to more directly explore the relationship between THC-induced DNA methylation changes and genes involved in the glutamatergic system and synaptic transmission, we used PPI databases (Lachmann and Ma'ayan, 2010) to assess the potential sharing of annotated mammalian PPIhubs by DMR genes and genes involved in the ‘regulation of synaptic plasticity' (n=146; GO:0048167) and ‘glutamatergic synaptic transmission' (n=79; GO:0035249). We found that physical protein–protein interaction networks from DMR genes were enriched for many of the same PPIhubs connected to synaptic plasticity/transmission gene sets (Figure 5b; Supplementary Table S5). For example, of the top 10 most significantly enriched PPIhubs, 7 were also significantly enriched for GO:0048167 and GO:0035249 gene sets (P<0.05). The most enriched hub for DMR-associated proteins, Dlg4, was also either the most significant or second most significant PPIhub in the other two gene set networks. Dlg4 encodes postsynaptic density protein 95 (Psd-95). The physical interaction network of Dlg4-associated genes from the three gene sets analyzed (n=35 genes) is shown in Figure 5b; in total there were 14 genes associated with THC-DMRs (9 hypermethylated; 5 hypomethylated) in the Dlg4 network.
Figure 5.
DMR-associated genes and genes involved in synaptic plasticity and synaptic transmission interact with common PPIhubs. (a) Heatmap showing Fisher's exact test P-values from PPIhub enrichment analyses (calculated by Lists2Networks (Lachmann and Ma'ayan, 2010)) using three gene sets: DMR-associated genes (n=492), and genes in GO categories GO:0048167 (‘regulation of synaptic plasticity' n=146) and GO:0035249 (‘glutamatergic synaptic transmission' n=79). Only PPIhubs for which a significant enrichment (P<0.05) was found for the DMR gene set are shown. (b) Protein interaction network including proteins corresponding to genes from DMR, GO:0048167, and GO:0035249 sets that interact with Dlg4, based on known interactions from rat, mouse, and human interaction databases (Berger et al, 2007; Lachmann and Ma'ayan, 2010). Proteins are colored according to whether they belong to one or more of the three gene sets. DMR, differentially methylated region.
DMR-Associated Genes in the Dlg4 Network Exhibit Differential mRNA Expression in the NAc Adult F1 Offspring from THC-Exposed Lines
We had previously found expression of Grin2a to be associated with the cross-generational effects of THC exposure in the NAc of a cohort of F1 adult male rats (Szutorisz et al, 2014). In the present study, we identified a significant hypomethylated DMR located within the first coding exon of Grin2a (Figure 2c). Reanalysis of an expanded male/female cohort revealed significant downregulation of Grin2a mRNA levels (P=0.049; Table 3), consistent with our previous findings, and suggestive of a possible link between observed methylation differences and gene expression. To further explore potential effects of differential methylation on gene transcript levels, we conducted mRNA expression analysis for Dlg4 and eight additional DMR-associated genes in the Dlg4 protein–protein interaction network, as well as three genes overlapped by top DMRs presented in Table 1 (Table 3). In total, 5 out of 10 genes in the Dlg4 network (Dlgap2, Kcna5, Begain, Grin2a, and Dlg4) showed significant differential expression between THC and VEH groups (P<0.05; Table 3). Of the three genes associated with top DMRs that were tested, Mta1 tended to have higher expression in THC animals, though this difference was not statistically significant (P=0.057; Table 3). These results indicate effects of parental germline THC exposure on the epigenetic state of gene loci related to synaptic plasticity and the glutamatergic pathway within the NAc of adult offspring.
Table 3. Analysis of mRNA Expression of DMR-Associated Genes.
| Gene | THC-associated expression change | % of VEH mean (THC) | ±SEM (THC) | P-valuec | DMR state | DMR location |
|---|---|---|---|---|---|---|
| Dlg4a | Upregulated | 113 | 0.045 | 0.044 | NA | NA |
| Dlgap2a | Upregulated | 115 | 0.038 | 0.015 | Hyper | Gene body; intron |
| Kcna5a | Upregulated | 121 | 0.06 | 0.017 | Hyper | Promoter region; exon |
| Begaina | Upregulated | 113 | 0.049 | 0.044 | Hyper | Gene body; exon |
| Grin2aa | Downregulated | 76 | 0.086 | 0.049 | Hypo | Gene body; exon |
| Dlgap3a | NS | 112 | 0.06 | 0.136 | Hyper | Gene body; exon |
| Scn5aa | NS | 85 | 0.069 | 0.136 | Hypo | Gene body; intron |
| Grik5a | NS | 108 | 0.037 | 0.179 | Hypo | Gene body; intron |
| Kcnj10a | NS | 106 | 0.027 | 0.208 | Hypo | Gene body; exon |
| Shank1a | NS | 94 | 0.058 | 0.522 | Hyper | Gene body; exon |
| Mta1b | NS | 121 | 0.042 | 0.057 | Hyper | Gene body; intron |
| Cdh15b | NS | 110 | 0.039 | 0.164 | Hyper | Gene body; exon |
| Ntrk2b | NS | 105 | 0.028 | 0.338 | Hypo | Gene body; intron |
Discussion
There is increasing interest in exploring the potential role of cross-generational epigenetic effects in mediating the risk of complex phenotypes, including those related to drug addiction and other neuropsychiatric conditions. The results of this study demonstrate that THC exposure before mating can elicit molecular disturbances leading to lasting cross-generational DNA methylation alterations in the NAc of adult, unexposed progeny. These DNA methylation changes were localized to clusters of CpGs occurring at 406 hypermethylated and 621 hypomethylated DMRs across the genome. Importantly, our use of stringent filtering criteria and a sliding-window-based analysis for DMR discovery limited the inclusion of false-positive CpGs. Thus, although on average the observed methylation differences were modest, these sites likely represent robust epigenetic changes given that we required multiple neighboring CpGs within DMRs to exhibit concordant methylation patterns (Table 1; Figure 2). Furthermore, the magnitude of observed methylation differences are likely consistent with those expected from a model of epigenetic cross-generational transmission, and are comparable to what has been observed by epigenome-wide association studies in complex traits (De Jager PL, 2014; Huynh et al, 2014; Pidsley et al, 2014). For example, alterations in DNA methylation at the human IGF2 locus have been associated with periconceptional famine and shown to persist into adulthood, with levels differing by only ~5% on average between famine-exposed and unexposed individuals (Heijmans et al, 2008).
It is also important to note that even subtle methylation differences between individuals have been associated with variation in mRNA and protein levels (Bakulski et al., 2012; Huynh et al, 2014). From the data presented here, however, it is not yet clear what molecular effects are associated with the identified DMRs. Building on expression analysis from our previous publication (Szutorisz et al, 2014), we showed that several DMR-associated genes tested were also differentially expressed between THC and VEH groups. It is interesting to note that three genes containing hypermethylated DMRs showed mRNA overexpression in the cohort examined (Dlgap, Begain, and Kcna5), whereas one gene (Grin2a) was associated with both hypomethylation and decreased expression status; in all four cases, DMRs were located within gene bodies, in which effects of methylation are less clearly defined (Deaton and Bird, 2011). Large-scale assessment of the broader mechanistic relationships between DNA methylation variation within THC-associated DMRs and gene regulation will be the subject of future analyses.
Although not directly tested, alternative effects of THC-associated DMRs on gene regulation are also likely. Notably, we observed a significant enrichment of DMRs in gene bodies, which, in addition to potential roles for regulating mRNA levels, could be suggestive of effects on gene splicing and isoform-specific regulation. There is also evidence that gene body methylation, particularly in CpGi's, could highlight secondary sites of transcription initiation (Deaton and Bird, 2011; Jones, 2012); in this context, ~47% of DMR-CpGs identified here within gene bodies also resided within CpGi's. In addition, it is important to consider that observed methylation signatures could mediate molecular processes in a more complex manner, for example, only in response to certain stimuli, in conjunction with regulatory proteins, or in a time-dependent manner at specific developmental stages. For example, a recent genome-wide DNA methylation study in schizophrenia showed that disease-associated changes were preferentially located at CpGs that exhibit dynamic variation during fetal development, suggesting that disease-associated methylation disturbances may exert effects during earlier neurodevelopmental periods prior to disease onset (Pidsley et al, 2014).
An additional outstanding question in the field concerns the mechanisms underlying the establishment and transmission of epigenetic marks across generations. Although it has been demonstrated that the reprogramming of DNA methylation occurs genome wide during ontogeny, some select loci including imprinted genes and classes of repeat/transposable elements are known to escape this process in part, suggesting that transgenerational epigenetic inheritance could be localized to these regions of the genome (Hackett et al, 2013; Lane et al, 2003). However, few epigenome-wide data sets of DMRs associated with models of cross-generational epigenetic inheritance have been generated and interrogated in the context of this idea. Interestingly, the cadherin gene Cdh15, containing a DMR exhibiting the second largest methylation difference in our data set, is a candidate imprinted gene in mice (Proudhon et al, 2012), as are two DMR-associated genes in the Dlg4 network exhibiting differential expression between THC and VEH groups (Begain and Dlgap2). Epigenetic changes of Dlgap2 (Chertkow-Deutsher et al, 2010) and Begain (Nagy et al, 2015) have already been implicated in neuropsychiatric phenotypes. However, a comparison of our gene set with a more comprehensive list of putative imprinted genes in rat, mouse, and human did not reveal enrichment for such loci. Nor did we observe evidence for preferential overlap of DMRs with annotated repeat elements in the rn4 genome, suggesting that alternate mechanisms likely underlie the maintenance of persistent methylation signatures in our identified DMRs.
Most intriguing from our analysis is the significant enrichments of DMRs within loci involved in a range of GO terms including genes localized to neuronal cellular components with prominent roles in the regulation of development, synaptic plasticity, and neurotransmission (Table 2; Figure 5). Importantly, many of these genes have functional relevance to behavioral and physiological traits we previously demonstrated to be impaired in this rat model (Szutorisz et al, 2014). This included striatal molecular disturbances in adult F1 progeny with parental THC exposure that affected several glutamate receptor subunits at both the mRNA and protein levels, and associated electrophysiological impairments related to dysregulation of synaptic plasticity (Szutorisz et al, 2014). Drawing potential links between DMRs and gene networks suspected to underlie phenotypes in this model, we showed that DMR-associated genes shared many common PPIhub proteins with gene sets specifically known to regulate synaptic plasticity and glutamatergic synaptic transmission (Figure 5). The most significant of these PPIhub enrichments involved Dlg4, encoding Psd-95, a membrane associated guanylate kinase scaffolding protein located in neural postsynaptic densities, involved in the regulation of dopamine–glutamate interactions. It associates with the NMDA subtype of GluR's and is required for synaptic plasticity associated with NMDA receptor signaling. Interestingly, it has recently been shown that epigenetic dysregulation of Dlg4 contributed to abnormal glutamatergic transmission and rewarding behavior induced by morphine conditioning (Wang et al, 2014), consistent with our observations of increased heroin self-administration in adult F1s in the model investigated here (Szutorisz et al, 2014). Although we did not identify a DMR within Dlg4 meeting our stringent criteria, we found that Dlg4 mRNA levels were statistically higher in F1 offspring of THC-exposed parents. In addition, four other DMR-associated genes in this interaction network were also differentially expressed, highlighting the importance of this pathway in our model.
In summary, this study provides the first set of data on cross-generational epigenomic alterations associated with THC exposure in the rat NAc, including DMRs localized to genes with important roles in neural function, complex psychiatric diseases, and addiction-related traits. Future work will require more rigorous comparisons between epigenomic and transcriptome alterations in order to address the mechanistic implications of these findings. The knowledge emerging from such investigations will likely contribute to the understanding of the heritability of epigenetic marks in relation to the consequences of parental THC exposure.
Funding And Disclosure
Our research was supported by NIH grants DA030359 and DA033660. The authors declare no conflict of interest. The authors declare that over the past three years AJS has received compensation from Guidepoint Global LLC.
Acknowledgments
We thank James Sperry, Jenna M. Carter, and Nayana D. Patel for technical help. ERRBS library preparation, sequencing, and data acquisition was carried out with the assistance of Alicia Alonso at the Weill Cornell Genomics Core Facility (New York, NY).
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary Material
References
- Akalin A, Garrett-Bakelman FE, Kormaksson M, Busuttil J, Zhang L, Khrebtukova I et al (2012. a). Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genet 8: e1002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A et al (2012. b). methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol 13: R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerman S (2014). Marijuana. Adolesc Med State Art Rev 25: 70–88. [PubMed] [Google Scholar]
- Bakulski KMDD, Sartor MA, Paulson HL, Konen JR, Lieberman AP, Albin RL et al (2012). Genome-wide DNA methylation differences between late-onset Alzheimer's disease and cognitively normal controls in human frontal cortex. J Alzheimers Dis 29: 571–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker C, Weigel D (2012). Epigenetic variation: origin and transgenerational inheritance. Curr Opin Plant Biol 15: 562–567. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300. [Google Scholar]
- Berger S, Posner J, Ma'ayan A (2007). Genes2Networks: connecting lists of gene symbols using mammalian protein interactions databases. BMC Bioinformatics 8: 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C (2010). Analysing and interpreting DNA methylation data. Nat Rev Genet 13: 705–719. [DOI] [PubMed] [Google Scholar]
- Brenet F, Moh M, Funk P, Feierstein E, Viale A, Socci N et al (2011). DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One 6: e14524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H, Bergman Y (2009). Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 10: 295–304. [DOI] [PubMed] [Google Scholar]
- Chertkow-Deutsher Y, Cohen H, Klein E, Ben-Shachar D (2010). DNA methylation in vulnerability to post-traumatic stress in rats: evidence for the role of the post-synaptic density protein Dlgap2. Int J Neuropsychopharmacol 13: 347–359. [DOI] [PubMed] [Google Scholar]
- De Jager PLSG, Lunnon K, Burgess J, Schalkwyk LC, Yu L, Eaton ML et al (2014). Alzheimer's disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci 17: 1156–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton A, Bird A (2011). CpG islands and the regulation of transcription. Genes Dev 25: 1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinieri JA, Hurd YL (2012). Rat models of prenatal and adolescent Cannabis exposure. Methods Mol Biol 829: 231–242. [DOI] [PubMed] [Google Scholar]
- Eden E, Lipson D, Yogev S, Yakhini Z (2007). Discovering motifs in ranked lists of DNA sequences. PLoS Comput Biol 3: e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z (2009). GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Nestler EJ (2013). Epigenetic mechanisms of drug addiction. Curr Opin Neurobiol 23: 521–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M (1987). CpG islands in vertebrate genomes. J Mol Biol 196: 261–282. [DOI] [PubMed] [Google Scholar]
- Guibert S, Forne T, Weber M (2012). Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res 22: 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Arcelus M, Ongen H, Lappalainen T, Montgomery SB, Buil A, Yurovsky A et al (2015). Tissue-specific effects of genetic and epigenetic variation on gene regulation and splicing. PLoS Genet 11: e1004958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA et al (2013). Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339: 448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Martienssen RA (2014). Transgenerational epigenetic inheritance: myths and mechanisms. Cell 157: 95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES et al (2008). Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA 105: 17046–17049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfer C (2014). Implications of Marijuana Legalization for Adolescent Substance Use. Subst Abus 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Wan J, Su Y, Song Q, Zeng Y, Nguyen HN et al (2013). DNA methylation presents distinct binding sites for human transcription factors. Elife 2: e00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd YL, Michaelides M, Miller ML, Jutras-Aswad D (2014). Trajectory of adolescent Cannabis use on addiction vulnerability. Neuropharmacology 76 Pt B: 416–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh JL, Garg P, Thin TH, Yoo S, Dutta R, Trapp BD et al (2014). Epigenome-wide differences in pathology-free regions of multiple sclerosis-affected brains. Nat Neurosci 17: 121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LD, O'Malley PM, Miech RA, Bachman JG, Schulenberg JE (2015) Monitoring the Future national survey results on drug use: 1975-2014: Overview, key findings on adolescent drug use. Institute for Social Research, The University of Michigan: Ann Arbor. [Google Scholar]
- Jones P (2012). Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13: 484–492. [DOI] [PubMed] [Google Scholar]
- Jurka J (2000). Repbase Update: a database and an electronic journal of repetitive elements. Trends Genet 16: 418–420. [DOI] [PubMed] [Google Scholar]
- Kent WJSC, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D (2002). The human genome browser at UCSC. Genome Res 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Volkow ND (2010). Neurocircuitry of addiction. Neuropsychopharmacology 35: 217–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applicationsBioinformatics 27: 1571–1572. [DOI] [PMC free article] [PubMed]
- Lachmann A, Ma'ayan A (2010). Lists2Networks: integrated analysis of gene/protein lists. BMC Bioinformatics 11: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J et al (2003). Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 35: 88–93. [DOI] [PubMed] [Google Scholar]
- Lange U, Schneider R (2010). What an epigenome remembers. Bioessays 32: 659–668. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Mathur BN, Lovinger DM (2012). Endocannabinoid-dopamine interactions in striatal synaptic plasticity. Front Pharmacol 3: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy C, Suderman M, Yang J, Szyf M, Mechawar N, Ernst C et al (2015). Astrocytic abnormalities and global DNA methylation patterns in depression and suicide. Mol Psychiatry 20: 320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidsley R, Viana J, Hannon E, Spiers HH, Troakes C, Al-Saraj S et al (2014). Methylomic profiling of human brain tissue supports a neurodevelopmental origin for schizophrenia. Genome Biol 15: 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudhon C, Duffie R, Ajjan S, Cowley M, Iranzo J, Carbajosa G et al (2012). Protection against de novo methylation is instrumental in maintaining parent-of-origin methylation inherited from the gametes. Mol Cell 47: 909–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2011). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAMHSA (2011) Results from the 2010 National Survey on Drug Use and Health: National Findings Office of Applied Studies, NSDUH Series H-41, HHS Publication No. (SMA) 11–4658, Rockville: MD, USA: MD, USA. [Google Scholar]
- Schmitt A, Malchow B, Hasan A, Falkai P (2014). The impact of environmental factors in severe psychiatric disorders. Front Neurosci 8: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P (1996–2010) RepeatMasker Open 3: 0.
- Spiers H, Hannon E, Schalkwyk LC, Smith R, Wong CC, O'Donovan MC et al (2015). Methylomic trajectories across human fetal brain development. Genome Res 25: 338–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szutorisz H, DiNieri JA, Sweet E, Egervari G, Michaelides M, Carter JM et al (2014). Parental THC exposure leads to compulsive heroin-seeking and altered striatal synaptic plasticity in the subsequent generation. Neuropsychopharmacology 39: 1315–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuesta LM, Zhang Y (2014). Mechanisms of epigenetic memory and addiction. EMBO J 33: 1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassoler FM, Sadri-Vakili G (2014). Mechanisms of transgenerational inheritance of addictive-like behaviors. Neuroscience 264: 198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Yan P, Hui T, Zhang J (2014). Epigenetic upregulation of PSD-95 contributes to the rewarding behavior by morphine conditioning. Eur J Pharmacol 732: 123–129. [DOI] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P et al (2010). The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38: W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson ME, Sengoku T (2013). Developmental regulation of neuronal genes by DNA methylation: environmental influences. Int J Dev Neurosci 31: 448–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Meltzer P, Davis S (2013). RCircos: an R package for Circos 2D track plots. BMC Bioinformatics 14: 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.