Abstract
CONNJUR Workflow Builder (WB) is an open-source software integration environment that leverages existing spectral reconstruction tools to create a synergistic, coherent platform for converting biomolecular NMR data from the time domain to the frequency domain. WB provides data integration of primary data and metadata using a relational database, and includes a library of pre-built workflows for processing time domain data. WB simplifies maximum entropy reconstruction, facilitating the processing of non-uniformly sampled time domain data. As will be shown in the paper, the unique features of WB provide it with novel abilities to enhance the quality, accuracy, and fidelity of the spectral reconstruction process. WB also provides features which promote collaboration, education, parameterization, and non-uniform data sets along with processing integrated with the Rowland NMR Toolkit (RNMRTK) and NMRPipe software packages. WB is available free of charge in perpetuity, dual-licensed under the MIT and GPL open source licenses.
Keywords: CONNJUR, data model, NUS, spectral reconstruction, software integration
Introduction
The application of the Fourier transform (FT) to NMR spectroscopy (Ernst & Anderson, 1966) has been revolutionary. FT-NMR experiments are collected in less time than continuous wave (CW), with higher sensitivity and allow the introduction of indirect time dimensions and multidimensional NMR (Jeener, 1971; Oschkinat et al. 1988; Clore & Gronenborn, 1994).
Despite the simplicity implied by its moniker, reconstructing a frequency domain spectrum from the time domain signal is a lengthy process in FT-NMR. Many mathematical operations are applied stepwise to the data in order to correct for artifacts introduced by the data collection hardware/software, remove unwanted signals, increase sensitivity and resolution, phase the spectrum so all signals appear absorptive, and correct for baseline distortions/offsets (Ernst et al. 1991; Hoch & Stern, 1996). Unwanted signals are commonly de-convolved from the time domain data as a first step, followed by linear prediction (LP) techniques for increasing sensitivity and resolution, apodization/windowing functions for improving line-shape, and zero-filling to improve digital resolution (Verdi et al. 2007). Baseline distortions are commonly corrected for by operations in either the time and frequency domains. Finally, the phase components of the signals are modified to provide purely absorptive line shapes. This process is undertaken for each of the various independent dimensions within a single multidimensional experiment.
Most of these operations must be parameterized with attention to the properties of the specific spectrum being reconstructed, including the digital resolution of the collected data, the relative frequencies of unwanted signals, the anticipated intensities of both signals and noise and the various modes of possible data collection. Some of this information is captured during data collection and stored as metadata with the time domain data. Other information is prior knowledge gained from experience with the spectrometer or from experience with the sample preparation and conditions. Finally, a few of the mathematical functions must be parameterized interactively, or by trial and error after examining the effect on the transformed data.
There exist a large number of software packages available for spectral reconstruction. The commercially available spectrometers all have such software: Delta (Delta), TopSpin (TopSpin), and VnmrJ (VnmrJ). There are also a large number of third-party software tools for reconstructing spectra off of the spectrometer: ACD NMR Processor (ACD/NMR Processor Academic Edition), Mnova (Mnova NMR lite), PROSA (Güntert, et al., 1992), SpinWorks (SpinWorks), SwaN-MR (SwaN-MR), PERCH NMR Software (PERCH Solutions), iNMR (iNMR), Felix (FelixNMR) & Azara (Azara). Most of these packages are used primarily for 1D and 2D NMR processing of small molecules and metabonomics analyses. Perhaps the most common spectral reconstruction software for multidimensional biomolecular NMR studies are NMRPipe (Delaglio et al 1995) and the Rowland NMR Toolkit (RNMRTK) (Hoch & Stern, 1985).
As mentioned previously, the mathematical operations for cleaning spectral data are typically applied stepwise; depending on the software utilized, the spectroscopist can either configure each mathematical function independently, keeping track of the cumulative effect of all the operations, or he/she can configure the expected appearance of the final spectrum and allow the software to track the interdependencies of the various mathematical functions. Due to the requirement of interactive inspection of the spectrum for proper phasing, each of these software packages includes a graphical display program to assist in determining the proper phase parameters.
The net result of this complex process is that while there exist several deterministic algorithms for conducting the Fourier Transform, there is no one deterministic algorithm which can optimally process all the multidimensional spectra commonly collected. There are many preferred strategies and schemes, often codified as processing scripts or workflows, and these scripts or schemes typically require human input for their parameterization. The spectroscopist is relied upon to decide which solvent signals are to be removed and by which method, how many time points can be reliably predicted through LP, how much windowing is required for optimal line-shape, and what magnitude of phase correction need be applied. Spectroscopists often share their processing schemes through online repositories (www.nanuc.ca, www.bmrb.wisc.edu) or attend workshops on this topic (www.nanuc.ca, connjur.uchc.edu/workshop2012).
An alternate approach to the FT process described above utilizes maximum entropy methods for reconstruction. Such methodology treats the reconstruction of a frequency spectrum as an inverse problem – rather than transforming time domain data directly to frequency, frequency data is calculated and optimized such that when transformed back into the time domain, it matches the collected data within the accuracy of the measurement. Such methodology eliminates the need for linear prediction and apodization, both of which are consequences of insufficient sampling in the time domain (Stern et al. 2002). It also allows for additional spectral improvements such as line-width and J-coupling deconvolution, to narrow NMR lineshapes and computationally collapse split peaks respectively (Stoven et al. 1997; Shimba et al. 2003).
Perhaps most importantly, the maximum entropy method reduces the need for uniformly sampled data. The Nyquist theorem (Nyquist 1928) dictates that the range of frequencies which can be differentiated, referred to as the spectral width, is inversely proportional to the delay between the sampled points. A small time increment must be used to distinguish a wide range of frequencies. Conversely, the spectral resolution – or the ability to distinguish signals with similar frequencies – is dependent on sampling out to long time delays. The consequence of these two competing demands - needing to sample with small time increments out to very long delays – results in a large number of total samples and a concomitantly long experimental collection time. This issue is particularly exacerbated with higher dimensionality experiments, as a large number of sample points collected along one dimension increases the total collection time multiplicatively with the other dimensions. The expansion in number of points is also exacerbated by higher magnetic fields, where the increased spectral dispersion necessitates shorter sampling intervals.
In recent years several solutions to this problem have been proposed, which amount to decreasing the experimental collection time by refraining from sampling all of the time points dictated by the Nyquist grid. For a review of the many proposed strategies, please see Gryk, et al., 2010. The promise of these various strategies is enormous. It has been demonstrated that similar quality spectra can be obtained by non-uniformly sampling as little as 33% of the data points per dimension (Gryk et al. 2010). This means that 3D and 4D experiments which used to take days can be collected in a few hours.
Unfortunately, there is one critical drawback to the technique. Incomplete sampling of the Nyquist grid results in sampling artifacts (appearing as phantom or aliased signals) when reconstructing the spectrum using conventional FT methods. There are many proposed solutions for removing these artifacts, some implemented within the FT realm (Kazimierczuk et al. 2006a; Kazimierczuk et al. 2006b; Kazimierczuk et al. 2012), others using non-FT methods such as maximum entropy reconstruction (Schmieder et al. 1993; Hoch and Stern 1996; Maciejewski et al. 2009). Regardless of the reconstruction method, the non-uniformity of the data samples makes the reconstruction process much more difficult and requires additional training and knowledge for the spectroscopist. The consequence of failure is dire, phantom signals which are not treated properly can nullify the entire analysis. The fear of such failure prevents most NMR groups in the world from adopting this important methodology.
An unfortunate aspect of maximum entropy reconstruction is that there are two variable parameters – def, and aim or lambda - which must be set correctly for optimal reconstructions. These parameters roughly correlate with estimates of signal intensity and the magnitude of the noise; but as such, they tend to vary from experiment to experiment, and from laboratory to laboratory, forcing the spectroscopist to re-determine these parameters for each spectrum. A semi-automated strategy has been published by our group for estimating these parameters and represents an important first step in automating spectral reconstruction (Mobli et al. 2007a; Mobli et al. 2007b).
The ongoing CONNJUR project is an effort to provide a software integration platform for biomolecular NMR (www.connjur.org). We had previously released a Java application for translating binary spectral data between the various software specific file formats (Nowling, et al, 2011). In this paper we describe a larger-scope software integration application – CONNJUR Workflow Builder (WB). WB utilizes the CONNJUR Spectrum Translator (ST) in addition to integrating the third-party tools NMRPipe and RNMRTK.
From the spectroscopist's perspective, the objective of WB is to create and execute processing workflows for transforming multidimensional time domain NMR data into frequency domain spectra, using NMRPipe and/or RNMRTK as the processing engines. This goal is aided through a graphical, virtual canvas on which the various mathematical operations (in the form of actors) are laid out and connected. WB handles all data and metadata management, as well as the actual invocation of the individual software tools. Due to the metadata management, the individual actors can be configured to analyze the current state of the data within the workflow and adapt accordingly. The sophisticated level of adaptive configuration possible is illustrated through the maximum entropy actor which enhances the semi-automated strategy to setting def, aim & lamda as proposed by Mobli, et al. (2007a). WB includes a library of workflows implementing typical reconstructions that work with the majority of standard pulsed experiments.
Materials and Methods
WB is written in Java and requires a Java Virtual Machine (JVM v. 1.7 or later) (www.Java.com). The JVM is multiplatform and typically comes preinstalled with many desktop computers. WB thus runs on platforms that support JVMs, including Macintosh, Windows, and Linux. However, WB makes use of several platform-dependent third party tools; on platforms not supported by these tools, WB will run, but will not be able to use the corresponding tools. Nevertheless, WB could be used to create a spectral reconstruction workflow even on a platform where the underlying tools do not run; the workflow can be saved to a networked database or exported as XML or NMR-STAR, and loaded into WB on a separate system for execution. For optimal use, a working MySQL installation is required; this may be local or accessed over the network. WB can be run without a database connection, but will not automatically manage and persist data.
WB has been developed using Eclipse (www.eclipse.org) as an integration development environment. A source code control system (Concurrent Versioning System, or CVS) (http://www.nongnu.org/cvs/) is used to track changes to the software and allow changes to be applied in a controlled manner. A comprehensive set of regression tests ensures the correctness of changes. An automated build occurs daily. The library of workflows was built using the WB snapshot build from 2014/09/22; data model version 7, ENC 2013; connected to MySQL 5.5.27 database; using NMRPipe 8.1, RNMRTK 3.1; on OS X 10.6.8.
Design overview
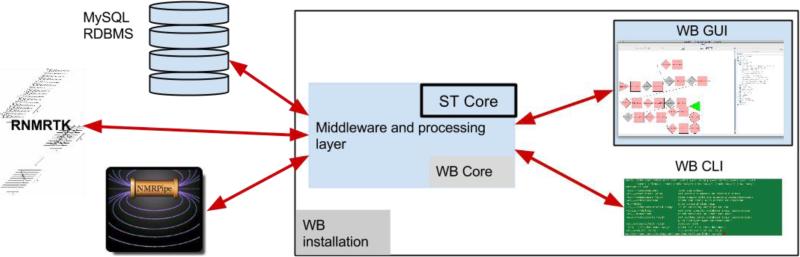
WB consists of a core program which integrates a graphical user interface (GUI) front-end, a MySQL Relational Database Management System (RDBMS) backend, a command line interface (CLI), and external tools. The core is composed of middleware which mediates the interactions between programs, and includes the ST core for data conversion. See Figure 1 for an overview of the design. While WB is a standalone project, it uses existing NMR tools such as NMRPipe (Delaglio et al. 2005) and the Rowland NMR Toolkit (Hoch & Stern, 1996) to perform data processing tasks. The GUI allows for interactive, graphical creation of processing workflows including functions such as zerofills, linear predictions, and apodizations. Pop-up windows provide information about the required and optional parameters for functions, as well as indicating the default and legal values. Workflows are created using actors; these contain domain logic and visual displays to assist the user in correctly parameterizing and processing their data by clearly indicating the available parameters and their domains.
Figure 1.
the WB design. The WB core includes the ST core, and a middleware and processing layer for software and data integration. A GUI for graphical user interaction, command line interface, external tools such as NMRPipe and RNMRTK, and a database backend for data persistence are all integrated through the WB Core. The WB installation includes WB Core, WB GUI, and the WB command line interface.
The database backend implements a data model of processing workflows. This includes the sequence of actors and their parameterizations, metadata, and the binary data (both time- and frequency-domain). It is optional to store the binary data in the database. The database is not required to run WB, as the application is capable of reading and writing processing workflows in XML and NMR-Star formats. However, running the application without a database prevents easy comparison, analysis, and recapitulation of workflows and data sets, removing one of the primary benefits of data integration, which is that the data is stored in a single, central and uniformly accessible location.
The middle layer mediates the interactions between external tools, the database, and the GUI. When an external tool such as NMRPipe is run, the processing layer sends the parameters received from the GUI to an external process, which calls the tool with the appropriate parameterization. Then the tool executes and its output is interactively read and sent to the processing layer to be passed along to the GUI for display to the user. When the tool finishes executing, the final output data set is read and sent to the GUI and the database (if necessary).
WB provides tool integration: while different tools use different formats for time- and frequency-domain data (Delaglio et al. 1995), WB is able to automatically and invisibly handle format conversions by using a data conversion library previously developed by the CONNJUR team for translating between spectral formats. This same library, when coupled with a command line interface, is released separately as CONNJUR ST (Nowling et al 2011).
WB provides spectral reconstruction features by wrapping and integrating the existing tools NMRPipe and RNMRTK. It can be extended to integrate additional tools. WB makes use of ST for handling time- and frequency-domain data formats and the various interconversions required. Thus, WB drives external tools by providing a link between the input data sources (whether files or the database), and the spectral processing tools, mediating the interactions and format conversions, including the syntax, semantics, and metadata of the formats involved. WB then monitors the progress of the tool during the course of execution, capturing reports and presenting them to the user, before collecting the output data set and reintegrating it into the CONNJUR system.
Results
Summary of features
WB overview
The core functionality WB provides is software integration of existing spectral reconstruction tools. WB also provides a graphical environment for building spectral reconstruction workflows. These workflows consist of sequences of actors which consume and produce time- and frequency-domain NMR data. The use of actors for scientific workflows has previously been applied and described (Altintas et al. 2004, Bowers et al. 2005). WB actors assist the user in correctly configuring and parameterizing the underlying NMR function, and use knowledge of the workflow context to determine whether an operation is valid and suggest likely parameter values. Actors (depicted as diamonds, as shown in Figure 2) implement tasks such as baseline correction and apodization, and are the smallest unit of logic within a WB spectral reconstruction. The correspondence between a single actor and a single function from a program such as NMRPipe or RNMRTK is not exact, as some NMRPipe and RNMRTK functions carry out multiple independent but related tasks.
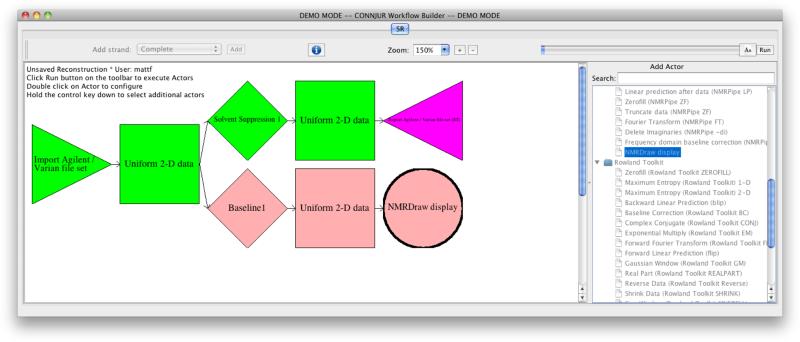
Figure 2.
A workflow shown in the WB GUI. The workflow includes an import actor (triangle pointing left-to-right), export actor (triangle pointing right-to-left), display actor (circle), and two processing actors (diamonds). Data sets are represented by squares. A branch is used to process the data in two different ways. The colors indicate progress: green means already successfully completed, pink means it has not been run yet, and purple means the actor has not been configured yet and so cannot run.
WB provides strands, which are reusable sequences of actors which perform a higher-level function, such as apodization, zero-filling, and Fourier transform of an indirect dimension. WB comes with four pre-built strands; ‘basic’ provides a rudimentary processing workflow; ‘complete’ applies all standard actors; and ‘linear prediction’ and ‘solvent suppression’ provide typical actors for indirect and direct dimensions respectively. A workflow is a complete sequence of actors that processes data from start to finish; multiple strands, many actors, or a combination of both may be used to build a workflow. A workflow that has been applied to a specific data set is known as a reconstruction. The entire set of actors shown in Figure 2 is a workflow, and applying that workflow creates a reconstruction. Actors, strands, workflows, reconstructions, primary data, and metadata are stored in the relational database. Workflows may contain branches in which multiple different results are calculated.
Strands and workflows are built in the GUI by placing visual representations of actors in logical sequences. Actors are then parameterized using interactive dialogs (Figure 5) which provide additional contextual information to the user. There are several types of actors: import actors are responsible for loading data from the file system or database and are represented by triangles pointing left-to-right; actors which take a data set as input and produce a new data set as output are represented as diamonds; export actors, which are responsible for writing data sets to the file system, are represented as triangles pointing right-to-left; and display actors, which load data sets into display programs such as NMRDraw, are represented as circles. Primary data are represented as squares, and lines are used to connect primary data to actors in sequence, showing the logical structure of the workflow. The dimensionality and uniformity/non-uniformity of each data set is displayed as well.
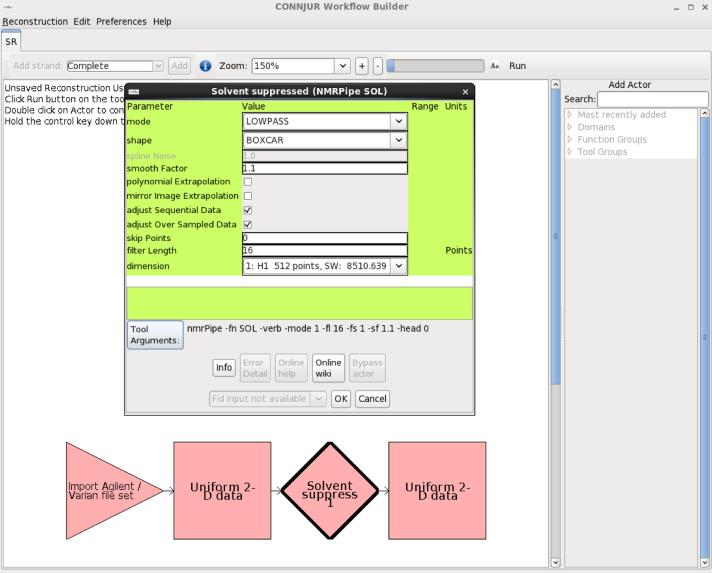
Figure 5.
a WB actor (Solvent Suppress) is parameterized by means of a popup menu. The menu provides widgets for each of the actor's parameters, ensures that the values are valid, and provides links to documentation that describe how to use and parameterize the tool.
A workflow typically begins with an import actor, which imports data from the file system. (Conversely, workflows can be applied to datasets already existing in the database.) The data may be uniform or non-uniform time-domain, or frequency-domain. Supported formats include Bruker, Varian/Agilent, RNMRTK and NMRPipe. The import actor allows the nucleus-channel assignment, and setting of sensitivity enhancement and negation of imaginaries which are performed upon import in the NMRPipe suite. If necessary, metadata may be corrected using a metadata correction actor, which shows the current values and allows updates. Figure 2 provides an example of an import actor, depicted as a triangle pointing from left to right.
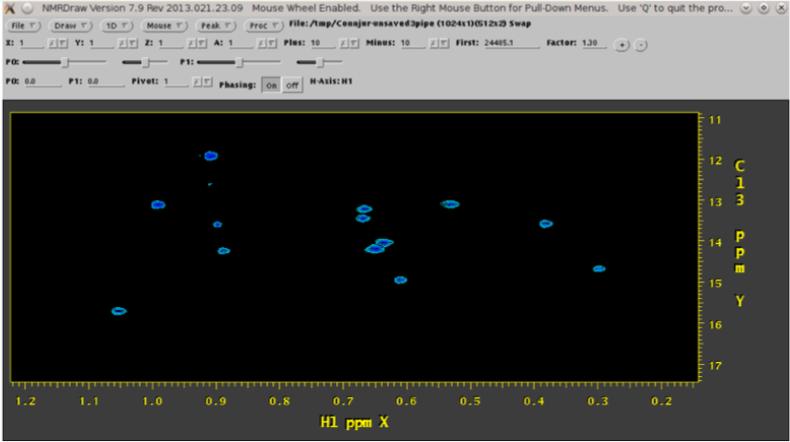
Actors are added by clicking on the icon representing the primary data which will serve as input, then selecting the desired actor from a list displayed on the right side of the screen. Only semantically appropriate actors can be applied; WB prevents inappropriate use of actors such as applying a time-domain process to frequency-domain data. At the end of an actor sequence, an export actor is placed to export to the file system. Display actors allow the time- and frequency-domain data to be viewed in the NMRDraw program; they may be placed as desired and are often useful for optimizing workflows by visualizing intermediate data sets. They are depicted as circles; see Figure 2 for an example. Branched workflows are created by adding multiple actors to the same primary data, as in Figure 2, creating a fork in the workflow. Branches allow comparison between two actor parameterizations. Comparisons of parameterizations are useful when dealing with tasks such as apodization which present inherent tradeoffs. Branches are also useful when it is necessary to process a spectrum completely in order to measure a parameter, which is then used to re-process the spectrum slightly differently. For example, when decoupling is not performed by the spectrometer, as in the Carbon dimension of Figure 3, it is possible to decouple in the reconstruction phase. This requires estimation of the J-coupling value. Maximum Entropy reconstruction allows deconvolution and removal of the splitting, as in Figure 4.
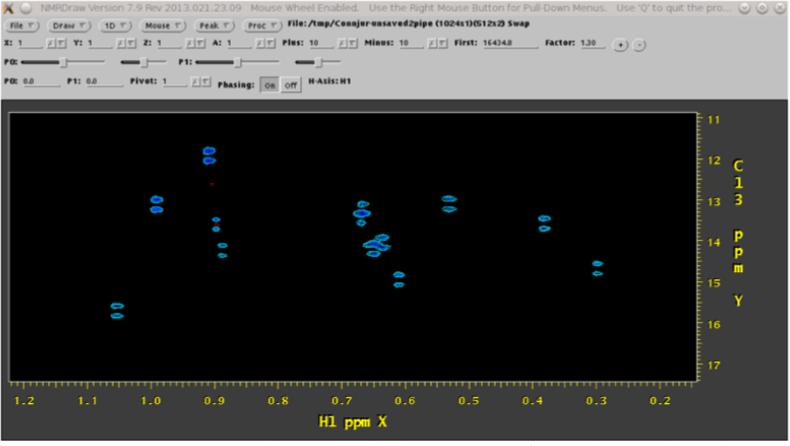
Figure 3.
a spectrum processed using standard Fourier transform techniques has splitting in the Carbon dimension, which is especially problematic when it causes overlap between splitting patterns. Screenshot is of NMRDraw (Delaglio, et al., 1995), a third-party tool initiated from within WB.
Figure 4.
a spectrum processed with Maximum Entropy techniques which allow J-decoupling; the peaks are not split in the Carbon dimension.
An actor's parameters are set using a dialog box which displays parameters, current values, value domain, and units for the actor's underlying function. WB performs domain checks to ensure that values are allowable, providing checkboxes and drop-down menus as appropriate if the domain is an enumeration of a small number of discrete values. To capitalize on a user's familiarity with existing tools, the commands passed to the tool that the actor utilizes are shown, including the command and function name, parameters and values. See Figure 5 for an example.
Workflows are executed using the ‘run’ button. WB loads the primary data and threads it through the sequence of actors, calling external tools as necessary to perform the semantic operations, checking preconditions and postconditions to ensure that clean execution environments are created beforehand, and the execution environments are cleaned up after execution. WB monitors the progress of the tools and reports this back to the user. If branched workflows are used, WB may utilize parallel execution in order to reduce overall execution time. During workflow execution, WB continuously provides feedback to the user through several mechanisms. The first is that the actor's and data set's display colors are updated to indicate progress, with different colors for ‘not run’, ‘successfully run’, ‘failed’, and ‘skipped’. The second is that the error and possibly standard output of the underlying tools is captured and displayed, indicating what is happening and the ramifications of parameter settings and actor choices.
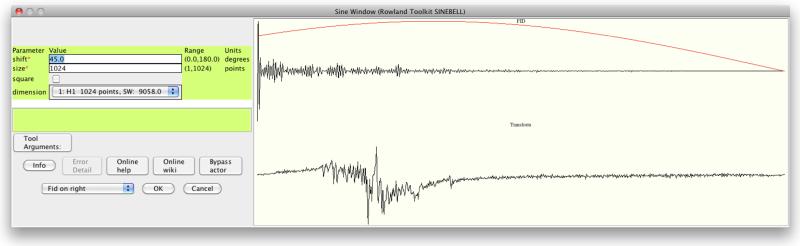
WB captures intermediate time-domain and frequency-domain data and metadata. These data may be stored in the relational database backend, in memory, or written to files as desired. Once captured, they can be inspected in order to verify that results are as expected. After a workflow has been executed, mousing over a data set shows the first FID. This is useful for understanding and debugging functions such as phasing and zero fills because it provides a quick visual cue that the intermediate results are reasonable, and shows what a workflow is doing. Correct parameterization of apodization actors, requiring a compromise between sensitivity and resolution, is assisted by interactive feedback; the first FID of the data set, the envelope of the apodization function, and their product are shown as parameters are set (Figure 6).
Figure 6.
interactive parameterization of sinebell apodization shows first data column (top right, black), function envelope (top right, red), and multiplication (bottom right).
Library of workflows
A set of workflows that operate on the majority of standard pulsed NMR experiments is bundled with WB. The workflows are included as part of the database install script, and so are installed at the same time as the database schema in MySQL. They are also included in XML and NMR-STAR format as tar files. To use one of these workflows, a new reconstruction is created; if using a workflow saved in the database, the “Reconstruction/Apply workflow” menu option is used; the “Import workflow” option is used to load workflows from XML and NMR-STAR files.
WB handles non-uniform time-domain data and automated maximum entropy
WB provides access to RNMRTK Maximum Entropy reconstruction of non-uniformly sampled data through a MaxEnt smart actor. This actor estimates the noise level, assisting the user in correctly setting the def, aim, and lambda parameters. Additionally, the user interface of WB presents the full set of parameters, along with standard default settings, to the user, making it clear what the values are, so that the user can peruse the RNMRTK documentation for additional clarification of appropriate parameter settings. When a workflow containing a MaxEnt actor is executed, WB uses ST to seamlessly provide format conversion, allowing addition of NMRPipe actors to the same workflow.
WB enhances the noise estimate of the automatic maximum entropy spectral reconstruction method published by Mobli, et. al. (2007a). The time domain data is conventionally Fourier transformed, and the spectrum is scanned to determine the range of values present, and a user-specified percentage of the values eliminated. The standard deviation of the remaining spectrum is taken to estimate noise. To eliminate the computationally intensive need to fully sort the spectrum data to determine which signals are included in the desired cutoff percentage, the calculation is approximated by a bucket sort (Cormen et al 2001). Values are placed unsorted into bins by their binary power and first two digits of the mantissa, and the set of bins that most closely approximates the specified user percentage is used.
Actors help users correctly parameterize workflows
WB actors provide useful feedback by analyzing the context in which they are placed in workflows. This context includes metadata and upstream actors, and is used to choose sensible parameter settings and decide if operations are applicable. Some actors, such as the one used for Maximum Entropy Reconstruction, use the full data set in order to provide this feedback.
Table 2 presents a partial list of actors implemented in WB. The actors are categorized by the NMR-domain task to be accomplished, and grouped into columns based on the tool to which the underlying function(s) belongs. Actors often correspond to a single function.
Table 2.
WB actors, grouped by underlying tool (NMRPipe, RNMRTK). Many functions are provided by both NMRPipe and RNMRTK, although both tools implement some functionality that the other does not.
| Task | NMRPipe | RNMRTK |
|---|---|---|
| resolution enhancement | ZF | zerofill |
| Apodization | GM, GMB, SINE | sinebell, em, gauss |
| solvent suppression | SOL | sstdc |
| phase correction | PS | phase |
| baseline correction | MED, POLY, BASE | bc |
| linear prediction | LP | blip, flip |
| Hilbert transform | HT | |
| Fourier transform | FT | fft |
| maximum entropy reconstruction | MEM | msa, msa2d |
WB was intended to focus on data integration and rely on third-party tools for NMR operations. However, tool idiosyncrasies such as sign conventions and defaults must be handled by WB to allow faithful interoperation. WB ensures that operations are consistent regardless of data format using a small set of additional actors implemented in Java. These include a circular shift for Bruker data, negation of imaginaries and sensitivity enhancement, metadata correction, and import and export. The additional ST actors reduce the burden of manually managing trivial and tedious details, while preventing mistakes. Table 3 presents a list of actors based on ST functions, which facilitate translation between formats and tools. Sensitivity enhancement and negation of imaginaries are typically performed upon import by NMRPipe but may also be performed in the middle of a processing pipeline. WB matches NMRPipe behavior by default.
Table 3.
WB actors providing access to ST translation features, implemented as semantic operations in ST. These tasks are often implicitly performed by NMRPipe and RNMRTK, or are necessary to correct for varying conventions. Therefore, these tasks are useful for translating between formats and tools.
| ST translation operation | NMRPipe | RNMRTK |
|---|---|---|
| time and frequency import/export | -in, -out | load*/put* |
| sensitivity enhancement | * | sefix1, sefix2 |
| metadata correction | N/A | N/A |
| circular shift | cs | rotate |
| negate imaginaries | sign | quadfix |
Export of workflows and reconstructions: XML and NMR-STAR files
While storing workflows in a centralized relational database greatly simplifies managing workflows and sharing them between users in the same research group or institution, WB also supports export of workflows in standard formats, facilitating exchange of workflows between fellow scientists using eXtensible Markup Language (XML), an industry standard format, as well as archival of workflows in the common NMR data repository of the BioMagResBank (BMRB), which uses the NMR-STAR format for data storage and dissemination.
XML is a textual format for data and markup that has been widely used for more than two decades. Tools for handling XML are available in most major languages. XML facilitates sharing data between computers and users, thereby promoting collaboration. XML has previously been used within the structural biology community by the PDB (Bernstein et al. 1977) and CCPN (Vranken et al. 2005), as well as by the systems biology community (Hucka et al. 2003). WB provides an XML schema and exports workflows according to this schema, against which the documents may be validated.
Command line interface
A console version of WB is available to facilitate validation and testing of actors and workflows. It is invoked from a shell and does not use a GUI. Monte Carlo methods are used to search for invalid actor configuration combinations so the configuration options can be corrected. The validation only attempts to identify which options the underlying processing tools – currently NMRPipe and Rowland Toolkit – accept as valid input. This interface provides both read and write access to the database and allows workflow execution from XML files. XML workflows may be saved to the database as well.
Teaching and workshops
WB was used as an integral component of two recent tutorial sessions on spectral reconstruction held at the University of Connecticut Health Center, in Farmington, CT. The first was in June 2012 and the second was in June 2014. WB was installed and set up with a MySQL database instance on several Linux virtual machine instances. Users then connected to the virtual machines through a VNC client-server instance. A sample workflow was demonstrated in order to showcase WB's features and proper use as well as to teach the attendees how to properly process a data set. This included topics such as metadata correctness. Documentation and tutorials from these workshops are freely available online (www.connjur.org).
Discussion and Conclusions
CONNJUR WB is a software integration environment, that is, it manages the configuration and operation of underlying, third party tools (currently NMRPipe, NMRDraw and the Rowland Toolkit). WB manages the syntax and semantics of tool operation; as well as the import, export, storage and translation of the underlying spectral data to be processed. WB can be used to generate processing workflows which are operationally identical to standard NMRPipe or RNMRTK processing scripts. However, by integrating ST, NMRPipe, and RNMRTK, WB allows for novel spectral reconstruction features difficult to reproduce with a shell script. For instance, since ST allows for facile translation between tool formats, a single workflow may easily combine NMRPipe and RNMRTK functions. This allows users familiar with NMRPipe functions, parameter names, and semantics to combine NMRPipe with maximum entropy reconstruction from RNMRTK: initially, only the new MaxEnt actor must be learned, while WB/ST handles the data translation behind the scenes.
WB's use of a MySQL relational database to store all results - including the time- and frequency-domain data, the metadata, and the workflows and parameterizations - frees the user from the tedium of manual data management. Instead, uniform access to the data is made possible. Furthermore, the semantics of these data are consistent in that a single medium is employed, and explicit due to the structure inherent in a database. This data integration facilitates software integration: the data is stored in a single location, with a single, uniform structure, regardless of the origin of the data. A programmer seeking to implement new functionality need therefore only deal with a single data API, reaping the benefits of integration with all formats supported by ST.
MySQL enables advanced querying and includes indexing for high performance. It uses a relational, non-hierarchical storage model in which data are stored in tables consisting of tuples of key-value pairs, which are linked to each other by means of relationships. This uniform model allows querying at any level of the data, and SQL's rich array of set operators allows for simple and powerful results to be obtained through a small amount of code. The structure and indexing capabilities optimize queries and data access, especially when compared to parsing textual or binary files.
Reproducibility, the ability to reach the same final result given a scientific procedure, is enhanced by explicit capture of the processing workflow, intermediates, and metadata. This is because the context of a result -- in this case, a frequency-domain spectrum -- is captured and made available. This in turn promotes collaboration and sharing of results and procedures; by collaborating and with access to the results of others’ work, the barriers to reusing valid scientific results are far lower; the effect is that progress is made more rapidly. WB supports export of workflows in the NMR-Star format, used by the BMRB, and in the industry-standard XML format. This enables NMR spectroscopists to exchange processing workflows, sharing techniques and results, and modifying those workflows as necessary to continually achieve better results.
WB's relational database backend provides a convenient platform for sharing data and experiments. Researchers with access to the same network can view, copy and modify other researcher's workflows and reconstructions. This was used during an NMR workshop; instructors were able to review and identify errors in student workflows without requiring direct access to the student workstations and interrupting their tutorial sessions.
WB's explicit recording of all relevant data is valuable for teaching the principles of spectral reconstruction as it makes the process tangible and concrete, so that it can be studied in detail. Successful reconstructions can be inspected to examine the effects of individual actors, while unsuccessful reconstructions can be inspected to learn how to fix incorrect implementations. WB provides a clean, simple solution for capturing primary data and metadata, including intermediates; as well as providing visualizations of that data to the user that clearly indicate what happened and why.
WB provides a high-level abstraction over existing tools
WB was designed with the goal of integrating existing functionality, not rewriting it. Popular tools such as NMRPipe have been in use for well over a decade. WB does not seek to supplant these tools, but rather to provide a safe and powerful abstraction over existing spectral reconstruction strategies. This allows WB's user to focus on the semantics of NMR and the goal of high-quality spectra, while WB handles trivial minutiae such as data format, layout, metadata syntax, and tool-specific details. In addition to saving time, this reduces the possibility of mistakes. The effort, tedium and slowness in the life cycle in changing, running and evaluating NMR signal processing scripts does not encourage experimentation and optimization; with its ease of use, forked workflows, and real time display of first fids, WB does. Its independent open source nature allows new processing techniques to be added without the overhead of developing a new graphical interface.
Smart actors
NMR functions used in spectral reconstruction are difficult to correctly parameterize, due both to inherent domain issues as well as to incidental ones. Examples of the former are the intricate knowledge needed of the tradeoff between sensitivity and resolution combined with knowledge of the signal decay and noise properties needed to correctly select and parameterize an apodization function; knowledge of the properties and restrictions of the Fast Fourier Transform (FFT) algorithm that are needed to correctly parameterize a zero-fill function, since the output needs to be of a size that is an integer power of 2. Examples of the latter include the knowledge of the semantics, parameter names, and allowable values of each of the functions provided by a concrete implementation of processing workflows such as NMRPipe and RNMRTK. WB addresses these issues by means of smart actors. Smart actors, as all other actors in WB, simplify the task of understanding and correctly parameterizing a function by intimately understanding the function which they wrap, and meaningfully presenting that understanding to the user. Furthermore, smart actors assist the user in coming up with useful parameter settings. An excellent example of this is the smart actor for Maximum Entropy reconstruction. As described in Mobli et al. (2007a, 2007b), correct parameterization requires an accurate estimate of the noise level. This actor provides this functionality, helping the user to parameterize the function. Parameters may be input using different units than accepted by the underlying tool, insulating the user from unnecessary knowledge of tool internals. Help buttons on actors take users directly to relevant online documentation, helping the user to understand the purpose and correct parameterization of processing functions.
Semantic and metadata correctness
WB promotes semantic correctness of processing workflows. Semantically correct workflows respect rules of the NMR domain. For instance, a Fourier Transform should not be applied twice along one dimension of a data set; frequency-domain data should not be apodized. WB implements semantic correctness by tracking the domain (time- or frequency-) of the data, and requiring that an actor be applied to data of the correct domain. The domain of each dimension is tracked and the applicability of actors enforced. Some actors, such as those for baseline correction and solvent suppression, may only be added to a workflow once. WB also distinguishes between uniform and nonuniform time domain data.
Metadata correctness is important to spectral processing correctness, because primary data cannot be correctly interpreted and used without correct and complete metadata. Incorrect metadata can silently cause problems whose root cause is not obvious. WB is designed to deal with metadata issues by allowing querying and modification as necessary, as well as saving metadata alongside binary data in the database. Tracking metadata also enables simple checks of consistency and semantic correctness of results.
Library of workflows provides common and general functionality
WB's built-in library of workflows provides spectroscopists with workflows for processing standard NMR experiments. These workflows simplify dealing with real data because they abstract over the actual layout of the data, eliminating a combinatorial explosion: a single workflow handles all possible choices of dimension-channel assignment and axis ordering, in contrast to processing scripts, which do not abstract over the data layout. When applying a workflow, the user must tell WB which dimension corresponds to which nucleus, after which WB automatically handles those settings for the rest of the workflow.
Interactive dialogs and immediate feedback facilitate experimentation and help novice users
WB's GUI places relevant information at the disposal of the user, enabling a more efficient process for reconstruction. In place of creating a single script containing numerous steps that is executed at the end, WB allows spectroscopists to use an incremental, step-wise process to create a reconstruction, with quality checking at each step. Spectroscopists can design one or two actions to be carried out on a data set, execute those actions, and verify the correctness of the results. If the results are incorrect, the location of the problem is readily apparent as the user knows exactly which action caused the problem and only needs to backtrack one or two steps and make a correction. This process means that errors are caught and corrected more quickly due to the continual focus on quality. An additional benefit is reaped from the ability to branch workflows, enabling the comparison of multiple actor parameterizations, reducing the tendency to settle for non-optimal parameter sets and supporting understanding of parameter choices. For many actors, optimal parameter values are not easy to find. Experimenting with several different values helps to find a suitable one.
The ability to mouse over a data set to show the first FID provides immediate feedback to the user, which is of immense help in understanding the effect of various processing functions, as well as determining whether there is an error and locating such an error. By looking just at the first FID, an experienced spectroscopist can quickly and easily identify such phenomena as phase errors, baseline distortions, improper apodization parameterization, likelihood of truncation artifact, zerofill, resolution, and sensitivity. The immediate feedback of the effect of an apodization function in the apodization actors is a further example of the value provided by interactive data visualizations.
Facilitation of processing of non-uniformly sampled data
Non-uniform sampling offers potential savings in spectrometer usage time and increases in resolution and sensitivity. Importantly, its benefits increase with the number of dimensions of the experiment. The difficulty is that once non-uniformly sampled data is collected, it may not be simply processed with the Fourier Transform, as is done with uniformly sampled data. While simple methods exist for converting non-uniformly sampled data to uniformly sampled data, allowing use of the Fourier transform, such transformations are non-optimal, producing artifacts in the final spectrum. In order to obtain a high-quality spectrum, advanced processing techniques such as Maximum Entropy reconstruction are required. However, implementations of the algorithm are difficult to integrate and difficult to correctly parameterize. WB addresses these problems by using ST for data integration and a smart actor to help the user correctly parameterize the function. This makes non-uniform reconstructions usable for a wider range of NMR spectroscopists.
WB is open source
All CONNJUR software programs, including WB, have been released under the standard MIT and GPL open source licenses and will continue to be made available under such licenses, free of charge, in perpetuity. CONNJUR is open source because we believe that open source software is beneficial to the community as it facilitates collaboration, continuity, quality, and openness and efficiency of development. Users are therefore encouraged to use the source code as they see fit, including patches, using the source for other purposes, and submitting modifications back to the CONNJUR team. CONNJUR's goal is to produce software of the highest quality and usefulness, and we believe that the community involvement enabled by open source licensing help ultimately results in better software which is more able to meet its users’ needs.
Table 1.
Summary of CONNJUR WB Features
| Feature | Significance |
|---|---|
| Rich data model of spectral reconstruction, metadata, and parameterization | Allows proper conversion of units and guided actor parameterization |
| Centralized data management provided by relational database using a single format | Data integration and advanced querying capabilities |
| Explicit capture of metadata | Enables reproducibility |
| Inspection and modification of metadata | Visibility of metadata helps to avoid common errors due to improper settings of metadata parameters |
| GUI environment for workflow building, execution, and analysis | Uniform interface to third-party tools, enhances discoverability of functions and parameters |
| Actors understand their context and assist the user | Simplified parameterization and protection from mistakes |
| Actors have access to metadata and logic making use of the metadata can be built in to actors | User has access to information about spectral characteristics before executing the workflow |
| Semantic constraints to ensure the validity of workflows | Reduces errors in workflow creation |
| Support for non-uniformly sampled data sets and Maximum Entropy reconstruction | Shortens data collection time, and decrease difficulty of data processing |
| Support for Bruker, Varian/Agilent, RNMRTK, and NMRPipe formats | Automatically handles translation between, to, and from various formats |
| Interactive, immediate feedback from actors and workflow -- first FIDs are displayed when mousing over data sets; in apodization, effect of function is shown while parameters are updated | Very quick to detect and address obvious errors and issues with parameterization |
| Export to XML and NMR-STAR formats | Enhances collaboration and facilitates sharing |
| Branched and parallel execution of workflows | Efficient reconstruction of spectra with multiple different parameter sets |
| Software integration of existing processing tools | Function and parameter names are unchanged, capitalizing on users’ familiarity with existing tools |
| Available for free under the standard MIT and GPL open source licenses | Source code may be inspected, modified, and redistributed |
Acknowledgements
This research was funded by United States National Institutes of Health grant GM-083072. The authors wish to thank Drs. Frank Delaglio, Jeffrey Hoch, Mark W. Maciejewski and Alan Stern for useful conversations and assistance with the operation of NMRPipe and the Rowland NMR Toolkit. The authors would also like to thank Dr. Eldon Ulrich for assistance with the NMR-STAR format and data dictionary.
Footnotes
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Altintas I, Berkley C, Jaeger E, Jones M, Ludascher B, Mock S. Scientific and Statistical Database Management, 2004. Proceedings. 16th International Conference on. IEEE; Jun, 2004. Kepler: an extensible system for design and execution of scientific workflows. pp. 423–424. [Google Scholar]
- ACD/NMR Processor Academic Edition 2014 Sep 17; Web. < http://www.acdlabs.com/resources/freeware/nmr_proc/>.
- Azara home page 2014 Sep 17; Web. < http://www2.ccpn.ac.uk/azara/>.
- Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Brice MD, Rodgers JR, Tasumi M. The protein data bank. European Journal of Biochemistry. 1977;80(2):319–324. doi: 10.1111/j.1432-1033.1977.tb11885.x. [DOI] [PubMed] [Google Scholar]
- Bowers S, Ludäscher B. Conceptual Modeling–ER 2005. Springer; Berlin Heidelberg: 2005. Actor-oriented design of scientific workflows. pp. 369–384. [Google Scholar]
- Clore GM, Gronenborn AM. Multidimensional heteronuclear nuclear magnetic resonance of proteins. Methods Enzymol. 1994;239:349–363. doi: 10.1016/s0076-6879(94)39013-4. [DOI] [PubMed] [Google Scholar]
- Cormen TH, Leiserson CE, Rivest RL, Stein C. Introduction to algorithms. Vol. 2. MIT press; Cambridge: 2001. pp. 531–549. [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Delta NMR Data Processing Software 2014 Sep 17; Web. < http://www.jeolusa.com/tabid/380/Default.aspx>.
- Ernst RR, Anderson WA. Application of Fourier transform spectroscopy to magnetic resonance. Review of Scientific Instruments. 1966;37(1):93–102. [Google Scholar]
- Ernst RR, Bodenhausen G, Wokaun A. Principles of nuclear magnetic resonance in one and two dimensions. 1991 (No. LRMB-BOOK-1991-001) [Google Scholar]
- Güntert P, Dötsch V, Wider G, Wüthrich K. Processing of multi-dimensional NMR data with the new software PROSA. J. Biomol. NMR. 1992;2:619–629. [Google Scholar]
- Gryk MR, Vyas J, Maciejewski MW. Biomolecular NMR data analysis. Progress in nuclear magnetic resonance spectroscopy. 2010;56(4):329. doi: 10.1016/j.pnmrs.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch JC, Stern AS. NMR data processing. Wiley-Liss; New York: 1996. [Google Scholar]
- Hoch JC, Stern A. The Rowland NMR Toolkit. Rowland Institute for Science Technical Memorandum, (18t) 1985 [Google Scholar]
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Wang J. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19(4):524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- iNMR . Sep 17, 2014. Web. < http://www.inmr.net/>. [Google Scholar]
- Jeener J. Oral presentation in Ampere International Summer School II. Basko Polje, Yugoslavia: 1971. 1971. [Google Scholar]; b Aue WP, Bartholdi E. Ernst. J. Chem. Phys. 64:2229. [Google Scholar]
- Kazimierczuk K, Misiak M, Stanek J, Zawadzka-Kazimierczuk A, Kozminski W. Generalized Fourier transform for non-uniform sampled data. Top. Curr. Chem. 2012;316:79–124. doi: 10.1007/128_2011_186. [DOI] [PubMed] [Google Scholar]
- Kazimierczuk K, Kozminski W, Zhukov I. Two-dimensional Fourier transform of arbitrarily sampled NMR data sets. J. Magn. Reson. 2006a;179:323–328. doi: 10.1016/j.jmr.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Kazimierczuk K, Zawadzka A, Kozminski W, Zhukov I. Random sampling of evolution time space and Fourier transform processing. J. Biomol. NMR. 2006b;36:157–168. doi: 10.1007/s10858-006-9077-y. [DOI] [PubMed] [Google Scholar]
- Mnova NMR Lite 2014 Sep 17; Web. < http://mestrelab.com/software/mnova/nmr-lite/>.
- Maciejewski MW, Qui HZ, Rujan I, Mobli M, Hoch JC. Nonuniform sampling and spectral aliasing. J. Magn. Reson. 2009;199:88–93. doi: 10.1016/j.jmr.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobli M, Maciejewski MW, Gryk MR, Hoch JC. An automated tool for maximum entropy reconstruction of biomolecular NMR spectra. Nat Methods. 2007a;4:3–4. doi: 10.1038/nmeth0607-467. [DOI] [PubMed] [Google Scholar]
- Mobli M, Maciejewski MW, Gryk MR, Hoch JC. Automatic maximum entropy spectral reconstruction in NMR. J. Biomol. NMR. 2007b;39:133–139. doi: 10.1007/s10858-007-9180-8. [DOI] [PubMed] [Google Scholar]
- Nowling RJ, Vyas J, Weatherby G, Fenwick MW, Ellis HJ, Gryk MR. CONNJUR spectrum translator: an open source application for reformatting NMR spectral data. Journal of biomolecular NMR. 2011;50(1):83–89. doi: 10.1007/s10858-011-9497-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyquist H. Certain topics in telegraph transmission theory. American Institute of Electrical Engineers, Transactions of the. 1928;47(2):617–644. [Google Scholar]
- Oschkinat H, Griesinger C, Kraulis PJ, Sørensen OW, Ernst RR, Gronenborn AM, Clore GM. Three-dimensional NMR spectroscopy of a protein in solution. Nature. 1988;332(6162):374–376. doi: 10.1038/332374a0. [DOI] [PubMed] [Google Scholar]
- PERCH Solutions 2014 Sep 17; Web. < http://new.perchsolutions.com/index.php?id=36>.
- Schmieder P, Stern AS, Wagner G, Hoch JC. Application of nonlinear sampling schemes to COSY-type spectra. J. Biomol. NMR. 1993;3:569–576. doi: 10.1007/BF00174610. [DOI] [PubMed] [Google Scholar]
- Shimba N, Stern AS, Craik CS, Hoch JC, Dötsch V. Elimination of 13Cα Splitting in Protein NMR Spectra by Deconvolution with Maximum Entropy Reconstruction. J. Am. Chem. Soc. 2003;125:2382–1. doi: 10.1021/ja027973e. [DOI] [PubMed] [Google Scholar]
- SpinWorks 2014 Sep 17; Web. < http://www.columbia.edu/cu/chemistry/groups/nmr/SpinWorks.html>.
- Stern AS, Li K-, Hoch JC. Modern Spectrum Analysis in Multidimensional NMR Spectroscopy: Comparison of Linear-Prediction Extrapolation and Maximum-Entropy Reconstruction. J. Am. Chem. Soc. 2002;124:1982–12. doi: 10.1021/ja011669o. [DOI] [PubMed] [Google Scholar]
- Stoven V, Annereau JP, Delsuc MA, Lallemand JY. A New N-Channel Maximum Entropy Method in NMR for Automatic Reconstruction of Decoupled Spectra and J-Coupling Determination. J. Chem. Inf. Comput. Sci. 1997;37:265–272. [Google Scholar]
- SwaN-MR 2014 Sep 17; Web < http://www.inmr.net/swan/>.
- TopSpin 2014 Sep 17; Web. < http://www.bruker.com/products/mr/nmr/nmr-software/software/topspin/overview.html>.
- Verdi KK, Ellis HJ, Gryk MR. Conceptual-level workflow modeling of scientific experiments using NMR as a case study. BMC Bioinformatics. 2007;8:31. doi: 10.1186/1471-2105-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VnmrJ 2014 Sep 17; Web. < http://www.chem.agilent.com/en-US/products-services/Software-Informatics/VnmrJ/Pages/default.aspx>.
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins: Structure, Function, and Bioinformatics. 2005;59(4):687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]