Abstract
Heterotrimeric G-proteins are composed of Gα, Gβ and Gγ subunits and regulate many fundamental processes in plants. Nonetheless plants have a considerably simpler repertoire of G-protein signaling components than metazoans. In animals, ligand binding to 7 transmembrane (7TM) cell surface receptors designated GPCRs leads to G protein activation, however, activation of the plant G protein complex is constitutive, therefore the exact role of plant 7TM proteins is unclear. MLOs are the best characterized 7TM plant proteins. While genetic ablation of either MLO2 or G proteins alters resistance to pathogens, it is unknown if G proteins directly couple signaling through MLO2. Here we exploited two well-documented phenotypes of Arabidopsis mlo2 mutants, broad-spectrum powdery mildew resistance and spontaneous callose deposition, to assess the relationship of MLO2 proteins to the G protein complex. Although our data reveal modulation of antifungal defence responses by Gβ and Gγ subunits, our findings are overall inconsistent with a role of MLO2 as a canonical GPCR. We discovered that mutants defective in the Gβ subunit show delayed accumulation of a subset of defence-associated genes following exposure to the microbe-associated molecular pattern (MAMP), flg22. Moreover Gβ mutants were found to be hypersusceptible to spray-inoculation with the bacterial pathogen, Pseudomonas syringae. In sum, our data do not support a function for MLO proteins as GPCRs, but unravel a role for Gβ and Gγ subunits as modulators of basal defence against biotrophic and hemibiotrophic phytopathogens.
Keywords: G-protein coupled receptor, heterotrimeric G-protein, MLO protein, plant immunity, powdery mildew
INTRODUCTION
Powdery mildew parasites are Ascomycete fungi that cause disease of a wide range of plant species. Successful pathogenesis depends on penetration into epidermal host cells by the obligate biotrophic phytopathogen, which requires the presence of members of the plant-specific seven transmembrane (7TM) domain MLO (Mildew Resistance Locus O) protein family. Recessive mutations of the founder of this gene family, barley MLO, confer durable and broad-spectrum powdery mildew resistance via early termination of fungal pathogenesis before successful penetration (Jørgensen, 1992; Büschges et al., 1997). Reminiscent of fully resistant barley mlo single mutants, Arabidopsis (Arabidopsis thaliana) triple mutants defective in the three co-orthologs MLO2, MLO6 and MLO12 are fully immune against the compatible powdery mildew fungi Golovinomyces orontii and Golovinomyces chicoracearum, indicating unequal genetic redundancy between the three Arabidopsis genes (Consonni et al., 2006). By contrast, mutant versions of MLO2 alone confer only partial resistance to these pathogens, a phenotype that is dependent on components of preinvasive antifungal defence (Consonni et al., 2006; Consonni et al., 2010).
Mutations in barley and Arabidopsis MLO genes result in additional developmentally-controlled pleiotropic phenotypes. The formation of spontaneous cell wall deposits containing callose, a β-(1,3) polyglucan, in mesophyll cells and early leaf chlorosis/necrosis occur during vegetative development in both barley and Arabidopsis mlo mutants (Wolter et al., 1993; Piffanelli et al., 2002; Consonni et al., 2006; Consonni et al., 2010). In Arabidopsis, spontaneous callose deposition in mlo2 mutants is mediated by PMR4/GSL5 (POWDERY MILDEW RESISTANT4/GLUCAN SYNTHASE-LIKE5)-activity, an enzyme also required for the biosynthesis of callose at wound sites and in local cell wall appositions (papillae) following pathogen attack (Jacobs et al., 2003; Nishimura et al., 2003; Consonni et al., 2010). Developmentally-controlled callose deposition and early leaf chlorosis/necrosis in mlo2 mutants were found to be fully dependent on functional salicylic acid (SA) biosynthesis and signalling, which in turns are not essential for mlo2-mediated powdery mildew resistance, demonstrating that the pleiotropic effects can be genetically uncoupled from mlo-based resistance (Consonni et al., 2006; Consonni et al., 2010).
mlo-mediated powdery mildew immunity is functionally conserved in various plant species (Jørgensen, 1992; Consonni et al., 2006; Bai et al., 2008; Humphry et al., 2011), other members of the Arabidopsis MLO protein family operate in processes unrelated to powdery mildew pathogenesis (Chen et al., 2009; Kessler et al., 2010). The biochemical functions of the plant-unique MLO proteins remain elusive. MLO proteins are plasma membrane-resident and constitute one of the largest heptahelical protein families in Arabidopsis with an extracellular N-terminal domain, and a cytoplasmic C-terminal domain (Devoto et al., 1999). Thus the domain architecture and subcellular localization of MLO proteins is reminiscent of the G-protein coupled receptor (GPCR) superfamily in metazoans (Temple and Jones, 2007; Oldham and Hamm, 2008). In animals, GPCRs activate heterotrimeric G-protein signalling, one of the most evolutionarily conserved signalling pathways in metazoans, by relying extracellular signals through intracellularly associated G-proteins (Oldham and Hamm, 2008).
The canonical GPCR-associated G-protein consists of three distinct subunits, Gα, Gβ and Gγ, which form a heterotrimeric complex in the inactive state. In animals, ligand binding to a GPCR induces the conversion of an inactive G-protein into its active conformation via exchange of GDP by GTP bound to Gα. As a result, Gα–GTP separates from the Gβγ dimer and both, Gα–GTP and the Gβγ dimer, can activate downstream effectors ultimately leading to a cellular response. The intrinsic hydrolytic GTPase activity of Gα recovers the GDP-bound state, which promotes reassociation of the complex into its inactive form. Regulator of G-protein Signaling (RGS ) proteins accelerate the GTPase activity of Gα. In animals, RGS proteins are cytoplasmic proteins that are recruited to the G protein complex to accelerate deactivation of signaling.
In comparison to metazoans, the set of known heterotrimeric G-protein signalling components in plants is much simpler. The fully sequenced genome of the reference plant A. thaliana codes for a single canonical Gα subunit (GPA1), one Gβ subunit (AGB1), three Gγ subunits (AGG1, AGG2 and AGG3) and a single RGS protein (RGS1) (Chen et al., 2003; Jones et al., 2003; Ullah et al., 2003; Trusov et al., 2007; Chakravorty et al., 2011). The plant RGS protein is unusual in that it contains an N-terminal 7TM domain. Despite its simplicity, the Arabidopsis heterotrimeric G-protein complex has been implicated in a broad range of biological processes, including seed germination, cell division, hormone sensitivity, sugar sensing, pathogen defence and abiotic stress responses (Assmann, 2002; Jones and Assmann, 2004; Perfus-Barbeoch et al., 2004; Chen, 2008). The unique property of the plant Gα subunit is spontaneous GTP loading, i.e. no canonical GPCR is needed for activation. Rather, the 7TM RGS regulates the activation state of the G protein complex (REF). However, other 7TM proteins may also be involved in G protein activation. With a total of 15 members the plant-specific MLO proteins define one of the largest families of 7TM domain proteins in Arabidopsis (Lu et al., 2009) and although no significant sequence similarities between mammalian GPCRs and MLO proteins exist, these plant-unique proteins may play a role in plant G protein activation (Devoto et al., 1999; Kim et al., 2002; Moriyama et al., 2006; Lu et al., 2009).
Previous combined pharmacological and genetic studies in barley did not provide any evidence for a function of heterotrimeric G-protein signalling in MLO-mediated powdery mildew pathogenesis (Kim et al., 2002). However, these analyses were based on single cell transient overexpression and dsRNAi-mediated gene silencing of barley Gα variants that were deduced from the animal or yeast system. Moreover, the specificity of the pharmaceutical agents is questioned and criticized (Fujisawa et al., 2001; Miles et al., 2004). In summary, the question of a GPCR role for the MLO proteins is unanswered.
Consequently, in this study we took a genetic approach to rigorously test a potential role of the MLO2 protein as a putative plant GPCR. Based on an informative set of G-protein signalling mutants and mlo2 G-protein double mutants, we assessed both powdery mildew resistance and developmental callose deposition as readouts for MLO2 function. Overall our data are inconsistent with a presumptive role for MLO2 as a canonical GPCR, but unravel a role for Gβ and Gγ subunits as modulators of basal defence against biotrophic and hemibiotrophic phytopathogens.
RESULTS
Gβ-deficient mutants allow enhanced host cell entry by adapted and non-adapted powdery mildew fungi
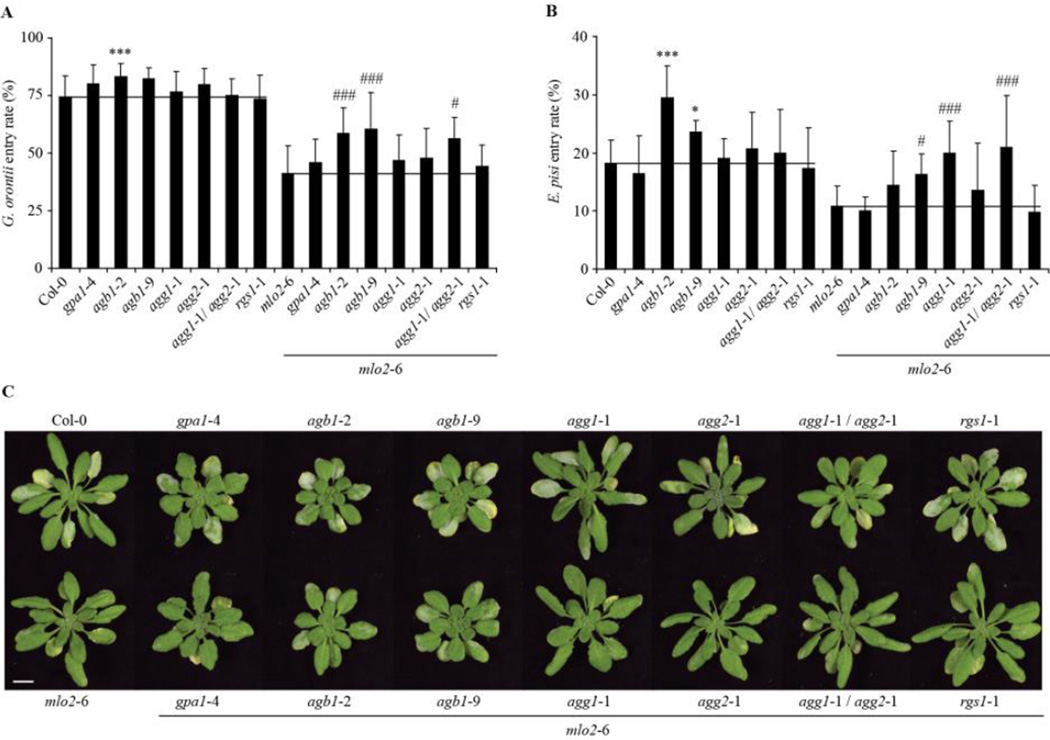
To investigate a potential molecular link between MLO2 and the heterotrimeric G-protein complex in powdery mildew susceptibility, we assessed the powdery mildew infection phenotype of Arabidopsis single mutants defective in each component of heterotrimeric G-protein signalling (Gα/GPA1, At2g26300; Gβ/AGB1, At4g34460; Gγ1/AGG1, At3g63420; Gγ2/AGG2, At3g22942 and RGS1, At3g26090) and the respective mlo2-6 double mutants. Given that Gγ1 and Gγ2 may act redundantly, the double mutant agg1-1/agg2-1 and the corresponding agg1-1,agg2-2,mlo2-6 triple mutant were also added to the analysis. Since mlo-mediated resistance operates early and primarily affects host cell penetration we focussed on this parameter for the assessment of susceptibility of the mutants to the powdery mildew disease. Quantitative analysis of fungal entry into plant epidermal leaf cells by the adapted powdery mildew fungus Golovinomyces orontii was performed at 48 h post inoculation (hpi) and revealed an increased penetration rate of the fungus on the Gβ null mutant, agb1-2 (74 ± 9 % for Col-0 versus 83 ± 6 % for agb1-2; Fig. 1A, P=). The independent EMS mutant allele, agb1-9, the gpa1-4 mutant, and the agg2-1 mutant also showed elevated fungal entry rates, although for these lines the data were not statistically significant (Fig. 1A). The increased susceptibility phenotype was more pronounced in the double mutants agb1-2/mlo2-6 and agb1-9/mlo2-6, where entry rates were 59 ± 11 % and 60 ± 16 % compared to 41 ± 12 % for the mlo2-6 single mutant (Fig. 1A, P= ). In agreement with this finding, the agb1-2/mlo2-6 and agb1-9/mlo2-6 double mutants showed enhanced powdery mildew disease symptoms at 7 d post inoculation (dpi) on (Fig. 1C, P=). Together these data suggest a role for the Gβ subunit, AGB1, in restricting host cell penetration of the adapted powdery mildew fungus G. orontii independently of the presence or absence of MLO2.
Figure 1.
Powdery mildew infection phenotypes of G-protein mutants and corresponding mlo2-6 double mutants. Powdery mildew inoculations were preformed with conidiospores of the respective fungus on rosette leaves of 4-week old plants of the indicated genotypes. Quantitative analysis of G. orontii (A) and E. pisi (B) entry rates scored at 48 hours and 7 d post inoculation, respectively. Results represent mean ± standard deviation of at least three independent experiments. Asterisks indicate a significant difference from Col-0 wild-type (*** P < 0.01, * < 0.05, Student’s t-test) and number signs indicate a significant difference from mlo2-6 mutant (### P < 0.01, # P < 0.05, Student’s t-test). (C) Macroscopic G. orontii infection phenotypes at 7 days post inoculation. Bar = 1cm
In the compatible interaction with G. orontii, high pathogenicity of the host-adapted fungus may mask a potential contribution of the G-protein complex to powdery mildew defence, especially in the presence of MLO2. Therefore, we additionally scored the penetration success of a less virulent, non-adapted powdery mildew fungus, the pea pathogen Erysiphe pisi. Consistent with the findings seen in the compatible interaction with G. orontii (see above; Fig. 1A), the agb1-2 and agb1-9 single mutants exhibited an enhanced frequency of successful entry by the non-adapted fungus (Fig. 1B). Similarly, agb1-2/mlo2-6 and agb1-9/mlo2-6 double mutants allowed significantly increased penetration by E. pisi (note that similar to the interaction with G. orontii entry levels of E. pisi are lower in the mlo2-6 double mutants; Consonni et al., 2006). These findings further corroborate a function of the Gβ subunit in pre-invasive defence against powdery mildew fungi independent of MLO2. While the single Gγ-deficient mutants as well as the agg1-1/agg2-1 double mutant did not display enhanced powdery mildew penetration phenotypes, the triple mutant agg1-1/agg2-1/mlo2-6 showed elevated plant cell entry by both fungal pathogens (Fig. 1A and B). Notably, at 7 dpi the triple mutant retained resistance to the compatible powdery mildew fungus, G. orontii, which was comparable to the mlo2-6 control plant (Fig. 1C, P=). Furthermore, the agg1-1/mlo2-6 double mutant exhibited enhanced penetration by the pea powdery mildew fungus, which was quantitatively similar to the triple mutant agg1-1/agg2-1/mlo2-6, but higher compared to the entry rates on the two agb1/mlo2-6 double mutants (Fig. 1B, P=). Interestingly, these effects were not observed upon infection with the adapted pathogen, G. orontii (Fig. 1A), which suggests differential requirements of G-protein complex components in mlo2-mediated defence against adapted and non-adapted powdery mildew fungi.
mlo2-mediated developmental callose deposition and salicylic acid accumulation are impaired in the Gγ1-deficient mutant
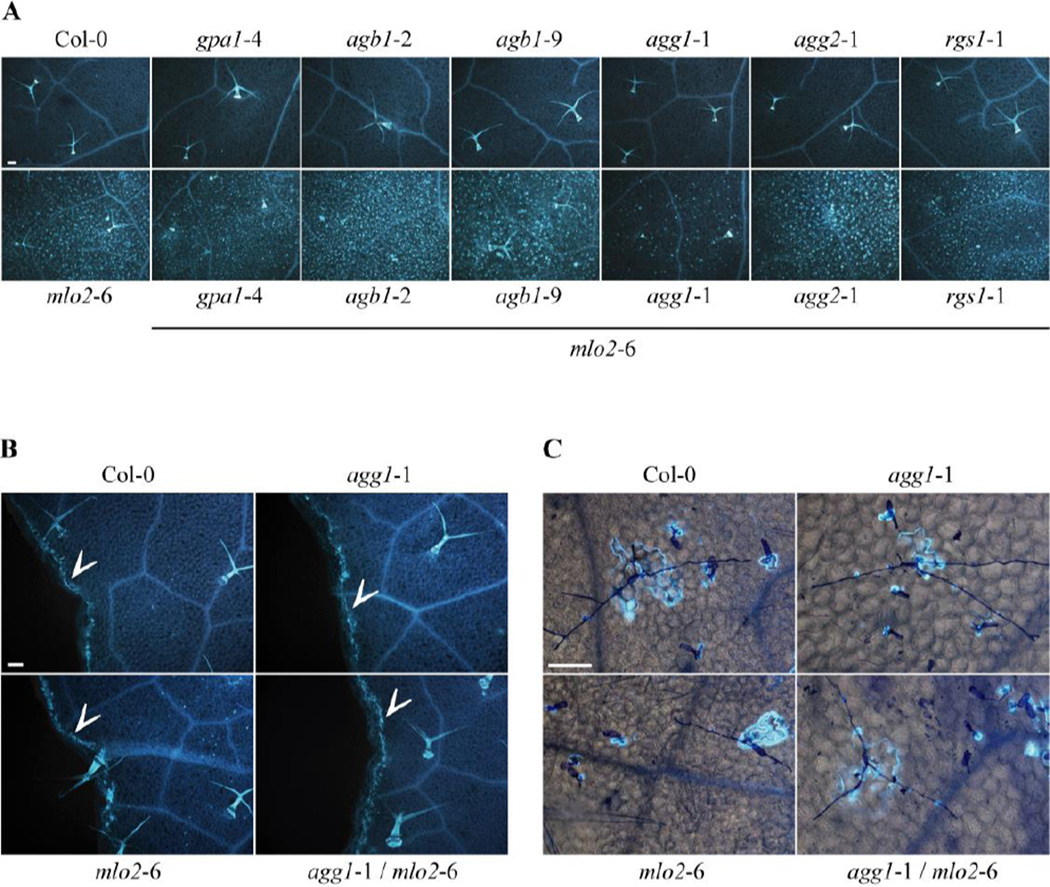
The biochemical function of MLO proteins during powdery mildew host cell invasion remains currently unknown. We thus cannot rule out the possibility that MLO proteins operate as G-protein-coupled receptors in normal plant physiology, but are co-opted for a G-protein-independent activity during fungal pathogenesis (Panstruga and Schulze-Lefert, 2003). In such a scenario, the contribution of G-proteins to MLO activity would remain undiscovered in powdery mildew infection assays. We thus sought for a readout of MLO2 function that is independent of the involvement of any pathogen. When grown under pathogen-free conditions, mlo2 mutant plants show spontaneous callose accumulation in leaf mesophyll cells. This phenotype is under developmental control and is detectable from six weeks onwards (Consonni et al., 2006; Consonni et al., 2010). To test whether heterotrimeric G-protein signalling contributes to this mlo2-associated but pathogen-independent process, callose deposition was analyzed in non-infected rosette leaves of 6-week old Col-0 control plants, G-protein single mutants (gpa1-4, agb1-2, agb1-9, agg1-1, agg2-1 , rgs1-1) and the respective mlo2-6 double mutant plants. Similar to the Col-0 wild-type, G-protein single mutants exhibited no callose deposition, whereas the majority of the corresponding mlo2-6 double mutants behaved like mlo2-6 control plants and developed numerous spontaneous callose deposits (Fig. 2A). Interestingly, the agg1-1/mlo2-6 double mutant showed markedly reduced callose accumulation, indicating an involvement of the Gγ1 subunit, AGG1, in the developmentally controlled biosynthesis of callose in the mlo2-6 mutant (Fig. 2A).
Figure 2.
Callose accumulation in rosette leaves. Callose was stained with aniline blue. (A) Representative micrographs demonstrating spontaneous callose deposition in leaves of 6-week old plants of the indicated genotypes grown in pathogen-free conditions. The experiment was repeated at least three times with similar results. Bar = 100 µm. (B) Leaves from 4-week old plants of the indicated genotypes injured with forceps showing callose deposition at wound sites (= arrowheads). The experiment was performed twice with similar results. Bar = 100 µm. (C) Four-week old leaves from plants of the indicated genotypes at 7 d post inoculation with the non-adapted powdery mildew fungus E. pisi exhibit callose deposition at sites of fungal interaction. Fungal structures were stained with Coomassie Brillant Blue. The experiment was performed once. Bar = 100 µm.
The formation of callose deposits in mlo2 mutants requires the PMR4/GSL5 callose synthase, an enzyme best known for its role in the biosynthesis of the β−(1,3) polyglucan at wound sites and in papillae following pathogen attack (Jacobs et al., 2003; Nishimura et al., 2003). To investigate if the Gγ1 subunit has a general function in callose biosynthesis and also contributes to wound and/or papillary callose depositions, we determined callose accumulation after leaf wounding and subsequent to inoculation with the non-adapted powdery mildew fungus, E. pisi. Compared to Col-0 and mlo2-6 control plants, wound- and pathogen-induced callose accumulation was unaltered in the agg1-1 single and the agg1-1/mlo2-6 double mutant (Fig. 2B and C). These findings indicate that the Gγ1 subunit is dispensable for localized PMR4/GSL5-generated callose formation in response to wounding and pathogen challenge. Instead, the data suggest a specific role for AGG1 in mlo2-dependent developmentally controlled deposition of the β−(1,3) polyglucan.
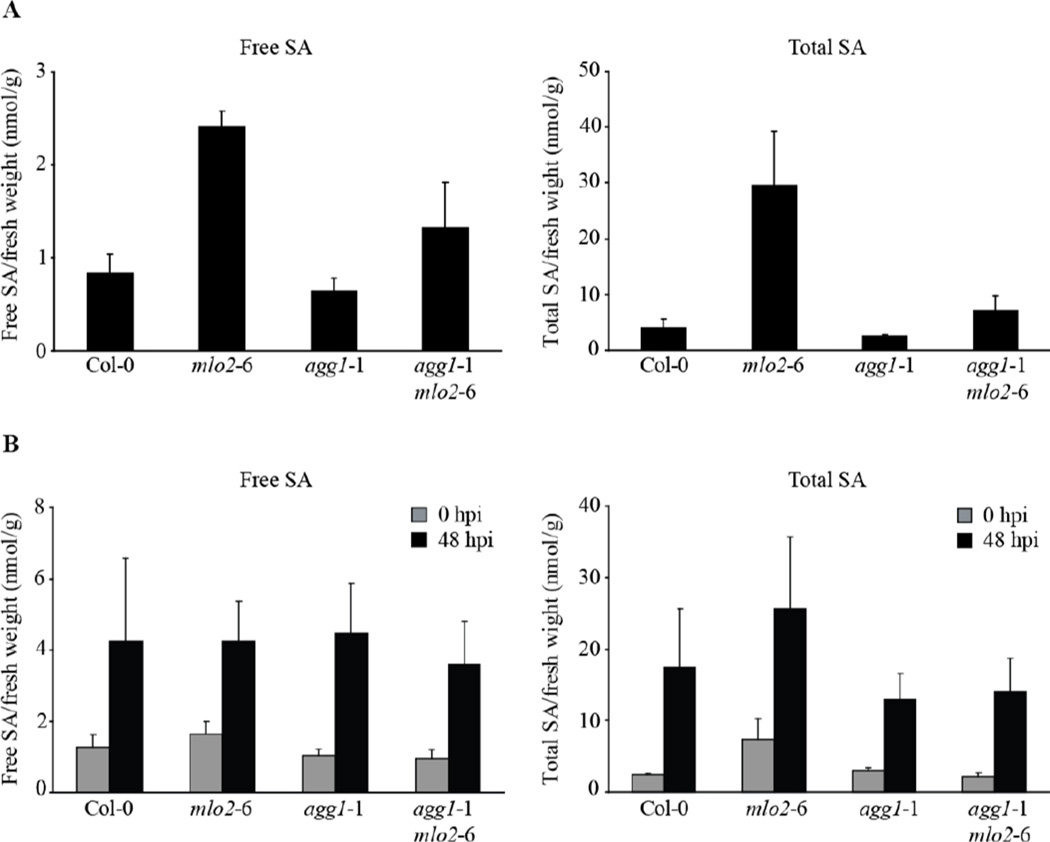
Spontaneous callose formation in the mlo2 mutant depends on functional salicylic acid (SA) biosynthesis and signaling, as double mutants defective in both MLO2 and components of the SA pathway are suppressed in developmentally controlled callose deposition (Consonni et al., 2006). Moreover, mlo2 mutants display elevated levels of free and total SA in the course of development. The timing of the increase in SA levels correlates with the onset of spontaneous callose deposition (from six weeks onwards; Connsoni et al., 2006), suggesting a molecular link between SA accumulation and callose deposition. We therefore reasoned that reduced SA levels might account for the impaired spontaneous callose accumulation in the agg1-1/mlo2-6 mutant and determined SA concentrations in uninfected 6-week old mutant plants. Consistent with previous results (Consonni et al., 2006), the mlo2-6 mutant displayed increased free (ca. 3-fold higher) and total (ca. 10-fold higher) SA levels compared to Col-0, while SA levels in the agg1-1 single mutant were indistinguishable from the wild-type (Fig. 3). In the agg1-1/mlo2-6 double mutant, concentrations of free SA were intermediate between Col-0 and the agg1-1 mutant, whereas levels of total SA were similar to Col-0 wild-type and thus strongly reduced compared to the mlo2-6 single mutant (Fig. 3). These findings suggest that the Gγ1 subunit, AGG1, positively contributes to developmentally-induced SA accumulation in the context of the mlo2-6 mutant, which explains the reduced number of callose deposits in the agg1-1/mlo2-6 double mutant. To investigate if the agg1 mutant has a general defect in SA accumulation and/or biosynthesis, we quantitatively assessed SA accumulation in Col-0 wild type plants, mlo2-6, agg1-1 and agg1-1/mlo2-6 mutants following G. orontii challenge at 48 hpi. Irrespective of the genotype, free and total SA concentrations were elevated to similar levels in response to the pathogen (Fig. 3B). In summary, these data indicate that the Gγ1 subunit is dispensable for G. orontii-triggered SA accumulation, but has a specific function in mlo2-mediated developmental SA accumulation.
Figure 3.
Accumulation of free and total salicylic acid (SA) in wild-type, mlo2-6, agg1-1 and agg1-1/mlo2-6 mutant plants. (A) SA levels were determined in rosette leaves of 6-week old plants grown under pathogen-free conditions. (B) SA concentrations were measured in rosette leaves of 4-week old plants at 48 h post inoculation with G. orontii. Results represent means ± SE of at least three independent experiments.
AGB1 and AGG1 are coexpressed with key components of antifungal defense
We recently proposed that several genes with a documented function in pre-invasive defense against powdery mildew fungi (e.g. MLO2, PEN1, PEN2, PEN3, SNAP33, VAMP722) show correlated transcript accumulation across a broad range of conditions and plant tissues (“coexpression”), together with a suite of additional genes, form a transcriptional regulon that defines a conserved functional module in plants (Humphry et al., 2010). Using a publicly accessible resource for the assessment of gene coexpression in Arabidopsis (ATTED-II; http://atted.jp/) we discovered that genes coexpressed with either AGB1 or AGG1 show a considerably higher overlap with genes found in this transcriptional regulon than genes coexpressed with either GPA1 or RGS1 (ca. 2–3-times as many; Table 1). A considerable overlap of genes present in the conserved defence regulon and genes coexpressed with either AGB1 and/or AGG1 hints at an authentic role for the Gβ and Gγ1 subunits in plant immunity. The fact that AGB1 and AGG1 are not integral parts of the previously identified conserved defence regulon suggests that these two G-protein subunits play a minor role in plant immunity. This is consistent with the weak but reproducible phenotypes observed with the respective mutants in our pathogen infection assays (Fig. 1A and B, Fig. 6).
Table 1.
Number of top 300 genes 1 co-expressed with G-protein components present in a previously described antifungal defence regulon 2.
| entire regulon (406 genes) |
co-expressed with MLO2/PEN1/ SNAP33 (107 genes) |
co-expressed PEN1/SNAP33/ VAMP722 (282 genes) |
co-expressed with PEN2/PEN3 (164 genes) |
|
|---|---|---|---|---|
| GPA1 | 31 | 15 | 23 | 9 |
| AGB1 | 91 | 32 | 75 | 23 |
| AGG1 | 90 | 46 | 62 | 34 |
| RGS1 | 43 | 18 | 39 | 11 |
Top 300 genes co-expressed with GPA1, AGB1, AGG1 or RGS1 were obtained from ATTED-II (http://atted.jp/)
see Humphry et al. (2010) for details on the defence refulon
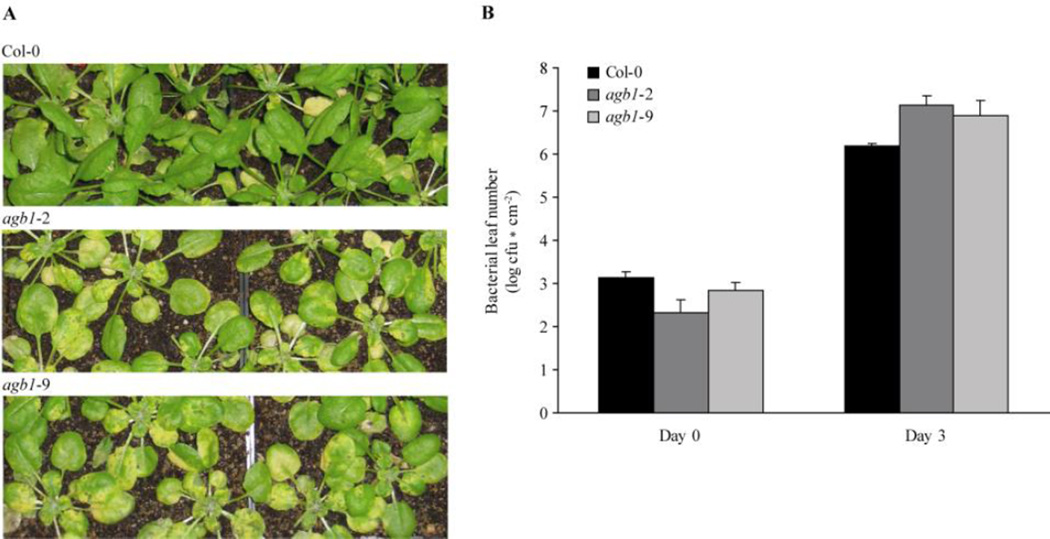
Figure 6.
Plants carrying agb1 mutations are more susceptible to Pst DC3000 bacteria. Four-week old plants were sprayed with Pst DC3000 bacteria at OD600 = 0.2. (A) Macroscopic disease symptoms of the indicated genotypes 3 days after Pst Dc3000 inoculation. (B) Pst DC3000 bacterial titers were determined 3 h (= Day 0) and 3 days after inoculation. A representative data set with mean ± standard deviation of one experiment is shown. The experiment was performed three times with similar results.
Gβ-deficient mutants display reduced flg22-triggered immune responses
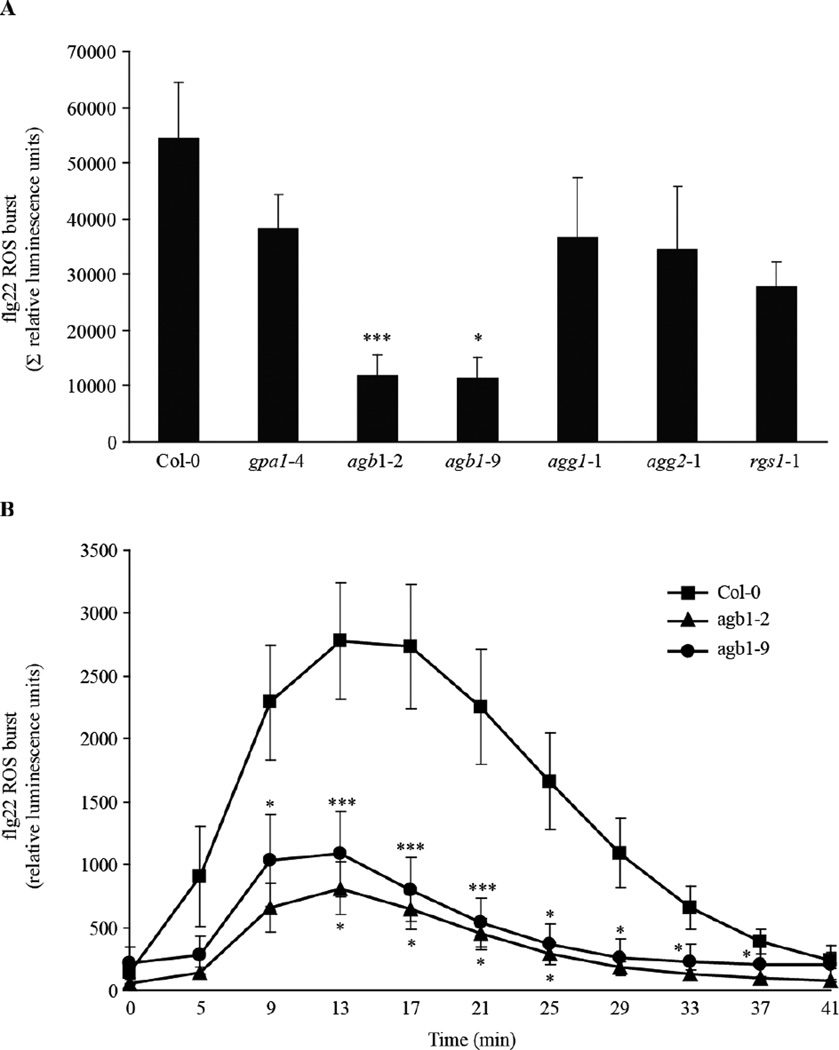
The presumed role of AGB1 as a peripheral part of the defence regulon prompted us to take a broader look at immune responses in the agb1 mutant. A previous study demonstrated a role for Arabidopsis AGB1 in MAMP-induced oxidative burst control (Ishikawa, 2009). However, in this work only a single AGB1 mutant allele was tested and no complementation line was analyzed. Using two independent agb1 mutants, agb1-2 and agb1-9, we confirmed reduced ROS production in response to flg22 treatment in these mutants, while ROS production was slightly but not statistically significantly reduced in mutants of other G-protein components (Fig. 4A and B). As previously shown by Ishikawa (2009), activation of the MAPK cascade upon MAMP treatment was not altered in the agb1-2 mutant.
Figure 4.
Flg22-induced ROS burst in G-protein single mutants. Leaf discs from 4-week old plants were treated with 1 µM flg22 and ROS formation was measured in a chemiluminescence assay. (A) Flg22-induced ROS production in the indicated genotypes is represented as the integrated area under the ROS curve measured during a time course of 41 min and is referred to as Σ relative luminescence units. (B) ROS burst in the indicated genotypes was measured for the indicated time points after flg22 application. Results are presented as mean ± SE of at least five independent experiments. Asterisks indicate a significant difference from Col-0 wild-type (*** P < 0.01, * < 0.05, Student’s t-test).
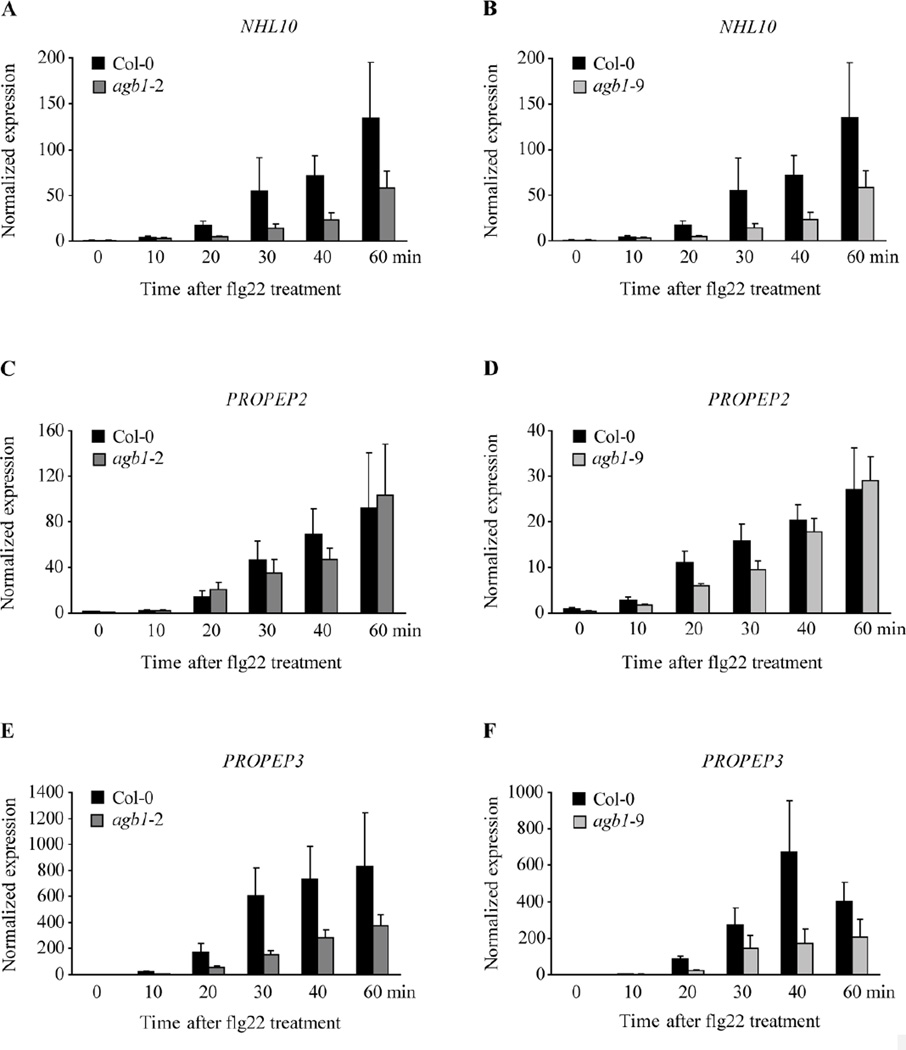
To elucidate a potential role of the Gβ subunit in MAMP-mediated transcriptional activation of defence genes, we studied transcript accumulation of the known MAMP-induced genes NHL10, PHI1, PROPEP2, PROPEP3 and WRKY22 over time after flg22 treatment as described (Huffaker et al., 2006; Lu et al., 2009; Boudsocq et al., 2010; Kwaaitaal et al., 2011). In Col-0 wild-type plants, NHL10, PROPEP2 and PROPEP3 transcript levels gradually increased within one h after flg22 application (Fig. 5). Flg22-induced transcript accumulation of these genes was markedly reduced in both agb1 mutants, agb1-2 and agb1-9, with a pronounced reduction in NHL10 and PROPEP3 expression levels and a minor decrease in PROPEP2 transcript accumulation compared to the wild-type (Fig. 5). Although the kinetics of flg22-induced defence gene activation and the extent of its reduction in agb1 mutant seedlings varied (c.f absolute expression values of the two sets of experiments), the expression level of these three genes was consistently decreased at 30 min after flg22 treatment in both agb1 mutant lines throughout multiple independent experiments (Table 2). Notably, WRKY22 and PHI1 transcript accumulation in response to flg22 was not altered in agb1 mutant plants compared to the wild-type (Fig. S1). Together these data suggest a function of the Gβ subunit in flg22-mediated transcriptional reprogramming of a subset of Arabidopsis defence genes including NHL10, PROPEP2 and PROPEP3. However, AGB1 seems dispensable for the expression of other flg22-induced defence genes such as WRKY22 and PHI1.
Figure 5.
Transcript levels of the MAMP-induced genes NHL10, PROPEP2 and PROPEP3 are reduced in agb1 mutants after flg22 treatment. Two-week old agb1-2 (A, C, E) and agb1-9 (B, D, F) seedlings were treated with 1 µM flg22 and sampled at the indicated time points after flg22 application. Transcript levels were determined by quantitative RT-PCR. (A, B) NHL10, (C, D) PROPEP2 and (E, F)PROPEP3 gene expression was normalized to the transcript levels of the reference gene At4g26420 and is presented relative to the transcript abundance of non-treated Col-0 wild-type at time point 0 minutes. A representative data set with mean ± standard deviation of three technical replicates per genotype and time point is shown. The experiment was performed three times with similar results.
Table 2.
Transcript levels of the MAMP-induced genes NHL10, PROPEP2 and PROPEP3 are reduced in agb1 mutants at 30 min after flg22 treatment in multiple experiments. 1
| Fold change agb1-2 / Col-0 at 30 min after flg22 treatment |
Fold change agb1-9 / Col-0 at 30 min after flg22 treatment |
|||||
|---|---|---|---|---|---|---|
| Experiment |
NHL10 | PROPEP2 | PROPEP3 | NHL10 | PROPEP2 | PROPEP3 |
| # 1 | 0.26 | 0.22 | 0.27 | 0.1 | 0.44 | 0.22 |
| # 2 | 0.28 | 0.56 | 0.36 | 0.24 | 0.32 | 0.53 |
| # 3 | 0.46 | 0.76 | 0.97 | 0.55 | 0.72 | 0.53 |
| # 4 | 0.63 | 1.16 | 0.76 | 0.61 | 0.6 | 0.67 |
| # 5 | 0.71 | 0.78 | - | 0.65 | 0.76 | - |
| mean ± SE | 0.47 ± 0.09 | 0.7 ± 0.15 | 0.59 ± 0.17 | 0.43 ± 0.11 | 0.57 ± 0.08 | 0.49 ± 0.1 |
Expression levels were determined by quantitative RT-PCR as described in Fig. 5.
The agg1-1 single mutation originates from the Arabidopsis accession Ws-0 and was subsequently introgressed into the Col-0 ecotype by eight backcrosses (Trusov et al., 2007) because the Ws-0 accession carries a natural mutation in the gene encoding the flagellin receptor, FLS2, making it insensitive to flg22 treatment (Gómez-Gómez and Boller, 2000). Sequencing of the FLS2 gene in the agg1-1/agg2-1 double mutant revealed the presence of the natural fls2 mutation in the double mutant but not in the backcrossed agg1-1 single mutant (data not shown). We therefore excluded the agg1-1/agg2-1 double mutant from the analysis of flg22-triggered effects.
Gβ-deficient mutants exhibit enhanced susceptibility to the virulent bacterial pathogen Pseudomonas syringae pv. tomato DC3000
Given that Gβ function is required for full expression of a subset of flg22-induced defence responses, we reasoned that agb1 mutants may be compromised in anti-bacterial defence. Previous studies did not reveal evidence for a role of the Gβ subunit in bacterial growth restriction (Trusov et al., 2006; Ishikawa, 2009), but these relied on experiments that were based on an inoculation method (syringe infiltration of bacterial cultures) that bypasses the first step of the genuine bacterial infection process. In nature, bacteria enter host plant leaves through wounds and openings such as stomata. In order to mimic the natural infection conditions, we performed bacteria spray inoculation on 4-week old agb1 mutant plants using the Pst DC3000 wild-type strain. At three days after spray inoculation with Pst DC3000 we observed a tendency towards enhanced macroscopic disease symptoms on the agb1 mutants, agb1-2 and agb1-9 (Fig. 6A). Quantification of bacterial growth revealed an increase in bacterial titers in both agb1 mutants (Fig. 6B). Although the differences in bacterial proliferation on the agb1 mutants compared to wild-type plants was not statistically significant, we observed the same tendency (increased bacterial titers in both agb1 alleles) in each of three independent experiments.
DISCUSSION
The MLO2 protein is not a GPCR in defense and callose deposition
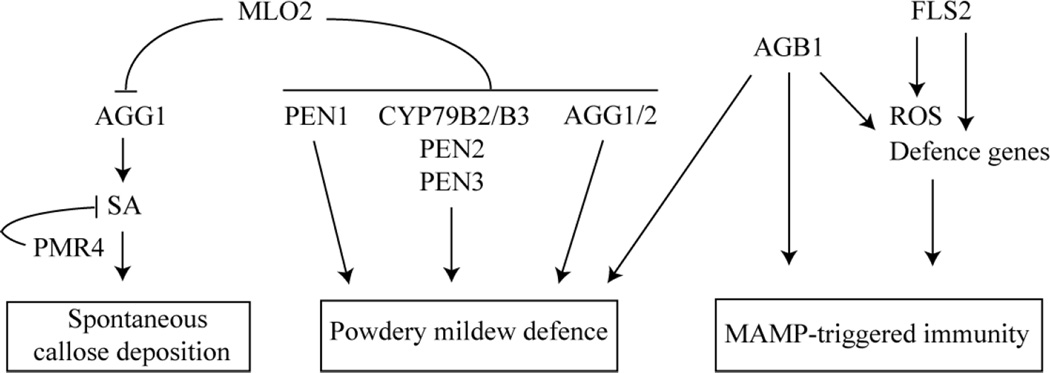
In the canonical model of heterotrimeric G-protein signalling, extracellular ligand binding to the GPCR induces activation of the intracellularly associated Gα subunit, resulting in the dissociation of Gα from the Gβγ dimer and subsequent downstream signalling (Oldham and Hamm, 2008). If we assumed that the MLO2 protein acts as a canonical plant GPCR, Arabidopsis mutants defective in the Gα and/or Gβ and Gγ subunit we would expect to phenocopy MLO2-deficient plants resulting in powdery mildew resistance. However, none of the Arabidopsis mutants lacking any of the known heterotrimeric G-protein components displayed a resistant powdery mildew infection phenotype similar to mlo2 plants (Fig. 1A and B). Instead we observed that Gβ- and Gγ-deficient mutants exhibited enhanced powdery mildew susceptibility, indicating an involvement of these G-protein components in defence against powdery mildew fungi (Fig. 1A, B and C). While elevated fungal entry rates on agb1 mutants occurred irrespective of the presence or absence of the MLO2 protein, enhanced invasiveness of the pathogens on agg mutants was dependent on loss of MLO2 function (Fig. 1 A and B). These findings suggest a role of Gβ in powdery mildew defence that is independent of MLO2, whereas the contribution of the Gγ subunits to powdery mildew immunity seems to be controlled by the MLO2 protein (see below, Fig. 7). We used mlo2-conditioned spontaneous callose deposition as a second, pathogen-independent readout of MLO2 function, which revealed, with the exception of the mlo2-6/agg1-1 double mutant (discussed further below), unaltered phenotypes in the G-protein single mutants and respective double mutants in combination with mlo2-6 (Fig. 2A). In sum, none of the observed phenotypes of the G-protein mutants is in agreement with a putative canonical GPCR function for MLO2. This is consistent with results obtained in the context of two other Arabidopsis MLO family members, MLO4 and MLO11. These MLO proteins co-function to control root thigmomorphogenesis, and it was recently found that MLO4 and MLO11 are unlikely to operate through heterotrimeric G-proteins in this process (Chen et al., 2009).
Figure 7.
Proposed model for the involvement of the heterotrimeric G-protein complex in powdery mildew defense, in MAMP-triggered immunity and in MLO2-mediated control of spontaneous callose deposition. The model integrates previous findings (Consonni et al. 2006, 2010) and data obtained in this study.
Although results obtained in this and previous studies (Kim et al., 2002; Chen et al., 2009) disagree with a GPCR function for MLO2 in powdery mildew pathogenesis, premature leaf senescence and root thigmomorphogenesis, this activity remains a formal possibility for other yet unknown MLO-dependent processes. Alternatively, MLO proteins may constitute heptahelical cell surface receptors that transmit extracellular signals through mechanisms that function independently of G-protein coupling (aka signaling ‘at zero G’) as has been proven for some GPCRs in Dictyostelium discoideum and mammalian cells (Brzostowski and Kimmel, 2001). Precedence in plants for such a scenario is provided by the putative Arabidopsis GPCR, GCR1, which shares sequence similarity (~ 20 %) with the D. discoideum CAR1 receptor and has been shown to act in both, G-protein-dependent and -independent pathways (Chen et al., 2004; Pandey and Assmann, 2004). However, kinetic studies demonstrated that the sole canonical Arabidopsis Gα subunit is in the activated GTP-bound state by default, suggesting that the heterotrimeric G-protein acts without cognate GPCRs in Arabidopsis (Johnston et al., 2007; Jones et al., 2011).
It is also conceivable that the function of MLO proteins is entirely unrelated to ligand binding but may involve transduction of signals perceived by other types of receptors or may serve a non-receptor function altogether. The Arabidopsis genome contains more than 600 receptor-like kinases (RLKs), which represent nearly 2.5 % of the annotated protein-coding genes (Shiu and Bleecker, 2001). Results from a recent study aiming at the elucidation of the Arabidopsis membrane interactome by applying the yeast split-ubiquitin technology suggest some MLO proteins interact with RLKs (Lalonde et al., 2010). In agreement with a presumptive function of MLO proteins in RLK-associated processes, MLO7/NORTIA, another member of the Arabidopsis MLO protein family, and FERONIA (FER), a RLK, both control pollen tube reception (Kessler et al., 2010). Moreover, fer and mlo7 as well as mlo2 mutants share several phenotypic similarities, suggesting that FER and various MLO proteins co-function to regulate pollen tube reception, powdery mildew pathogenesis and maybe other yet unknown processes. However, a direct physical interaction between MLO proteins and FER has not been found as yet (S. Kessler, personal communication).
The Gβ subunit acts in defense against (hemi-) biotrophic pathogens
A number of early pharmacological analyses originally suggested a role for the heterotrimeric G-protein in plant defence (Legendre et al., 1992; Vera-Estrella et al., 1994; Beffa et al., 1995; Gelli et al., 1997; Mahady et al., 1998; Rajasekhar et al., 1999; Han and Yuan, 2004). Meanwhile genetic studies with Arabidopsis G-protein mutants provided conclusive evidence for a function of heterotrimeric G-protein signalling in plant immunity. Arabidopsis mutants lacking AGB1 display increased susceptibility to the necrotrophic fungi Plectosphaerella cucumerina, Alternaria brassicicola, Fusarium oxysporum and Botrytis cinerea, indicating a role for Gβ in defence against these pathogens (Llorente et al., 2005; Trusov et al., 2006; Trusov et al., 2007). In contrast, previous analyses failed to support an involvement of Gβ in defence against pathogens with a (hemi-) biotrophic life style such as P. syringae bacteria or the oomycete Hyaloperonospora arabdopsidis (Llorente et al., 2005; Trusov et al., 2006).
In the present study we found that Arabidopsis Gβ-deficient mutants allow higher host cell entry by the biotrophic powdery mildew fungi, G. orontii and E. pisi, indicating a role for Gβ in pre-invasive defence against these obligate biotrophic pathogens (Fig. 1A, B and C). Moreover, upon inoculation of P. syringae via spray inoculation, a procedure that more closely resembles the conditions of natural bacterial infection, we also observed increased bacterial susceptibility on Gβ-deficient mutants pointing to an additional role of Gβ in bacterial growth restriction (Fig. 6A and B). Thus, data acquired in this study for the first time support a function of the Gβ subunit in Arabidopsis defence against pathogens with a (hemi-) biotrophic life style (Fig. 7). Given that Gβ-dependent processes contribute to defence against a broad spectrum of plant pathogens with various life styles, Gβ likely either plays a role in basal plant immune mechanisms or indirectly affects plant immunity. While coexpression data favour the first possibility (Table 1), recent publications revealed a novel function for Gβ in the regulation of cell wall architecture, which may support the second scenario (Klopffleisch et al., 2011; Delgado-Cerezo et al., 2012). It is well established that alterations in Arabidopsis cell wall composition can lead to modified immune responses to various plant pathogens (Ellis and Turner, 2001; Vogel et al., 2002; Vogel et al., 2004; Hernández-Blanco et al., 2007; Cantu et al., 2008). Therefore, loss of Gβ-regulated cell wall modifications may be the cause for the altered plant disease responses.
The Gγ subunits are required for mlo2-mediated powdery mildew resistance
Arabidopsis mlo2-based powdery mildew resistance relies on functional SNARE protein-dependent secretory defence and the biosynthesis and extrusion of indolic secondary metabolites, as mutations in components of these two independent defence pathways compromise powdery mildew immunity in the mlo2 mutant background (Consonni et al., 2006; Consonni et al., 2010). Similarly, mlo2-mediated resistance to the powdery mildew fungi G. orontii and E. pisi was compromised by mutations in the Gγ subunits, AGG1 and AGG2, indicating that Gγ functions are also required for mlo2-mediated powdery mildew resistance (Fig. 1A and B, Fig. 7). Notably, mlo2 resistance against G. orontii, was only compromised when both genes, AGG1 and AGG2, were disrupted, indicating that both Gγ subunits have complementary functions in defence against the adapted powdery mildew pathogen (Fig. 1A). In comparison to the previously described suppressors of mlo2 resistance (Consonni et al., 2006; Consonni et al., 2010), we found that elevated G. orontii entry rates in the agg1-1/agg2-1/mlo2-6 triple mutant were intermediate between Col-0 wild-type and mlo2 control plants, indicating a less pronounced role for the Gγ subunits in defence against the compatible powdery mildew fungus. By contrast, mlo2-mediated resistance against the pea powdery mildew fungus, E. pisi, was impaired in the absence of the Gγ1 subunit only (Fig. 1B). This finding indicates that defence against the non-adapted pathogen in the mlo2 genotype is selectively mediated by Gγ1, while the Gγ2 subunit seems to be dispensable in this context. In contrast to the previously identified suppressors of mlo2 resistance, mutations in AGG1 (and/or AGG2) do not affect resistance against E. pisi in the presence of MLO2 (Fig. 1B), suggesting that AGG1 and AGG2 operate in a pathway that is unrelated to SNARE protein-mediated secretion and indolic secondary metabolite biosynthesis. Irrespective of the underlying mechanism these data indicate differential requirements of Gγ1 and Gγ2 in mlo2-mediated defence against adapted and non-adapted powdery mildew fungi. Selective functionality of the Gγ subunits was previously described in defence against the necrotrophic fungus Alternaria brassicicola (Trusov et al., 2007).
The Gγ1 subunit is required for mlo2-mediated developmental callose deposition and salicylic acid accumulation
We showed that function of Gγ1, but not Gγ2, is necessary for the developmentally-controlled callose deposition in mlo2 mutant plants (Fig. 2A, Fig. 7). The formation of callose deposits in mlo2 plants requires the PMR4/GSL5 callose synthase (Consonni et al., 2010). We conclude that loss of Gγ1 functions does not directly affect PMR4/GSL5-dependent callose biosynthesis, as synthesis of papillary and wound callose, which also requires PMR4/GSL5 (Jacobs et al., 2003; Nishimura et al., 2003), was not altered in the agg1-1/mlo2-6 double mutant (Fig. 2B and C). These data indicate that Gγ1 functions upstream of PMR4/GSL5-dependent callose deposition (Fig. 7). As spontaneous callose formation in mlo2 mutant plants is linked to functional SA signalling and increased SA accumulation (Consonni et al., 2010), we speculate that reduced SA level accounts for the impaired spontaneous callose deposition in agg1-1/mlo2-6 mutant plants. In fact, levels of free and total SA were markedly decreased in the double mutant (Fig. 3). By contrast, SA accumulation triggered by powdery mildew inoculation was not altered in the agg1-1/mlo2-6 mutant compared to control plants (Fig. 3B). Together these data indicate a positive regulatory role of Gγ1 in SA signalling or SA biosynthesis that is specific for developmentally-controlled callose deposition in mlo2 mutant plants (Fig. 7).
The Gβ subunit mediates flg22-triggered immune responses
Perception of MAMPs by the corresponding PRRs is associated with the activation of diverse physiological responses that are thought to contribute to robust immunity. However, mechanisms by which MAMP-induced responses are achieved are largely unknown. Increasing evidence points to a role of heterotrimeric G-protein signalling in the integration of MAMP-perception into downstream responses (Zhang et al., 2008; Ishikawa, 2009; Zhang et al., 2011). A previous study revealed a role for AGB1 in MAMP-induced oxidative burst control, positioning Gβ-mediated signalling early after MAMP perception upstream of RbohD (respiratory burst oxidase homologs D) activity, the NADPH oxidase essential for MAMP-induced ROS production (Zhang et al., 2007; Ishikawa, 2009). Using two independent agb1 mutants, agb1-2 and agb1-9, we corroborated reduced ROS production in response to flg22 treatment in these mutants (Fig. 4, Fig. 7). Residual ROS spiking in agb1 mutants suggests the involvement of additional components in flg22-triggered oxidative burst control.
Although flg22-induced ROS production was reduced in Gβ-deficient mutants, MAMP-triggered activation of MAPK cascades was not affected, indicating that Gβ-dependent signalling is not required for MAPK activity (Ishikawa, 2009). Together these results suggest that either both events occur independently of each other or that MAPKs act upstream of Gβ-mediated ROS formation. The relationship between ROS and MAPK cascades in plant defence is controversial; while some results favour the action of MAPKs upstream of ROS formation (Zhang et al., 2007), other data contradict this hypothesis (Lu et al., 2009; Boudsocq et al., 2010).
Data obtained in this study support a role for Gβ in flg22-mediated transcriptional reprogramming. Interestingly, Gβ seems to regulate the transcript accumulation of a subset of defence marker genes (NHL10, PROPEP2, PROEP3), whereas it does not affect the expression of other defence-related genes (WRKY22, PHI1) (Fig. 5, Supplemental Fig. 1). These findings indicate the existence of Gβ-dependent and Gβ-independent pathways regulating transcriptional reprogramming in response to MAMP treatment. This concept is in agreement with the recently proposed model of MAPK- and CDPK-specific activation of MAMP-triggered defence gene expression (Boudsocq et al., 2010). In summary, results from this and previous studies demonstrate that despite the simple repertoire of heterotrimeric G-protein components found in plants, signalling processes using the heterotrimeric G-protein are highly complex
MATERIAL AND METHODS
Plant material and growth conditions
Arabidopsis (Arabidopsis thaliana) wild-type Col-0, agb1-2 (Ullah et al., 2003), agg1-1 (Trusov et al., 2007), agg2-1 (Trusov et al., 2007), agg1-1/agg2-1 (Trusov et al., 2007), gpa1-4 (Jones et al., 2003), mlo2-6 (Consonni et al., 2006), agb1-9 (REF) and rgs1-1 (Chen et al., 2003) mutants were described G-protein mlo2 double and triple mutants were generated by crossing mlo2-6 plants with the corresponding G-protein mutants. Homozygous insertion mutants were selected by PCR using T-DNA- and gene-specific primer sets as described on the T-DNA Express webpage (http://signal.salk.edu/cgi-bin/tdnaexpress). Primer sequences are available on request. Homozygous agb1-9 mutants were identified by cleaved amplified polymorphic sequence (dCAPS) analysis using the dCAPS markers: 5’-GCTATGCGAGCAACAACACTTGCTACGCTT-3’ and 5’-CTGACAACCCCAAACAGC-TT-3’ followed by a DdeI digestion.
For flg22-triggered gene expression assays seedlings were pre-grown on ½ MS agar plates with 0.25 % sucrose for 5 d and subsequently on analogous liquid medium for additional 5–7 d under a 10/14 h light/dark cycle at 21°C and 70 % relative humidity. For all other experiments plants were soil grown under controlled conditions under a 10/14 h light/dark cycle at 23°C and 65 % relative humidity.
Phytopathogens and infection assays
The powdery mildew isolates of G. orontii and E. pisi kept at the Max-Planck Institute for Plant Breeding Research were used in this study (Lipka et al., 2005) and powdery mildew infection assays were performed as described previously (Consonni et al., 2006). The bacterial strain used was P. syringae pv. tomato DC3000 and bacteria spray infection assays were conducted as described before (Heidrich et al., 2011).
Analysis of callose deposition
For visualization of callose, rosette leaves were detached, cleared with ethanol/acidic acid (3/1 (v/v)) and subsequently stained for 24 h with 0.01 % aniline blue in 150 mM KH2PO4 (pH 5.8). Callose deposits were visualized by epifluorescence microscopy using an UV filter set. For the analysis of spontaneous callose depositon 6-week old plants were used, for the analysis of wound-induced callose, leaves from 4-week old plants were squeezed with forceps and collected after 24 h. For visualization of papillary callose, 4-week old plants were inoculated with E. pisi and leaves were harvested at 7 dpi.
Quantification of SA
SA quantification was done essentially as previously described (Straus et al., 2010).
ROS analysis
ROS assays were performed as described (Gómez-Gómez et al., 1999)with the following modifications. Leaf discs (5 mm diameter) excised from 4-week old plants were incubated over night in water before they were transferred into microtiter plates containing 50 µl water. ROS production was triggered by the addition of 1 µM flg22 (peptide, QRLSTGSRINSAKDDAAGLQIA, synthesized by Centic Biotec, Weimar, Germany), applied in a reaction mixture containing 50 µl water, 20 µM luminol (Fluka) and 1 µg horseradish peroxidase (Sigma-Aldrich). Luminescence was measured in a Centro LB 960 microplate luminometer (Berthold Technologies, Bad Wildbad, Germany). Flg22-mediated ROS production (relative luminescence units) in Figure 4A is represented as the integrated area under the ROS curve measured during a time course of 41 min and is referred to as Σ relative luminescence units. Figure 4B displays the kinetic of flg22-mediated ROS production during a time course of 41 min.
Gene expression analysis
The treatment of seedlings with flg22, the extraction of total RNA, cDNA synthesis, and quantitative RT-PCR were performed essentially as described previously (Kwaaitaal et al., 2011). To quantify transcripts of the indicated genes the following forward and reverse primers were used: 5’-GAGCTGAAGTGGCTTCCATGAC-3’ and 5’-GGTCCGACATACCCATGATCC-3’ for At4g26420 (Czechowski et al., 2005); 5’-TTCCTGTCCGTAACCCAAAC-3’ and 5’-CCCTCGTAGTAGGCATGAGC-3’ for NHL10 (At2g35980) (Boudsocq et al., 2010); 5’-TTGGTTTAGACGGGATGGTG-3’ and 5’-ACTCCAGTACAAGCCGATCC-3’ for PHI1 (At1g35140) (Boudsocq et al., 2010); 5’-AGAAAAGCCTAGTTCAGGTCGTC-3’ and 5’-CTCCTTATAAACTTGTATTGCCGC-3’ for PROPEP2 (At5g64890); 5’-GTTCCGGTCTCGAAAGTTCATC-3’ and 5’-TGAACTCTAATTGTGTTTGCCTCC-3’ for PROPEP3 (At5g64905) and 5’-CATCCGATCAACAGACGAGTAAAT-3’ and 5’-AAATTCGTCGGCTGAAGTCAC-3’ for WRKY22 (At4g01250) (Lu et al., 2009).
Supplementary Material
Acknowledgments
We thank Anja Reinstädler for technical assistance in plant genotyping and SA measurements.
This work was supported by grants of the Deutsche Forschungsgemeinschaft (grant no. PA861-6/1) to R.P. and the Max-Planck Society to J.L.
LITERATURE CITED
- Assmann SM. Heterotrimeric and unconventional GTP binding proteins in plant cell signaling. Plant Cell. 2002;14:S355–S373. doi: 10.1105/tpc.001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Pavan S, Zheng Z, Zappel NF, Reinstädler A, Lotti C, De Giovanni C, Ricciardi L, Lindhout P, Visser R, Theres K, Panstruga R. Naturally occurring broad-spectrum powdery mildew resistance in a central American tomato accession is caused by loss of Mlo function. Mol Plant Microbe Interact. 2008;21:30–39. doi: 10.1094/MPMI-21-1-0030. [DOI] [PubMed] [Google Scholar]
- Beffa R, Szell M, Meuwly P, Pay A, Vogeli-Lange R, Metraux JP, Neuhaus G, Meins F, Nagy F. Cholera toxin elevates pathogen resistance and induces pathogenesis-related gene expression in tobacco. EMBO J. 1995;14:5753–5761. doi: 10.1002/j.1460-2075.1995.tb00264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng S-H, Sheen J. Differential innate immune signalling via Ca2+ sensor protein kinases. Nature. 2010;464:418–422. doi: 10.1038/nature08794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzostowski JA, Kimmel AR. Signaling at zero G: G-protein-independent functions for 7-TM receptors. Trends Biochem Sci. 2001;26:291–297. doi: 10.1016/s0968-0004(01)01804-7. [DOI] [PubMed] [Google Scholar]
- Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J, Töpsch S, Vos P, Salamini F, Schulze-Lefert P. The barley Mlo gene: a novel control element of plant pathogen resistance. Cell. 1997;88:695–705. doi: 10.1016/s0092-8674(00)81912-1. [DOI] [PubMed] [Google Scholar]
- Cantu D, Vicente AR, Labavitch JM, Bennett AB, Powell ALT. Strangers in the matrix: plant cell walls and pathogen susceptibility. Trends Plant Sci. 2008;13:610–617. doi: 10.1016/j.tplants.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Chakravorty D, Trusov Y, Zhang W, Acharya BR, Sheahan MB, McCurdy DW, Assmann SM, Botella JR. An atypical heterotrimeric G-protein γ-subunit is involved in guard cell K+-channel regulation and morphological development in Arabidopsis thaliana . Plant J. 2011;67:840–851. doi: 10.1111/j.1365-313X.2011.04638.x. [DOI] [PubMed] [Google Scholar]
- Chen J-G. Heterotrimeric G-protein signaling in Arabidopsis: puzzling G-protein-coupled receptor. Plant Signal Behav. 2008;3:1042–1045. doi: 10.4161/psb.3.12.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-G, Pandey S, Huang J, Alonso JM, Ecker JR, Assmann SM, Jones AM. GCR1 can act independently of heterotrimeric G-protein in response to brassinosteroids and gibberellins in Arabidopsis seed germination. Plant Physiol. 2004;135:907–915. doi: 10.1104/pp.104.038992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP. A seven-transmembrane RGS protein that modulates plant cell proliferation. Science. 2003;301:1728–1731. doi: 10.1126/science.1087790. [DOI] [PubMed] [Google Scholar]
- Chen Z, Noir S, Kwaaitaal M, Hartmann HA, Wu M-J, Mudgil Y, Sukumar P, Muday G, Panstruga R, Jones AM. Two seven-transmembrane domain MILDEW RESISTANCE LOCUS O proteins cofunction in Arabidopsis root thigmomorphogenesis. Plant Cell. 2009;21:1972–1991. doi: 10.1105/tpc.108.062653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consonni C, Bednarek P, Humphry M, Francocci F, Ferrari S, Harzen A, Ver Loren van Themaat E, Panstruga R. Tryptophan-derived metabolites are required for antifungal defense in the Arabidopsis mlo2 mutant. Plant Physiol. 2010;152:1544–1561. doi: 10.1104/pp.109.147660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consonni C, Humphry ME, Hartmann HA, Livaja M, Durner J, Westphal L, Vogel J, Lipka V, Kemmerling B, Schulze-Lefert P, Somerville SC, Panstruga R. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat Genet. 2006;38:716–720. doi: 10.1038/ng1806. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Cerezo M, Sánchez-Rodríguez C, Escudero V, Miedes E, Fernández PV, Jordá L, Hernández-Blanco C, Sánchez-Vallet A, Bednarek P, Schulze-Lefert P, Somerville S, Estevez JM, Persson S, Molina A. Arabidopsis heterotrimeric G-protein regulates cell wall defense and resistance to necrotrophic fungi. Mol Plant. 2012;5:98–114. doi: 10.1093/mp/ssr082. [DOI] [PubMed] [Google Scholar]
- Devoto A, Piffanelli P, Nilsson I, Wallin E, Panstruga R, von Heijne G, Schulze-Lefert P. Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J Biol Chem. 1999;274:34993–35004. doi: 10.1074/jbc.274.49.34993. [DOI] [PubMed] [Google Scholar]
- Ellis C, Turner JG. The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell. 2001;13:1025–1033. doi: 10.1105/tpc.13.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa Y, Kato H, Iwasaki Y. Structure and function of heterotrimeric G proteins in plants. Plant Cell Physiol. 2001;42:789–794. doi: 10.1093/pcp/pce111. [DOI] [PubMed] [Google Scholar]
- Gelli A, Higgins VJ, Blumwald E. Activation of plant plasma membrane Ca2+-permeable channels by race-specific fungal elicitors. Plant Physiol. 1997;113:269–279. doi: 10.1104/pp.113.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Felix G, Boller T. A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana . Plant J. 1999;18:277–284. doi: 10.1046/j.1365-313x.1999.00451.x. [DOI] [PubMed] [Google Scholar]
- Han R-B, Yuan Y-J. Oxidative burst in suspension culture of Taxus cuspidataInduced by a laminar shear stress in short-term. Biotech Progress. 2004;20:507–513. doi: 10.1021/bp034242p. [DOI] [PubMed] [Google Scholar]
- Heidrich K, Wirthmueller L, Tasset C, Pouzet C, Deslandes L, Parker JE. Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science. 2011;334:1401–1404. doi: 10.1126/science.1211641. [DOI] [PubMed] [Google Scholar]
- Hernández-Blanco C, Feng DX, Hu J, Sánchez-Vallet A, Deslandes L, Llorente F, Berrocal-Lobo M, Keller H, Barlet X, Sánchez-Rodríguez C, Anderson LK, Somerville S, Marco Y, Molina A. Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell. 2007;19:890–903. doi: 10.1105/tpc.106.048058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Pearce G, Ryan CA. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Nat Acad Sci USA. 2006;103:10098–10103. doi: 10.1073/pnas.0603727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphry M, Bednarek P, Kemmerling B, Koh S, Stein M, Göbel U, Stüber K, Piślewska-Bednarek M, Loraine A, Schulze-Lefert P, Somerville S, Panstruga R. A regulon conserved in monocot and dicot plants defines a functional module in antifungal plant immunity. Proc Nat Acad Sci USA. 2010;107:21896–21901. doi: 10.1073/pnas.1003619107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphry M, Reinstädler A, Ivanov S, Bisseling T, Panstruga R. Durable broad-spectrum powdery mildew resistance in pea er1 plants is conferred by natural loss-of-function mutations in PsMLO1 . Mol Plant Path. 2011;12:866–878. doi: 10.1111/j.1364-3703.2011.00718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa A. The Arabidopsis G-protein beta-subunit is required for defense response against Agrobacterium tumefaciens . Biosci Biotech Biochem. 2009;73:47–52. doi: 10.1271/bbb.80449. [DOI] [PubMed] [Google Scholar]
- Jacobs AK, Lipka V, Burton RA, Panstruga R, Strizhov N, Schulze-Lefert P, Fincher GB. An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell. 2003;15:2503–2513. doi: 10.1105/tpc.016097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CA, Taylor JP, Gao Y, Kimple AJ, Grigston JC, Chen J-G, Siderovski DP, Jones AM, Willard FS. GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proc Nat Acad Sci USA. 2007;104:17317–17322. doi: 10.1073/pnas.0704751104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Assmann SM. Plants: the latest model system for G-protein research. EMBO Reports. 2004;5:572–578. doi: 10.1038/sj.embor.7400174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Ecker JR, Chen J-G. A reevaluation of the role of the heterotrimeric G protein in coupling light responses in Arabidopsis. Plant Physiol. 2003;131:1623–1627. doi: 10.1104/pp.102.017624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Temple BRS, Jones AM, Dohlman HG. Functional reconstitution of an atypical G protein heterotrimer and regulator of G protein signaling protein (RGS1) from Arabidopsis thaliana . J Biol Chem. 2011;286:13143–13150. doi: 10.1074/jbc.M110.190355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen IH. Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica. 1992;63:141–152. [Google Scholar]
- Kessler SA, Shimosato-Asano H, Keinath NF, Wuest SE, Ingram G, Panstruga R, Grossniklaus U. Conserved molecular components for pollen tube reception and fungal invasion. Science. 2010;330:968–971. doi: 10.1126/science.1195211. [DOI] [PubMed] [Google Scholar]
- Kim MC, Panstruga R, Elliott C, Muller J, Devoto A, Yoon HW, Park HC, Cho MJ, Schulze-Lefert P. Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature. 2002;416:447–451. doi: 10.1038/416447a. [DOI] [PubMed] [Google Scholar]
- Klopffleisch K, Phan N, Augustin K, Bayne RS, Booker KS, Botella JR, Carpita NC, Carr T, Chen J-G, Cooke TR, Frick-Cheng A, Friedman EJ, Fulk B, Hahn MG, Jiang K, Jorda L, Kruppe L, Liu C, Lorek J, McCann MC, Molina A, Moriyama EN, Mukhtar MS, Mudgil Y, Pattathil S, Schwarz J, Seta S, Tan M, Temp U, Trusov Y, Urano D, Welter B, Yang J, Panstruga R, Uhrig JF, Jones AM. Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Mol Syst Biol. 2011;7 doi: 10.1038/msb.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwaaitaal M, Huisman R, Maintz J, Reinstädler A, Panstruga R. Ionotropic glutamate receptor (iGluR)-like channels mediate MAMP-induced calcium influx in Arabidopsis thaliana . Biochem J. 2011;440:355–365. doi: 10.1042/BJ20111112. [DOI] [PubMed] [Google Scholar]
- Lalonde S, Sero A, Pratelli Rj, Pilot G, Chen J, Sardi MI, Parsa SA, Kim D-Y, Acharya BR, Stein EV, Hu H-C, Villiers F, Takeda K, Yang Y, Han YS, Schwacke R, Chiang W, Kato N, Loqu D, Assmann SM, Kwak JM, Schroeder J, Rhee SY, Frommer WB. A membrane protein / signaling protein interaction network for Arabidopsis version AMPv2. Frontiers Physiol. 2010;1 doi: 10.3389/fphys.2010.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre L, Heinstein PF, Low PS. Evidence for participation of GTP-binding proteins in elicitation of the rapid oxidative burst in cultured soybean cells. J Biol Chem. 1992;267:20140–20147. [PubMed] [Google Scholar]
- Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D, Llorente F, Molina A, Parker J, Somerville S, Schulze-Lefert P. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science. 2005;310:1180–1183. doi: 10.1126/science.1119409. [DOI] [PubMed] [Google Scholar]
- Llorente F, Alonso-Blanco C, Sánchez-Rodriguez C, Jorda L, Molina A. ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina . Plant J. 2005;43:165–180. doi: 10.1111/j.1365-313X.2005.02440.x. [DOI] [PubMed] [Google Scholar]
- Lu G, Wang Z, Jones A, Moriyama E. 7TMRmine: a web server for hierarchical mining of 7TMR proteins. BMC Genomics. 2009;10:275. doi: 10.1186/1471-2164-10-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Tintor N, Mentzel T, Kombrink E, Boller T, Robatzek S, Schulze-Lefert P, Saijo Y. Uncoupling of sustained MAMP receptor signaling from early outputs in an Arabidopsis endoplasmic reticulum glucosidase II allele. Proc Nat Acad Sci USA. 2009;106:22522–22527. doi: 10.1073/pnas.0907711106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahady GB, Liu C, Beecher CWW. Involvement of protein kinase and G proteins in the signal transduction of benzophenanthridine alkaloid biosynthesis. Phytochem. 1998;48:93–102. doi: 10.1016/s0031-9422(97)00823-6. [DOI] [PubMed] [Google Scholar]
- Miles GP, Samuel MA, Jones AM, Ellis BE. Mastoparan rapidly activates plant MAP kinase signaling independent of heterotrimeric G proteins. Plant Physiol. 2004;134:1332–1336. doi: 10.1104/pp.103.037275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama E, Strope P, Opiyo S, Chen Z, Jones A. Mining the Arabidopsis thaliana genome for highly-divergent seven transmembrane receptors. Genome Biol. 2006;7:R96. doi: 10.1186/gb-2006-7-10-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura MT, Stein M, Hou B-H, Vogel JP, Edwards H, Somerville SC. Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science. 2003;301:969–972. doi: 10.1126/science.1086716. [DOI] [PubMed] [Google Scholar]
- Oldham WM, Hamm HE. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol. 2008;9:60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- Pandey S, Assmann SM. The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein alpha subunit GPA1 and regulates abscisic acid signaling. Plant Cell. 2004;16:1616–1632. doi: 10.1105/tpc.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panstruga R, Schulze-Lefert P. Corruption of host seven-transmembrane proteins by pathogenic microbes: a common theme in animals and plants? Microbes Infect. 2003;5:429–437. doi: 10.1016/s1286-4579(03)00053-4. [DOI] [PubMed] [Google Scholar]
- Perfus-Barbeoch L, Jones AM, Assmann SM. Plant heterotrimeric G protein function: insights from Arabidopsis and rice mutants. Cur Opin Plant Biol. 2004;7:719–731. doi: 10.1016/j.pbi.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Piffanelli P, Zhou F, Casais C, Orme J, Jarosch B, Schaffrath U, Collins NC, Panstruga R, Schulze-Lefert P. The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 2002;129:1076–1085. doi: 10.1104/pp.010954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekhar VK, Lamb C, Dixon RA. Early events in the signal pathway for the oxidative burst in soybean cells exposed to avirulent Pseudomonas syringaepv glycinea . Plant Physiol. 1999;120:1137–1146. doi: 10.1104/pp.120.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S-H, Bleecker AB. Plant receptor-like kinase gene family: diversity, function and signaling. Sci. STKE. 2001 doi: 10.1126/stke.2001.113.re22. [DOI] [PubMed] [Google Scholar]
- Straus MR, Rietz S, Ver Loren van Themaat E, Bartsch M, Parker JE. Salicylic acid antagonism of EDS1-driven cell death is important for immune and oxidative stress responses in Arabidopsis. Plant J. 2010;62:628–640. doi: 10.1111/j.1365-313X.2010.04178.x. [DOI] [PubMed] [Google Scholar]
- Temple BRS, Jones AM. The plant heterotrimeric G-protein complex. Ann Rev Plant Biol. 2007;58:249–266. doi: 10.1146/annurev.arplant.58.032806.103827. [DOI] [PubMed] [Google Scholar]
- Trusov Y, Rookes JE, Chakravorty D, Armour D, Schenk PM, Botella JR. Heterotrimeric G proteins facilitate Arabidopsis resistance to necrotrophic pathogens and are involved in jasmonate signaling. Plant Physiol. 2006;140:210–220. doi: 10.1104/pp.105.069625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov Y, Rookes JE, Tilbrook K, Chakravorty D, Mason MG, Anderson D, Chen J-G, Jones AM, Botella JR. Heterotrimeric G protein gamma subunits provide functional selectivity in Gbetagamma dimer signaling in Arabidopsis. Plant Cell. 2007;19:1235–1250. doi: 10.1105/tpc.107.050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H, Chen J-G, Temple B, Boyes DC, Alonso JM, Davis KR, Ecker JR, Jones AM. The β-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell. 2003;15:393–409. doi: 10.1105/tpc.006148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera-Estrella R, Higgins VJ, Blumwald E. Plant defense response to fungal pathogens (II. G-protein-mediated changes in host plasma membrane redox reactions) Plant Physiol. 1994;106:97–102. doi: 10.1104/pp.106.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Schiff C, Somerville SC. PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell. 2002;14:2095–2106. doi: 10.1105/tpc.003509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Somerville CR, Somerville SC. Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J. 2004;40:968–978. doi: 10.1111/j.1365-313X.2004.02264.x. [DOI] [PubMed] [Google Scholar]
- Wolter M, Hollricher K, Salamini F, Schulze-Lefert P. The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defence mimic phenotype. Mol Gen Genet. 1993;239:122–128. doi: 10.1007/BF00281610. [DOI] [PubMed] [Google Scholar]
- Zhang J, Shao F, Li Y, Cui H, Chen L, Li H, Zou Y, Long C, Lan L, Chai J, Chen S, Tang X, Zhou J-M. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe. 2007;1:175–185. doi: 10.1016/j.chom.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Zhang W, He SY, Assmann SM. The plant innate immunity response in stomatal guard cells invokes G-protein-dependent ion channel regulation. Plant J. 2008;56:984–996. doi: 10.1111/j.1365-313X.2008.03657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Jeon BW, Assmann SM. Heterotrimeric G-protein regulation of ROS signalling and calcium currents in Arabidopsis guard cells. J Exp Bot. 2011;62:2371–2379. doi: 10.1093/jxb/erq424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.