Abstract
Despite recent clinical advances in immunotherapy, a fraction of cancer patients fails to respond to these interventions. Evidence from preclinical mouse models as well as clinical samples has provided evidence that the extent of activated T cell infiltration within the tumor microenvironment is associated with clinical response to immunotherapies including checkpoint blockade. Therefore, understanding the molecular mechanisms mediating the lack of T cell infiltration into the tumor microenvironment will be instrumental for the development of new therapeutic strategies to render those patients immunotherapy responsive. Recent data have suggested that major sources of intersubject heterogeneity include differences in somatic mutations in specific oncogene pathways between cancers of individual subjects and also environmental factors including commensal microbial composition. Successful identification of such causal factors should lead to new therapeutic approaches that may facilitate T cell entry into noninflamed tumors and expand the fraction of patients capable of responding to novel immunotherapies.
1. RATIONALE FOR STUDYING REGULATION OF THE T CELL-INFLAMED TUMOR MICROENVIRONMENT PHENOTYPE
Interrogation of the tumor microenvironment in human melanoma patients was initially pursued to identify mechanisms that might explain clinical response vs resistance to cancer vaccines in individual patients. To this end, baseline biopsies of melanoma metastases were analyzed by transcriptional profiling and confirmatory assays. This analysis revealed two major subsets of tumor microenvironment that were largely characterized by the presence or absence of a gene signature indicative of a T cell-inflamed tumor microenvironment. The T cell-inflamed subset of tumors showed presence of T cell transcripts, chemokines that likely mediate effector T cell recruitment, macrophage activation markers, and a type I IFN transcriptional profile (Harlin et al., 2009). Immunohistochemistry confirmed the presence of CD8+ T cells, macrophages, and some B cells in these tumors. Interestingly, patients with clinical benefit to these vaccines exclusively showed the T cell-inflamed tumor microenvironment phenotype. Thus, the ability of cells within the tumor microenvironment to produce chemokines and recruit activated T cells appears to be instrumental for clinical benefit. Similar results have been observed in patients treated with high-dose IL-2 (Sullivan et al., 2009) and also with the anti-CTLA-4 mAb ipilimumab (Hamid et al., 2011).
The question arises as to why tumors infiltrated with T cells are not rejected spontaneously. More detailed analysis of the T cell-inflamed subset of tumors revealed the presence of transcripts encoding indoleamine-2, 3-dioxygenase (IDO), PD-L1, and FoxP3, all markers of immune-inhibitory pathways. IHC confirmed that PD-L1 and IDO protein expression, and also nuclear FoxP3+CD4+ cells, were found within T cell-inflamed tumors in the same region as CD8+ T cells. Mouse mechanistic studies confirmed that CD8+ T cells were required for the upregulation of all of these three factors within the tumor microenvironment. For PD-L1 and IDO induction, the requisite factor produced by the CD8+ T cells was interferon (IFN)-γ. For FoxP3+ Tregs, production of the chemokine CCL22 was identified, which mediated Treg recruitment into tumor sites (Spranger et al., 2013). Together, these data suggest that the involvement of these three immune-inhibitory mechanisms in T cell-inflamed tumors is driven by the activated CD8+ T cells themselves, and likely explains failed spontaneous tumor elimination.
Importantly, clinical response with anti-PD-1 mAb, which is blocking PD-L1/PD-1 interactions directly within the tumor microenvironment, was found to occur almost exclusively in patients with preexisting T cell infiltrates in the region of PD-L1 upregulation (Topalian et al., 2012). Following anti-PD-1 administration, these CD8+ T cells seemed to proliferate and expand to penetrate throughout the tumor, an event associated with tumor regression (Tumeh et al., 2014). These observations are consistent with preclinical data indicating that tumor regression upon checkpoint blockade was almost completely mediated by reactivation of CD8+ T cells directly within the tumor site to be able to proliferate and produce IL-2 (Spranger et al., 2014).
Based on the observation that multiple immune regulatory pathways appear to be operational within the same tumor microenvironment, combination immunotherapies are being pursued. In preclinical models, concurrent doublets of anti-CTLA-4± anti-PD-L1± an IDO inhibitor were found to be synergistic in the B16 melanoma model in vivo (Spranger et al., 2014). Interestingly, each of these combination therapies involved reacquisition of IL-2 production and proliferation by CD8+ T cells directly within the tumor microenvironment. In support of the importance of reactivation of T cells already present at the tumor site, therapeutic effects were preserved even in the presence of FTY720 blockade, which prevents exit of new T cells from lymph nodes. All three of these combination doublets are being tested clinically, and the combination of anti-CTLA-4+ anti-PD-1 was recently approved by the FDA for the treatment of patients with advanced melanoma (Larkin et al., 2015). With these ongoing successes in hand, increased attention is being invested into studying the non-T cell-inflamed tumor microenvironment, as a strategy to identify mechanisms of primary resistance to these successful immunotherapies.
2. MOLECULAR AND CELLULAR DRIVERS OF THE T CELL-INFLAMED TUMOR MICROENVIRONMENT
One step toward understanding what is failing to occur in the non-T cell-inflamed tumor microenvironment subset has been to investigate the mechanistic steps involved in successful generation of a T cell-inflamed phenotype when it does occur (Fig. 1). A critical process in spontaneous T cell priming against tumor-associated antigens involves the recruitment and activation of Batf3-lineage dendritic cells (DCs), expressing the markers CD8α or CD103 in the mouse (Edelson et al., 2010; Engelhardt et al., 2012; Fuertes et al., 2011; Hildner et al., 2008). By using knockout mice deficient for the transcription factor Batf3, it has been proven that lack of this particular subset of DCs is resulting in a significant reduction of systemic activation antitumor CD8+ T cells (Fuertes et al., 2011). In addition, successful priming of antitumor immune responses requires this subset of DCs to receive signaling by type I IFNs (IFN-α/β), which are induced to be produced by host cells in the presence of tumor challenge (Fuertes et al., 2011). In concordance with these preclinical findings, analysis of human melanoma samples showed that the absence of type I interferons was also associated with the absence of T cells from the tumor microenvironment (Fuertes, Woo, Burnett, Fu, & Gajewski, 2013).
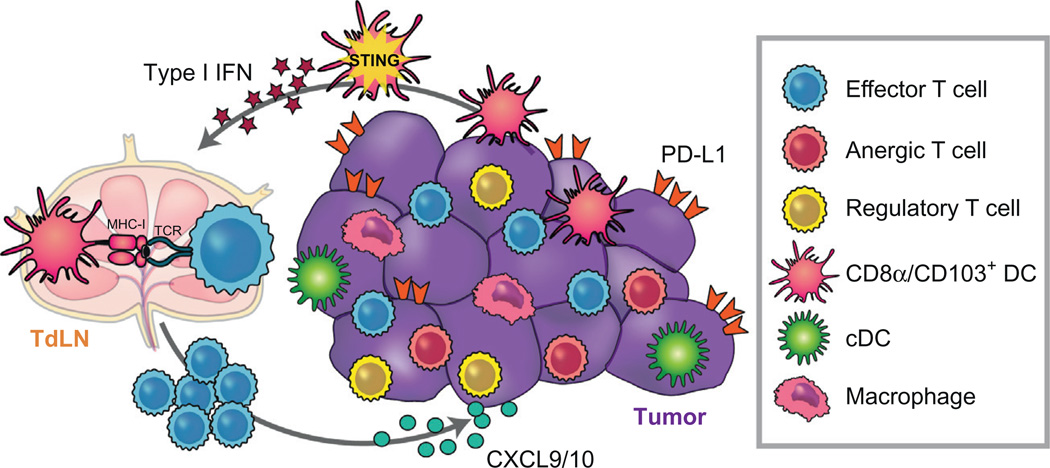
Figure 1.
Processes involved in generation of the T cell-inflamed tumor microenvironment phenotype. Innate immune sensing of tumors appears to be dominantly mediated by the host STING pathway, which results in type I IFN production and activation of Batf3-lineage DCs. This DC subset cross-presents antigens to CD8+ T cells, which once activated can be recruited into the inflamed tumor microenvironment and contribute to tumor control. The same STING pathway engagement likely leads to production of CXCL9 and CXCL10, which are the key chemokines that support effector T cell recruitment.
The critical involvement of host type I IFNs and Batf3-lineage DCs raised the next level question as to what tumor-derived factors might be responsible for inducing the requisite type I IFN production by host immune cells. Using a series of gene-targeted mice deficient in specific innate immune sensing pathways, recent evidence has implicated the DNA-sensing cGAS/STING pathway as critical for the spontaneous priming of antitumor T cells in vivo (Woo et al., 2014). Consistent with these results, intratumoral injection of STING agonists has resulted in profound immune-mediated tumor elimination in preclinical models in vivo (Corrales et al., 2015). Thus, one strategy with the potential to initiate de novo immune responses in non-T cell-inflamed tumors is through the use of STING agonists.
Once effector T cells are activated differentiated in peripheral lymphoid organs, the presence of appropriate chemokines within the tumor microenvironment is critical for those T cells to traffic into tumor sites. Gene expression profiling of human melanoma metastases identified a range of chemokines associated with CD8+ T cell presence (Erdag et al., 2012; Harlin et al., 2009; Salerno, Olson, McSkimming, Shea, & Slingluff, 2014). Recent work has indicated that the CXCR3/CXCL9/10 chemokine axis is mandatory for T cell entry into the tumor microenvironment (Mikucki et al., 2015). Interestingly, one consequence of STING pathway activation in DCs is production of CXCL9 and CXCL10 (Woo et al., 2014), suggesting that properly activated DCs within the tumor microenvironment might also contribute to effector T cell recruitment. As mentioned earlier, once activated T cells successfully traffic into the tumor site, if antigen-expressing tumor cells are not immediately eradicated, immune-inhibitory mechanisms become upregulated that counteract chronic inflammation. Thus, immunotherapies aiming to block the function of those pathways are expected to be preferentially active in tumors displaying the T cell-inflamed phenotype. Uncovering molecular causes for absence of the T cell-inflamed tumor microenvironment should therefore enable new interventions to be developed that can expand the proportion of patients with immunotherapy-responsive tumors.
3. TUMOR-INTRINSIC ONCOGENE PATHWAYS MEDIATING THE NON-T CELL-INFLAMED TUMOR MICROENVIRONMENT
3.1 Tumor-Intrinsic WNT/β-Catenin Pathway Activation is Causal for T Cell Exclusion in Melanoma
The possibility that somatic differences at the level of the tumor cells themselves might explain the lack of a T cell-inflamed tumor microenvironment in a subset of cases was recently pursued through gene expression profiling of 266 individual melanoma metastases in concert with exome sequencing of the same tumors (Spranger, Bao, & Gajewski, 2015). Exome-sequencing data revealed that gain of function mutations in the β-catenin gene were found in seven tumors (14%) and loss-of-function mutations in negative regulators of the β-catenin pathway (APC, Axin1, TCF1) were identified in an additional ten of the non-T cell-inflamed tumors (23%). In addition, gene expression analysis of defined β-catenin target genes revealed that 48% of the non-T cell-inflamed tumors showed evidence for activation of the WNT/β-catenin pathway. Additional analysis of those β-catenin target genes showed a negative correlation with CD8α expression in the tumor, whereas PD-L1 expression showed an expected positive correlation with CD8α (Spranger et al., 2013). Through confirmatory immunohistochemistry, high β-catenin protein expression was associated with the absence of CD8+ T cells within the tumor, indicating a significant correlation between activation of the WNT/β-catenin pathway and a non-T cell-inflamed tumor microenvironment (Spranger et al., 2015).
To interrogate the mechanistic relevance of tumor-intrinsic β-catenin signaling in controlling the host immune response to melanoma, genetically engineered mice were developed using a tamoxifen-regulated Cre-driven by the tyrosinase promoter in combination with active Braf (BrafV600E) and conditional PTEN deletion (PTEN−/−), with or without a conditional active β-catenin mutant (CAT-STA) (Bosenberg et al., 2006; Damsky et al., 2011; Dankort et al., 2009). Melanomas arising from BrafV600E/PTEN−/− mice did have a modest T cell infiltrate at baseline as analyzed by flow cytometry and immunohistochemistry. However, melanomas comprising BrafV600E combined with active β-catenin (BrafV600E/CAT-STA) completely lacked a T cell infiltrate. Moreover, addition of PTEN deletion (BrafV600E/PTEN−/−/CAT-STA) to accelerate tumor growth resulted in melanomas also lacking a T cell infiltrate. These results demonstrate that activation of the β-catenin pathway within melanoma tumor cells can dominantly exclude T cells from the tumor microenvironment.
The step at which tumor-intrinsic β-catenin activation might be antagonizing antitumor T cell responses was pursued by integrating Cre-inducible expression of the model antigen SIY (SIYRYYGL) (Cheung, Dupage, Dong, Chen, & Jacks, 2008). The use of this model antigen allowed the use of adoptive transfer of SIY-specific 2C TCR-transgenic T cells in order to evaluate endogenous T cell activation. Indeed, mice with SIY+ tumors driven by mutated Braf, PTEN deletion, and active β-catenin (BrafV600E/PTEN−/−/CAT-STA) showed absence of spontaneous T cell activation, while tumors without active β-catenin showed normal T cell activation measured by CFSE dilution. In-depth analysis revealed a complete lack of Batf3-lineage DCs expressing the surface markers CD103 or CD8α in tumors expressing β-catenin. This was correlated with lack of expression of CCL4, a major chemokine responsible for CD103+ DC recruitment. Mechanistically, CCL4 expression in β-catenin-expressing tumor cells was suppressed by the transcriptional repressor ATF3. Consistent with the lack of baseline T cell infiltration, combination immunotherapy with αCTLA-4 and αPD-L1 showed no therapeutic benefit in mice-bearing BrafV600E/PTEN−/−/CAT-STA tumors, while reduction in tumor outgrowth was observed for BrafV600E/PTEN−/− tumors. Therefore, these collective data identify the WNT/β-catenin pathway as the first defined tumor-intrinsic oncogene pathway that can prevent the induction of anti-tumor T cell responses, excluding T cell from the tumor microenvironment and resulting in resistance to immunotherapy (Fig. 2) (Spranger et al., 2015).
Figure 2.
Model for how tumor-intrinsic β-catenin signaling mediates exclusion of T cells from the tumor microenvironment. Activation of the β-catenin pathway in tumor cells results in induction of ATF3, which serves as a transcriptional repressor for CCL4 gene expression. Lack of CCL4 accounts for failed recruitment of CD103+ and CD8α+ DCs into the tumor microenvironment, thereby preventing cross-priming of host CD8+ T cells.
Collectively, these data suggest a new model in which tumor-intrinsic β-catenin activation mediates immune evasion from an antitumor immune response through tumor immune avoidance. Although the data supporting this notion were obtained with β-catenin activation beginning early at the stage of tumor initiation, it is plausible that this process of immune avoidance could occur at any given time in tumor development if mutations activating the β-catenin pathway become acquired. In concordance with contemporary thinking around immune evasion (the three E hypothesis) (Dunn, Bruce, Ikeda, Old, & Schreiber, 2002), in which the tumor escapes from the immune system by elimination of immunogenic tumor cells with subsequent upregulation of immune-inhibitory mechanisms that suppress the function of residual T cells, we propose that tumor escape also can occur through selection of WNT/β-catenin-positive tumor cells. This new mechanism would result in T cell exclusion from the tumor microenvironment and could represent the second major tumor phenotype observed clinically (non-T cell-inflamed). An additional prediction of this model is that patients who encounter extreme immune selection (eg, following immunotherapies) might show acquisition of β-catenin pathway activation as a potential secondary escape mechanism. This possibility is testable clinically by analyzing tumors before successful treatment vs after secondary progression.
3.2 Other Candidate Oncogene Pathways That May Contribute to Immune Exclusion
It is very conceivable that additional molecular oncogenic pathways could result in similar effects to limit the accumulation of T cells within the tumor microenvironment. In particular since 48% of the non-T cell-inflamed melanomas showed evidence of WNT/β-catenin activation, the remaining 52% might have other molecular aberrations resulting in a similar functional outcome. One potential candidate pathway is mutant p53. In preclinical mouse models, restoration of intact p53 signaling has been found to increased recruitment and activation of innate immune cells (Xue et al., 2007). Additionally, restoration of p53 wildtype signaling was sufficient to induce tumor regression, accompanied with increased expression of proinflammatory cytokines, in a murine liver carcinoma model. In related work, activation and recruitment of natural killer (NK) cells were responsible for tumor regression in response to reexpression of wildtype p53 (Iannello, Thompson, Ardolino, Lowe, & Raulet, 2013). Mechanistically, NK cell recruitment was found to be driven by CCL2, which was expressed at higher levels in tumor cells after p53 wildtype restoration. In concordance with those findings in preclinical models, a recent study analyzing triple negative breast cancer indicated a correlation between mutant p53 and the absence of T cells in the tumor microenvironment (Quigley et al., 2015). Therefore, p53 signaling at steady state may enhance recruitment of innate immune cells as well as their activation, suggesting that mutations in the p53 pathway could contribute to the non-T cell-inflamed phenotype.
Another conceivable oncogenic pathway that could contribute to T cell exclusion is activation of STAT3 signaling. In transplantable tumor models, constitutively active STAT3 signaling results in decreased expression of proinflammatory mediators, while a dominant negative variant of STAT3 resulted in augmented expression of proinflammatory factors (Burdelya et al., 2005; Wang et al.,2004), including the chemokines CCL5 and CXCL10. Furthermore, recent studies using a carcinogen-induced lung cancer model as well as a genetically induced prostate cancer model have provided in vivo support for these notions (Ihara et al., 2012; Toso et al., 2014). Abolishing STAT3 signaling using a conditional knockout mouse model resulted in increased antitumor immune responses. This was associated with increased T cell accumulation and functionality within the tumor microenvironment, as well as increased expression of the chemokines CCL5 and CXCL10. Thus, activation of STAT3 signaling may represent another oncogenic pathway able to mediate reduction of immune cell recruitment into tumor sites.
The NFκB signaling pathway represents another important candidate that could impact on host immune responses. In general, cancer cell-intrinsic activation of this pathway has been associated with tumor progression (Baldwin, 2012; Basseres & Baldwin, 2006). Focusing on a more immunologically relevant aspect of this pathway, augmented NFκB signaling in hepatocellular carcinoma cells triggered by immune-derived TNF has been shown to promote tumor progression (Pikarsky et al., 2004). These data suggest that NFκB signaling in tumors could contribute to tumor growth despite immune system interactions. On the other hand, increased production of tumor-derived chemokines has been associated with activation of NFκB, an observation which could have a positive effect on T cell infiltration (Greten et al., 2004). This notion is also supported by studies on hyperactivation of NFκB within the tumor microenvironment, which has been reported to be associated with increased chemokine expression and recruitment of activated T cells (Muthuswamy et al., 2012). Therefore, the impact of tumor-intrinsic NFκB activation on the host immune response might depend on the cellular context of the tumor, and its role might be dependent on the type of immune infiltrate that develops. Additional cancer type-specific studies using GEM model systems will be required to fully determine the effects of NFκB activation on the immune response within the tumor microenvironment.
The PI3K/PTEN/AKT pathway is an additional interesting candidate to be considered as potentially impacting host immune responses. Studies of inflammation-induced cancer progression have investigated activation of the PI3K signaling, either through activating mutations in PIK3CA or loss-of-function mutations in PTEN. These studies have given indications that an increased accumulation of tumor-associated macrophages results, which in turn can contribute to an immune-suppressive microenvironment (Coussens, Zitvogel, & Palucka, 2013; Schmid et al., 2011). The accumulation of macrophages and differentiation into M2-like macrophages has been associated with increased tumor-derived production of TNF, IL-6, CSF-1 VEGF-A, and IL-8 (Bronte & Murray, 2015), which all can support tumor growth, either directly or indirectly. In contrast, recent reports analyzing tumor types which are not associated with inflammation-induced progression including triple negative breast cancer, have indicated that expression of PTEN was associated with the absence of T cells as well as low PD-L1 expression in the tumor microenvironment (Mittendorf et al., 2014), arguing that loss of PTEN expression (and constitutive PI3K activation) is associated with presence of T cells in the tumor microenvironment. Therefore, like NFκB signaling, the impact of tumor-intrinsic PI3K signaling on host T cell function will require further mechanistic studies in well-defined mouse models.
4. ENVIRONMENTAL FACTORS INFLUENCING THE TUMOR MICROENVIRONMENT: THE HOST MICROBIOTA
In addition to tumor-intrinsic oncogenic pathways that could impact on the nature of the host immune response against tumors, another source of intersubject heterogeneity that could theoretically regulate antitumor immunity is the commensal microbiota. A growing body of data has implicated the composition of intestinal commensal bacteria as a major environmental factor that varies between individuals, with the ability to impact systemic immunity. Commensal bacteria have been shown to direct differentiation of T cells leading to expansion of specific molecular subsets, thereby influencing systemic inflammatory processes that involve these T cell differentiation states (Hooper, Littman, & Macpherson, 2012; Ivanov & Honda, 2012). Other studies have highlighted a role for commensal bacteria in modulating the activation state of innate antigen-presenting cells (APCs), thereby impacting priming of systemic immune responses (Abt et al., 2012; Ganal et al., 2012).
While these studies establish a role for commensal bacteria in modulating systemic immune function in general, a potential impact of gut commensal microbes on systemic antitumor immune responses has only begun to be investigated. Two seminal studies showed that the therapeutic effect of chemotherapy was facilitated by the presence of commensal microbes. In one model, treatment with oral antibiotics reduced the therapeutic effect of oxaliplatin or the response to anti-IL-10R + CpG-ODN as an immunotherapy. Mechanistically, the effect was mapped to the level of inflammatory cytokine production by myeloid cells (Iida et al., 2013). In a second model, bacterial translocation from the gut into peripheral organs was implicated in the therapeutic effect of cyclophosphamide, as treatment with antibiotics reduced the generation of T helper 17 (Th17) that were involved with anti-tumor efficacy in this system (Viaud et al., 2013). In cancer models associated with inflammation, commensal bacteria also have been reported to promote tumor growth. In a recent study, TLR5-mediated sensing of gut commensals was shown to increase systemic levels of IL-6, leading to recruitment of MDSCs and γδ suppressor T cells into the tumor microenvironment, in a p53/K-ras-driven tumor model (Rutkowski et al., 2015). These data begin to highlight a role for commensal bacteria in shaping antitumor immune responses, with implications on cancer progression and treatment.
Whether commensal microbiota in fact influence spontaneous immune responses against tumors was initially investigated by comparing subcutaneous B16.SIY melanoma growth in genetically similar C57BL/6 mice derived from two different mouse facilities, Jackson Laboratory (JAX) and Taconic Farms (TAC), which have been shown to differ in their commensal microbes (Ivanov et al., 2009). These experiments revealed that C57BL/6 mice colonized by different microbial communities exhibited significant differences in B16.SIY melanoma growth rate and associated tumor antigen-specific T cell responses. Specifically, tumors in mice obtained from TAC grew more aggressively, accompanied by weaker antitumor T cell responses. Cohousing prior to tumor inoculation ablated the differences in tumor growth and tumor-specific immune responses between the two mouse populations, arguing for an environmental influence. Upon cohousing, TAC mice acquired the JAX phenotype, suggesting that JAX mice may harbor commensal microbes that dominantly facilitate improved antitumor immunity (Sivan et al., 2015).
In a more direct approach to assess the involvement of commensal microbes in modulating the antitumor immune response, the effects of fecal material transfer between the two mouse populations were investigated. This approach showed that prophylactic transfer of JAX fecal material into TAC recipients was sufficient to delay tumor growth and to enhance induction and infiltration of tumor-specific CD8+ T cells, supporting a microbe- or microbial product-derived effect that was consistent with the effects observed upon cohousing. Administration of JAX fecal material to TAC mice-bearing established tumors showed a therapeutic effect that was comparable to treatment with systemic αPD-L1 monoclonal antibodies (mAbs), and combination treatment with both JAX feces and αPD-L1 mAbs was able to further enhance the tumor-specific T cell response, leading to near complete tumor elimination. Consistent with these results, αPD-L1 therapy alone was significantly more efficacious in JAX mice compared to TAC mice, which paralleled improved antitumor T cell responses. These data indicated that the commensal microbial composition can influence spontaneous antitumor immunity as well as response to immunotherapy with αPD-L1 mAb (Sivan et al., 2015).
Identification of the bacterium contained within JAX feces that elicited improved antitumor immune responses was pursued using 16S ribosomal RNA (rRNA) sequencing of fecal bacterial content collected over time from mice that were subjected to administration of fecal permutations. This approach revealed that administration of JAX fecal material to TAC mice induced a gradual, but consistent change in microbial community diversity, such that TAC mice became very similar to JAX mice in microbial composition. Comparative in-depth analyses of bacterial taxa abundance between the different fecal permutation groups pointed to Bifidobacterium breve and Bifidobacterium longum as being significantly associated with increased tumor antigen-specific T cell responses. Administration of a cocktail of Bifidobacterium species, which included B. breve and B. longum, to TAC mice-bearing established tumors led to an increase in Bifidobacterium levels in the fecal material of these mice, assessed by both 16S rRNA sequencing and quantitative PCR. This increase was accompanied by significantly improved tumor control and robust tumor-specific T cell responses in comparison to non-Bifidobacterium treated counterparts, either when Bifidobacterium was given alone or in combination with αPD-L1 mAbs, in a CD8+ T cell-dependent manner (Sivan et al., 2015).
Mechanistically, CD8+ SIY-specific 2C TCR Tg T cells exposed to tumors in JAX and Bifidobacterium-treated TAC mice produced markedly greater IFN-γ in both the tumor draining lymph node and the spleen, relative to TAC tumor-bearing mice, consistent with analyses of the endogenous T cell response, and pointed to an improvement in immune responses upstream of T cells, at the level of host DCs (Fig. 3). Indeed, DCs isolated from early tumors of JAX and Bifidobacterium-fed TAC showed increased maturation and expression of genes critical for antitumor responses including those involved in CD8+ T cell activation and costimulation (Bak et al., 2012; Mackey et al., 1998; Scholer, Hugues, Boissonnas, Fetler, & Amigorena, 2008), DC maturation (Pan et al., 2004; Pettit et al., 1997), antigen processing and cross presentation (Compeer, Flinsenberg, van der Grein, & Boes, 2012; Jancic et al., 2007; Stober, Brode, White, Popoff, & Blackwell, 2007), chemokine-mediated recruitment of immune cells to the tumor microenvironment (Kabashima et al., 2007; Nukiwa et al., 2006; Zhang et al., 2003), and type I IFN signaling (Fuertes et al., 2011; Woo et al., 2014). In vitro, purified DCs isolated from lymphoid organs of JAX and Bifidobacterium-fed TAC mice elicited significantly elevated levels of T cell IFN-γ production in naïve CD8+ SIY-specific 2C TCR Tg T cells. Taken together, these data suggest that commensal Bifidobacterium-derived signals modulate the activation of DCs in the steady state, which in turn supports improved effector function of tumor-specific CD8+ T cells (Sivan et al., 2015).
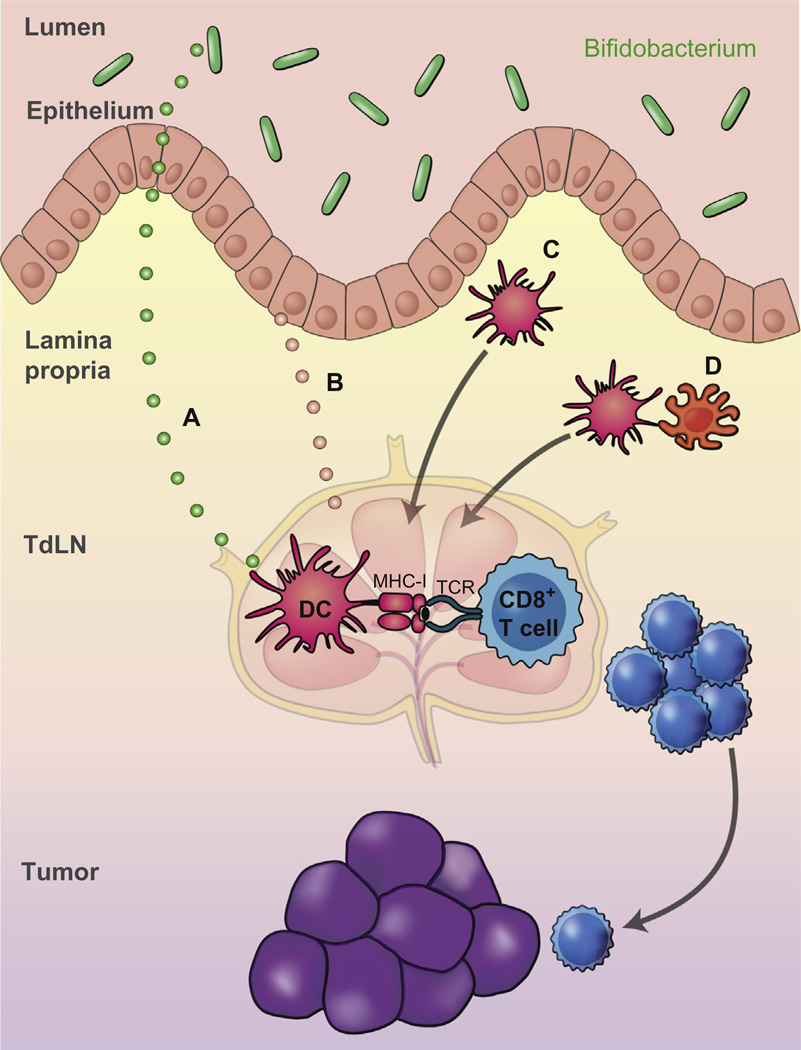
Figure 3.
Potential mechanisms for Bifidobacterium-mediated improvement in antitumor immune responses. (A) and (B) A systemic factor might be released by Bifidobacterium (A) or by host cells, such as intestinal epithelial cells (B) that promotes global DC preactivation. (C) Altered DCs or DC precursors might be preactivated locally in the intestinal lamina propria and disseminate to distant sites. (D) A host cell, altered by crosstalk with Bifidobacterium, might interact with DCs to modulate DC function.
These data support a model wherein commensal species belonging to the Bifidobacterium genus are involved in enhancing antitumor immunity in vivo, through calibration of DC activation, ultimately leading to improved T cell priming and increased CD8+ T cell infiltration into distant tumor sites. An additive effect is observed when Bifidobacterium colonization is combined with antibodies targeting the αPD-1/αPD-L1 axis, likely due to targeting two distinct phases of the antitumor immune response—DC-mediated priming of CD8+ T cells and reactivation of tumor-infiltrating CD8+ T cells (Spranger et al., 2014), respectively. Given that these beneficial effects were observed in multiple tumor settings and that alteration of innate immune function was observed, this improved antitumor immunity is likely occurring in an antigen-independent fashion. Mechanistically, Bifidobacterium may colonize a specific compartment within the gut that enables it to interact with host cells that are critical for modulating DC function, or to release soluble factors that disseminate systemically leading to improved DC function (Fig. 3).
Based on this model, we propose that one source of intersubject heterogeneity with regard to spontaneous antitumor immunity and therapeutic effects of immune-based interventions may be the composition of gut microbes, which could be manipulated for therapeutic benefit. Importantly, commensal bacteria species other than Bifidobacterium may also have the capability to regulate antitumor immunity, either positively or negatively. Similar analyses can be performed in humans using 16S rRNA sequencing of stool samples from patients receiving checkpoint blockade or other immunotherapies, to identify commensals associated with clinical benefit. It is therefore conceivable that clinical activity of these agents might similarly be improved through rational modulation of the commensal microbiota.
5. GERMLINE GENETIC DIFFERENCES AS AN ADDITIONAL SOURCE OF INTERPATIENT HETEROGENEITY
A third source of intersubject heterogeneity that could impact on the magnitude and quality of endogenous immune responses against tumors is germline polymorphisms in immune regulatory genes. Spontaneous antitumor immune responses have similarities to autoimmunity, and autoimmune diseases have been linked to a wide range of heritable genetic variants. The pursuit of single-nucleotide polymorphisms (SNPs) that might be associated with the degree of T cell infiltration into tumors or clinical response to immunotherapies is just in its infancy. The first germline polymorphism investigation described in this regard is a SNP in the gene encoding the chemokine receptor CCR5. Indeed, a CCR5 polymorphism was identified to be associated with clinical response to high-dose IL-2 (Bedognetti et al., 2013). More recently, a polymorphism in the IRF5 gene was identified that was associated with clinical benefit in a cohort of patients treated with T cell adoptive transfer (Uccellini et al., 2012), suggesting a link to the type I IFN pathway. A comprehensive analysis of germline SNPs in concert with the degree of T cell infiltration within the tumor microenvironment and/or clinical response to contemporary immunotherapies, such as anti-PD-1, is warranted.
6. CONCLUSIONS AND IMPLICATIONS
Novel immunotherapies, such as blocking Abs against PD-1, are showing clinical activity in a broad range of human cancers. Combination therapies targeting two immune regulatory checkpoints concurrently are expected to improve clinical benefit even further, particularly in patients showing the T cell-inflamed tumor microenvironment phenotype. Gaining additional clinical benefit with immunotherapies in patients with the non-T cell-inflamed tumor microenvironment will require a deep mechanistic understanding of tumor-intrinsic and host-derived processes that restrict generation of a spontaneous antitumor T cell response. Interrogation of these possibilities in patients has become technically feasible using modern genomics approaches. Analysis of tumor biopsies by gene expression profiling and exome sequencing, of germline SNPs from blood cells, and of commensal microbiota differences through 16S rRNA sequencing can be performed with respect to clinical response to defined immunotherapies. An integrated bioinformatics approach may also reveal functional interactions between these different axes. Identified resistance pathways that are validated mechanistically in preclinical models can be considered as targets for new therapeutic development. Recent data have identified tumor-intrinsic β-catenin pathway activation, as well as absence of host Bifidobacterium colonization, as two candidates ripe for forward translation back into the clinic. Similar analyses in patients bearing the full range of tumor types being treated with anti-PD-1 or other active immunotherapies should be performed in prospective studies, to identify the spectrum of regulatory mechanisms that might be manipulated therapeutically.
REFERENCES
- Abt MC, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. http://dx.doi.org/10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak SP, et al. Differential requirement for CD70 and CD80/CD86 in dendritic cell-mediated activation of tumor tolerized CD8 T cells. Journal of Immunology. 2012;189:1708–1716. doi: 10.4049/jimmunol.1201271. http://dx.doi.org/10.4049/jimmunol.1201271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin AS. Regulation of cell death and autophagy by IKK and NF-kappaB: Critical mechanisms in immune function and cancer. Immunological Reviews. 2012;246:327–345. doi: 10.1111/j.1600-065X.2012.01095.x. http://dx.doi.org/10.1111/j.1600-065X.2012.01095.x. [DOI] [PubMed] [Google Scholar]
- Basseres DS, Baldwin AS. Nuclear factor-kappaB and inhibitor of kappaB kinase pathways in oncogenic initiation and progression. Oncogene. 2006;25:6817–6830. doi: 10.1038/sj.onc.1209942. http://dx.doi.org/10.1038/sj.onc.1209942. [DOI] [PubMed] [Google Scholar]
- Bedognetti D, et al. CXCR3/CCR5 pathways in metastatic melanoma patients treated with adoptive therapy and interleukin-2. British Journal of Cancer. 2013;109:2412–2423. doi: 10.1038/bjc.2013.557. http://dx.doi.org/10.1038/bjc.2013.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosenberg M, et al. Characterization of melanocyte-specific inducible Cre recombinase transgenic mice. Genesis. 2006;44:262–267. doi: 10.1002/dvg.20205. http://dx.doi.org/10.1002/dvg.20205. [DOI] [PubMed] [Google Scholar]
- Bronte V, Murray PJ. Understanding local macrophage phenotypes in disease: Modulating macrophage function to treat cancer. Nature Medicine. 2015;21:117–119. doi: 10.1038/nm.3794. http://dx.doi.org/10.1038/nm.3794. [DOI] [PubMed] [Google Scholar]
- Burdelya L, et al. Stat3 activity in melanoma cells affects migration of immune effector cells and nitric oxide-mediated antitumor effects. Journal of Immunology. 2005;174:3925–3931. doi: 10.4049/jimmunol.174.7.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AF, Dupage MJ, Dong HK, Chen J, Jacks T. Regulated expression of a tumor-associated antigen reveals multiple levels of T-cell tolerance in a mouse model of lung cancer. Cancer Research. 2008;68:9459–9468. doi: 10.1158/0008-5472.CAN-08-2634. http://dx.doi.org/10.1158/0008-5472.CAN-08-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compeer EB, Flinsenberg TWH, van der Grein SG, Boes M. Antigen processing and remodeling of the endosomal pathway: Requirements for antigen cross-presentation. Frontiers in Immunology. 2012;3 doi: 10.3389/fimmu.2012.00037. http://dx.doi.org/10.3389/fimmu.2012.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales L, et al. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Reports. 2015;11:1018–1030. doi: 10.1016/j.celrep.2015.04.031. http://dx.doi.org/10.1016/j.celrep.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Zitvogel L, Palucka AK. Neutralizing tumor-promoting chronic inflammation: A magic bullet? Science. 2013;339:286–291. doi: 10.1126/science.1232227. http://dx.doi.org/10.1126/science.1232227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damsky WE, et al. beta-Catenin signaling controls metastasis in Braf-activated Pten-deficient melanomas. Cancer Cell. 2011;20:741–754. doi: 10.1016/j.ccr.2011.10.030. http://dx.doi.org/10.1016/j.ccr.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankort D, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nature Genetics. 2009;41:544–552. doi: 10.1038/ng.356. http://dx.doi.org/10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: From immunosurveillance to tumor escape. Nature Immunology. 2002;3:991–998. doi: 10.1038/ni1102-991. http://dx.doi.org/10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- Edelson BT, et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha + conventional dendritic cells. The Journal of Experimental Medicine. 2010;207:823–836. doi: 10.1084/jem.20091627. http://dx.doi.org/10.1084/jem.20091627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt JJ, et al. Marginating dendritic cells of the tumor microenvironment cross-present tumor antigens and stably engage tumor-specific T cells. Cancer Cell. 2012;21:402–417. doi: 10.1016/j.ccr.2012.01.008. http://dx.doi.org/10.1016/j.ccr.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdag G, et al. Immunotype and immunohistologic characteristics of tumor-infiltrating immune cells are associated with clinical outcome in metastatic melanoma. Cancer Research. 2012;72:1070–1080. doi: 10.1158/0008-5472.CAN-11-3218. http://dx.doi.org/10.1158/0008-5472.CAN-11-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes MB, Woo SR, Burnett B, Fu YX, Gajewski TF. Type I interferon response and innate immune sensing of cancer. Trends in Immunology. 2013;34:67–73. doi: 10.1016/j.it.2012.10.004. http://dx.doi.org/10.1016/j.it.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes MB, et al. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8{alpha} + dendritic cells. The Journal of Experimental Medicine. 2011;208:2005–2016. doi: 10.1084/jem.20101159. http://dx.doi.org/10.1084/jem.20101159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganal SC, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37:171–186. doi: 10.1016/j.immuni.2012.05.020. http://dx.doi.org/10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- Greten FR, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. http://dx.doi.org/10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Hamid O, et al. A prospective phase II trial exploring the association between tumor microenvironment biomarkers and clinical activity of ipilimumab in advanced melanoma. Journal of Translational Medicine. 2011;9:204. doi: 10.1186/1479-5876-9-204. http://dx.doi.org/10.1186/1479-5876-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlin H, et al. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Research. 2009;69:3077–3085. doi: 10.1158/0008-5472.CAN-08-2281. http://dx.doi.org/10.1158/0008-5472.CAN-08-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildner K, et al. Batf3 deficiency reveals a critical role for CD8alpha + dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. http://dx.doi.org/10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. http://dx.doi.org/10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannello A, Thompson TW, Ardolino M, Lowe SW, Raulet DH. p53-Dependent chemokine production by senescent tumor cells supports NKG2D-dependent tumor elimination by natural killer cells. The Journal of Experimental Medicine. 2013;210:2057–2069. doi: 10.1084/jem.20130783. http://dx.doi.org/10.1084/jem.20130783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara S, et al. Inhibitory roles of signal transducer and activator of transcription 3 in antitumor immunity during carcinogen-induced lung tumorigenesis. Cancer Research. 2012;72:2990–2999. doi: 10.1158/0008-5472.CAN-11-4062. http://dx.doi.org/10.1158/0008-5472.CAN-11-4062. [DOI] [PubMed] [Google Scholar]
- Iida N, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. http://dx.doi.org/10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Honda K. Intestinal commensal microbes as immune modulators. Cell Host & Microbe. 2012;12:496–508. doi: 10.1016/j.chom.2012.09.009. http://dx.doi.org/10.1016/j.chom.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. http://dx.doi.org/10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancic C, et al. Rab27a regulates phagosomal pH and NADPH oxidase recruitment to dendritic cell phagosomes. Nature Cell Biology. 2007;9:367–378. doi: 10.1038/ncb1552. http://dx.doi.org/10.1038/ncb1552. [DOI] [PubMed] [Google Scholar]
- Kabashima K, et al. CXCL12-CXCR4 engagement is required for migration of cutaneous dendritic cells. The American Journal of Pathology. 2007;171:1249–1257. doi: 10.2353/ajpath.2007.070225. http://dx.doi.org/10.2353/ajpath.2007.070225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. The New England Journal of Medicine. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. http://dx.doi.org/10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey MF, et al. Dendritic cells require maturation via CD40 to generate protective antitumor immunity. Journal of Immunology. 1998;161:2094–2098. [PubMed] [Google Scholar]
- Mikucki MFD, Matsuzaki J, Skitzki J, Gaulin N, Muhitch J, Ku A, et al. Non-redundant requirement for CXCR3 signaling during tumoricidal T cell trafficking across tumor vascular checkpoints. Nature Communications. 2015;6:7458. doi: 10.1038/ncomms8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittendorf EA, et al. PD-L1 expression in triple-negative breast cancer. Cancer Immunology Research. 2014;2:361–370. doi: 10.1158/2326-6066.CIR-13-0127. http://dx.doi.org/10.1158/2326-6066.CIR-13-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuswamy R, et al. NF-kappaB hyperactivation in tumor tissues allows tumor-selective reprogramming of the chemokine microenvironment to enhance the recruitment of cytolytic T effector cells. Cancer Research. 2012;72:3735–3743. doi: 10.1158/0008-5472.CAN-11-4136. http://dx.doi.org/10.1158/0008-5472.CAN-11-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nukiwa M, et al. Dendritic cells modified to express fractalkine/CX3CL1 in the treatment of preexisting tumors. European Journal of Immunology. 2006;36:1019–1027. doi: 10.1002/eji.200535549. http://dx.doi.org/10.1002/eji.200535549. [DOI] [PubMed] [Google Scholar]
- Pan J, et al. Interferon-gamma is an autocrine mediator for dendritic cell maturation. Immunology Letters. 2004;94:141–151. doi: 10.1016/j.imlet.2004.05.003. http://dx.doi.org/10.1016/j.imlet.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Pettit AR, et al. Nuclear localization of RelB is associated with effective antigen-presenting cell function. Journal of Immunology. 1997;159:3681–3691. [PubMed] [Google Scholar]
- Pikarsky E, et al. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–466. doi: 10.1038/nature02924. http://dx.doi.org/10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- Quigley D, et al. Lymphocyte invasion in IC10/basal-like breast tumors is associated with wild-type TP53. Molecular Cancer Research: MCR. 2015;13:493–501. doi: 10.1158/1541-7786.MCR-14-0387. http://dx.doi.org/10.1158/1541-7786.MCR-14-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski MR, et al. Microbially driven TLR5-dependent signaling governs distal malignant progression through tumor-promoting inflammation. Cancer Cell. 2015;27:27–40. doi: 10.1016/j.ccell.2014.11.009. http://dx.doi.org/10.1016/j.ccell.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salerno EP, Olson WC, McSkimming C, Shea S, Slingluff CL., Jr T cells in the human metastatic melanoma microenvironment express site-specific homing receptors and retention integrins. International Journal of Cancer. Journal international du cancer. 2014;134:563–574. doi: 10.1002/ijc.28391. http://dx.doi.org/10.1002/ijc.28391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MC, et al. Receptor tyrosine kinases and TLR/IL1Rs unexpectedly activate myeloid cell PI3kgamma, a single convergent point promoting tumor inflammation and progression. Cancer Cell. 2011;19:715–727. doi: 10.1016/j.ccr.2011.04.016. http://dx.doi.org/10.1016/j.ccr.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholer A, Hugues S, Boissonnas A, Fetler L, Amigorena S. Intercellular adhesion molecule-1-dependent stable interactions between T cells and dendritic cells determine CD8+ T cell memory. Immunity. 2008;28:258–270. doi: 10.1016/j.immuni.2007.12.016. http://dx.doi.org/10.1016/j.immuni.2007.12.016. [DOI] [PubMed] [Google Scholar]
- Sivan A, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. http://dx.doi.org/10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523(7559):231–235. doi: 10.1038/nature14404. http://dx.doi.org/10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- Spranger S, et al. Up-regulation of PD-L1, IDO, and tregs in the melanoma tumor microenvironment is driven by CD8+ T cells. Science Translational Medicine. 2013;5:200ra116. doi: 10.1126/scitranslmed.3006504. http://dx.doi.org/10.1126/scitranslmed.3006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spranger S, et al. Mechanism of tumor rejection with doublets of CTLA-4, PD-1/PD-L1, or IDO blockade involves restored IL-2 production and proliferation of CD8(+) T cells directly within the tumor microenvironment. Journal for Immunotherapy of Cancer. 2014;2:3. doi: 10.1186/2051-1426-2-3. http://dx.doi.org/10.1186/2051-1426-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stober CB, Brode S, White JK, Popoff J, Blackwell JM. Slc11a1, formerly Nramp1 is expressed in dendritic cells and influences major histocompatibility complex class II expression and antigen-presenting cell function. Infection and immunity. 2007;75:5059–5067. doi: 10.1128/IAI.00153-07. http://dx.doi.org/10.1128/iai.00153-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan RJ, et al. A single center experience with high-dose (HD) IL-2 treatment for patients with advanced melanoma and pilot investigation of a novel gene expression signature as a predictor of response. Journal of Clinical Oncology. 2009;27:9003. [Google Scholar]
- Topalian SL, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. The New England Journal of Medicine. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. http://dx.doi.org/10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toso A, et al. Enhancing chemotherapy efficacy in Pten-deficient prostate tumors by activating the senescence-associated antitumor immunity. Cell Reports. 2014;9:75–89. doi: 10.1016/j.celrep.2014.08.044. http://dx.doi.org/10.1016/j.celrep.2014.08.044. [DOI] [PubMed] [Google Scholar]
- Tumeh PC, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. http://dx.doi.org/10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uccellini L, et al. IRF5 gene polymorphisms in melanoma. Journal of Translational Medicine. 2012;10:170. doi: 10.1186/1479-5876-10-170. http://dx.doi.org/10.1186/1479-5876-10-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaud S, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–976. doi: 10.1126/science.1240537. http://dx.doi.org/10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nature Medicine. 2004;10:48–54. doi: 10.1038/nm976. http://dx.doi.org/10.1038/nm976. [DOI] [PubMed] [Google Scholar]
- Woo SR, et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41:830–842. doi: 10.1016/j.immuni.2014.10.017. http://dx.doi.org/10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. http://dx.doi.org/10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. New England Journal of Medicine. 2003;348:203–213. doi: 10.1056/NEJMoa020177. http://dx.doi.org/10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]