Abstract
Genome architecture varies considerably among eukaryotes in terms of both size and structure (e.g. distribution of sequences within the genome, elimination of DNA during formation of somatic nuclei). The diversity in eukaryotic genome architectures and the dynamic processes that they undergo are only possible due to the well-developed nature of an epigenetic toolkit, which likely existed in the Last Eukaryotic Common Ancestor (LECA). This toolkit may have arisen as a means of navigating the genomic conflict that arose from the expansion of transposable elements within the ancestral eukaryotic genome. This toolkit has been coopted to support the dynamic nature of genomes in lineages across the eukaryotic tree of life. Here we highlight how the changes in genome architecture in diverse eukaryotes are regulated by epigenetic processes by focusing on DNA elimination, genome rearrangements, and adaptive changes to genome architecture. The ability to epigenetically modify and regulate genomes has contributed greatly to the diversity of eukaryotes observed today.
Epigenetic mechanisms regulate gene expression, modify genome structures, silence mobile genetic elements, and are widespread among eukaryotes, suggesting that at least some were present in the last eukaryotic common ancestor [LECA; 1,2–4]. For example, the RNAi pathway that is involved in the post-transcriptional regulation of transposable elements (TEs) also plays a role in guiding large-scale chromatin remodeling processes such as de novo DNA methylation in plants [5,6] and diatoms [7], as well as in modifying histones [8,9]. Evidence for transgenerational epigenetic inheritance, a concept the emerged from Barbara McClintock’s discovery of the impact of transposable elements (TEs) on phenotypes in corn, is now well established in plants and animals where it often involves chromatin modifications [10]. While less is known about microeukaryotic lineages, there is a growing body of literature suggesting that epigenetic processes underlie the structure and function of genomes in diverse lineages.
One hypothesis for the proliferation of epigenetic mechanisms in eukaryotes is that they evolved first to manage genome conflict that resulted from the expansion of TEs and then became coopted for other uses [11]. Silencing of TEs can be done post-transcriptionally or through heterochromatin formation targeting mobile elements [12,13], and both require epigenetic mechanisms that are now deployed more generally throughout the genome. As described below, several eukaryotic lineages have managed to reduce the negative impact of TEs through developmentally regulated genome rearrangements, which include the loss of ‘germline-specific’ genome sequences during the generation of somatic nuclei [14]. Other lineages have coopted epigenetic mechanisms to regulate gene expression and nuclear architecture [15,16].
Here we describe the links between epigenetic mechanisms and the diversity of genome architectures in lineages from across the eukaryotic tree of life. Available data are most abundant for plants, animals and fungi, and we discuss only select data from these multicellular lineages as reviews exist to cover many topics within these clades [17–19]. Data from the rest of the eukaryotic tree of life are patchy, and come largely from model lineages (e.g. ciliates), and parasites and pathogens (e.g. Entamoeba, Plasmodium, Phytophthora). We are confident that examples of the roles of epigenetic processes in shaping genomes will only expand as poorly-sampled lineages receive greater scrutiny. We also believe that the value of this review includes highlighting the exceptions to biological principles (e.g. the concept of a static genome within species) that emerge from studies of diverse eukaryotic lineages.
Diversity of eukaryotic genome contents
Understanding the impact of epigenetic processes in eukaryotes requires an appreciation of the tremendous variation in size and content of eukaryotic genomes [11]. This is perhaps best exemplified by the C-value paradox whereby genome size is highly variable and does not obviously correlate with any measure of complexity, particularly in eukaryotes [11,20,21]. Among eukaryotes, size variation can be extreme with genomes ranging from only 2.3 Mbp in the microsporidian fungus Encephalitozoon intestinalis (Opisthokonta; Fungi) [22], 3 Gbp in Homo sapiens (Opisthokonta; Metazoa) [23], to over 20 Gbp in the gymnosperm Pinus taeda (Loblolly pine; Plantae [24]) and an estimated 670 Gbp in the Amoeba dubia (Amoebozoa) [25]. Variation in the number of TEs is one factor that contributes to variation in genome sizes, with the proportion of transposable elements comprising more than 50% of the genome content in some lineages [11]. Transposable elements are rare in other lineages including the ancient-asexual Bdelloid rotifers (Opisthokonta; Metazoa) [26] and the somatic macronuclei of ciliates (SAR) [27] where they comprise less than 10% of the genome.
DNA elimination in establishing somatic genomes
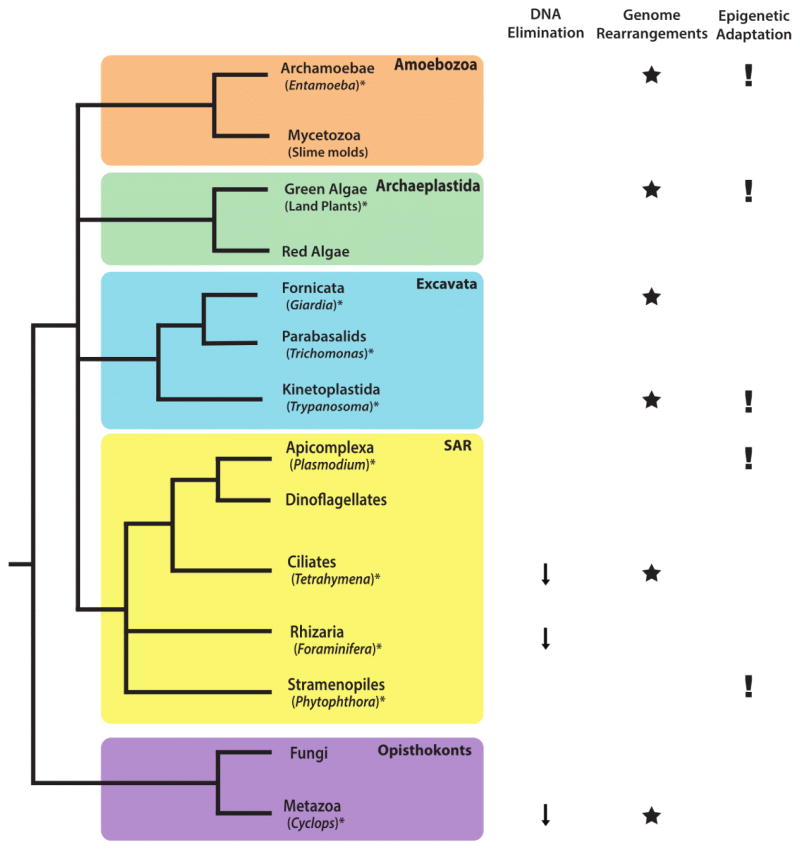
One example of epigenetic control of eukaryotic genome structure can be seen in the purging of portions of the genome during the development of somatic nuclei. This distinction between germline and somatic nuclei defines both animals and ciliates, and is also found in a subset of foraminifera (Figure 1) [28].
Figure 1.
Distribution of epigenetic processes across the eukaryotic tree of life. These exemplar epigenetically regulated processes are widespread across eukaryotes. Organisms denoted with ‘*’ are discussed in this review.
Distinct germline and somatic genomes in animals
Beyond simply differing between haploid and diploid, multiple non-sister animal lineages generate somatic genomes with distinct contents that often includes reduced levels of TEs and other repetitive elements (Figure 1)[14]. During early animal development, the germline genome is physically sequestered into specialized tissues where it often remains heavily heterochromatinized for much of the life cycle [29,30]. The loss of germline-specific DNA, also described as chromatin diminution, has been documented in a diversity of non-monophyletic animal lineages [14] and molecular details have been worked out in ascarid worms [31], copepods [32], and in early-diverging vertebrates (i.e. hagfish and lampreys) [33–35]. In copepods, for example, the zygotic genome expands through successive rounds of endoreplication and/or TE proliferation [32,36,37], which is then followed by large-scale elimination of germline-limited sequences [37]. In Cyclops kolensis (Opisthokonta), the genome is amplified from ~ 1 Gbp up to ~75 Gbp [37]. Recently, Sun et al. [36] sequenced portions of both the somatic and germline genomes of Mesocyclops edax (Opisthokotna) revealing that TEs are rare in the somatic genome, and younger (i.e. less degenerate) TEs appear to be more effectively eliminated (absent) from the somatic genome [36]. Given the broad distribution of examples of DNA elimination during the formation of somatic nuclei in lineages across the animal tree of life [14], we suspect that this process may be even more widespread and may have evolved as a means of managing the genome conflict introduced by the invasion of TEs.
Distinct germline and somatic genomes in ciliates
Ciliates are marked by the presence of distinct germline and somatic genomes within a shared cytoplasm. Because of mechanistic similarities in some elements of chromosome processing, Klobutcher and Herrick [38] argued that nuclear dualism in ciliates arose as a means of eliminating TEs from the somatic genome (Figure 1; SAR). The somatic macronucleus harbors gene-rich chromosomes that are the result from developmentally regulated genome processing following conjugation (i.e. sex). These processes include DNA elimination, genome rearrangements and genome amplification [39,40]. In contrast, the germline micronucleus is enriched in repetitive regions that interrupt gene-coding regions [27,39]. Many of these repetitive regions harbor signatures of TEs, suggesting that an ancient proliferation of TEs was counterbalanced by the evolution/cooption of mechanisms for DNA elimination of germline-limited sequences during somatic development [38,41]. For example, a domesticated PiggyBac transposase (i.e. PiggyMAC) is responsible for excision of germline-limited DNA, effectively deleting TEs from the somatic genome.
The molecular mechanisms behind genome reduction have been worked out in some ciliate lineages and involve a suite of epigenetic players [42–44]. In the model ciliate Tetrahymena thermophila (SAR), which only eliminates ~30% of its germline genome, small RNAs are enriched in germline specific sequences and are believed to serve as scan RNAs during the development of the somatic nucleus [45]. In contrast, the ciliate Stylonychia lemnae (SAR), which eliminates >90% of its germline genome, small RNAs appear to target somatic sequences to be kept [46]. These same small RNAs also contribute to heterochromatin formation, by guiding repressive histone modifications [44] and DNA methylation [47] in regions to be eliminated.
Transposable elements, epigenetics, and the potential for adaptation
The idea that epigenetic mechanisms evolved at least in part as a means of silencing transposable elements is well-established and has been reviewed elsewhere [19,48,49]. Some well documented examples of epigenetic silencing of transposable elements include: RNA-directed de novo DNA methylation in plants and diatoms [50,51], repeat-induced point mutations in fungi [52], and small RNA guided transposon silencing in animals [53]. Despite the ability of diverse eukaryotes to effectively ‘purge’ or silence TEs throughout development, TEs and their associated processing/silencing in genomes can also play an adaptive role [10,11,54] and perhaps even influence patterns of speciation [55]. For example, cell-to-cell heterogeneity and life stage specific control of gene expression – both of which are categorized as stochastic developmental variation –are underlain by epigenetic modifications to chromatin and have been argued to be adaptive in lineages as diverse as bacteria, yeast, animals, plants, apicomplexa, ciliates, green algae, slime molds, and choanoflagellates [56–59]. The broad distribution of stochastic developmental variation among lineages of bacteria, archaea and eukaryotes suggests that this phenomenon may have been present in the last universal common ancestor [LUCA; 58].
Epigenetic mechanisms and expansive TE burden in plants
The prevalence of TEs in plants led to the concept that a diverse epigenetic toolkit evolved for genome defense from TEs and viruses [60], and that this toolkit has become part of an adaptive, TE-mediated response to stress [61,62]. The diverse suite of epigenetic mechanisms in plants can been attributed to the large portion of genomes comprised of both functional TEs and repetitive elements (i.e. degraded TEs; >80% in some plants such as Zea mays; Plantae [63]). Silencing of TEs in plants occurs through RNA-directed DNA methylation, where transcribed TEs are processed into the small RNAs that guide their own de novo methylation [64,65]. During non-stressed growth, epigenetic proteins ensure the maintenance of heterochromatin and genomic stability in the vast TE rich chromosomal regions [66,67].
Evidence for the adaptive impact of TEs in adaptive responses in plants has emerged in recent decades. Upon abiotic stress in Arabidopsis (Plantae), TE activity increases measurably, leading to distinct changes in genome organization through both homologous recombination and copy number variation of TEs and protein coding genes [62,68]. Interestingly, these effects are heritable through multiple generations of progeny, suggesting the possibility that this response is adaptive [62,68,69]. For example, increased rates of homologous recombination are heritable in Nicotiana tabacum (tobacco; Plantae), where stress induces global changes in hypermethylation of DNA and loci-specific hypomethylation that allows for recombination [70]. It is possible that the impacts of genome rearrangement are adaptive to some individuals due to beneficial changes in gene regulation or even gene copy number (Figure 1).
Epigenetic modifications of genome structures in eukaryotic parasites
We focus on the role of epigenetics in parasites to exemplify processes in eukaryotic microbes, largely due to the lack of data in non-parasitic lineages; we do recognize that data are beginning to emerge from lineages such as dinoflagellates, stramenopiles and other marine algae [71–73]. Epigenetic mechanisms play a role in phenotypic plasticity and in the ability of parasites to modify host physiology and behavior [74–77]. Moreover, mechanisms like pathogen-induced chromatin modifications also play a role in bacterial disease [74], suggesting that they may be very ancient.
The apicomplexan parasite Plasmodium falciparum (Figure 1; SAR), the causative agent of malaria, relies on epigenetic mechanisms to regulate the transcription of genes necessary for its varying life cycle stages [56,74,78–80]. Transitions between life cycle stages in Plasmodium is in part driven by post-translational modifications of histones [56] and in part by large scale reorganization of nuclear architecture [79]. Plasmodium falciparum also differentially modifies the expression of the var genes that underlie antigenic variation through epigenetic modification of histones in small chromatin domains; the var genes are located in subtelomeric regions and their expression is regulated both by localized modification of chromatin and position within the nucleus [56]. Epigenetic mechanisms in the apicomplexan Toxoplasma gondii (Figure 1; SAR) have evolved to alter the behavior of one of their hosts, the rat, to make it less fearful of cats, which are the final hosts for the parasite [77].
Life cycle variation is also epigenetically regulated in the parasite Giardia intestinalis (Figure 1; Excavata) [81]. Changes in histone acetylation correspond to transition from free-living to encysted states [81]. Another interesting feature about the structure of the G. intestinalis genome is the restriction of active retrotransposons to subtelomeric regions [82]. The variation in the number of retrotransposons (and their recombination) may contribute to the variable karyotypes observed among strains of Giardia [82–84]. These homologous regions could allow for recombination in the absence of traditional meiosis, providing Giardia with an alternative means to generate genomic diversity after the fusion of its two nuclei [84,85].
Another disease-causing group of Excavata, the kinetoplastids (e.g. Leishmania and Trypanosoma; Figure 1; Excavata), also deploy epigenetic mechanisms in causing disease (e.g. Leishmaniasis, African sleeping sickness) and evading host immune systems. The genus Trypanosoma relies on epigenetic modification of VSG (variable surface glycoprotein) genes to evade host immune systems [75], including inducing homologous recombination of VSG genes nestled in subtelomeric regions. Similar to the var genes in Plasmodium, changes in nuclear position of the active VSG gene initiate changes in chromatin structure (e.g. chromatin condensation) that lead to differential and mono-allelic VSG expression [15]. Beyond altering their own genome, the parasite Leishmania donovani (the causative agent of leishmaniasis) is able to induce epigenetic modifications in host macrophages that allow for the successful invasion by the parasite [76].
Epigenetics may also underlie karyotype variation in the genus Entamoeba (Figure 1; Amoebozoa), which includes Entamoeba histolytica, the causative agent of dysentery [86]. As in Giardia, karyotype variation may be generated by recombination between transposable elements within the genome, and may contribute to the ability of Entamoeba to escape host immune systems [86]. Adding a further layer of complexity, differential methylation of TEs in Entamoeba has been linked to varying levels of virulence [75,87]. Together, these data indicate the role the epigenetic toolkit plays in virulence of this human pathogen.
Genome architecture also drives patterns of substitutions in the genomes of some eukaryotic lineages. Oomycetes and some filamentous fungi (Figure 1; SAR; Stramenopiles and Opisthokonta; Fungi respectively) have managed to physically partition their genomes into core regions with greater conservation that are interrupted by gene-poor plastic regions[88–90]. This is most apparent in Phytophthora infestans, the causative agent in the Irish potato famine, whose 240 Mbp genome is divided unevenly as the regions of conserved ‘house-keeping’ genes that comprise about 25% of the total genome size. The gene-poor regions that comprise the bulk of the P. infestans genome are rich in mobile and repetitive elements and are associated with pathogenicity and epigenetic silencing [89]. This division of function within the P. infestans genome behaves almost as two functionally and spatially distinct genomes, and is determined by epigenetic mechanisms. RNAi-mediated heterochromatin formation not only controls the activity of mobile elements but also has major impacts on the transcription of nearby effector genes (more than half of all effector genes in P. infestans are within <2kb of a TE) where increasing proximity can alter an effector gene’s transcription due to the spreading of heterochromatin from targeted loci [91,92]. The combination of complex epigenetic silencing and the evolutionary impacts of the repetitive genome on gene evolution (e.g. copy number variation, and recombination) contribute to the incredible virulence of the pathogenic oomycetes.
Perspective
Epigenetic mechanisms that regulate transposable elements as part of genome defense have been coopted and contribute to the development of diversity across the eukaryotic tree of life. Eukaryotes share a core epigenetic toolkit (though individual components vary among lineages) comprised of proteins and RNAs that regulate histone and DNA modifications, and that enable RNA scanning mechanisms. These epigenetic processes have expanded among eukaryotic lineages and have enabled eukaryotes to explore diverse genomic landscapes. The resulting epigenetic toolkit provides the basis for the dynamic processes that have contributed to the overall diversity and success of eukaryotic lineages.
Glossary
- Endoreplication
Replication of the genome without any following cell division that leads to changes in ploidy
- Heterochromatin
Tightly packed chromatin that blocks transcription from occurring and is associated with histone modifications
- Histone modification
Post-transcriptional modifications of the histone proteins at varying amino acid residues. The most well known are histone methylation and acetylation, which are often generalized to be repressive and activating modifications, respectively
- Macronucleus
Somatic and transcriptionally active nucleus in ciliates. Contains streamlined chromosomes that leack centromeric sequences and are often gene-rich. In some ciliate lineages, processing of germline chromosomes leads to macronuclei with chromosomes coding for single-genes and that can be highly amplified
- Micronucleus
The germline nucleus in ciliates that is heterochromatinized and has a more traditional genome architecture (e.g. long chromosomes with centromeric sequences). Micronuclear genomes also contain transposable element sequences that sometimes interrupt protein-coding genes
- Stochastic developmental variation
Seemingly random changes in phenotype such as heterogeneity in gene expression among cells. Stochastic developmental variation provides populations with genetic diversity that may allow exploration of adaptive landscapes
- Transposable elements
Regions of DNA that are capable of changing their position in the genome
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shabalina SA, Koonin EV. Origins and evolution of eukaryotic RNA interference. Trends in Ecology & Evolution. 2008;23:578–587. doi: 10.1016/j.tree.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parfrey LW, Lahr DJG, Katz LA. The dynamic nature of eukaryotic genomes. Molecular Biology and Evolution. 2008;25:787–794. doi: 10.1093/molbev/msn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oliverio AM, Katz LA. The dynamic nature of genomes across the tree of life. Genome Biology and Evolution. 2014;6:482–488. doi: 10.1093/gbe/evu024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cerutti H, Casas-Mollano JA. On the oridin and functions of RNA-mediated silencing: from protists to man. Current Genetics. 2006;50:81–99. doi: 10.1007/s00294-006-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wassenegger M, Heimes S, Riedel L, Sanger HL. Rna-Directed De-Novo Methylation of Genomic Sequences in Plants. Cell. 1994;76:567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 6.Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJM. Targets of RNA-directed DNA methylation. Current Opinion in Plant Biology. 2007;10:512–519. doi: 10.1016/j.pbi.2007.06.007. [DOI] [PubMed] [Google Scholar]
- *7.Veluchamy A, Lin X, Maumus F, Rivarola M, Bhavsar J, Creasy T, O’Brien K, Sengamalay NA, Tallon LJ, Smith AD, et al. Insights into the role of DNA methylation in diatoms by genome-wide profiling in Phaeodactylum tricornutum. Nature Communications. 2013;4 doi: 10.1038/ncomms3091. Provides evidence for de novo RNA-directed DNA methylation, which has previously been found only in plants, suggesting this process may be a prevalent way of combating TE expansion in eukaryotes. [DOI] [PubMed] [Google Scholar]
- 8.Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Current Biology. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 10.Heard E, Martienssen RA. Transgenerational epigenetic inheritance: myths and mechanisms. Cell. 2014;157:95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **11.Fedoroff NV. Presidential address. Transposable elements, epigenetics, and genome evolution. Science. 2012;338:758–767. doi: 10.1126/science.338.6108.758. Synthesizes evidence on the role of TEs in shaping the evolution of genomes, including hypothezing that epigenetic mechanisms in eukaryotes have evolved from pathways to silence TEs. Provides examples from plants to document phenomena. [DOI] [PubMed] [Google Scholar]
- 12.Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Research. 2007;35:5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D-melanogaster germline. Current Biology. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- 14.Wang JB, Davis RE. Programmed DNA elimination in multicellular organisms. Current Opinion in Genetics & Development. 2014;27:26–34. doi: 10.1016/j.gde.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *15.Landeira D, Navarro M. Nuclear repositioning of the VSG promoter during developmental silencing in Trypanosoma brucei. J Cell Biol. 2007;176:133–139. doi: 10.1083/jcb.200607174. Large scale changes in the nuclear position of loci result in the rapid activation of genes and aid in host immune evasion in T. brucei. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espada J, Esteller M. Epigenetic control of nuclear architecture. Cellular and Molecular Life Sciences. 2007;64:449–457. doi: 10.1007/s00018-007-6358-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diez CM, Roessler K, Gaut BS. Epigenetics and plant genome evolution. Current Opinion in Plant Biology. 2014;18:1–8. doi: 10.1016/j.pbi.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 18.Feng SH, Jacobsen SE, Reik W. Epigenetic Reprogramming in Plant and Animal Development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nature Reviews Genetics. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 20.Gregory TR. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biological Reviews. 2001;76:65–101. doi: 10.1017/s1464793100005595. [DOI] [PubMed] [Google Scholar]
- 21.Cavaliersmith T. Nuclear volume control by nucleoskeletal DNA, selection for cell-volume and cell-growth rate, and solution of DNA C-value paradox. Journal of Cell Science. 1978;34:247–278. doi: 10.1242/jcs.34.1.247. [DOI] [PubMed] [Google Scholar]
- 22.Corradi N, Pombert JF, Farinelli L, Didier ES, Keeling PJ. The complete sequence of the smallest known nuclear genome from the microsporidian Encephalitozoon intestinalis. Nature Communications. 2010;1 doi: 10.1038/ncomms1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morton NE. Parameters of the Human Genome. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:7474–7476. doi: 10.1073/pnas.88.17.7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wegrzyn JL, Liechty JD, Stevens KA, Wu LS, Loopstra CA, Vasquez-Gross H, Dougherty WM, Lin BY, Zieve JJ, Martinez-Garcia PJ, et al. Unique Features of the Loblolly Pine (Pinus taeda L) Megagenome Revealed Through Sequence Annotation. Genetics. 2014;196:891. doi: 10.1534/genetics.113.159996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friz CT. The biochemical composition of the free-living Amoebae Chaos chaos, Amoeba dubia and Amoeba proteus. Comp Biochem Physiol. 1968;26:81–90. doi: 10.1016/0010-406x(68)90314-9. [DOI] [PubMed] [Google Scholar]
- 26.Arkhipova I, Meselson M. Deleterious transposable elements and the extinction of asexuals. Bioessays. 2005;27:76–85. doi: 10.1002/bies.20159. [DOI] [PubMed] [Google Scholar]
- 27.Coyne RS, Lhuillier-Akakpo M, Duharcourt S. RNA-guided DNA rearrangements in ciliates: Is the best genome defence a good offence? Biology of the Cell. 2012;104:309–325. doi: 10.1111/boc.201100057. [DOI] [PubMed] [Google Scholar]
- 28.Katz LA. Evolution of nuclear dualism in ciliates: a reanalysis in light of recent molecular data. Int J Syst Evol Microbiol. 2001;51:1587–1592. doi: 10.1099/00207713-51-4-1587. [DOI] [PubMed] [Google Scholar]
- 29.Maatouk DM, Kellam LD, Mann MRW, Lei H, Li E, Bartolomei MS, Resnick JL. DNA methylation is a primary mechanism for silencing postmigratory primordial germ cell genes in both germ cell and somatic cell lineages. Development. 2006;133:3411–3418. doi: 10.1242/dev.02500. [DOI] [PubMed] [Google Scholar]
- 30.Robert VJP, Sijen T, van Wolfswinkel J, Plasterk RHA. Chromatin and RNAi factors protect the C-elegans germline against repetitive sequences. Genes & Development. 2005;19:782–787. doi: 10.1101/gad.332305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bachmann-Waldmann C, Jentsch S, Tobler H, Muller F. Chromatin diminution leads to rapid evolutionary changes in the organization of the germ line genomes of the parasitic nematodes A. suum and P. univalens. Molecular and Biochemical Parasitology. 2004;134:53–64. doi: 10.1016/j.molbiopara.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 32.Drouin G. Chromatin diminution in the copepod Mesocyclops edax: diminution of tandemly repeated DNA families from somatic cells. Genome. 2006;49:657–665. doi: 10.1139/g06-022. [DOI] [PubMed] [Google Scholar]
- 33.Smith JJ, Antonacci F, Eichler EE, Amemiya CT. Programmed loss of millions of base pairs from a vertebrate genome. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11212–11217. doi: 10.1073/pnas.0902358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kohno S, Kubota S, Nakai Y. Chromatin diminution and chromosome elimination in hagfishes. Biology of Hagfishes. 1998:81–100. [Google Scholar]
- 35.Nakai Y, Kubota S, Kohno S. Chromatin Diminution and Chromosome Elimination in 4 Japanese Hagfish Species. Cytogenetics and Cell Genetics. 1991;56:196–198. doi: 10.1159/000133087. [DOI] [PubMed] [Google Scholar]
- 36.Sun C, Wyngaard G, Walton DB, Wichman HA, Mueller RL. Billions of basepairs of recently expanded, repetitive sequences are eliminated from the somatic genome during copepod development. Bmc Genomics. 2014;15 doi: 10.1186/1471-2164-15-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wyngaard GA, Rasch EM, Connelly BA. Unusual augmentation of germline genome size in Cyclops kolensis (Crustacea, Copepoda): further evidence in support of a revised model of chromatin diminution. Chromosome Research. 2011;19:911–923. doi: 10.1007/s10577-011-9234-3. [DOI] [PubMed] [Google Scholar]
- **38.Klobutcher LA, Herrick G. Developmental genome reorganization in ciliated protozoa: The transposon link. Progress in Nucleic Acid Research and Molecular Biology. 1997;5656:1–62. doi: 10.1016/s0079-6603(08)61001-6. This paper proposes that eliminated portions of the germline genome in ciliates are derived from transposable elements and argues that the mechanisms for supressing TEs have been coopted to shape ciliate genome development and evolution. [DOI] [PubMed] [Google Scholar]
- 39.Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jahn CL, Klobutcher LA. Genome remodeling in ciliated protozoa. Annual Reviews in Microbiology. 2002;56:489–520. doi: 10.1146/annurev.micro.56.012302.160916. [DOI] [PubMed] [Google Scholar]
- 41.Klobutcher LA, Herrick G. Developmental genome reorganization in ciliated protozoa: The transposon link. Progress in Nucleic Acid Research and Molecular Biology. 1997;5656:1–62. doi: 10.1016/s0079-6603(08)61001-6. [DOI] [PubMed] [Google Scholar]
- 42.Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- 43.Chalker DL, Meyer E, Mochizuki K. Epigenetics of Ciliates. Cold Spring Harbor Perspectives in Biology. 2013;5 doi: 10.1101/cshperspect.a017764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, Taverna SD, Muratore TL, Shabanowitz J, Hunt DF, Allis CD. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes & Development. 2007;21:1530–1545. doi: 10.1101/gad.1544207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mochizuki K, Gorovsky MA. Small RNAs in genome rearrangement in Tetrahymena. Current Opinion in Genetics & Development. 2004;14:181. doi: 10.1016/j.gde.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 46.Chalker DL, Yao MC. DNA Elimination in Ciliates: Transposon Domestication and Genome Surveillance. In: Bassler BL, Lichten M, Schupbach G, editors. Annual Review Genetics. Vol. 45. 2011. pp. 227–246. [DOI] [PubMed] [Google Scholar]
- 47.Bracht JR, Perlman DH, Landweber LF. Cytosine methylation and hydroxymethylation mark DNA for elimination in Oxytricha trifallax. Genome Biology. 2012;13 doi: 10.1186/gb-2012-13-10-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lisch D. Epigenetic Regulation of Transposable Elements in Plants. Annual Review of Plant Biology. 2009;60:43–66. doi: 10.1146/annurev.arplant.59.032607.092744. [DOI] [PubMed] [Google Scholar]
- 49.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 50.Saze H, Tsugane K, Kanno T, Nishimura T. DNA Methylation in Plants: Relationship to Small RNAs and Histone Modifications, and Functions in Transposon Inactivation. Plant and Cell Physiology. 2012;53:766–784. doi: 10.1093/pcp/pcs008. [DOI] [PubMed] [Google Scholar]
- 51.Rogato A, Richard H, Sarazin A, Voss B, Navarro SC, Champeimont R, Navarro L, Carbone A, Hess WR, Falciatore A. The diversity of small non-coding RNAs in the diatom Phaeodactylum tricornutum. Bmc Genomics. 2014;15 doi: 10.1186/1471-2164-15-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galagan JE, Selker EU. RIP: the evolutionary cost of genome defense. Trends in Genetics. 2004;20:417–423. doi: 10.1016/j.tig.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 53.Halic M, Moazed D. Transposon Silencing by piRNAs. Cell. 2009;138:1058–1060. doi: 10.1016/j.cell.2009.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lai JS, Li YB, Messing J, Dooner HK. Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:9068–9073. doi: 10.1073/pnas.0502923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belyayev A. Bursts of transposable elements as an evolutionary driving force. J Evol Biol. 2014;27:2573–2584. doi: 10.1111/jeb.12513. [DOI] [PubMed] [Google Scholar]
- 56.Cortes A, Crowley VM, Vaquero A, Voss TS. A View on the Role of Epigenetics in the Biology of Malaria Parasites. Plos Pathogens. 2012;8 doi: 10.1371/journal.ppat.1002943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Levy SF, Ziv N, Siegal ML. Bet Hedging in Yeast by Heterogeneous, Age-Correlated Expression of a Stress Protectant. Plos Biology. 2012;10 doi: 10.1371/journal.pbio.1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vogt G. Stochastic developmental variation, an epigenetic source of phenotypic diversity with far-reaching biological consequences. Journal of Biosciences. 2015;40:159–204. doi: 10.1007/s12038-015-9506-8. [DOI] [PubMed] [Google Scholar]
- **59.Rouxel T, Grandaubert J, Hane JK, Hoede C, van de Wouw AP, Couloux A, Dominguez V, Anthouard V, Bally P, Bourras S, et al. Effector diversification within compartments of the Leptosphaeria maculans genome affected by Repeat-Induced Point mutations. Nature Communications. 2011;2 doi: 10.1038/ncomms1189. Describes how an epigenetic defence against TE proliferation has been coopted to increase the diversity of effector genes, improving the organism’s pathogenicity. This paper provides a detailed discussion on the history of TE proliferation and how selection has retained their presence when in close proximity to genes involved in pathogenicity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *60.Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity (vol 15, 394, 2014) Nature Reviews Genetics. 2014;15 doi: 10.1038/nrg3683. Provides thorough insight into the recently described de novo DNA methylation pathway found in plants, evaluting/assessing? divergent paralogs of core eukaryotic proteins. [DOI] [PubMed] [Google Scholar]
- 61.Bond DM, Finnegan EJ. Passing the message on: inheritance of epigenetic traits. Trends Plant Sci. 2007;12:211–216. doi: 10.1016/j.tplants.2007.03.010. [DOI] [PubMed] [Google Scholar]
- *62.Molinier J, Ries G, Zipfel C, Hohn B. Transgeneration memory of stress in plants. Nature. 2006;442:1046–1049. doi: 10.1038/nature05022. This study demonstrated that epigenetic memory of stress is heritable across numerous generations. Several generations of offspring from lines of stressed plants responded to stress by increasing their rates of homologous recombination. [DOI] [PubMed] [Google Scholar]
- 63.Tenaillon MI, Hufford MB, Gaut BS, Ross-Ibarra J. Genome Size and Transposable Element Content as Determined by High-Throughput Sequencing in Maize and Zea luxurians. Genome Biology and Evolution. 2011;3:219–229. doi: 10.1093/gbe/evr008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity (vol 15, 394, 2014) Nature Reviews Genetics. 2014:15. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 65.Law JA, Vashisht AA, Wohlschlegel JA, Jacobsen SE. SHH1, a Homeodomain Protein Required for DNA Methylation, As Well As RDR2, RDM4, and Chromatin Remodeling Factors, Associate with RNA Polymerase IV. Plos Genetics. 2011;7 doi: 10.1371/journal.pgen.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stancheva I. Caught in conspiracy: cooperation between DNA methylation and histone H3K9 methylation in the establishment and maintenance of heterochromatin. Biochemistry and Cell Biology-Biochimie Et Biologie Cellulaire. 2005;83:385–395. doi: 10.1139/o05-043. [DOI] [PubMed] [Google Scholar]
- 67.Zilberman D, Henikoff S. Silencing of transposons in plant genomes: kick them when they’re down. Genome Biology. 2004;5 doi: 10.1186/gb-2004-5-12-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.DeBolt S. Copy Number Variation Shapes Genome Diversity in Arabidopsis Over Immediate Family Generational Scales. Genome Biology and Evolution. 2010;2:441–453. doi: 10.1093/gbe/evq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tricker PJ. Transgenerational inheritance or resetting of stress-induced epigenetic modifications: two sides of the same coin. Frontiers in Plant Science. 2015;6 doi: 10.3389/fpls.2015.00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *70.Kathiria P, Sidler C, Golubov A, Kalischuk M, Kawchuk LM, Kovalchuk I. Tobacco Mosaic Virus Infection Results in an Increase in Recombination Frequency and Resistance to Viral, Bacterial, and Fungal Pathogens in the Progeny of Infected Tobacco Plants. Plant Physiology. 2010;153:1859–1870. doi: 10.1104/pp.110.157263. This paper provides evidence for increased recombination (genomic instability) in plants stressed by numerous pathogens. Also, progeny displayed elevated rates of homologous recombination in pathogen resistance genes and have a shorted delay in response to pathogens that had stressed their parents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin S. Genomic understanding of dinoflagellates. Res Microbiol. 2011;162:551–569. doi: 10.1016/j.resmic.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 72.Lopez-Gomollon S, Beckers M, Rathjen T, Moxon S, Maumus F, Mohorianu I, Moulton V, Dalmay T, Mock T. Global discovery and characterization of small non-coding RNAs in marine microalgae. BMC Genomics. 2014;15:697. doi: 10.1186/1471-2164-15-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maumus F, Rabinowicz P, Bowler C, Rivarola M. Stemming epigenetics in marine stramenopiles. Curr Genomics. 2011;12:357–370. doi: 10.2174/138920211796429727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gomez-Diaz E, Jorda M, Angel Peinado M, Rivero A. Epigenetics of Host-Pathogen Interactions: The Road Ahead and the Road Behind. Plos Pathogens. 2012;8 doi: 10.1371/journal.ppat.1003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Croken MM, Nardelli SC, Kim K. Chromatin modifications, epigenetics, and how protozoan parasites regulate their lives. Trends in Parasitology. 2012;28:202–213. doi: 10.1016/j.pt.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marr AK, MacIsaac JL, Jiang RW, Airo AM, Kobor MS, McMaster WR. Leishmania donovani Infection Causes Distinct Epigenetic DNA Methylation Changes in Host Macrophages. Plos Pathogens. 2014;10 doi: 10.1371/journal.ppat.1004419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *77.Hari Dass SA, Vyas A. Toxoplasma gondii infection reduces predator aversion in rats through epigenetic modulation in the host medial amygdala. Mol Ecol. 2014;23:6114–6122. doi: 10.1111/mec.12888. Toxoplasma infections drive a reduction in DNA methylation in the rat medial amygdala. The impact is a dramatic switch in rat behavior, resulting in ‘fearless’ rats that ultimately aids Toxoplasma transmission into cats, which host the sexual cycle of the parasites. [DOI] [PubMed] [Google Scholar]
- 78.Salcedo-Amaya AM, Hoeijmakers WA, Bartfai R, Stunnenberg HG. Malaria: could its unusual epigenome be the weak spot? Int J Biochem Cell Biol. 2010;42:781–784. doi: 10.1016/j.biocel.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 79.Ay F, Bunnik EM, Varoquaux N, Vert J-P, Noble WS, Le Roch KG. Multiple dimensions of epigenetic gene regulation in the malaria parasite Plasmodium falciparum. Bioessays. 2015;37:182–194. doi: 10.1002/bies.201400145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Deshmukh AS, Srivastava S, Dhar SK. Plasmodium falciparum: epigenetic control of var gene regulation and disease. Sub-Cellular Biochemistry. 2013;61:659–682. doi: 10.1007/978-94-007-4525-4_28. [DOI] [PubMed] [Google Scholar]
- 81.Sonda S, Morf L, Bottova I, Baetschmann H, Rehrauer H, Caflisch A, Hakimi M-A, Hehl AB. Epigenetic mechanisms regulate stage differentiation in the minimized protozoan Giardia lamblia. Molecular Microbiology. 2010;76:48–67. doi: 10.1111/j.1365-2958.2010.07062.x. [DOI] [PubMed] [Google Scholar]
- 82.Arkhipova IR, Morrison HG. Three retrotransposon families in the genome of Giardia lamblia: Two telomeric, one dead. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:14497–14502. doi: 10.1073/pnas.231494798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Le Blancq SM, Adam RD. Structural basis of karyotype heterogeneity in Giardia lamblia. Molecular and Biochemical Parasitology. 1998;97:199–208. doi: 10.1016/s0166-6851(98)00150-9. [DOI] [PubMed] [Google Scholar]
- 84.Poxleitner MK, Carpenter ML, Mancuso JJ, Wang CJR, Dawson SC, Cande WZ. Evidence for karyogamy and exchange of genetic material in the binucleate intestinal parasite Giardia intestinalis. Science. 2008;319:1530–1533. doi: 10.1126/science.1153752. [DOI] [PubMed] [Google Scholar]
- 85.Ramesh MA, Malik SB, Logsdon JM. A phylogenomic inventory of meiotic genes: Evidence for sex in Giardia and an early eukaryotic origin of meiosis. Current Biology. 2005;15:185–191. doi: 10.1016/j.cub.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 86.Weedall GD, Hall N. Evolutionary genomics of Entamoeba. Res Microbiol. 2011;162:637–645. doi: 10.1016/j.resmic.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *87.Kumari V, Sharma R, Yadav VP, Gupta AK, Bhattacharya A, Bhattacharya S. Differential distribution of a SINE element in the Entamoeba histolytica and Entamoeba dispar genomes: role of the LINE-encoded endonuclease. BMC Genomics. 2011;12:267. doi: 10.1186/1471-2164-12-267. Comparative genome analysis of between pathogenic E. histolytica and non-pathogenic E. dispar suggests that the distribution of TEs may play a role in pathogenicity, supposedly due to the impact of their silencing on nearby loci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gijzen M. Runaway repeats force expansion of the Phytophthora infestans genome. Genome Biology. 2009;10 doi: 10.1186/gb-2009-10-10-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Haas BJ, Kamoun S, Zody MC, Jiang RHY, Handsaker RE, Cano LM, Grabherr M, Kodira CD, Raffaele S, Torto-Alalibo T, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 90.Raffaele S, Kamoun S. Genome evolution in filamentous plant pathogens: why bigger can be better. Nature Reviews Microbiology. 2012;10:417–430. doi: 10.1038/nrmicro2790. [DOI] [PubMed] [Google Scholar]
- 91.van West P, Shepherd SJ, Walker CA, Li S, Appiah AA, Grenville-Briggs LJ, Govers F, Gow NAR. Internuclear gene silencing in Phytophthora infestans is established through chromatin remodelling. Microbiology-Sgm. 2008;154:1482–1490. doi: 10.1099/mic.0.2007/015545-0. [DOI] [PubMed] [Google Scholar]
- *92.Vetukuri RR, Asman AK, Jahan SN, Avrova AO, Whisson SC, Dixelius C. Phenotypic diversification by gene silencing in Phytophthora plant pathogens. Communicative & integrative biology. 2013;6:e25890–e25890. doi: 10.4161/cib.25890. This study shows how the local release of TE silencing in Oomycetes can rapidly alter the active pathogenic effector genes and putatively allows for quick shifts in their preferred hosts. [DOI] [PMC free article] [PubMed] [Google Scholar]