Abstract
In the human body, there are 10 bacterial cells for every one human cell. This fact highlights the importance of the National institutes of Health’s initiative to map the human microbiome. The Human Microbiome Project was the first large-scale mapping of the human microbiome of 5 body sites: GI tract, mouth, vagina, skin and nasal cavity using culture-independent methods. The bladder was not originally tested because it was considered to be sterile and there were complexities regarding sample collection. Over the last couple years our team along with other investigators have shown that a urinary microbiome exists and for most individuals it plays a protective role.
Keywords: Urinary microbiome, urgency incontinence, bacteria
Introduction
With the aid of interdisciplinary research, clinicians are learning more about the complex roles that microbial communities (microbiota) play within each organ system. The interactions between these microbiota and their host, and the relationship between these interactions and organ health and disease are complex. For example, we know that the bacterial species Clostridium difficile and Helicobacter pylori live in the gut, contributing to gastrointestinal health. With fluctuations in the host environment, however, these bacteria may cause severe illness in the form of C. diff colitis or H. pylori peptic ulcers. Understanding such symbiotic, yet potentially pathologic, relationships has been an ongoing journey for researchers; the more we learn, the more we realize we do not fully understand. As the Loyola Urinary Education and Research Collaborative (LUEREC), a interdisciplinary team that includes clinicians, clinical microbiologists, and basic scientists, works to understand the microbiota of the bladder and their impact on wellness and illness, we appreciate the scientists who paved the way for our current discoveries and recognize the value of new technologies that permit us to make new discoveries.
Historical perspective
The origin of the “urine is sterile” dogma dates to the mid-1800s and the attempt to understand germ theory by the earliest bacteriologists, including Louis Pasteur, Joseph Lister and William Roberts [2–4], who showed that a vial of urine in a sealed container did not turn cloudy, in contrast to a vial of urine exposed to air or with added tap water. The conclusion was that “…fresh and healthy urine is perfectly free from bacteria or other minute organisms” [3, 4]. In other words, the dogma originated in an era when all bacteria were considered pathogens and microbiology was in its infancy. Yet, the dogma persists.
In the 1950s, Edward Kass, an infectious disease physician at Harvard Medical School, established a threshold for infection to detect patients with pyelonephritis [6]. After analyzing the urine of symptomatic versus asymptomatic women in pregnancy or with diabetes or a cystocele, Kass determined that 105 colony forming units per milliliter (CFU/ml) was the dividing line between contamination and infection in most populations [7]. Since then, this standard culture method has been adopted to include lower urinary tract infections, despite several studies that have since provided evidence that the ≥105 CFU/mL threshold is insufficient to detect significant bladder infections [9–13].
There were those who went against the dogma and as a result improved patient outcomes. In the mid-1800s, Lister developed aseptic catheterization to prevent contamination of a sterile body site. Aseptic catheterization soon became standard of practice for urethral catheterization [3]. However, in the 1960s, the urologist Jack Lapides suggested that intermittent catheterization did not have to be a sterile procedure performed exclusively by medical professionals. It was hypothesized that intraluminal urinary tract pressure results in local ischemia of the bladder wall, resulting in bladder distention, thereby causing urinary tract infections (UTI) in spinal cord injury patients. He taught a clean technique in which a patient, after washing his or her hands, would insert a catheter for voiding. Lapides’ research in neurogenic bladder patients challenged the idea that UTIs were caused by instrumentation. Clean intermittent catheterization not only improved continence, but decreased rates of UTI and pyelonephritis in these patients [3, 14]. His work demonstrated that UTIs could be prevented by, rather than be the result of, catheterization.
If instrumentation is not always the cause of an infection, could there be more to the bladder environment than previously thought? Rosalind Maskell, a clinician directing a clinical microbiology laboratory in England, thought the answer might be ‘yes.’ Maskell noticed that patients with UTI symptoms, but negative standard urine cultures, contained slow growing organisms that required different growth conditions than those in standard method [15, 16]. She concluded that standard urine culture was insufficient to diagnose many urinary disorders, and she urged general practitioners to collaborate with microbiologists to understand urinary disorders [15, 16]. Her advice was repudiated or ignored. It was not until culture-independent methods were developed that we began to understand that the slow-growing organisms revealed by Maskell’s experiments were just the tip of the iceberg.
The Human Microbiome Project (HMP)
Only recently has it become accepted that most human body sites are colonized with bacteria, and that the majority of those bacteria are non-pathogenic. The first large-scale mapping of the human microbiome using culture-independent methods was the Human microbiome project (HMP). In the human body, there are 10 bacterial cells for every one human cell. However, medical studies have traditionally focused on disease-causing bacteria. Therefore, very little was known about the natural residents, or commensals, of the human body. Initially, the HMP characterized the gut microbiome because samples from this heavily colonized (high biomass) environment are easy to collect, significant diversity exists between individuals [17], and differences had previously been observed in health and disease states [18]. Following the initial phase, the project was expanded to sample the microbiome of 300 “normal” individuals at a total of five body sites: GI tract, mouth, vagina, skin and nasal cavity, using culture-independent methods, such as metagenomic and amplicon sequencing [19].
Sequencing Technologies
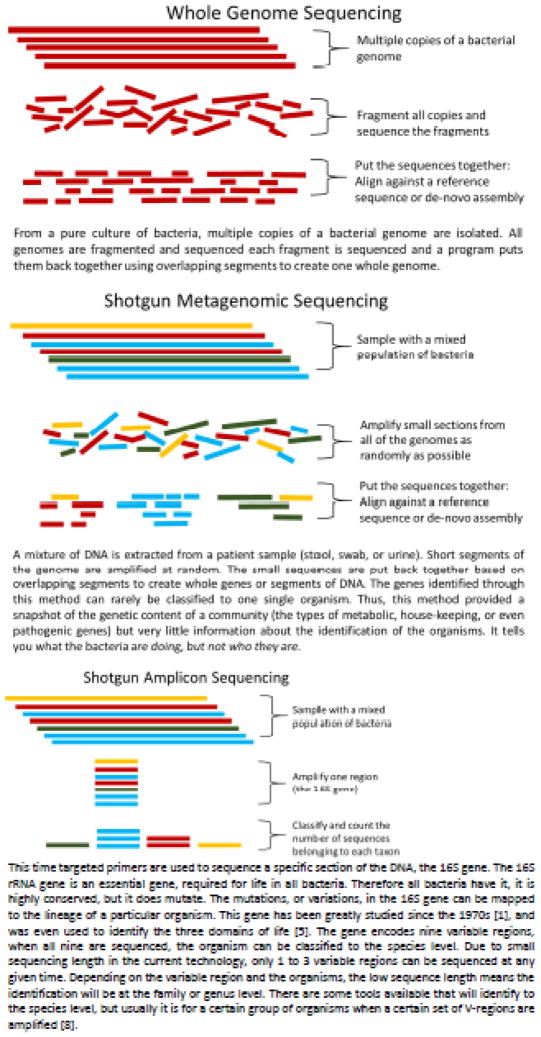
Most current high throughput DNA sequencing technologies can only produce relatively short sequence reads (~250–400 base pairs). Because they use massively parallel sequencing approaches and thus generate large numbers of sequences, these technologies (often called Next Generation Sequencing or NGS) allow for large coverage of a genome or great depth of a single genomic site. Sequencing strategies fall into two categories: whole genome and metagenome. Whole genomes sequencing starts with an isolated pure culture, while metagenomics is performed on patient samples with mixed populations, such as a swab, fecal material, or urine. Each sequencing strategy uses a different technique to turn the large amounts of short sequences generated by NGS into useful information (Figure 1).
Figure 1.
Sequencing Technologies
The most common technique to classify bacteria within a population is 16S rRNA gene amplicon sequencing (Figure 1C). The 16S rRNA gene is used because it is highly conserved [20, 5, 1] among bacteria, a direct result of its critical cellular role, but is lacking in other domains of life. Within the gene, some stretches do evolve. Thus, conserved regions are interspersed with hypervariable regions. These hypervariable regions contain specific polymorphisms that can be used for measuring evolutionary distance and thus phylogenetic relatedness (20). All 9 known hypervariable regions (V1–V9) of the 16S rRNA gene are proven to contain sufficient polymorphisms for species level classification. However, sequencing one hypervariable region is only sufficient to achieve taxonomic classification to the family or genus level.
Much of the HMP research has relied on 16S rRNA amplicon sequencing to obtain a first glimpse of bacteria present in different niches, at different times, and between different disease states. Amplicon sequencing relies on polymerase chain reaction (PCR) amplification. This method is not quantitative; different sequences amplify at different rates; thus, the number of sequence reads obtained from different samples cannot be directly compared. Instead, researchers compare these data qualitatively, looking at the relative amounts of classified bacteria, rather than the numbers of raw sequences.
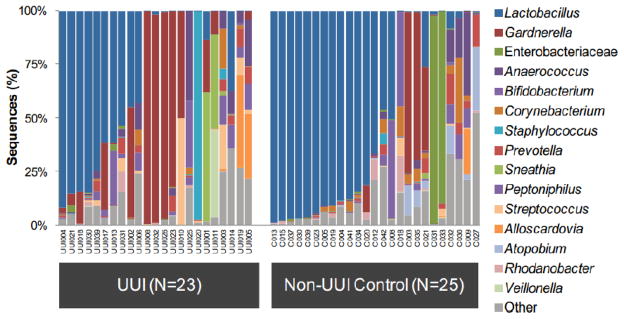
A relative abundance graph (Figure 2) depicts the bacterial community structure in terms of percentage of total classified reads (y-axis) per sample collected (x-axis). Each color represents a different taxonomic classification. These graphs can be shown at any taxonomic level (Phylum, Class, Order, Family, or Genus) or combination of levels. For example, the previously published data in Figure 2 represents catheterized urines sequenced from women with urgency urinary incontinence or from continent controls [21]. Some urine samples were dominated by the genera Lactobacillus (blue) or Gardnerella (red) or by the family Enterobacteriaceae (green), which includes Escherichia, Klebsiella, and Proteus. The V4 region does not contain enough variability to distinguish between members of the Enterobacteriaceae family. In other samples, no bacterium dominated; these were termed “diverse.” In this way, one graph can display large amounts of taxonomic information concerning the qualitative differences between samples.
Figure 2. Relative Abundance of urgency urinary incontinence (UUI) patients versus continent controls.
Here is shown the bacterial community structure in terms of percentage of total classified reads (y-axis) per sample collected (x-axis) from previously published data [21]. Each color represents a different taxonomic classification at the Family or Genus level. Some urine samples were dominated by Lactobacillus (blue) or Gardnerella (red) or by the family Enterobacteriaceae (green), which includes Escherichia, Klebsiella, and Proteus. In other samples, no bacterium dominated; these were termed “diverse.” The UUI cohort has more individuals in the Gardnerella urotype and tends to be more diverse overall than the continent controls.
Results of the HMP
Due to complex community structures, and variation between individuals, the data obtained through HMP was more complex that initially expected and a new set of analyses had to be developed. One qualitative approach was the identification of community types; they are called enterotypes in the gut [22], community state types (CSTs) in the vagina [23], and urotypes in the urine [21]. To form community types, bacterial profiles are clustered together based on taxonomic similarity. Those clusters can then be tested for their statistical association between individuals and diseases. Using this method, different enterotypes were found to be correlated to one’s diet, obesity, or Crohn’s disease [24, 22, 25].
The HMP resulted in a large increase in our understanding of the vaginal microbiome. It is well documented that the vagina of reproductive age women is highly colonized with bacteria (high biomass) and that the vaginal microbiome tends to fall into one of five CSTs: CST 1, 2, 3, and 5 are dominated by different species of Lactobacillus (L. crispatus, L. gasseri, L. iners, and L. jensenii, respectively) [23]. In contrast, CST 4 is dominated by a diverse group of anaerobes, including Anaerococcus, Peptoniphilus, Prevotella, Streptococcus, Atopobium, and Gardnerella [23].
The bladder is not sterile
The bladder was not originally tested in the HMP, primarily because it was considered sterile. Therefore, the study of the bladder microbiome is years behind the study of other human niches. As Maskell demonstrated, bacteria have not been detected in the bladder primarily because culture techniques were developed for fast growing aerobic uropathogens, such as uropathogenic E. coli. However, the paradigm is changing [26].
Two other major hurdles prevented the bladder microbiome field from moving forward: first, bacteria from the bladder had to be distinguished from vulvo-vaginal contamination; second, the detected DNA had to be shown to originate from live bacteria. To overcome the first hurdle, LUEREC researchers sampled urine directly from the bladder using suprapubic aspiration (SPA). SPA collection bypasses the vagina and thus the samples could not be contaminated by vulvo-vaginal flora. Using 16S amplicon sequencing, the bacterial DNA in SPA samples was compared to the bacterial DNA in urine from the same individuals obtained by transurethral catheter (TUC). TUC samples were also compared to voided urine and vaginal swabs. SPA and TUC urine had similar profiles, and were distinct from voided urine and vaginal swabs. Since SPA sampling bypasses the vagina, it was concluded that the bladder contained bacterial DNA and that TUC also samples the bladder environment [27]. Since TUC is a more feasible urine collection method, currently it is sampling method of choice [28, 29].
To overcome the second hurdle, LUEREC researchers showed that the detected bacterial DNA represents living bacteria, instead of DNA from dead cells or free floating DNA. The previous study had provided taxonomic identification of the 16S sequences detected in the bladder [27]. Most of the detected bacteria were cultivatable, but not under standard urine culture conditions. Therefore, an enhanced quantitative urine culture (EQUC) protocol was developed. Unlike the standard urine culture protocol, which plates 1μl urine on blood and MacConkey agar with incubation in air for 24 hours, EQUC plates a greater amount of urine (100μl) on a variety of media with incubation under diverse atmospheric conditions for 48 hours [30]. The result is that many organisms detected by sequencing can be grown by culture. The development of EQUC and other enhanced culture protocols [29] showed that the DNA detected by sequencing represent live bacteria.
The female urinary microbiota in relation to health and disease
Now that we know the female urinary microbiota (FUM) exist, the next obvious issue is whether they contribute to health and disease. A few key things are known. First, the FUM are distinct between overactive bladder (OAB) patients and continent controls [21, 31] (Figure 2). Overall, the OAB cohorts tend to be more diverse by culture and sequencing, and have fewer Lactobacillus-dominant profiles. This is reminiscent of the vaginal microbiome story. However, it is unclear how much of the difference in diversity is due to the age, and therefore hormone status, of the individual versus the disease. Currently, studies are underway to address this concern.
Furthermore, the FUM profile, or phenotype, of a patient’s bladder microbiome could be predictive of anticholinergic (solifenacin) treatment response [31]. Less diverse FUM (mean of 3 unique organisms by EQUC) correlate with robust treatment response, whereas FUM with more diverse cultivatable organisms tend to require either an increased dose or do not respond to the medication. All of these data show that there is a correlation between the FUM and important factors associated with OAB syndrome; however, it is still unclear if these correlations are simple biomarkers or play a pivotal role in the etiology, progression, or resolution of the disease.
Finally, there is evidence to suggest the presence of protective FUM. Notice that most of the organisms discussed so far are not commonly thought of as uropathogens. In fact, the most common genus, Lactobacillus, is considered beneficial in many other areas of the body, including the gut and vagina. In a study of women undergoing instrumentation, it was found that detection of urinary bacteria correlated with greater symptom resolution and a decreased incidence of post-instrumentation UTIs [32], suggesting that certain FUM profiles can be beneficial or protective.
Future directions
If a bladder microbiome exists, how can we tell if it is friend or foe? Most likely, it will not be from urine cultures alone since we cannot identify bacterial virulence factors with this information. Lower urinary tract symptoms such as urinary urgency, frequency and incontinence can be a starting point. We hope to identify the bacteria that cause specific symptoms including an inflammatory response in the host. Many of us hypothesize that, similar to other bacterial niches, the inflammatory response to the same bacteria in the bladder may differ based on host factors.
Likewise, we hope to elucidate the factors that contribute to a “healthy” (asymptomatic) bladder. Since the incidence of UTI in women increases after menopause, age and estrogen are thought to play significant roles. It is more than likely that the FUM is altered with age. Other microbial niches, such as the vaginal tract, are greatly altered in the absence of estrogen. Given that estrogen alters the bladder epithelium [33, 34], and that estrogen improves the incidence of UTIs [35–37] and urgency urinary incontinence (UUI) [38–40], it is very likely that the microbiome will be altered following menopause which could be a contributing factor to many lower urinary tract symptoms associated with age.
At this time, all of our work has been done with catheterized urine samples to avoid obtaining the urethral, periurethral, vaginal and perineal microbiome that becomes part of the voided urine specimen. It will be critical that we develop methods that allow us to use voided urine specimens since large-scale multi-age group studies cannot be encumbered by needing to obtain catheterized urine specimens. In addition, the data generated from our work on the urinary microbiome requires the development of large databases and the expertise to statistically analyze these data.
Summary
A urinary microbiome exists and for most individuals it plays a protective role. Changes in the urinary microbiome appear to be a factor for some individuals who have the symptom of urgency urinary incontinence. The urinary microbiome may also be predictive of anticholinergic (solifenacin) treatment response. Our future work is based on better understanding what lifestyle and host factors can contribute to developing and maintaining a healthy urinary microbiome.
Footnotes
Conflict of Interest
Drs. Thomas-White, Brady, and Wolfe declare that they have no conflict of interest. Dr. Mueller reports consultancy fees and grants from Astellas.
Compliance with Ethics Guidelines
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Contributor Information
Krystal Thomas-White, Department of Microbiology and Immunology, Stritch School of Medicine, Loyola University, Chicago, Illinois, USA.
Megan Brady, Departments of Obstetrics & Gynecology and Urology, Loyola University Medical Center, Maywood, Illinois, USA.
Alan J. Wolfe, Department of Microbiology and Immunology, Stritch School of Medicine, Loyola University, Chicago, Illinois, USA
Elizabeth R. Mueller, Email: emuelle@lumc.edu, Associate Professor, Departments of Obstetrics and Gynecology and Urology, Division of Female Pelvic Medicine and Reconstructive Surgery, Stritch School of Medicine, Loyola University Chicago, 2160 South First Avenue, Maywood, IL 60153, Phone: (708) 216-3326, FAX: (708) 216-4305
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Van de Peer Y, Chapelle S, De Wachter R. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic acids research. 1996;24(17):3381–91. doi: 10.1093/nar/24.17.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duclaux E. Pasteur: The History of the Mind. Philidelphia and London: W.B. Saunders Company; 1920. [Google Scholar]
- 3.Bloom DA, McGuire EJ, Lapides J. A brief history of urethral catheterization. The Journal of urology. 1994;151(2):317–25. doi: 10.1016/s0022-5347(17)34937-6. [DOI] [PubMed] [Google Scholar]
- 4.Roberts W. On the Occurrence of Micro-Organisms in Fresh Urine. Br Med J. 1881;2(1085):623–5. doi: 10.1136/bmj.2.1085.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proceedings of the National Academy of Sciences of the United States of America. 1977;74(11):5088–90. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kass EH. Pyelonephritis and bacteriuria. A major problem in preventive medicine. Ann Intern Med. 1962;56:46–53. doi: 10.7326/0003-4819-56-1-46. [DOI] [PubMed] [Google Scholar]
- 7.Kass EH. Asymptomatic infections of the urinary tract. Transactions of the Association of American Physicians. 1956;69:56–64. [PubMed] [Google Scholar]
- 8.Fettweis JM, Serrano MG, Sheth NU, Mayer CM, Glascock AL, Brooks JP, et al. Species-level classification of the vaginal microbiome. BMC genomics. 2012;13(Suppl 8):S17. doi: 10.1186/1471-2164-13-S8-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stamm W, Counts G, Running K, Fihn S, Turck M, Holmes K. Diagnosis of coliform infection in acutely dysuric women. The New England journal of medicine. 1982;307(8):463–8. doi: 10.1056/NEJM198208193070802. [DOI] [PubMed] [Google Scholar]
- 10.Stark RP, Maki DG. Bacteriuria in the Catheterized Patient. New England Journal of Medicine. 1984;311(9):560–4. doi: 10.1056/NEJM198408303110903. [DOI] [PubMed] [Google Scholar]
- 11.Lipsky BA, Ireton RC, Fihn SD, Hackett R, Berger RE. Diagnosis of bacteriuria in men: specimen collection and culture interpretation. J Infect Dis. 1987;155(5):847–54. doi: 10.1093/infdis/155.5.847. [DOI] [PubMed] [Google Scholar]
- 12.Thomas MH, Pacita LR, Marsha EC, Ann ES. Voided Midstream Urine Culture and Acute Cystitis in Premenopausal Women. New England Journal of Medicine. 2013;369 doi: 10.1056/NEJMoa1302186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooton TM, Roberts PL, Cox ME, Stapleton AE. Voided midstream urine culture and acute cystitis in premenopausal women. The New England journal of medicine. 2013;369(20):1883–91. doi: 10.1056/NEJMoa1302186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14•.Lapides J, Diokno AC, Silber SJ, Lowe BS. Clean, intermittent self-catheterization in the treatment of urinary tract disease. 1972. The Journal of urology. 2002;167(2 Pt 2):1131–3. discussion 4 This landmark paper describes how intermittent self-cathaterization decreased the incidence of UTIs. [PubMed] [Google Scholar]
- 15.Maskell R, Pead L, Allen J. The puzzle of “urethral syndrome”: a possible answer? Lancet. 1979;1(8125):1058–9. doi: 10.1016/s0140-6736(79)92953-2. [DOI] [PubMed] [Google Scholar]
- 16.Maskell RM. The natural history of urinary tract infection in women. Medical hypotheses. 2010;74(5):802–6. doi: 10.1016/j.mehy.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 17.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–3. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 19.Group NHW, Peterson J, Garges S, Giovanni M, McInnes P, Wang L, et al. The NIH Human Microbiome Project. Genome research. 2009;19(12):2317–23. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dubnau D, Smith I, Morell P, Marmur J. Gene conservation in Bacillus species. I. Conserved genetic and nucleic acid base sequence homologies. Proceedings of the National Academy of Sciences of the United States of America. 1965;54(2):491–8. doi: 10.1073/pnas.54.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21•.Pearce MM, Hilt EE, Rosenfeld AB, Zilliox MJ, Thomas-White K, Fok C, et al. The Female Urinary Microbiome: a Comparison of Women with and without Urgency Urinary Incontinence. mBio. 2014;5(4) doi: 10.1128/mBio.01283-14. This manuscript describes a difference in the microbiome of women with urinary incontinence compared to continent controls both by culture and sequencing methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, et al. Vaginal microbiome of reproductive-age women. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4680–7. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–8. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quince C, Lundin EE, Andreasson AN, Greco D, Rafter J, Talley NJ, et al. The impact of Crohn’s disease genes on healthy human gut microbiota: a pilot study. Gut. 2013;62(6):952–4. doi: 10.1136/gutjnl-2012-304214. [DOI] [PubMed] [Google Scholar]
- 26•.Whiteside SA, Razvi H, Dave S, Reid G, Burton JP. The microbiome of the urinary tract-a role beyond infection. Nature reviews Urology. 2015;12(2):81–90. doi: 10.1038/nrurol.2014.361. This review article lays out the literature on the bladder microbiome in men and women. [DOI] [PubMed] [Google Scholar]
- 27•.Wolfe AJ, Toh E, Shibata N, Rong R, Kenton K, Fitzgerald M, et al. Evidence of uncultivated bacteria in the adult female bladder. Journal of clinical microbiology. 2012;50(4):1376–83. doi: 10.1128/JCM.05852-11. This article shows why trasurethral cathaterization is the collection method of choice and how it compares to aspirated and voided urine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fouts DE, Pieper R, Szpakowski S, Pohl H, Knoblach S, Suh MJ, et al. Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. Journal of translational medicine. 2012;10:174. doi: 10.1186/1479-5876-10-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khasriya R, Sathiananthamoorthy S, Ismail S, Kelsey M, Wilson M, Rohn JL, et al. Spectrum of bacterial colonization associated with urothelial cells from patients with chronic lower urinary tract symptoms. Journal of clinical microbiology. 2013;51(7):2054–62. doi: 10.1128/JCM.03314-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30••.Hilt EE, McKinley K, Pearce MM, Rosenfeld AB, Zilliox MJ, Mueller ER, et al. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. Journal of clinical microbiology. 2014;52(3):871–6. doi: 10.1128/JCM.02876-13. This paper defines the expanded quantative urine culture (EQUC) technique and how it relates to sequencing results. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31•.Thomas-White KJ, Hilt EE, Fok C, Pearce MM, Mueller ER, Kliethermes S, et al. Incontinence medication response relates to the female urinary microbiota. International urogynecology journal. 2015 doi: 10.1007/s00192-015-2847-x. This recent research article has shown a correlation between the microbiome of women with UUI and their response to the anticholinergic medication solifenacin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearce MM, Zilliox MJ, Thomas-White KJ, Richter HE, Nager CW, Visco AG, et al. The female urinary microbiota in urgency urinary incontinence. American journal of obstetrics and gynecology. 2015 doi: 10.1016/j.ajog.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luthje P, Brauner H, Ramos NL, Ovregaard A, Glaser R, Hirschberg AL, et al. Estrogen supports urothelial defense mechanisms. Science translational medicine. 2013;5(190):190ra80. doi: 10.1126/scitranslmed.3005574. [DOI] [PubMed] [Google Scholar]
- 34.Wang C, Symington JW, Ma E, Cao B, Mysorekar IU. Estrogenic modulation of uropathogenic Escherichia coli infection pathogenesis in a murine menopause model. Infection and immunity. 2013;81(3):733–9. doi: 10.1128/IAI.01234-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raz R. Urinary tract infection in postmenopausal women. Korean journal of urology. 2011;52(12):801–8. doi: 10.4111/kju.2011.52.12.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raz R, Colodner R, Rohana Y, Battino S, Rottensterich E, Wasser I, et al. Effectiveness of estriol-containing vaginal pessaries and nitrofurantoin macrocrystal therapy in the prevention of recurrent urinary tract infection in postmenopausal women. Clin Infect Dis. 2003;36(11):1362–8. doi: 10.1086/374341. [DOI] [PubMed] [Google Scholar]
- 37.Raz R, Stamm WE. A controlled trial of intravaginal estriol in postmenopausal women with recurrent urinary tract infections. The New England journal of medicine. 1993;329(11):753–6. doi: 10.1056/NEJM199309093291102. [DOI] [PubMed] [Google Scholar]
- 38.Nelken RS, Ozel BZ, Leegant AR, Felix JC, Mishell DR., Jr Randomized trial of estradiol vaginal ring versus oral oxybutynin for the treatment of overactive bladder. Menopause. 2011;18(9):962–6. doi: 10.1097/gme.0b013e3182104977. [DOI] [PubMed] [Google Scholar]
- 39.Serati M, Salvatore S, Uccella S, Cardozo L, Bolis P. Is there a synergistic effect of topical oestrogens when administered with antimuscarinics in the treatment of symptomatic detrusor overactivity? European urology. 2009;55(3):713–9. doi: 10.1016/j.eururo.2008.06.051. [DOI] [PubMed] [Google Scholar]
- 40.Tseng LH, Wang AC, Chang YL, Soong YK, Lloyd LK, Ko YJ. Randomized comparison of tolterodine with vaginal estrogen cream versus tolterodine alone for the treatment of postmenopausal women with overactive bladder syndrome. Neurourology and urodynamics. 2009;28(1):47–51. doi: 10.1002/nau.20583. [DOI] [PubMed] [Google Scholar]