Abstract
Several human polyomaviruses including JCV, BKV and TSV are associated with diseases, particularly in immunosuppressed patients. While the large T antigen (LT) encoded by the monkey polyomavirus SV40 is well studied, and possesses intrinsic ATPase and DNA helicase activities, the LTs of the human polyomaviruses are relatively uncharacterized. In order to evaluate whether these enzymatic activities, which are required for viral DNA replication, are conserved between polyomaviruses, we performed a comparative study using the LTs from JCV, TSV and SV40. The ATPase and DNA helicase activities and the interaction with the cellular tumor suppressor p53 were assayed for the purified Zn-ATPase domains of the three LTs. We found that all Zn-ATPases were active ATPases. The Zn-ATPase domains also functioned as DNA helicases, although the measured kinetic constants differed among the three proteins. In addition, when tested against four small molecule ATPase inhibitors, the Zn-ATPase domains of TSV was more resistant than that of SV40 and JCV. Our results show that, while LTs from JCV and TSV share the core ATPase and DNA helicase activities, they possess important functional differences that might translate into their respective abilities to infect and replicate in hosts.
Keywords: Polyomavirus, ATPase, DNA helicase, Protein–protein interaction, p53
Introduction
The polyomaviruses are double stranded DNA viruses with a genome size of about 5 kb. Recent efforts in virus discovery have resulted in a rapid expansion of the number of known polyomaviruses. There are currently over 90 species of polyomaviruses and 65 complete reference polyomavirus genomes in the NCBI database. Polyomaviruses infect a wide range of vertebrate hosts from birds and rodents to humans. The polyomaviruses depend on the host machinery for replication. To this end, all polyomaviruses encode a major regulatory protein, the large tumor antigen (LT)1, which establishes complicated interactions with various host proteins to coordinate the production of viral progeny.
Simian virus 40, i.e. SV40, is the founding member of the Polyomaviridae. The LT of SV40 has been studied extensively using structural biology, biochemistry, cell culture and animal models [1,2]. During a productive life cycle the LT is expressed from the early region of the SV40 genome. By releasing activator E2Fs from the Rb repressor complexes and activating E2F dependent gene transcription the LT drives the infected cell into S phase in order to hijack the cellular DNA replication/repair machinery for synthesis of progeny viral genomes. Moreover, LT is the master protein that directs the replication of the viral genome by recognizing and binding the replication origin within the viral genome, recruiting cellular DNA replication factors, assembling into homododecamers, and unwinding the viral DNA genome.
SV40 LT consists of four folded domains (Fig. 1A). The DnaJ domain has a co-chaperone activity required to stimulate ATP hydrolysis by cellular Hsc70 and to subsequently disrupt Rb–E2F complexes [3–8]. The origin-binding domain (OBD) of the SV40 LT specifically recognizes and makes contacts with the GAGGC pentanucleotides within the replication origin [9,10]. Both the Zn-binding and ATPase domains contribute to hexamerization of the LT. The ATPase domain of SV40 LT contains functional motifs characteristic of the AAA+ superfamily (ATPases Associated with various cellular Activities) [11], which include the Walker A box (Gly-Pro-Ile-Asp-Ser-Gly-Lys-Thr, GPIDSGKT) [12], the Walker B box (Leu-Val-Val-Phe-Glu-Asp, LVVFED) [13], and the Arg finger that are essential for ATP binding and hydrolysis. Using the energy produced by ATP hydrolysis, SV40 LT functions as an SF3 DNA helicase and unwinds the entire viral genome to provide templates for DNA replication [14–16].
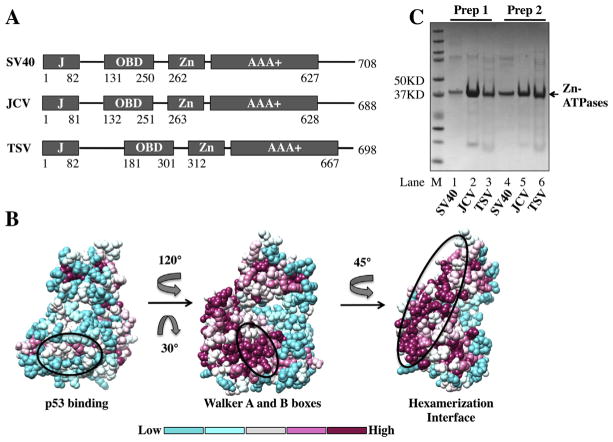
Fig. 1.
Structure, modeling and purification of the Zn-ATPase domains of SV40, JCV and TSV. (A) Diagrams of the domain structure of LTs. The fourfolded domains in LTs [DnaJ (J), origin binding (OBD), Zn-binding (Zn) and AAA+ ATPase (AAA+)] are represented by boxes, the linkers between domains are shown as lines. Amino acid numbers are listed for each domain. (B) Sequence conservation of the Zn-ATPase domains of LTs mapped onto the solved SV40 LT structure (PDB identifier_ 1SVM). The level of conservation is shown in colors. Three views are shown. Left_ the least conserved side of LTs. The oval indicates the p53-binding surface of SV40 LT. Middle_ the highly conserved Walker boxes (within the oval). Right_ the residues (within the oval) involved in interaction with neighboring LT monomer. The direction and degree of rotation are indicated. The bar at the bottom indicates the levels of conservation from low (left) to high (right). (C) Evaluation of two independent purified preparations of Zn-ATPases. The purified proteins were visualized by coomassie staining after SDS-PAGE and are indicated by arrows. SV40, lanes 1 and 4; JCV, lanes 2 and 5; TSV, lanes 3 and 6; and M, protein ladder.
The functions of SV40 LT are facilitated and modulated in a highly sophisticated fashion through interactions with various cellular targets and/or post-translational modifications. The His-Pro-Asp (HPD) motif in the DnaJ domain and the LXCXE (Leu-X-Cys-X-Glu) motif in the flexible linker downstream of the DnaJ domain interact with the cellular components Hsc70 and pRb proteins respectively [17]. These interactions are required to disrupt the Rb–E2F complexes noted above. Residues in the ATPase domain mediate LT binding to the cellular tumor suppressor protein p53 [18]. This interaction stabilizes p53 protein, but at the same time masks its DNA binding motif, thus inactivating p53 target gene transcription and blocking p53 dependent apoptosis [19,20]. Various motifs in the OBD and the Zn-ATPase domains of SV40 LT coordinate interactions with cellular replicative factors, including RPA, alpha-primase and Topoisomerase I, and facilitate DNA replication [21–24].
Although the LT of SV40 has been well characterized, the biochemical activities of human polyomaviruses LTs are less understood. JCV (John Cunningham Virus) and TSV (Trichodysplasia Spinulosa associated virus) have been shown to cause progressive multifocal leukoencephalopathy and trichodysplasia spinulosa, respectively, in immune-compromised patients [25,26]. To address whether LTs from human polyomaviruses share the core enzymatic activities, we decided to characterize the ATPase and DNA helicase activities of JCV and TSV LTs in direct comparison with SV40 LT.
Experimental procedures
Cloning
The domain boundaries for the Zn-ATPase domains of JCV and TSV LTs were determined by ClustalW alignment of JCV or TSV LT against SV40 LT (Figs. 1A and 2). The coding sequences (CDS) of Zn-ATPase domains were amplified by PCR from the following constructs: pWZL-SV40 early region [27], pLenti6.3-JCV early region [28] and pUC19-TSV [26]. The PCR products were cloned into pET151 using the Champion pET Directional TOPO Expression Kit (Life Technologies, Carlsbad CA). All constructs were verified by DNA sequencing to contain the matching corresponding reference sequences (SV40 NC001669, JCV NC_001699, pUC19-TSV NC_014361) in the NCBI database. The final constructs allow expression of each recombinant Zn-ATPase fused to a 6X-His tag at the amino-terminus. The construct containing the GST-p53 DNA binding domain (DBD, aa. 92-292) was described previously [29].
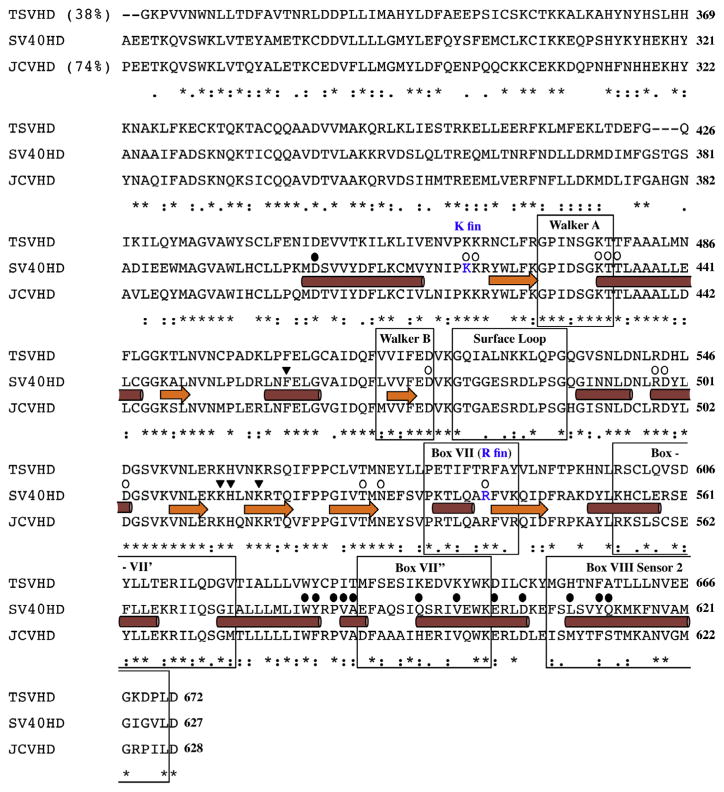
Fig. 2.
Sequence alignment of SV40, JCV and TSV Zn-ATPase domains. The alignment was performed using ClustalW. The α-helices are indicated by cylinders and the β-strands are indicated by arrows based on the solved SV40 structure (PDB identifier_ 1SVM). Known functional motifs (Walker A box and Walker B box) and subdomain components (surface loop, box VII, box VII′ box VII″ and box VIII sensor 2) are highlighted in labeled boxes. The SV40 lysine finger (K Fin) and arginine finger (R Fin) are indicated. Several groups of critical residues are highlighted on their tops: ATP/ADP binding residues, open circles; DNA binding residues in the central channel, inverted triangles; and p53 binding residues, filled circles. Global percent identity compared to SV40 Zn-ATPase domains is shown in parentheses.
Protein expression and purification
Plasmids containing the CDS for Zn-ATPases and p53 DBD were transformed into Escherichia coli BL21 star cells (Life Technologies, Carlsbad CA). Expression of recombinant protein was induced with 0.4 mM IPTG for 24 h at room temperature.
For purification of the Zn-ATPases, bacterial pellets containing the recombinant proteins were resuspended and lysed in non-denaturing buffer (20 mM HEPES, pH 7.2, 400 mM NaCl, 0.1% NP40, 5 mM imidazole, 10 mM beta-mercaptoethanol, 5% glycerol and protease inhibitors). All following steps were carried out at 4 °C. The lysate was clarified by centrifugation at 15,000g for 30 min and the supernatant was incubated with ProBond™ Nickel-chelating resin (Life Technologies, Carlsbad CA) for batch purification. After washing the resin with the non-denaturing lysis buffer containing increasing amounts of imidazole (20 mM twice and 30 mM once), the recombinant protein was eluted with 200 mMimidazole in the same buffer. The 6X-His tag was removed through cleavage with a 6xHis-tagged TEV protease at room temperature followed by a second round of incubation with Nickel chelating resin, which removed the uncleaved His-tagged recombinant proteins, TEV protease and the free 6X-His tag. For the helicase assay, the proteins were dialyzed into binding buffer (25 mMTris HCl, pH 7.5, 125 mMNaCl, 1 mMDTT and 5% glycerol), and then further purified using a S6 ion exchange column. The Zn-ATPases were present in the unbound fractions.
Bacterial pellets containing GST-human p53 DNA binding domain (DBD, amino acid 92–292) were lysed in GST purification buffer (25 mM Tris HCl, pH 8.0, 250 mM NaCl, 1 mM DTT and 5% Glycerol) and batch purified using glutathione beads (Pierce, Thermo Scientific). The beads were washed extensively with the lysis buffer and then used for pull-down assays as described below. The full-length SV40 LT was purified from SF9 cells infected with recombinant baculovirus containing the early region of SV40 as described previously [30].
ATPase assay
ATPase assays were performed as previously described [30]. Briefly, 1.2 μM of the purified Zn-ATPases were mixed with 10 μM of unlabeled ATP and 1 μCi of γ-32P-ATP (3000 Ci/mmol, 10 mCi/ml, PerkinElmer, Waltham, MA) in a reaction buffer (25 mM HEPES, pH 7.3, 10 mM MgCl2, 20 mM KCl, 0.05% NP40, 0.1 mM EDTA, 1 mM DTT). When ssDNA stimulation of ATPase activity was assessed, 60-mer poly-dT oligos (IDT DNA, Coralville, IA) were added to the reaction to a final concentration of 0.37 μM, and the reactions were incubated at room temperature. Aliquots of 2 μl were removed at specific time points and the reaction was stopped by immediately mixing with 4 μl of 0.75 M KH2PO4 buffer. A total of 1 μl aliquots of these samples was spotted on TLC plates (Thermo Fisher Scientific, Waltham, MA), which were then developed in a chamber using 0.75 M KH2PO4 buffer as the solvent. The results were visualized using FLA5100 fluorescent image analyzer and quantified using Multigauge software.
For enzyme turnover measurement, the amount of unlabeled ATP was increased to 50 μM and the amounts of the Zn-ATPases were lowered to 0.6 μM. When the effects of the inhibitor compounds were tested in the ATPase assays, the inhibitors were pre-incubated with the purified Zn-ATPases in the reaction buffer for 30 min at 4 °C before ATP and γ-32P-ATP were added to the system. DMSO, the solvent for the inhibitors, was used as a negative control. The complete reaction was incubated at room temperature for 1 h, and the reaction was stopped and resolved by TLC as described above.
DNA helicase assay
The DNA helicase assay was carried out based on a previously published protocol with modifications [30]. The purified Zn-ATPases were tested by their abilities to unwind a partially double-stranded DNA substrate. To generate the substrate, the −40 M13 primer (GTTTTCCCAGTCACGACGTTGTAAAA) was annealed to M13mp18 (+) DNA (NEB, Ipswich, MA), and then labeled with α-32P-dATP by primer extension using Klenow DNA polymerase (NEB, Ipswich, MA). For a 20 μl annealing reaction, 0.6 pmole of M13mp18 single stranded DNA was mixed with the −40 M13 primer at a 1–5 molar ratio in annealing buffer (70 mM Tris–HCl, pH 7.6, 50 mM NaCl, 10 mM MgCl2, 5 mM DTT). The reaction was heated to 90 °C for 90 sec, followed by an incubation at 65 °C for 5 min, and then slowly cooled to room temperature. Primer extension was carried out by combining the 20 μl annealed DNA with the labeling mixture (1X NEB buffer 2, 0.4 mM dGTP, 0.4 mM dTTP, 50 μCi of α-32P-dATP and 10 units of Klenow enzyme) to a final volume of 50 μl. The reaction was incubated at 30 °C for 30 min followed by addition of 0.2 mM dATP and another incubation at 30 °C for 30 min. The labeled substrate was purified through ethanol precipitation and 3 rounds of Centri-Spin 20 columns.
For the helicase assay, various amounts of purified Zn-ATPases were mixed with the substrate (1 μl of purified substrate/20 μl reaction) in helicase buffer (20 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 50 mM NaCl, 0.1 mg/ml bovine serum albumin, 1 mM DTT and 5% glycerol) freshly supplemented with an ATP regeneration system containing 2 mM ATP, 40 mM creatine phosphate (Roche Applied Science, Indianapolis, IN) and 82 μg/ml creatine kinase (Roche Applied Science, Indianapolis, IN). All reactions were set up on ice, and then incubated at 37 °C for 1 h. The reactions were terminated by adding 4 μl of 5X stop buffer (10 mM EDTA pH 8.0, 0.5% SDS, 0.1% bromophenol blue and 50% glycerol). The quenched reactions were resolved on a 12% polyacrylamide gel in TBE. The gel was dried and images were captured with an FLA5100 fluorescent image analyzer.
Pull-down assays
To measure the interaction between the Zn-ATPases and p53, 400 μg of purified SV40, JCV or TSV Zn-ATPases were mixed with purified GST-p53 DBD immobilized on glutathione beads at a 1–2 molar ratio in 1.5 ml of binding buffer (25 mM HEPES, pH7.2, 150 mM NaCl, 8% glycerol and 1 mM DTT). The mixture was incubated at 4 °C and kept in suspension through constant rotation for 2 h. The supernatant was then removed and the beads were washed three times using PBS. Bound proteins were eluted with 10 mM reduced glutathione in PBS and the eluate was resolved by SDS–PAGE. The gel was stained with Coomassie blue and the identities of the bound proteins were determined based on their molecular weights.
Results
Differential ATPase activities of the purified Zn-ATPase domains from SV40, JCV and TSV
The domain structures of SV40, JCV and TSV LTs are very similar with respect to both length and sequence identity. The Zn-ATPase domains of all three LTs are highly conserved (Figs. 1 and 2). Taking advantage of the solved structure of the SV40 Zn-ATPase domain [31,32], models of high confidence for the domains from JCV and TSV were generated through homology modeling by BioSerf v2.0 [33]. The overall architecture and the secondary structure components of both models are in good agreement with the solved SV40 Zn-ATPase domain structure. To further determine sequence conservation among LTs, we manually selected 76 reference sequences of LTs from the Uniprot database. Using ConSurf server, position specific amino acid conservation scores were calculated based on the multiple sequence alignment of the above LTs [34,35]. The conservation scores were then mapped onto the solved SV40 LT structure and visualized using Chimera (Fig. 1B). Examples shown in Fig. 1B reveal the variations in surface conservation across LTs. Whereas the p53 binding interface of SV40 LT (Fig. 1B, left) is less conserved, both Walker A and B boxes, as well as the hexamerization interface residues are highly conserved among LTs (Fig. 1B middle and right), indicating the ATPase and DNA helicase activities of all LTs are likely to be common.
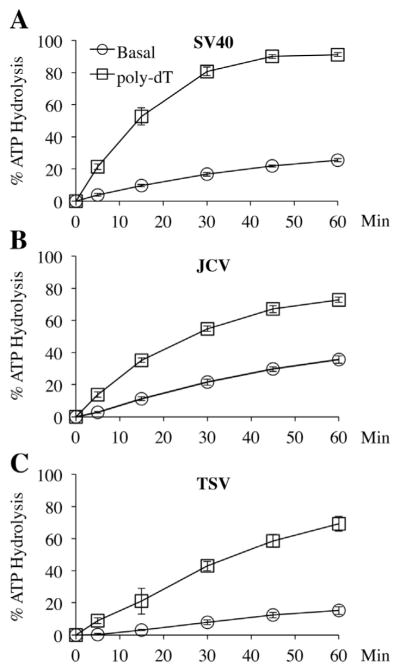
Two independent preparations of the Zn-ATPase domains were verified by SDS–PAGE and Coomassie staining (Fig. 1C). Based on results from a steady-state ATPase assay (Fig. 3), the JCV Zn-ATPase showed the highest basal level ATPase activity, i.e., roughly 35% of input ATP was hydrolyzed at the 60-min time point while SV40 helicase converted about 25% of the ATP substrate at the same time point. The TSV helicase was the slowest ATPase, which only hydrolyzed 15% of ATP after 60 min. It was previously shown that ssDNA stimulates the ATPase activity of SV40 LT [30]. Our results show that, similar to the SV40 Zn-ATPase domain, the ATPase activities of the Zn-ATPase domains from both JCV and TSV are stimulated by poly-dT ssDNA.
Fig. 3.
Basal and ssDNA-stimulated ATPase activities of Zn-ATPase domains. Percentage of ATP hydrolysis (Y-axis) at various time points (X-axis) were plotted for the basal hydrolysis rate and the rate after addition of poly-dT ssDNA. Each data point was the average of three measurements. The error bars represent standard deviation. Similar results were produced using two independent preparations of each Zn-ATPase. (A) SV40 Zn-ATPase; (B) JCV Zn-ATPase; (C) TSV Zn-ATPase.
A more accurate comparison of the ATPase activities was obtained by measuring the enzyme turnover numbers (kcat) for these proteins. The turnover numbers of basal and poly-dT stimulated ATPase activities for each Zn-ATPase domains are summarized in Table 1. The basal level turnover numbers of the three Zn-ATPases correlated with the ATP hydrolysis results across several time points (Fig. 3), i.e., the JCV Zn-ATPase domain had the highest kcat of 0.63 s−1, followed by SV40 Zn-ATPase (kcat = 0.54 s−1). The TSV Zn-ATPase domain was the least efficient in catalyzing ATP hydrolysis with a kcat of 0.22 s−1. However, both TSV and SV40 Zn-ATPase domains responded similarly to poly-dT stimulation. The fold stimulations (ratio of stimulated over basal ATP hydrolysis) for the SV40 and TSV enzymes are 6.6 and 6.8, respectively. The JCV Zn-ATPase domain only showed stimulation of 3.7-fold. Overall, the JCV Zn-ATPase domain exhibited the highest basal level ATPase activity but the lowest response to poly-dT stimulation. On the other hand, the same protein from TSV was the least efficient ATP hydrolyzing enzyme at the basal level but responded to poly-dT stimulation as well as the SV40 counterpart.
Table 1.
The ATPase turnover numbers of Zn-ATPases. The enzyme turnover numbers of both basal level and ssDNA stimulated ATPase activities were measured as the number of ATP molecules hydrolized per LT hexamer per second for the Zn-ATPases of SV40, JCV and TSV LTs and the results are summarized in the table. Fold stimulation by ssDNA was calculated for each Zn-ATPase.
| kcat s−1_ Basal | kcat s−1_ Poly-dT | Fold stimulation | |
|---|---|---|---|
| SV40 | 0.54 ± 0.04 | 3.6 ± 0.28 | 6.6 |
| JCV | 0.63 ± 0.06 | 2.3 ± 0.27 | 3.7 |
| TSV | 0.22 ± 0.04 | 1.5 ± 0.3 | 6.8 |
Small molecule inhibitors of LT ATPase activity also differentially affect the SV40, JCV and TSV Zn-ATPase domain activities (Fig. 4). Four inhibitors were tested: MAL2-11B, hexachlorophene, bithionol and MAL2-11B tetrazole. Hexachlorophene and bithionol were identified as SV40 LT ATPase inhibitors in a high throughput screen that sought small molecule inhibitors of SV40 LT using a colorimetric reporter assay [36,37]. In contrast, MAL2-11B is a derivative of a chaperone modulator that similarly inhibited SV40 LT ATPase activity but also abrogated the propagation of SV40 as well as BKV in infected mammalian cells [38]. The MAL-211B tetrazole is an analog of MAL2-11B, which showed more potent inhibition of the ATPase activity of SV40 LT and also inhibited SV40 and BKV replication in cell culture [39].
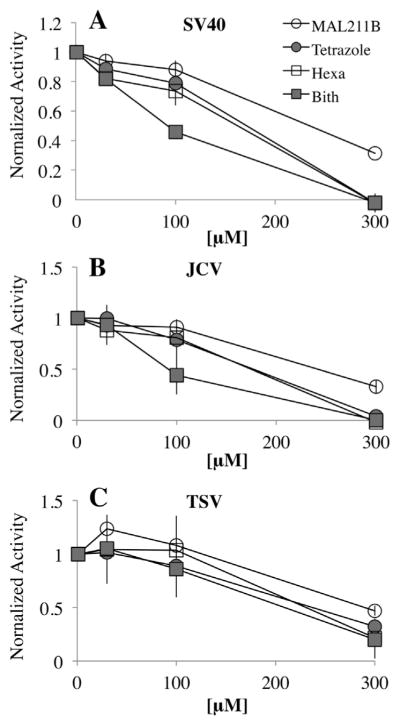
Fig. 4.
Effects of inhibitors on the basal ATPase activities. Inhibitors were tested across three concentrations: 30, 100 and 300 μM. ATP hydrolysis with inhibitors was normalized against the DMSO control and plotted against the corresponding concentrations. Y-axis, normalized ATP hydrolysis; X-axis, concentrations of the inhibitors. The point “0” indicates the DMSO control (no inhibitor) with 100% ATPase activities of the Zn-ATPases. Each data point was the average of three measurements. The error bars represent standard deviation. Similar results were produced using two independent preparations of each Zn-ATPase. (A) SV40 Zn-ATPase; (B) JCV Zn-ATPase; (C) TSV Zn-ATPase.
For each purified Zn-ATPase domain, the effects of the four compounds were tested individually at three concentrations. DMSO was used as a negative control. To compare the inhibitory effects among the different compounds, ATP hydrolysis activities in the presence of the inhibitors were normalized against the DMSO control. Therefore, the ATPase activities of the no compound controls are “1”. In general bithionol showed the strongest inhibition of all three Zn-ATPase domains and MAL2-11B was the least effective inhibitor (Fig. 4). Moreover, both the SV40 and JCV Zn-ATPase domains reacted to the compounds in a similar manner. In contrast, although the basal level ATPase activity of TSV Zn-ATPase domain was the lowest, the protein showed higher resistance to all tested inhibitors. Indeed, none of the inhibitors was able to completely abolish the TSV ATPase activity. On the other hand 300 μM of MAL2-11B tetrazole, hexachlorophene and bithionol completely abolished the ATPase activities of the purified SV40 and JCV Zn-ATPase domains. The combined data suggest that the homologous domains possess subtle yet conformational differences that result in distinguishable biochemical properties.
The DNA unwinding activities of the SV40, JCV and TSV Zn-ATPase domains
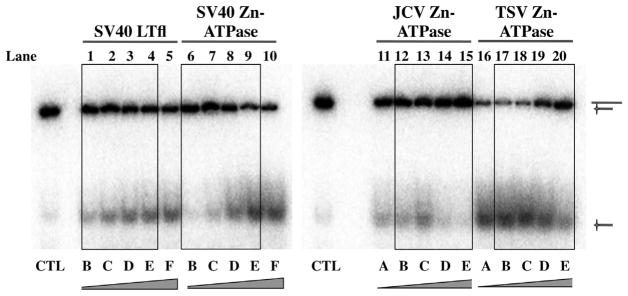
To further examine the distinguishing features between the different Zn-ATPase domains we performed DNA helicase assays. The concentration of substrate was held constant while the amount of the purified Zn-ATPase domains was altered. Active unwinding of the substrate is shown by accumulation of radiolabeled oligonucleotide at the bottom of the gels. Full-length SV40 LT purified from a baculovirus expression system was used as a positive control and as expected increased unwinding corresponded to a rise in protein concentration (Fig. 5, lanes 1–5). The purified SV40 Zn-ATPase domain also showed increased helicase activity at higher concentrations (lanes 6–10). However, the helicase activity of JCV Zn-ATPase domain was lower overall, and peaked at a concentration of 120 nM (lane 13). In contrast, the TSV Zn-ATPase domains displayed the most potent DNA helicase activity at lower concentrations (lanes 16–18). The robust DNA unwinding of TSV Zn-ATPase was maintained as a plateau over a wide range of protein concentrations, from 30 to 120 nM, but decreased at higher concentrations (Fig. 5, lanes 19 and 20). Similar trends of the helicase activities of the SV40, JCV and TSV were reproduced using a second independent preparation for each enzyme (data not shown).
Fig. 5.
DNA helicase activities of SV40, JCV and TSV Zn-ATPase domains. Full length SV40 LT (lanes 1–5) was used as a positive control. DNA helicase activities of purified SV40 (lanes 6–10), JCV (lanes 11–15) and TSV (lanes 16–20) Zn-ATPases are shown. Bands corresponding to the substrate (upper band) and the displaced radiolabeled oligo product (lower band) are indicated with schematics on the right. Lane CTL, control without protein. Increasing concentrations of the proteins are indicated at the bottom of the gel panel. The boxes highlight the results for the same concentration range. (A) 30 nM; (B) 60 nM; (C) 120 nM; (D) 240 nM; (E) 480 nM; and (F) 960 nM.
The Zn-ATPase domains of SV40 and JCV but not TSV interact directly with the DNA binding domain of p53
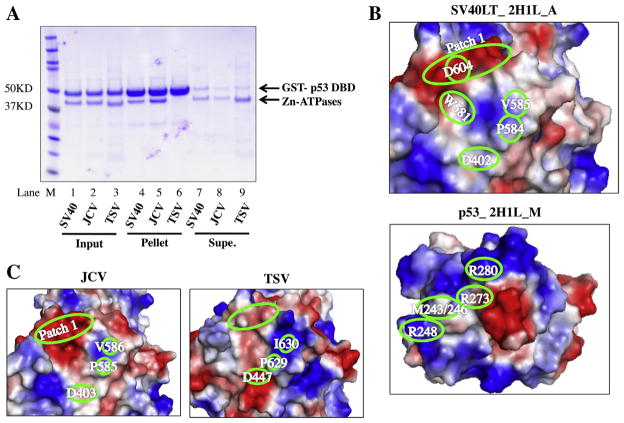
SV40 LT binds to p53 in order to block activation of p53 target gene transcription and the subsequent apoptosis. Data from both structural and mutational analyses indicate a complex LT–p53 interaction interface that involves multiple residues within the ATPase domain of SV40 LT and p53 DBD [40,29]. Sequence conservation analysis suggests residues involved in the p53 binding as determined in SV40 LT are less conserved among LTs (Fig. 1B, left). To determine whether the Zn-ATPase domains interact with p53 we performed an in vitro pull-down assay using the purified Zn-ATPases and a GST fusion containing the DBD of p53. In these experiments, the purified GST-p53 DBD immobilized on glutathione beads served as the bait, and the SV40 Zn-ATPase domain was used as a positive control. The amount of input protein and the molar ratio between the bait (GST-p53) and the prey (i.e., the purified Zn-ATPase domains) were consistent for all three reactions (Fig. 6A, input, lanes 1–3). Binding to GST-p53 was observed for the SV40 and JCV Zn-ATPase domains but binding between the same domain from TSV and the p53 DBD was absent (lanes 4–6). As anticipated, a greater amount of the TSV Zn-ATPase domain was instead present in the supernatant (lane 9). These data suggest that the TSV Zn-ATPase domain is unable to form a stable complex with p53.
Fig. 6.
The LT–p53 interaction. (A) Results of in vitro pull-down assay between Zn-ATPase domains and GST-p53. Protein inputs (lanes 1–3), bound proteins eluted from the pellets (lanes 4–6) and supernatants after binding (lanes 7–9) were resolved on SDS–PAGE and visualized by Coomassie staining. Bands corresponding to GST-p53 DBD and Zn-ATPases are indicated on the right of the gel panel. Lane M, protein ladder. (B) The binding interfaces of SV40 LT (PDB identifier_ 2H1L, chain A) and p53 (PDB identifier_ 2H1L, chain M) based on solved co-complex. In this representation, regions consist of acidic side chains are colored red, whereas basic regions are colored in blue. The intensity of the colors correlates with the degree of electropositivity (blue) and electronegativity (red). (C) Candidate p53 binding interfaces of JCV and TSV on their structure models. For B and C, critical binding residues and regions are highlighted in green ovals.
The p53 binding residues in SV40 as well as their counterparts (candidate p53 binding residues) in JCV and TSV are highlighted in the sequence alignment in Fig. 2 and summarized in Table 2 for comparison. We then compared the candidate p53 binding residues in the JCV and TSV Zn-ATPase domains with the known p53 binding residues in SV40 LT. Only three of the residues are identical among all three proteins (Fig. 2, Table 2): D402, W581 and P584 (SV40 LT amino acid numbers). Eight of the JCV LT candidate p53 binding residues are the same as the aligned SV40 residues. Two of the five variations are of similar chemical properties, i.e. M610 in JCV versus L609 in SV40 (both hydrophobic), and S614 in JCV versus Q613 in SV40 (both polar). In contrast, only four residues from the candidate p53-binding surface of TSV match the aligned SV40 residues. Seven of the nine variations between TSV and SV40 involve amino acids with dramatic differences in chemical properties.
Table 2.
The LT–p53 interface residues. The amino acids involved in direct interaction between SV40 LT (left, column_ SV40) and p53 (right, column_ p53) are summarized. The corresponding homologous residues in JCV (column_ JCV) and TSV (column_ TSV) are also listed. Sequence variations among the three LTs are represented as following: no filling_ same as SV40 LT; light gray filled_ different amino acid with similar chemical properties; dark gray filled_ amino acid with different chemical properties.
| SV40 | JCV | TSV | p53 |
|---|---|---|---|
| D402 | D403 | D447 | R248, R280 |
| W581 | W582 | W626 | M246 |
| P584 | P585 | P629 | M243 |
| Y582 | F583 | Y627 | M246 |
| V585 | V586 | I630 | M246 |
| A586 | A587 | T631 | P177 |
| Q593 | H594 | K638 | R181 |
| V597 | V598 | K642 | H178 |
| E601 | E602 | D646 | N239 |
| D604 | D605 | C649 | R273 |
| L609 | M610 | H654 | M246 |
| Y612 | F613 | F657 | M246 |
| Q613 | S614 | A658 | N288 |
To compare the candidate p53 binding interfaces in JCV and TSV against the known p53 interface of SV40 LT, we generated the surface electrostatic potential of the interface residues in SV40 LT and p53 using PyMOL to visualize the surface charges (Fig. 6B). The candidate interface residues in the JCV and TSV Zn-ATPase domains were also represented in a similar fashion (Fig. 6C) using their modeled structures. The positions of SV40 residues D402, V585, D604 are indicated on the binding surfaces. Patch 1 was previously described as a cluster of conserved charged residues (E598/601/606, D604 and K605) located on the surface of SV40 LT [40]. It has been shown mutations targeting residues within Patch 1 abolished p53 binding. Homologous residues between the JCV and TSV LTs are also highlighted in Fig. 6C. Overall, the predicted JCV interface closely resembles the actual p53-binding surface in SV40 LT: both the electrostatic properties and the positions of critical residues are highly consistent between the SV40 and JCV proteins. On the contrary, the candidate p53 binding surface in TSV differs dramatically from the p53 interface of SV40 LT. Compared to Patch 1 in SV40 LT, the homologous region in TSV LT shows a significant reduction in negative charge, likely due to the dissimilarity between C649 in TSV and D604 in SV40. The positively charged area in TSV LT is also expanded and the intensity of the positive charge is increased. TSV LT K638 and K642 are located within this area and most likely contribute to the electrostatic surface properties. Together, the candidate p53 interacting residues in TSV are distinct from those found in JCV and SV40. Moreover, although the size and the shape of the candidate p53-interacting interfaces in both JCV and TSV LTs are similar to the p53 binding surface of SV40 LT, the electrostatic potentials of the TSV, but not JCV candidate interface shows dramatic dissimilarity when compared with the p53 binding interface in SV40 LT. This dissimilarity is likely responsible for its failure to interact with the p53 DBD.
Discussion
The LT of SV40 functions as a replicative helicase [16,41], and the ATPase and DNA helicase activities of SV40 LT are essential for viral DNA replication. Although these activities have only been extensively studied for SV40 LT, it has been assumed that all LTs possess these same core enzymatic activities based on sequence conservation. Despite the high level conservation, sequence variations do exist across the LTs, and may underlie distinct functional properties. In this study, we have compared the ATPase and DNA helicase activities of LTs from JCV and TSV with the well-characterized LT of SV40.
We expected both LTs of JCV and TSV to function as an ATPase and DNA helicase because of the presence of the conserved Walker A and B boxes, as well as the Lys finger. Interactions between these motifs and ATP/ADP have been confirmed by crystal structures of SV40 LT – ATP/ADP co-complexes [31]. Their functional importance was also verified in mutagenesis studies of SV40 LT [42,43]. Our results confirmed that the purified Zn-ATPase domains are active ATPases and DNA helicases as expected. Interestingly, the three Zn-ATPase domains differed kinetically and displayed unique functional properties in several tests, including their basal and ssDNA (poly-dT)-stimulated ATPase activities, effects of inhibitors on basal ATP hydrolysis, and DNA unwinding. Since these differences in activities are consistent between two independent preparations of the proteins, they are most likely to be intrinsic instead of random variations produced by suboptimal protein isolates.
Previous biochemical studies showed that SV40 LT has an intrinsically weak ATPase activity that can be stimulated upon addition of ssDNA [43,30]. Our study showed similar results for all three tested Zn-ATPase domains. In contrast to the purified Zn-ATPase proteins from SV40 and JCV, the TSV Zn-ATPase domain was a slower ATP hydrolyzing enzyme at the basal level and formed ADP with less than 50% efficiency compared to the JCV and SV40 Zn-ATPase domains (Fig. 3, Table. 1). However, the Zn-ATPase domain from TSV was activated by ssDNA as efficiently as when the same region from SV40 was examined, while the JCV Zn-ATPase domain was less stimulated under the same conditions (Fig. 3, Table 1). These results suggest that SV40, JCV and TSV LTs may differ in their abilities to bind ssDNA.
All three Zn-ATPase domains were active DNA helicases but their unwinding kinetics was different. TSV showed the most robust unwinding activities at the lower protein concentrations tested (Fig. 5); the JCV Zn-ATPase domain only showed trace amounts of unwinding at the 30 and 60 nM concentration and SV40 Zn-ATPase domain tested in parallel displayed no unwinding at 30 nM. The lower basal ATPase activity of TSV Zn-ATPase seems to contradict these results. Even with the addition of ssDNA, the stimulated ATPase activity of the TSV Zn-ATPase domain never reached the same levels as that of the SV40 enzyme. It is possible that the robust DNA helicase activity of the TSV Zn-ATPase domain may be achieved by a higher affinity to the dsDNA substrate and/or more efficient assembly of hexamers on the substrate DNA.
We suggest that sequence variations in the functional motifs within the Zn-ATPase domains from SV40, JCV and TSV are responsible for their distinct kinetics in ATPase and DNA helicase activities. The sequence alignment in Fig. 2 shows that JCV and SV40 LTs have an identical Walker A motif, whereas the Walker A motif of TSV LT varies from that of SV40 and JCV by only one residue, an Asp, that is changed to an Asn. However, each LT has a unique Walker B motif: although the Glu and the following Asp within the Walker B box (consensus in polyomavirus LTs as hhhh(Glu/Asp)Asp, h stands for any hydrophobic residue) are conserved in all three LTs, the first and the third hydrophobic residues do vary. All additional motifs except the Arg and Lys fingers contain variations. For example, sensor 2 and a surface loop with unknown function show higher level of variations among the three proteins.
Efforts have been made to identify inhibitors with therapeutic potential in treating diseases caused by polyomavirus infection. Several inhibitors, including MAL2-11B, bithionol, hexachlorophene and MAL2-11B tetrazole were identified by our ongoing analyses of structure–activity relationships and via a high throughput screen using SV40 LT [36,37,39,44]. MAL2-11B and the tetrazole inhibited the ATPase activity of SV40 LT and inhibited SV40 and BKV DNA replication in cell culture. Hexachlorophene and bithionol also inhibited SV40 LT ATPase activity and inhibited the replication of SV40 but a continued analysis of these FDA-approved compounds was thwarted by their relatively high toxicity. Based on sequence conservation among LTs we predicted that inhibitors targeting SV40 LT might have similar effects on the Zn-ATPase domains from the closely related polyomaviruses such as JCV and BKV. Therefore, we compared the effects of these inhibitors on basal ATPase activities of the SV40, JCV and TSV Zn-ATPase domains. While the SV40 and JCV enzymes responded very similarly to the inhibitors, the TSV Zn-ATPase domain was more resistant to all four compounds (Fig. 4). These results suggest that the inhibitors may be most effective on LTs that are closely related, i.e., SV40 and JCV, but not TSV LT. Therefore, attempts to identify specific inhibitors for TSV or other polyomavirus that differ most from SV40 in similar screens should be carried out using the corresponding LT.
LTs from several polyomaviruses including SV40, JCV and BKV are found in complex with cellular p53 [28,45,46]. However, a direct interaction with p53 has only been confirmed for SV40 LT [29]. Our in vitro pull-down assay confirmed the direct interaction between SV40 Zn-ATPase and p53. Moreover, the purified p53 DBD also precipitated the JCV Zn-ATPase domain, suggesting that the two proteins directly interact and form a stable complex. The TSV Zn-ATPase domain, on the other hand, failed to precipitate p53 (Fig. 6A). Besides the conservation in most of the critical p53 binding residues, the surface electrostatic map of the potential p53-binding interface of JCV LT agrees well with the actual binding surface of SV40 LT. In contrast, the TSV LT contains more variations in its candidate p53 binding residues. The surface electrostatic map of TSV LT candidate p53 binding interface also suggests major differences from that of SV40 LT. We suggest that these variations in TSV LT fail to establish proper contact with p53, and are therefore responsible for its lack of p53 interaction. Furthermore, we expect LTs from other polyomaviruses closely related to SV40, such as BKV and SA12 [47,48], to directly interact with p53 in a similar fashion, whereas LTs more distant from SV40 LT should be less likely to bind p53 directly.
Our comparative study using purified versions of the Zn-ATPase domains of SV40, JCV and TSV LTs shows that these proteins vary in both enzymatic activities and protein–protein interactions with p53. LTs are highly sophisticated molecular machines capable of coordinating activities at multiple levels, including binding of ATP/ADP and a dsDNA substrate, the formation of hexamers and dodecamers, the hydrolysis of ATP and release of ADP, and the coupling of energy from ATP hydrolysis with conformational changes to power unwinding of duplex DNA. Each of these steps can be subjected to regulation during infection to achieve a productive viral life cycle. Understanding the regulatory mechanisms exerted by different LTs will not only advance our knowledge in polyomavirus biology, but also provide broad insight on the functions of ATPases and helicases and augment our ability to better inhibit the replication of these disease-causing viruses.
Acknowledgments
This work was supported by National Institutes of Health (R21 AI109339) and the Pittsburgh Center for Kidney Research (GM75061 and DK79307).
We thank Dr. Andrew VanDemark and his lab members for scientific and technical advice and for sharing equipments and reagents.
We thank Dr. Mariet C.W. FeltKamp for providing the construct containing the TSV genome.
We thank Dr. Xiaojiang Chen (University of Southern California) for providing the GST-p53 construct used for the pull-down assay.
Footnotes
Abbreviations used: SV40, Simian virus 40; JCV, John Cunningham Virus; BKV, BK virus; SA12, Simian agent 12; LT, large tumor antigen; pRb, Retinoblastoma protein; p53, tumor protein p53.
References
- 1.An P. Annu Rev Microbiol. 2012;66:213–236. doi: 10.1146/annurev-micro-092611-150154. [DOI] [PubMed] [Google Scholar]
- 2.Cheng J, DeCaprio JA, Fluck MM, Schaffhausen BS. Semin Cancer Biol. 2009;19(4):218–228. doi: 10.1016/j.semcancer.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Srinivasan A, McClellan AJ, Vartikar J, Marks I, Cantalupo P, Li Y, Whyte P, Rundell K, Brodsky JL, Pipas JM. Mol Cell Biol. 1997;17(8):4761–4773. doi: 10.1128/mcb.17.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sullivan CS, Cantalupo P, Pipas JM. Mol Cell Biol. 2000;20(17):6233–6243. doi: 10.1128/mcb.20.17.6233-6243.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zalvide J, Stubdal H, DeCaprio JA. Mol Cell Biol. 1998;18(3):1408–1415. doi: 10.1128/mcb.18.3.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stubdal H, Zalvide J, Campbell KS, Schweitzer C, Roberts TM, DeCaprio JA. Mol Cell Biol. 1997;17(9):4979–4990. doi: 10.1128/mcb.17.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelley WL, Georgopoulos C. Proc Natl Acad Sci USA. 1997;94(8):3679–3684. doi: 10.1073/pnas.94.8.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fewell SW, Pipas JM, Brodsky JL. Proc Natl Acad Sci USA. 2002;99(4):2002–2007. doi: 10.1073/pnas.042670999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLucia AL, Lewton BA, Tjian R, Tegtmeyer P. J Virol. 1983;46(1):143–150. doi: 10.1128/jvi.46.1.143-150.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bochkareva E, Martynowski D, Seitova A, Bochkarev A. EMBO J. 2006;25(24):5961–5969. doi: 10.1038/sj.emboj.7601452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wendler P, Ciniawsky S, Kock M, Kube S. Biochim Biophys Acta. 2012;1823(1):2–14. doi: 10.1016/j.bbamcr.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 12.Walker JE, Saraste M, Runswick MJ, Gay NJ. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanson PI, Whiteheart SW. Nat Rev Mol Cell Biol. 2005;6(7):519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 14.Hickman AB, Dyda F. Curr Opin Struct Biol. 2005;15(1):77–85. doi: 10.1016/j.sbi.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Singleton MR, Dillingham MS, Wigley DB. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 16.Stahl H, Droge P, Knippers R. EMBO J. 1986;5(8):1939–1944. doi: 10.1002/j.1460-2075.1986.tb04447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim HY, Ahn BY, Cho Y. EMBO J. 2001;20(1–2):295–304. doi: 10.1093/emboj/20.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linzer DI, Levine AJ. Cell. 1979;17(1):43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 19.Pipas JM, Levine AJ. Semin Cancer Biol. 2001;11(1):23–30. doi: 10.1006/scbi.2000.0343. [DOI] [PubMed] [Google Scholar]
- 20.Sheppard HM, Corneillie SI, Espiritu C, Gatti A, Liu X. Mol Cell Biol. 1999;19(4):2746–2753. doi: 10.1128/mcb.19.4.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waga S, Bauer G, Stillman B. J Biol Chem. 1994;269(14):10923–10934. [PubMed] [Google Scholar]
- 22.Arunkumar AI, Klimovich V, Jiang X, Ott RD, Mizoue L, Fanning E, Chazin WJ. Nat Struct Mol Biol. 2005;12(4):332–339. doi: 10.1038/nsmbXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou B, Arnett DR, Yu X, Brewster A, Sowd GA, Xie CL, Vila S, Gai D, Fanning E, Chen XS. J Biol Chem. 2012;287(32):26854–26866. doi: 10.1074/jbc.M112.363655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khopde S, Simmons DT. J Virol. 2008;82(3):1136–1145. doi: 10.1128/JVI.01314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calabrese LH, Molloy E, Berger J. Nat Rev Rheumatol. 2014 doi: 10.1038/nrrheum.2014.167. [DOI] [PubMed] [Google Scholar]
- 26.van der Meijden E, Janssens RW, Lauber C. PLoS Pathog. 2010;6(7):e1001024. doi: 10.1371/journal.ppat.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao JJ, Gjoerup OV, Subramanian RR, Cheng Y, Chen W, Roberts TM, Hahn WC. Cancer Cell. 2003;3(5):483–495. doi: 10.1016/s1535-6108(03)00088-6. [DOI] [PubMed] [Google Scholar]
- 28.Seneca NT, Saenz Robles MT, Pipas JM. Virology. 2014;468–470C:47–56. doi: 10.1016/j.virol.2014.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lilyestrom W, Klein MG, Zhang R, Joachimiak A, Chen XS. Genes Dev. 2006;20(17):2373–2382. doi: 10.1101/gad.1456306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castellino AM, Cantalupo P, Marks IM, Vartikar JV, Peden KW, Pipas JM. J Virol. 1997;71(10):7549–7559. doi: 10.1128/jvi.71.10.7549-7559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gai D, Zhao R, Li D, Finkielstein CV, Chen XS. Cell. 2004;119(1):47–60. doi: 10.1016/j.cell.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 32.Li D, Zhao R, Lilyestrom W, Gai D, Zhang R, DeCaprio JA, Fanning E, Jochimiak A, Szakonyi G, Chen XS. Nature. 2003;423(6939):512–518. doi: 10.1038/nature01691. [DOI] [PubMed] [Google Scholar]
- 33.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Nucleic Acids Res. 2013;41(Web Server issue):W349–57. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. Nucleic Acids Res. 2005;33(Web Server issue):W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. Nucleic Acids Res. 2010;38(Web Server issue):W529–33. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seguin SP, Ireland AW, Gupta T, Wright CM, Miyata Y, Wipf P, Pipas JM, Gestwicki JE, Brodsky JL. Antiviral Res. 2012;96(1):70–81. doi: 10.1016/j.antiviral.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seguin SP, Evans CW, Nebane-Akah M, McKellip S, Ananthan S, Tower NA, Sosa M, Rasmussen L, White EL, Maki BE, Matharu DS, Golden JE, Aube J, Brodsky JL, Noah JW. J Biomol Screen. 2012;17(2):194–203. doi: 10.1177/1087057111421630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wright CM, Chovatiya RJ, Jameson NE, Turner DM, Zhu G, Werner S, Huryn DM, Pipas JM, Day BW, Wipf P, Brodsky JL. Bioorg Med Chem. 2008;16(6):3291–3301. doi: 10.1016/j.bmc.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huryn DM, Brodsky JL, Brummond KM, Chambers PG, Eyer B, Ireland AW, Kawasumi M, Laporte MG, Lloyd K, Manteau B, Nghiem P, Quade B, Seguin SP, Wipf P. Proc Natl Acad Sci USA. 2011;108(17):6757–6762. doi: 10.1073/pnas.1015251108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahuja D, Rathi AV, Greer AE, Chen XS, Pipas JM. J Virol. 2009;83(17):8781–8788. doi: 10.1128/JVI.00621-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiekowski M, Schwarz MW, Stahl H. J Biol Chem. 1988;263(1):436–442. [PubMed] [Google Scholar]
- 42.Huang H, Zhao K, Arnett DR, Fanning E. J Biol Chem. 2010;285(43):33475–33484. doi: 10.1074/jbc.M110.156240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greenleaf WB, Shen J, Gai D, Chen XS. J Virol. 2008;82(12):6017–6023. doi: 10.1128/JVI.00387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wright CM, Seguin SP, Fewell SW, Zhang H, Ishwad C, Vats A, Lingwood CA, Wipf P, Fanning E, Pipas JM, Brodsky JL. Virus Res. 2009;141(1):71–80. doi: 10.1016/j.virusres.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trowbridge PW, Frisque RJ. Virology. 1993;196(2):458–474. doi: 10.1006/viro.1993.1502. [DOI] [PubMed] [Google Scholar]
- 46.Harris KF, Christensen JB, Imperiale MJ. J Virol. 1996;70(4):2378–2386. doi: 10.1128/jvi.70.4.2378-2386.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cantalupo P, Doering A, Sullivan CS, Pal A, Peden KW, Lewis AM, Pipas JM. J Virol. 2005;79(20):13094–13104. doi: 10.1128/JVI.79.20.13094-13104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cunningham TP, Pipas JM. J Virol. 1985;54(2):483–492. doi: 10.1128/jvi.54.2.483-492.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]