Abstract
Background: Research studies exploring the determinants of disease require sufficient statistical power to detect meaningful effects. Sample size is often increased through centralized pooling of disparately located datasets, though ethical, privacy and data ownership issues can often hamper this process. Methods that facilitate the sharing of research data that are sympathetic with these issues and which allow flexible and detailed statistical analyses are therefore in critical need. We have created a software platform for the Virtual Pooling and Analysis of Research data (ViPAR), which employs free and open source methods to provide researchers with a web-based platform to analyse datasets housed in disparate locations.
Methods: Database federation permits controlled access to remotely located datasets from a central location. The Secure Shell protocol allows data to be securely exchanged between devices over an insecure network. ViPAR combines these free technologies into a solution that facilitates ‘virtual pooling’ where data can be temporarily pooled into computer memory and made available for analysis without the need for permanent central storage.
Results: Within the ViPAR infrastructure, remote sites manage their own harmonized research dataset in a database hosted at their site, while a central server hosts the data federation component and a secure analysis portal. When an analysis is initiated, requested data are retrieved from each remote site and virtually pooled at the central site. The data are then analysed by statistical software and, on completion, results of the analysis are returned to the user and the virtually pooled data are removed from memory.
Conclusions: ViPAR is a secure, flexible and powerful analysis platform built on open source technology that is currently in use by large international consortia, and is made publicly available at [ http://bioinformatics.childhealthresearch.org.au/software/vipar/ ].
Keywords: ViPAR, data sharing, data federation, data pooling
Key Messages
ViPAR provides a solution for analysing disparately located datasets, without the need for permanent storage of all data in one location.
ViPAR facilitates and encourages collaborations by providing an easy-to-use, yet sophisticated, technology platform for data sharing to work from.
ViPAR enables all standard analyses (pre-configured for R, SAS, Stata) to be conducted on the federated data, without the user needing to have access to individual-level data.
ViPAR provides a platform for centralized data management (including data dictionary creation) and analyses for consortia, whereas data contributors maintain control of their own datasets remotely.
Introduction
The notion of sharing data from different scientific studies and experiments is one of the pillars of the modern scientific method. The complexity surrounding sharing data has been highlighted in numerous recent commentaries, 1–3 with particular focus on the need for the development of methods to facilitate the sharing of data in a secure manner that is sympathetic to privacy, ethical and other constraints that may exist.
To harness the power of results from multiple studies or data sources, statistical techniques such as meta-analyses 4 are commonly used as the only mechanism available to combine data from research studies where the presence of ethical, legal, privacy or technical limitations prevents the sharing of individual-level data.
The transfer of data between study sites is typically conducted using either physical media (e.g. CDs), electronic media (e.g. e-mail) or newer ‘cloud’ technologies (e.g. DropBox). However, many of these methods do not satisfy privacy, ethical and legal restrictions nor data security concerns surrounding the transfer, storage, location and analysis of the study data. Database federation techniques offer a viable solution to this problem by permitting controlled access to datasets located and managed in disparate locations without the need for permanent storage at a single location. 5,6 In this scenario, each study site retains control of their own data in separate databases at their respective site. A central analysis site (which may or may not be located at one of the study data sites) hosts an informatics platform that contains no study data itself but is able to connect to, view and interrogate the data held in each of the separate sites as if the data existed at the central site. When an analysis is to be conducted, data are retrieved from each study site and temporarily pooled at the central site until completion of the analysis, after which they are deleted. Although ethical consent and strong collaboration are still essential for this method to succeed, the ability to maintain local control of study data, and the absence of permanency in pooling cross-site data for analysis, create an appealing alternative to meta-analysis and similar summary techniques.
Navigating the complexities of ethical approval for access to data from multiple sources is only one part of the problem. In order to effectively analyse data under a federated model it is essential that it be harmonized appropriately. 7 In the simplest case this can be achieved in a prospective manner where data are collected in the same way across all sites following a defined protocol. The situation becomes more complicated, however, when data are harmonized retrospectively. This requires a more coordinated approach where overlapping variables across sites need to be identified and methods may need to be developed to resolve and harmonize problematic variables such as those with incompatible categories, varying metrics or different methods of measurement. Approaches exist to facilitate the process of data harmonization such as the DataSHaPER platform. 7,8
Here we present a software platform for the Virtual Pooling and Analysis of Research data, referred to as ViPAR. The platform is based on data federation and represents a secure, flexible solution for the management and analysis of harmonized research data stored in disparate locations, while preserving and respecting privacy and other restrictions on the source data. We describe the data model and technical features, and compare and contrast the ViPAR system with two similar technologies. We also demonstrate the utility of ViPAR as part of a multi-site international research consortium. ViPAR is made freely available to the research community and is available for download at the project’s homepage [ http://bioinformatics.childhealthresearch.org.au/software/vipar/ ], with the source code available at [ https://gitlab.com/kim.carter/ViPAR ].
Methods
Motivation
The development of ViPAR stems from our involvement with the International Collaboration for Autism Registry Epidemiology (iCARE—see Use Case section in Supplementary Methods , available as Supplementary data at IJE online). A key requirement of this project was to design a system that facilitated the combined analysis of large datasets from six international sites. Importantly, such a system needed to ensure that all sites retained management and ownership of their data. It was quickly apparent that access to these data involved complex ethical and legal limitations; however, all sites were able to obtain ethical approval for ‘virtual pooling’ where data from a remote site could exist temporarily at another site for the purposes of an analysis. In addition to creating a system to implement the virtual pooling concept, we also needed to accommodate the variety of statistical software in use by iCARE analysts, incorporate features that fostered the collaborative nature of the project and also ensure that the cost of implementation was minimized. No existing system fulfilled all these requirements.
Federated platform design
The ‘hub-and-spoke’ design is one of the most popular topologies for data warehousing and enterprise integration in modern computer systems. 9,10 In a traditional hub-and-spoke system, the central ‘hub’ is the main data storage location, with the communication and flow of data moving down the ‘spoke’ from systems at invariably remote sites, permanently to the hub. This is akin to how many researchers and consortia analyse datasets from disparate locations, namely by merging and transferring separate datasets into a single master dataset—assuming that arrangements (e.g. memorandums of understanding and ethics approvals) are in place to even allow data to be housed at a central location. Prior to any data being loaded into systems that are based on such a model, a coordinated approach should be performed to create a data dictionary that defines common and overlapping data from all contributing sites. In addition, any derived variables and their standardized unit measures and metrics should be specified, along with how missing data should be represented. This type of process is commonplace within multi-site consortia, and typically involves surveying the data at each site, developing the data dictionary to which the data is harmonized and then applying quality control testing to ensure that the harmonized data are still a true representation of the source data.
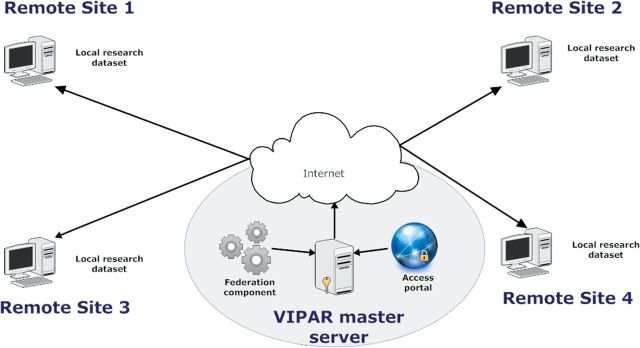
Database federation techniques provide centralized, transparent access to datasets solely located and managed in remote locations, without the need for permanent pooling at a single location. 5,6 ViPAR has been designed and built with these data federation principles in mind, where remote sites permanently house and manage their own harmonized datasets (see Supplementary material , available as Supplementary data at IJE online), and a centralized access portal and federation component provide transparent access to all of the sites and enable virtual pooling of the data therein. The federated design topology for ViPAR is illustrated in Figure 1 , and is described in detail in the following sections and in the Supplementary material .
Figure 1.
ViPAR topology. A typical multi-site ViPAR configuration where a ViPAR master server (VMS) is linked to a number of remote sites. Each remote site stores and maintains their research data. Users of the ViPAR system access the web-based analytical portal where they can initiate analyses. During an analysis, the federation component retrieves data from the remote sites into RAM on the VMS where they are analysed and removed without ever permanently being stored.
The ViPAR data model
ViPAR has been designed to allow researchers to securely and flexibly bring research data together, in a way that allows individual sites to retain control of data while also providing a mechanism for virtual (non-permanent) pooling with data from other sites. The database model is illustrated in Supplementary Figure 1 and described in more detail in the Supplementary materials (available as Supplementary data at IJE online). In brief, the model comprises components for secure data storage by site, secure data access from a set of data-contributing sites by authorized users and a web-based environment to administer analytical projects and to perform statistical data analyses. Access to the pooled study data is in the form of analytical projects, where data for subsets of variables from a defined data dictionary are made available for analysis to a set of researchers within the context of a specific research question. The results of analyses are available within the web-based environment and can be optionally shared with other researchers.
Results
Summary of the ViPAR federated infrastructure and its components
The hardware and software technologies underlying ViPAR are described in detail in the Supplementary materials (available as Supplementary data at IJE online) and summarized in Table 1 . The structure of a typical ViPAR setup is illustrated in Figure 1 . Each data-contributing site houses a local ViPAR database (LVD) on a physical or virtual server located at the site itself. Each LVD runs a MySQL database server for data storage and an SSH server for secure, encrypted data communication to the master site. A central analysis site houses the ViPAR master server (VMS), which hosts two key components of the infrastructure, namely the ViPAR daemon and the ViPAR web-based analysis portal (VWAP). The ViPAR daemon ( Box 1 ) is responsible for logging, controlling access to statistical packages, and encompasses the data federation component that governs the integration and virtual pooling of site data connected to each LVD using a secure SSH connection. The VWAP is the interface through which analyses, data management and other research and administrative activities are conducted. Details on how to define and prepare a data dictionary for ViPAR, along with details on how to load data into the system, are provided in the Supplementary material and are described in detail in the ViPAR manual available from the project website.
Table 1.
Summary of hardware and software requirements for the key ViPAR server components
| ViPAR Master Server (VMS) | Local ViPAR Database (LVD) Server | |
|---|---|---|
| Hardware | Physical or virtual server with at least two CPU cores, 8GB of RAM and 50GB of disk space | Physical or virtual server with at least 1 CPU core, 1GB of RAM and 5 GB of disk space |
| Software (pre-installed) | Perl 5.10 or greater, OpenSSH 5.4 or greater, MySQL 5.5 or greater, Apache webserver, R statistical software | OpenSSH 5.4 or greater, MySQL 5.5 or greater |
| Software (optional) | OpenSSL server-side certificates, SAS, STATA, denyhosts | denyhosts |
Box 1. Glossary of informatics terms used throughout the main text
SSH : Secure Shell, enables encrypted transfer of information between otherwise insecure platforms.
Daemon : a program that runs in the background and manages tasks such as logging and handling data.
MySQL Database : a popular platform used to manage the efficient storage and retrieval of data.
SSL Certification : a standard technology used to securely identify and encrypt data flow between two entities (e.g. one computer or individual to another computer).
FIFO : a method to allow two programs to temporarily communicate with each other without the need for an intermediary file.
Database Federation : allows remotely housed datasets to be transparently accessed from a central location.
ViPAR Web Analysis Portal (VWAP) —analytical interface
The primary interface to the ViPAR system is a web-based portal called the VWAP, which operates on top of the underlying federated infrastructure described previously. Once a user has successfully logged into the portal, they are presented with a welcome page displaying current analysis projects within any study to which they have access, information on other projects within the system,and details on the connection status of each site's LVD. The project management interface provides access to the three most commonly used features within the VWAP, namely starting a new analysis, viewing outputs from completed analyses and managing code libraries. These interfaces are illustrated in Figures 2 and 3 and in Supplementary Figure 2 (available as Supplementary data at IJE online) and are described as follows.
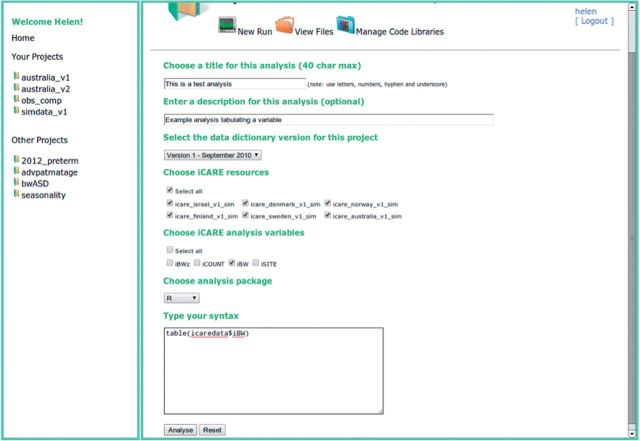
Figure 2.
VWAP analysis interface. Screenshot of browsing the VWAP analysis interface. Here the analyst has provided some simple syntax in the R language to provide summary information for the single selected variable across all selected resources.
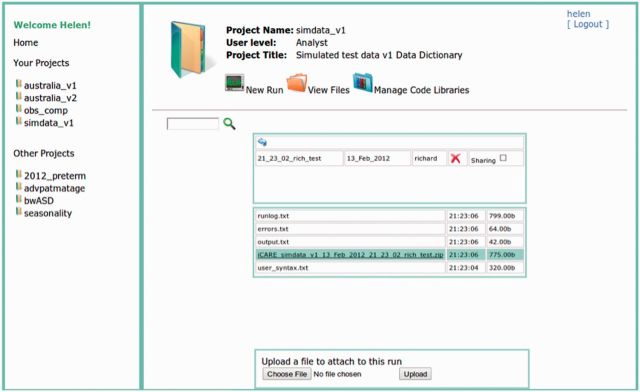
Figure 3.
VWAP file manager. Screenshot of browsing the VWAP file manager. Here the output files resulting from a single analysis are displayed. Users can download files individually or all at once in the provided ZIP file. Optionally users can upload files to associate with an analysis. In addition there are options for deleting the results of an analysis and for sharing the results with other users of the system.
Figure 2 shows the analysis interface, where a user can initiate an analysis run by first choosing from the subset of variables and sites made available to them within a particular analytical project and providing the required analysis syntax to be used in a text field on the interface. When a new analysis is submitted, the following process is enacted:
The Perl data federation component retrieves data for the selected variables from each of the requested site LVDs over the encrypted SSH connections. Data may be retrieved in serial (one site after another) or parallel (all sites at the same time) depending on how ViPAR has been configured; however, this part is transparent to the submitting user.
The data from each site are read into the VMS's memory and virtually pooled. These data are never committed to disk or permanently stored on the server at any point. The virtually pooled dataset is passed to the requested statistical package using a FIFO, a computational technique for passing data from one program to another without the need for an intermediary file (see ‘named pipe’ in Supplementary material and Box 1). ViPAR has been tested with both open-source (R) and commercially available (SAS, STATA) statistical software.
The file manager, illustrated in Figure 3 , marks an analysis task as either ‘running’, ‘completed’ or ‘failed’ (if an error has occurred). On completion, all results and logs are made available for download in the file manager, and the user is notified by e-mail.
To assist the analyst, a code library feature ( Supplementary Figure 2 , available as Supplementary data at IJE online) allows commonly used program codes (e.g. custom analysis functions) to be uploaded, stored and reused in multiple analyses within a single project. As multiple researchers may be involved in the same project, they too can access any shared program code as well as view the results of analyses conducted by other researchers within the same project. Additional administration features are available through the VWAP to users with administration-level user accounts. These administration interfaces simplify the management and creation of users and projects, as well as facilitate the creation of data dictionary versions and corresponding database tables on the LVDs and the setup and connection of new LVDs to the system. Examples of these interfaces are illustrated in Supplementary Figures 3 and Supplementary Data (available as Supplementary data at IJE online).
We provide a detailed use case of ViPAR in the Supplementary section of this manuscript as part of the International Collaboration for Autism Registry Epidemiology (iCARE) 11 .
Summary of data security and implications
All data transfers within the ViPAR system are encrypted using enterprise-grade technologies. Access to the VWAP is password protected and, if additional security is required, the portal can be further restricted through the use of SSL security certificates for both the server and the clients. At both the front end and behind the scenes, different user levels are defined to restrict and control access to data, and all interactions with the system are logged. We recommend that all research data within the ViPAR system are anonymized to help ensure the privacy of individuals. A detailed technical description of the security measures and implications is provided in the Supplementary material , available as Supplementary data at IJE online.
Discussion
ViPAR provides a secure platform for the analysis of multi-site research data when such data cannot be permanently stored or retained outside national, state or other borders. ViPAR is built on open-source technology, and contrary to the notion that federated techniques are expensive to implement, 6 we provide a free solution that with some informatics assistance can be rolled out across any number of research projects.
Whereas ViPAR has been designed to be compatible with ethico-legal restrictions, it should be noted that although analysis data are never committed to disk in any way and never permanently exist outside international or national borders, data do leave the LVD to be virtually pooled in a temporary way. Data are pooled in the random access memory (RAM) at the VMS at the time of analysis, and are then removed. We encourage that this point be made up front in data access and relevant ethics applications to ensure that this ‘virtual pooling’ principle is understood by all necessary parties. It is important to establish all the required privacy and confidentiality rules at national and international level before selecting the analytical method of choice. At the present moment we are not aware of any jurisdiction where virtual pooled data has a specific legal status. Virtually pooling data is possible when approval is given by local ethics committees to share site-specific data between partners. We can however confirm that the ViPAR virtual pooling approach has been approved for use by ethics committees across multiple international jurisdictions (covering Australia, the UK, the Nordic countries, the USA and Israel) for use in projects funded by Autism Speaks and the NIH. We believe this demonstrates and should give reassurance as to the viability and practicality of using this approach across jurisdictions. In addition to ViPAR’s aforementioned data security safeguards (e.g. encryption of data in transit, data access restriction within projects, variable granularity within data dictionaries) it is also useful to keep in mind that the virtual pooling approach within ViPAR can serve to restrict the re-identification of individuals, particularly those with extreme values for certain variables (e.g. very tall/short individuals). These individuals are much more likely to be re-identified within a single site study dataset than when diluted within a larger virtually pooled multi-site dataset that may well contain other individuals with similar extreme values. This concept may be more compliant with juristictional data protection and privacy laws. Ultimately though, the legalities and protections on any dataset reside in the jurisdiction where the data reside.
In the current implementation of ViPAR, the system does not actively monitor statistical syntax provided by researchers (though the syntax is always captured and saved). Using relevant syntax, a researcher may try to write study data to disk on the VMS. However, other users within the system will be able to see from the saved syntax and files that there has been inappropriate use of the system and can act accordingly. We are currently looking into methods to further enhance data security while maintaining the analytical flexibility of the system, such as analysing each line of analysis code that passes through the system for potential inappropriate use or placing limits (e.g. on file sizes) on the download of data through the file management interface.
The ViPAR system was originally developed for the purpose of analysing autism data in a collaboration involving data from six countries (Sweden, Denmark, Norway, Finland, Israel, and Australia), the iCARE project. Within the iCARE project, the ViPAR system handles almost 10 million records from across the aforementioned international sites. For a simple analysis with serial data retrieval, these data are pooled and analysed within 3 min (faster with parallel retrieval), which we believe clearly demonstrates that the system can handle large datasets. However, in some cases, care should be taken to assess whether particular data and analyses should be conducted using ViPAR, as the time taken for very large amounts of data to be sent across the internet from an LVD to the VMS may hinder the speed of analysis and may increase pressure on computer resources at the VWAP. For example, large genome-wide association studies can involve data from thousands of individuals for millions of genetic variants, and analysing data of this type within a single analysis run is not what this system was ideally designed for, although individual variants could be analysed within the system. Similarly, whereas ViPAR was primarily designed for virtual pooling and analysis of subject-specific data, the technique can be applied to other forms of data (e.g. clinical or genetic).
Finally, we would like to note that although ViPAR was designed for use in large multi-site collaborative projects, it can equally be used to connect databases in the same country or institute, or even on the same computer. For example, the VMS could also be the host for all LVDs on the same machine at the same site. We believe this design flexibility is a great strength of ViPAR, and lends itself to the heterogeneous nature of IT systems and research collaborations across sites around the world.
Comparison with existing/alternate methods
There are few other methods and tools that compare to the automated and flexible analyses that are possible through the ViPAR system. The GenomEUTwin 12 project stores epidemiological data for around 600 000 twins from across Europe and Australia together with genotypic data for a subset. The project also describes a data federation approach for the management and pooling of datasets in a hub-and-spoke network architecture similar to ViPAR, although a software package was never released. One key feature that distinguishes the GenomeEUTwin method from ViPAR is that, whereas data are stored and maintained at data-contributing sites, for project-based analyses the data are extracted and physically stored in a document/file for input to a statistical package. In ViPAR, however, named pipes are used to ensure that data never exist permanently outside the site of origin.
DataSHIELD 13,14 is a statistical method, implemented in the statistical software R, for individual- level meta-analysis developed as part of the Maelstrom Research project [ https://www.maelstrom-research.org/ ]. This method also adopts a hub-and-spoke architecture, where the spokes are data-contributing sites (DCs) and the hub is the analytical centre (AC). In the DataSHIELD method, no study data ever leave the DC, temporarily or otherwise. In an iterative process, DataSHIELD pools site-specific statistics rather than site-specific raw data. DataSHIELD may be more appropriate than ViPAR in collaborations where ethics strongly prohibit the movement of data away from a particular site at any time, since ViPAR analysis data do temporarily leave a data-contributing site. DataSHIELD is implemented in the software R only, and is currently limited to analyses conducted within a generalized linear model (GLM) framework. ViPAR, in comparison, does not have such limitations. It is even possible to add DataSHIELD as an analysis option within ViPAR, which gives ViPAR users an option for a more stringent method of analysis while providing DataSHIELD users with access to the collaborative project management interface and other benefits of the VWAP. We note that both the ViPAR and DataSHIELD methods rely on having cleaned, harmonized data at all sites involved.
Conclusion
We have created a secure and cost-effective solution to the key problem that often limits data sharing and analysis across research collaborations. ViPAR avoids the problems associated with physical pooling of data from several sites by using database federation techniques. It has been successfully used within two large international projects, enabling new insight into risk factors for autism. 11,15 ViPAR is flexible and scaleable, and is made freely available to the research community where it can be implemented into other research or data-sharing collaborations.
Funding
This work was supported by Autism Speaks [grant numbers: 6230, 6246–6249, 6251, 6295]; the Government of Western Australia Centres of Excellence programme [to K.W.C. and R.W.F.]; and the McCusker Charitable Foundation Bioinformatics Centre [to K.W.C. and R.W.F.]. The Open Access fee was funded by the Mike Schon-Hegrad Incentive Award (Telethon Kids Institute) and the McCusker Charitable Foundation Bioinformatics Centre.
Supplementary Material
Author contributions
R.W.F. and K.W.C. conceived, designed and implemented the ViPAR system and wrote the manuscript. iCARE Investigators (M.B., K.W.C., R.W.F., M.G., T.K.G., R.G., N.G., G.H., M.H., C.M.H., A.L., H.L., E.T.P., A.R., S.S., D.E.S., A.S., C.S., A.S., P.S., E.S) contributed to the development of the ViPAR system and critically reviewed the manuscript. iCARE Site IT Teams (J.H., S.N., G.P., L.S., C.S., A.S.V., Z.Y.) contributed to the development of the ViPAR system and managed and supported the site IT infrastructure critical to its function.
Conflict of interest: The authors have declared that no competing interests exist.
References
- 1. Boulton G, Rawlins M, Vallance P, Walport M . Science as a public enterprise: the case for open data . Lancet 2011. ; 377 : 1633 – 35 . [DOI] [PubMed] [Google Scholar]
- 2. Ross JS, Krumholz HM . Ushering in a new era of open science through data sharing: the wall must come down . JAMA 2013. ; 309 : 1355 – 56 . [DOI] [PubMed] [Google Scholar]
- 3. Walport M, Brest P . Sharing research data to improve public health . Lancet 2011. ; 377 : 537 – 39 . [DOI] [PubMed] [Google Scholar]
- 4. Glass GV . Primary, secondary, and meta-analysis of research . Educational Researcher 1976. ; 5 : 3 – 8 . [Google Scholar]
- 5. Haas LM, Lin ET, Roth MA . Data integration through database federation . IBM Syst J 2002. ; 41 : 578 – 96 . [Google Scholar]
- 6. Akula SP, Miriyala RN, Thota H, Rao AA, Gedela S . Techniques for integrating -omics data . Bioinformation 2009. ; 284 – 86 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fortier I, Doiron D, Little J, et al. . Is rigorous retrospective harmonization possible? Application of the DataSHaPER approach across 53 large studies . Int J Epidemiol 2011. ; 40 : 1314 – 28 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fortier I, Burton PR, Robson PJ, et al. . Quality, quantity and harmony: the DataSHaPER approach to integrating data across bioclinical studies . Int J Epidemiol 2010. ; 39 : 1383 – 93 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ariyachandra T, Watson HJ . Which data warehouse architecture is most successful? . Business Intelligence Journal 2006. ; 11 : 4 . [Google Scholar]
- 10. Sen A, Sinha AP . A comparison of data warehousing methodologies . Commun ACM 2005. ; 48 : 79 – 84 . [Google Scholar]
- 11. Schendel DE, Bresnahan M, Carter KW, et al. . The International Collaboration for Autism Registry Epidemiology (iCARE): multinational registry-based investigations of autism risk factors and trends . J Autism Dev Disord 2013. ; 43:2650 – 63 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Muilu J, Peltonen L, Litton JE . The federated database - a basis for biobank-based post-genome studies, integrating phenome and genome data from 600 000 twin pairs in Europe . Eur J Hum Genet 2007 Jul15 : 718 – 23 . [DOI] [PubMed] [Google Scholar]
- 13. Gaye A, Marcon Y, Isaeva J, et al. . DataSHIELD: taking the analysis to the data, not the data to the analysis . Int J Epidemiol 2014. ; 43:1929 – 44 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wolfson M, Wallace SE, Masca N, et al. . DataSHIELD: resolving a conflict in contemporary bioscience - performing a pooled analysis of individual-level data without sharing the data . Int J Epidemiol 2010. ; 39 : 1372 – 82 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sandin S, Schendel D, Magnusson P, et al. . Autism risk associated with parental age and with increasing difference in age between the parents . Mol Psychiatry 2015. , June 9. doi: 10.1038/mp.2015.70. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.